Abstract
Centrioles are intriguing cylindrical organelles composed of triplet microtubules. Proteomic data suggest that a large number of proteins besides tubulin are necessary for the formation and maintenance of a centriole's complex structure. Expansion of the preexisting centriole proteome from the green alga Chlamydomonas reinhardtii revealed additional human disease genes, emphasizing the significance of centrioles in normal human tissue homeostasis. We found that two classes of ciliary disease genes were highly represented among the basal body proteome: cystic kidney disease (especially nephronophthisis) syndromes, including Meckel/Joubert-like and oral-facial-digital syndrome, caused by mutations in CEP290, MKS1, OFD1, and AHI1/Jouberin proteins and cone-rod dystrophy syndrome genes, including UNC-119/HRG4, NPHP4, and RPGR1. We further characterized proteome of the centriole (POC) 1, a highly abundant WD40 domain-containing centriole protein. We found that POC1 is recruited to nascent procentrioles and localizes in a highly asymmetrical pattern in mature centrioles corresponding to sites of basal-body fiber attachment. Knockdown of POC1 in human cells caused a reduction in centriole duplication, whereas overexpression caused the appearance of elongated centriole-like structures. Together, these data suggest that POC1 is involved in early steps of centriole duplication as well as in the later steps of centriole length control.
INTRODUCTION
Centrioles are barrel-shaped structures composed of nine triplet microtubules. They are necessary for recruitment of pericentriolar material (PCM) to form a complete centrosome and they act as basal bodies during the formation of cilia and flagella. In most quiescent cells, centrioles move to and dock on the apical plasma membrane during ciliogenesis and provide a template for the extension of doublet microtubules, which make up the ciliary axoneme (Ringo, 1967; Sorokin, 1968; Snell et al., 1974; Vorobjev and Chentsov, 1982; Dawe et al., 2007). Centrioles that template cilia are known as basal bodies, and the proteins that compose them have received increased attention in recent years because of their role in ciliary diseases. Ciliary diseases, or ciliopathies, result in symptoms ranging from obesity and retinal degeneration to polydactyly and cystic kidneys (Pazour and Rosenbaum, 2002; Afzelius, 2004; Badano et al., 2006; Yoder, 2007; Marshall, 2008). Ciliopathies arise from mutations in not only ciliary genes but also in genes encoding proteins within the basal body (Ansley et al., 2003; Keller et al., 2005; Marshall, 2008). Proteomic analyses of centrioles from a number of diverse organisms reveal the presence of ciliary disease genes (Keller et al., 2005; Broadhead et al., 2006; Kilburn et al., 2007). Because having a properly anchored basal body is required for a functional cilium, mutations in any protein involved in the formation and maintenance of centriole or basal body integrity have the potential to lead to ciliary diseases.
The structure of the centriole is complex and highly precise, with centriole length tightly controlled, but the molecular mechanisms governing centriole assembly, length control, and maturation into basal bodies remain mysterious. Genetic screens in Chlamydomonas, Drosophila, and Caenorhabditis elegans have given clues to how centrioles assemble by providing mutants that act as premature stops in the centriole assembly pathway and have provided insight into such questions as how the ninefold symmetry of centrioles is established, what other tubulin isoforms are necessary for triplet microtubule formation, and how initial steps of centriole assembly progress in various species (Dutcher and Trabuco, 1998; Dutcher et al., 2002; Dammermann et al., 2004; Bettencourt-Dias et al., 2005; Delattre et al., 2006; Pelletier et al., 2006; Hiraki et al., 2007; Nakazawa et al., 2007). Although these studies reveal crucial steps in centriole assembly at the ultrastructural scale, we have only just begun to learn how steps in basal body assembly are reflected in individual protein recruitment events. Detailed localizations and determination of the order of assembly of particular centriole proteins are pertinent for a detailed depiction of how centrioles form and duplicate once per cell cycle, but thus far few centriole proteins have been characterized in detail.
In this report, we have expanded the Chlamydomonas centriole proteome based on new genomic data and the identification of additional centriole proteins. To begin to learn how the centriole proteome is put together, we investigated proteome of the centriole (POC) 1, one of the most abundant proteins from our centriole proteome, and we found it to be a proximal and very early marker of centriole duplication. Additionally, POC1 has a unique localization on intact mature centrioles, being found to colocalize with attachment points of multiple distinct fiber systems that contact the centriole/basal body. This is the first protein to date to have been localized to both early duplicating centrioles and to places of centriole fiber attachment, indicating that POC1 may be involved in multiple distinct aspects of centriole biology. Furthermore, knockdown of POC1 in human U2OS cells prevented overduplication of centrioles, whereas overexpression of POC1 caused the appearance of numerous elongated centriole-like structures. Based on these results, we suggest that POC1 is involved in the early stages of centriole duplication and also plays a role in the enigmatic process of centriole length control.
MATERIALS AND METHODS
Human Cell Culture
HeLa and U2OS cells were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum as described in Keller et al. (2005).
Generation of Green Fluorescent Protein (GFP)-expressing Cell Lines
The C-terminally tagged POC1B-GFP (Keller et al., 2005) was transfected with Lipofectamine 2000 according to manufacturers guidelines (Invitrogen). Using a limited dilution method in the presence of 500 μg/ml Geneticin (Invitrogen), stable clones expressing the POC1B–GFP fusion protein were isolated. Multiple independent clones were analyzed. The centrioles in each clone had no differences in known centriole markers compared with parental cell lines.
POC1A cDNA was cloned by reverse transcription-polymerase chain reaction (PCR) from a cDNA library constructed from unsynchronized HeLa cells. The cDNA was then subcloned into the pEGFP-C1 vector (Clontech, Mountain View, CA), which added a C-terminal GFP tag. Cells were transiently transfected with Lipofectamine 2000 according to manufacturer's guidelines (Invitrogen).
Human Cell Fixation and Immunofluorescence
Cells were grown on coverslips in 12- or 24-well plates, fixed, and visualized according to Keller et al. (2005). Cells were stained with either anti-γ-tubulin (GTU-88; Sigma-Aldrich, St. Louis, MO), 1:100; centrin-2 (a generous gift from M. Bornens, Institut Curie, Paris, France) 1:2000; acetylated-tubulin (clone 6;11B-1; Sigma-Aldrich), 1:500; polyglutamylated-tubulin ([B3] ab11324; Abcam, Cambridge, MA), 1:1000, or GFP (11 814 460 001; Roche Diagnostics, Indianapolis, IN), 1:250. The following secondary antibodies from Jackson ImmunoResearch Laboratories (West Grove, PA) were used at 1:1000: fluorescein isothiocyanate-conjugated AffiniPure goat anti-mouse immunoglobulin G (IgG) (115-095-003), tetramethylrhodamine B isothiocyanate (TRITC)-conjugated AffiniPure goat anti-mouse IgG (115-025-003), or TRITC-conjugated AffiniPure goat anti-rabbit IgG (111-025-144). Cells were then stained with 4,6-diamidino-2-phenylindole (DAPI) for 5 min and mounted with Mowiol mounting media. DeltaVision deconvolution fluorescence microscopy was used with PlanApo 60× and 100× objectives (Olympus, Tokyo, Japan), and 0.2-μm steps in the z-axis were used to make quick projections of deconvolved images. Intensity plots were made by choosing a region of interest followed by use of DeltaVision 3D graph data inspector software (Applied Precision, Issaquah, WA). Centriole lengths were measured using the Distance tool with standard two point settings.
To facilitate the visualization of GFP localization to centrioles, U2OS cells were occasionally treated with aphidicolin (3.2 μg/ml) for 50–72 h to induce S phase arrest and an accompanying overduplication of centrioles.
RNA Interference (RNAi)
Synthetic siRNA oligonucleotides were obtained from QIAGEN (Valencia, CA). Transfection of small interfering RNAs (siRNAs) by using HiPerFect Transfection Reagent (QIAGEN) was performed according to manufacturer's instructions. QIAGEN′s thoroughly tested and validated AllStars Negative Control siRNA was used as a negative control. The following predesigned siRNAs were ordered and used from QIAGEN: Hs_WDR51A_2, Hs_WDR51A_4, Hs_WDR51B_2, and WDR51B_4. Coverslips were fixed and stained as above after 55 h of treatment. For S phase-arrested cells, aphidicolin (3.2 μg/ml) was added to cells 2 h after siRNA transfection. Cells were examined 55 h later.
Human Cell Transient Transfection and Overexpression
HeLa and/or U2OS cells were seeded the day before transfection onto coverslips. The day of transfection, cells were transfected with the following constructs: full-length POC1A-Cherry, POC1B-Cherry, POC1A-WD40-GFP, POC1A-WD40-Cherry, POC1B-WD40-Cherry, POC1A-Cterm-Cherry, or POC1B-Cterm-Cherry. For S phase-arrested cells, aphidicolin (3.2 μg/ml) was added to cells 2 h after transfection, and cells were examined 55–72 h later. Mammalian cDNAs from the human open reading frame collection in the form of Gateway entry vectors were purchased from Open Biosystems (Huntsville, AL): POC4 (CV030533), POC6 (CV027317), POC7 (CV025021), POC8 (CV023168), POC9 (CV-029168), POC17 (CV023450), Rib43A (CV023821), CCT3 (CV027136), basal body proteins with up-regulated genes (BUG) 5 (CV027392), BUG7 (CV025099), and BUG22 (CV-026100). The following cDNAs in the form of Gateway entry vectors were purchased from GeneCopoeia (Germantown, MD): POC2 (GC-E0364), POC3 (GC-T7752), POC11 (GC-V0935), POC20 (GC-F0087), DIP13 (GC-Q0661), Hsp90 (GC-M0233), BUG11 (GC-U0139), BUG30 (GC-E0925), and BUG32 (GC-V1413).
Chlamydomonas Cell Culture and Immunofluorescence
Chlamydomonas reinhardtii wild-type (strains cc125 and cc124), basal body-deficient strains bld2 (cc478), uni3 (cc2508), and bld10 (cc4076), and temperature-sensitive flagellar assembly mutant strain fla10 (fla10-1 allele, cc1919) were obtained from the Chlamydomonas Genetics Center (Duke University, Durham, NC). Cells were grown and maintained in TAP media (Harris, 1989). Growth was at 25°C with continuous aeration and constant light except for the fla10 mutant, which was grown at 34°C as the restrictive temperature.
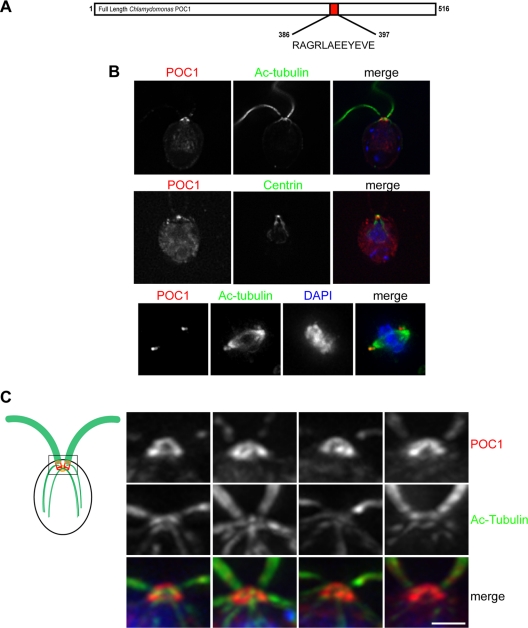
To study the localization and properties of Chlamydomonas POC1, a peptide antibody against the following peptide was raised and affinity purified: RAGRLAEEYEVE (Bethyl Laboratories, Montgomery, TX). This peptide was designed using Invitrogen antigen design tool (www.invitrogen.com) and the Open Biosystems antigenicity prediction tool (www.openbiosystems.com). The peptide is located within the last 150 amino acids (aa) of Chlamydomonas POC1, but it does not overlap with the C-terminal POC1 domain and does not share homology with any other organisms (Figure 3A). The antibody detects a single polypeptide and specific antibody staining in vivo was abolished after incubation with the peptide used for antibody preparation (Supplemental Figure S3).
Figure 3.
Chlamydomonas POC1 immunofluorescence. (A) Illustration depicting the position and sequence of the POC1 peptide that was used to create the Chlamydomonas POC1 antibody. Numbers represent amino acid positions. (B) POC1 (red) localizes to the basal bodies of wild-type Chlamydomonas cells, as shown by colocalization with acetylated tubulin antibody (top; green). Merged image is with nuclear stain (blue, DAPI). The middle panel indicates that POC1 (red) colocalizes with centrin (green) antibody only at the basal body. POC1 is absent from the centrin-based nuclear-attachment fibers. The bottom panel demonstrates that POC1 (red) costains with acetylated tubulin (green) to mark two centrioles at each pole of the mitotic spindle. Merged image is stained with a nuclear stain (blue, DAPI). (C) POC1 localizes to centrioles in a distinct pattern. Illustration of Chlamydomonas basal bodies (red), flagella (green), and inner rootlet microtubules (smaller green lines). The boxed region represents the area of high magnification in the next panels. Merged images demonstrate that POC1 (red) forms a cylindrical structure at the base of the flagella (green). Bar, 1 μm.
Chlamydomonas immunofluorescence followed the standard procedure of Cole et al. (1998). Cells were allowed to adhere to polylysine-coated coverslips before fixation in cold methanol for 5 min. Coverslips were then transferred to a solution of 50% methanol:50% TAP for an additional 5 min. After fixation, cells were blocked in 5% bovine serum albumin (BSA), 1% fish gelatin, and 10% normal goat serum in phosphate-buffered saline (PBS). Cells were then incubated in primary antibodies overnight: anti-POC1, 1:200, anti-acetylated tubulin (T6793; Sigma-Aldrich), 1:500; anti-Bld10p (a generous gift from M. Hirono, University of Tokyo, Tokyo, Japan), 1:100; and anti-centrin (a generous gift from J. Salisbury, Mayo Clinic, Rochester, MN). Secondary antibodies used are as stated above.
Chlamydomonas Flagellar Manipulations
Flagellar splay assays were conducted according to Johnson (1998). After fixation, coverslips were blocked in 5% BSA, 1% fish gelatin, and 10% normal goat serum in PBS. Coverslips were then incubated in primary antibodies: anti-POC1, 1:200 and acetylated-tubulin (T6793; Sigma-Aldrich), 1:500 overnight. Secondary antibodies used are as stated above.
To test whether POC1 is a component of the intraflagellar transport (IFT) machinery, the fla10-ts mutant was temperature shifted from 25°C to 34°C for 45 min. It has been shown previously that this mutant stops IFT (Kozminski et al., 1995) and loses IFT proteins from its flagella (Cole et al., 1998) within 100 min after shifting to the nonpermissive temperature. Cells were stained with POC1, 1:200 and IFT172.1, an IFT complex B protein, 1:200 (a generous gift from D. Cole, University of Idaho, Moscow, ID).
Quantitative PCR
All quantitative PCR was performed according to Keller et al. (2005).
Western Blot
For POC1 Western blots, purified basal bodies were mixed 1:1 with sample buffer and loaded onto a 10% polyacrylamide gel. Blots were probed with the Chlamydomonas POC1 peptide antibody at a concentration of 1:250 followed by staining with a rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (1:20,000; Jackson ImmunoResearch Laboratories). Human cells were grown in six-well dish and treated with 3.2 μg/ml aphidicolin for 55–72 h to achieve S phase arrest. Cells were then collected in wash buffer (10 mM HEPES, 1% Triton X, and 1 mM phenylmethylsulfonyl fluoride) and centrifuged for 15 min at 24,000 × g. Pellets were then resuspended in sample buffer and equal amounts of protein were loaded onto a 10% polyacrylamide gel. Blots were probed with an anti-GFP antibody (11 814 460 001; Roche Diagnostics), 1:250 followed by staining with a mouse HRP-conjugated secondary antibody (Jackson ImmunoResearch Laboratories; 1:20,000).
Fluorescence Intensity Quantification
All images were scaled identically to maintain quantitative information. Pixel boxes (50 × 50) surrounding each basal body image were then analyzed. Background was estimated from the average intensity of pixels on the edge of the box. A threshold was then applied such that all pixels with intensity less than half the dynamic range of the image were set to zero. The total intensity of the remaining pixels was added and then the estimated background contribution was subtracted.
Immunoelectron Microscopy
All immunoelectron microscopy (immuno-EM) was done as described in Geimer and Melkonian (2005), with the following changes. Serial sections (50 nm) were cut with an Ultracut UCT microtome (Leica Microsystems, Vienna, Austria) and collected on piliform-coated gold gilded copper grids. Incubation with POC1 primary antibody was done for 90 min at room temperature at a dilution of 1:250 followed by goat anti-rabbit IgG labeled with 15-nm colloidal gold (Jackson ImmunoResearch Laboratories). Sections were imaged with a CEM 902 transmission electron microscope (Carl Zeiss, Oberkochen, Germany) operated at 80 kV by using SO-163 EM film (Eastman Kodak, Rochester, NY).
RESULTS
Extending the Chlamydomonas Centriole Proteome
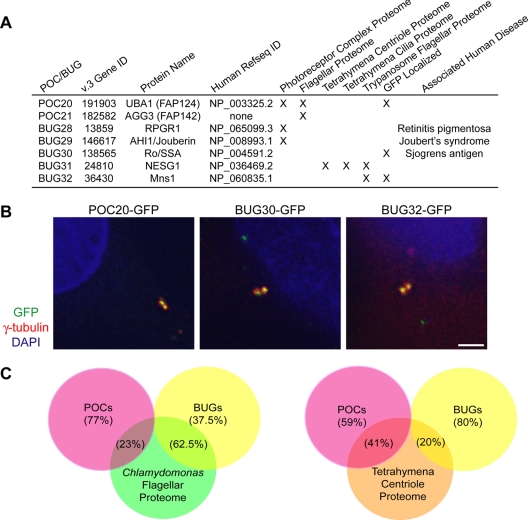
The original centriole proteome from Chlamydomonas reinhardtii (Keller et al., 2005) was determined by using multidimensional protein identification technology, a mass spectrometry-based method in which complex mixtures of proteins can be analyzed without prior electrophoretic separation (Washburn et al., 2001). This method was combined with matching individual peptides to predicted gene models in the Chlamydomonas genome sequence (Keller et al., 2005). However, after the time of the first centriole proteome study, the Chlamydomonas genome has been refined and extensively reannotated (Merchant et al., 2007), with many new and revised gene–model predictions added. We therefore reanalyzed our initial mass spectrometry data by using the new Chlamydomonas version 3.0 gene model predictions to search for additional centriole proteins. This analysis identified seven additional candidate centriole proteins (Figure 1A). Six of these proteins have human orthologues, three of which are associated with known human diseases, revalidating the importance of the centriole proteome.
Figure 1.
Expansion of the Chlamydomonas centriole proteome reveals additional human disease genes. (A) Table of newly discovered POCs and BUGs. Version 3 gene identification numbers are as specified in version 3.0 of the Chlamydomonas genome sequence, available at the Joint Genomes Institute website at http://genome.jgi-psf.org/Chlre3/Chlre3.home.html. Table also indicates protein name, human Refseq ID numbers, localization of proteins to other proteomes of interest, GFP localization to centrioles in human cells, and any associated human diseases. (B) Localization of GFP-fusion proteins corresponding to human homologues of Chlamydomonas centriole candidate proteins (POC20/FAP124, BUG30/Ro/SSA, and BUG21/Mns1) after transient transfection into HeLa cells. γ-Tubulin antibody stain shows centrosomes; DAPI stain shows DNA. Each GFP-fusion protein colocalizes to a pair of dots within the centrosome in HeLa cells, confirming centriolar localization. Bar, 5 μm. (C) Venn diagrams illustrating the overlap of POC and BUG proteins with the Chlamydomonas flagellar proteome (Pazour et al., 2005) and with the Tetrahymena centriole proteome (Kilburn et al., 2007). Percentage of POC and BUG proteins overlapping with the specified proteomes are indicated.
To select candidate proteomic hits that were most likely to encode centriole-related proteins, we characterized the novel proteins as either core components of the centriole (POCs) or as basal body proteins with up-regulated genes (BUGs) (Keller et al., 2005). The POC category indicates proteins that are conserved in species with cilia or else orthologues of proteins obtained in a human centrosome protein analysis (Andersen et al., 2003). The BUG category indicates proteins encoded by genes whose expression (as judged by quantitative real-time PCR and/or microarray analysis) increases during flagellar assembly, implicating them in the process of ciliogenesis. Figure 1A demonstrates that five of the seven novel centriole proteins have genes that are up-regulated during flagellar regeneration and are thus annotated as BUGs (BUG28–BUG32). A full list of cross-validated POCs and BUGs from the Chlamydomonas centriole proteome is in Supplemental Table 1. Two proteins, POC13 and BUG6, were not reannotated in the version 3.0 genome and are indicated as such in Supplemental Table 1.
To validate centriole proteins, we constructed C-terminal GFP fusion proteins and analyzed their localizations during transient transfections in both HeLa and U2OS cells. We previously published the localizations of four centriole proteins from the centriole proteome in human cells (POC1, BUG21/PACRG, POC12/MKS1, and BUG14) (Keller et al., 2005). We have now succeeded in localizing 28 of the POCs and BUGs at the centriole in our ongoing effort to verify our cross-validated groups of centriole proteins (Supplemental Table 1). POC20/FAP124, BUG30/Sjogrens autoantigen (Ro/SSA), and BUG32 are among the new proteins in the centriole proteome that we have shown to be centriolar by GFP localization in human cells (Figure 1B). Costaining with γ-tubulin to mark the centrosome reveals that each of the examined proteins localizes to a pair of dots representing the centrioles embedded in a matrix of PCM. The results of the GFP localizations for all POCs are summarized in Supplemental Figure S1. Results of GFP localizations for all BUGs are summarized in Supplemental Figure S2. Furthermore, other groups have demonstrated centriole localizations for additional POCs and BUGs, in both mammalian cells and in other organisms. BUG28/RPGR1 has been demonstrated to localize to centrioles and basal bodies in cells with primary cilia (Shu et al., 2005); and recently, POC10/NPHP-4 has also been shown to localize to the transition zone, a distal modification of the basal body that is required for ciliogenesis, in C. elegans sensory cilia (Jauregui et al., 2008). Additionally, both POC5 and POC19 localize to centrioles in mammalian cells (Azimzadeh and Bornens, personal communication). Many proteins from the centriole proteome did not have a mammalian homologue, which prevented us from checking their localization by using human GFP-tagged constructs. Overall, the localization data confirm the validity of our centriole proteome data and allow us to expand our centriole proteome by seven proteins.
Due to the large number of cilia and centriole proteomes that have been published since our original centriole proteome, we sought to examine which of the POCs and BUGs are also found in the proteomes from mouse photoreceptor complexes, Chlamydomonas flagella, Tetrahymena centrioles and cilia, and Trypanosome flagella (Pazour et al., 2005; Kilburn et al., 2007; Smith et al., 2005; Broadhead et al., 2006; Liu et al., 2007). The number of proteins overlapping between POCs and BUGs and these other proteomes is shown in Supplemental Table 1. It is noteworthy that compared with the flagellar proteome from Chlamydomonas, the majority of overlap occurs within the BUGs, corresponding to genes up-regulated during flagellar assembly, compared with the POCs whose genes are not up-regulated during flagellar assembly (Figure 1C). We hypothesize that these BUG proteins are components of a common structural motif based on microtubule doublets that is shared between centrioles and flagella. In contrast, POCs have a larger overlap than BUGs compared with the Tetrahymena centriole proteome suggesting that POCs do indeed constitute core structural components of the centriole (Figure 1C).
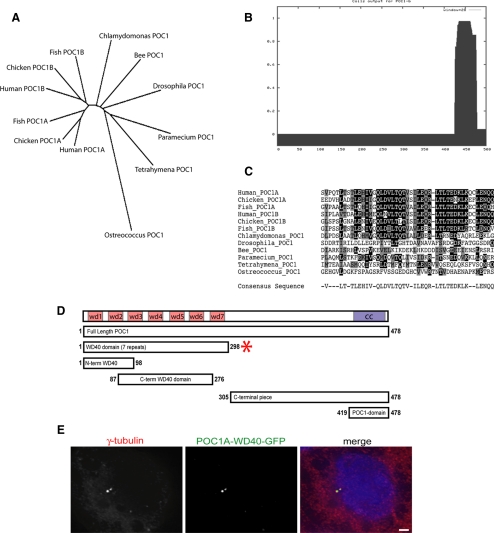
POC1 Is a Conserved Protein That Uses WD40 Repeats to Localize to Centrioles
The kidney and retinal disease gene products outlined above must act in the context of the overall complex structure of the basal body. A key goal at present is thus to know how all basal body proteins fit into this structure. As a first step toward this goal, we have focused on POC1, which is the most abundant centriole protein besides tubulin and tektin, based on spectral counts in the basal body proteome (Keller et al., 2005). Because tubulin and tektin are both centriole structural proteins (Hinchcliffe and Linck, 1998), we reasoned that POC1 might also be an important part of the centriole structure. The POC1 protein is evolutionarily conserved in all organisms from Chlamydomonas to humans, excluding C. elegans, which has a highly unusual centriole structure that is shorter than normal centrioles. All organisms with standard triplet microtubule-containing centrioles in at least part of their life cycle have a POC1 gene (Figure 2A). There is a gene duplication of POC1 in all vertebrates, and we refer to the two paralogues in humans as POC1A (NP_056241.2) and POC1B (NP_758440.1). POC1 has seven WD40 repeats in the N-terminal half of the protein, and the last 50 or so amino acids form a coiled coil based on the COILS server prediction (Figure 2B). Sequence alignment of the last 50 amino acids of POC1 reveals a novel consensus sequence we have termed the POC1 domain (Figure 2C).
Figure 2.
POC1 is a conserved centriole protein that is recruited to the centriole through the N-terminal WD40 repeat domain. (A) Phylogenetic unrooted tree showing that POC1 is conserved from Chlamydomonas to humans. The POC1 protein underwent a gene duplication in vertebrates as shown. (B) Coiled-coil prediction analysis of POC1 illustrates that there is a coiled-coil domain at the C terminus of the protein (COILS server; ch.EMBnet.org). (C) The POC1 domain is a conserved sequence at the C terminus of all conserved POC1 sequences. The POC1 consensus sequence is indicated at the bottom of the sequence alignment. (D) POC1 domain architecture. The WD40 repeat domain contains seven repeats (labeled wd1–wd7) and is sufficient for localization to centrioles in human cells (depicted by red star). Numbers represent amino acid positions. Coiled-coil domain indicated by purple box. (E) POC1-WD40-GFP (green; middle) colocalizes with γ-tubulin (red; left) in transiently transfected HeLa cells. Merged image with nuclear stain (right; blue, DAPI). Bar, 5 μm.
We confirmed previously that POC1B is a centriolar protein in HeLa cells (Keller et al., 2005). We cloned the POC1A gene from HeLa cell cDNA by reverse transcription and verified that it is also centriolar by POC1A–GFP transfection (Supplemental Figure S1). To identify what part of POC1 is necessary for centriolar localization, we constructed C-terminally tagged GFP pieces of POC1A and POC1B (Figure 2D). Our results indicate that besides full-length protein, the WD40 domain at the N terminus (aa 1-298) is sufficient for targeting POC1 localization to centrioles in human cells (Figure 2E). Sequences including only parts of the WD40 domain (aa 1-98 and aa 87-276) are not sufficient for POC1 localization to the centriole, but a comprehensive analysis of exactly which elements of the WD40 domain are or are not required has not been attempted. The conserved POC1 domain consensus sequence is neither necessary nor sufficient for centriole localization; therefore, we hypothesize it may be involved in interactions with other proteins.
POC1 Localizes to Basal Bodies in Human Cells and in Chlamydomonas
We found that POC1 remains localized to centrioles when they become basal bodies in ciliated human cells (Supplemental Figure S5). To more carefully explore POC1 localization within the basal body, we turned to Chlamydomonas in which the basal body cytology is extremely well defined (Ringo, 1967; O'Toole et al., 2003; Geimer and Melkonian, 2005). The Chlamydomonas POC1 peptide antibody (Figure 3A) stains the basal bodies of wild-type cells (Figure 3B). The POC1-positive spots colocalize with both acetylated-tubulin and centrin (Figure 3B), indicating that POC1 is a specific marker for centrioles in Chlamydomonas. The antibody seems to be specific to POC1 and only recognizes one polypeptide (Supplemental Figure S3). This polypeptide has the predicted molecular mass of POC1 (54.4 kDa). As a further control for antibody specificity, we have shown that preincubation with a POC1 peptide used for antibody production abolishes all basal body and flagellar staining, demonstrating that the antibody is specific to the POC1 protein in Chlamydomonas (Supplemental Figure S3).
During spindle formation, POC1 exclusively stains centrioles near the spindle poles. More specifically, POC1 was visualized in two spots at either pole of the spindle representing the four centrioles that are present in wild-type Chlamydomonas spindles (Figure 3B). At higher magnification, POC1 seems to stain the entire basal body from which acetylated tubulin-positive cilia extend (Figure 3C).
We also observed a faint punctate staining along the length of the flagella. Interestingly, the flagellar staining was dramatically increased in flagella that had become detached from the cell body (data not shown). Similar epitope behavior was reported with antibodies to the axonemal microtubule-associated protein BUG21/PACRG (Ikeda et al., 2007), suggesting that the POC1 epitope may be inaccessible in attached flagella.
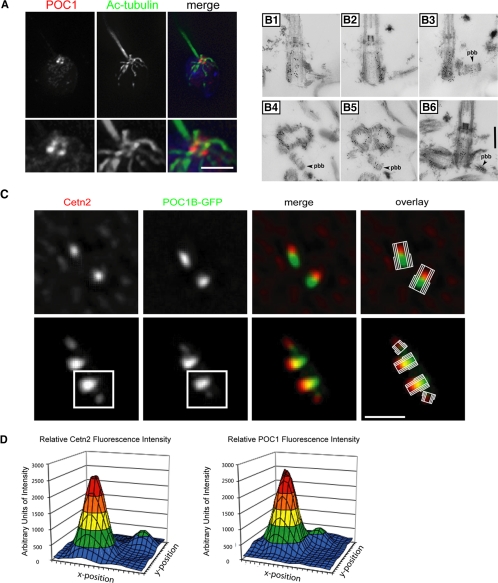
POC1 Localizes to Newly Duplicating Centrioles
We are particularly interested in investigating proteins involved in the early steps of centriole assembly because these may give clues to the unexplained process of centriole duplication. We used both the Chlamydomonas POC1 antibody and human POC1-GFP constructs to ask whether POC1 localizes to newly duplicating daughter centrioles in addition to fully developed mother centrioles. In Chlamydomonas, careful examination demonstrated that POC1 localizes to four distally located spots in wild-type cells (Figure 4A). This is reminiscent of both Bld10p and Vfl1p localizations, both of which show two to four dots at the base of the flagella in positions that resemble the basal bodies and probasal bodies (proBBs) (Silflow et al., 2001; Matsuura et al., 2004). To confirm the POC1 proBB localization in Chlamydomonas, we performed immuno-EM on isolated nucleoflagellar apparatuses (NFAps), which are cytoskeletal complexes containing the basal bodies, proBBs, axonemes, rootlet microtubules, and other fibrous structures tightly associated with the basal bodies (Wright et al., 1985). Gold particles conjugated to the secondary antibody were found associated not only with mature mother centrioles (Figure 4, B1–B3) but also with proBBs (Figure 4, B3, B5, and B6). POC1 is therefore a novel proBB protein in Chlamydomonas and part of a very small group of proteins that are known to be present in daughter proBBs and are likely essential for proper centriole assembly.
Figure 4.
POC1 localizes to newly duplicating centrioles in both Chlamydomonas and in human cells. (A) Chlamydomonas POC1 (red) localizes to both mature basal bodies and proBBs, indicated by the appearance of four dots at the base of the flagella (green) as shown in the higher magnification images. Bar, 1 μm. (B) Immuno-EM of Chlamydomonas basal body sections. Cells were labeled with anti-POC1 antibody and gold-conjugated secondary antibodies. Localization of POC1 to probasal bodies is highlighted with arrowheads. B1–3 are serial sections as are B4 and B5. Bar, 250 nm. (C) Immunofluorescence of stably expressing human POC1-GFP (green). HeLa cells with centrosomes stained with Cetn2 (red). Merged images show two mature centrioles (top) and two mature centrioles along with their newly duplicating daughter centrioles from their proximal ends (bottom). Overlay images demonstrate proximal and distal ends of the mature centrioles along with newly duplicating daughter centrioles. Bar, 1 μm. (D) Fluorescence intensity quantification of boxed images in C. Large peak represents mature basal body; smaller peak represents newly duplicating daughter centriole. Note the smaller peak in the Cetn2 graph is shifted farther (more distal) from the mature centriole than the smaller peak in the POC1 graph.
Because Chlamydomonas POC1 localizes to both mother and daughter centrioles, we wanted to further investigate human POC1 localization. We looked at both POC1A-GFP and POC1B-GFP constructs that had been transiently and stably transfected into HeLa and U2OS cells. Costaining with the early centriole marker, centrin 2 (Cetn2) revealed POC1 localization to both mother centrioles and newly forming daughter centrioles (Figure 4C). Centrin is known to localize to the distal lumen of mature centrioles (Azimzadeh and Bornens, 2007), in contrast POC1 localizes along the entire length of the microtubule-based centriole structure, consistent with the localization in Chlamydomonas. To further clarify POC1 position within the centriole, we examined centrioles during S phase before procentriole elongation. These images indicate that POC1 localizes to the whole centriole, whereas centrin occupies only the distal ends in both mature centrioles and in procentrioles (Figure 4C). Relative intensity plots were constructed to demonstrate that POC1 and centrin are slightly shifted from one another and to show that the mature mother centriole has higher fluorescence intensity then the daughter procentriole (Figure 4D). POC1 is therefore a component of both mother and daughter centrioles in both Chlamydomonas and in human cells.
POC1 Recruitment in Centriole Mutants with Ultrastructural Defects
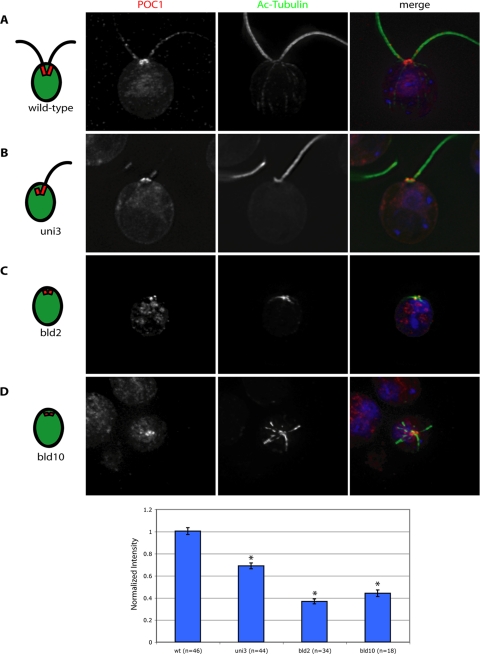
To investigate when POC1 becomes recruited to centrioles during the assembly process, we took advantage of previously described Chlamydomonas centriole mutants that block specific steps in the centriole assembly pathway. Mutations in δ-tubulin (UNI3) and ε-tubulin (BLD2) cause basal bodies to have doublet and singlet microtubules, respectively, unlike the wild-type triplet microtubules (Goodenough and StClair, 1975; O'Toole et al., 2003). Centrioles in bld2 mutants are also much shorter than wild-type centrioles, suggesting a defect in centriole length control (Goodenough and StClair, 1975). Additionally, the bld10 mutant has been reported to completely lack centrioles; however, on very rare occasions cells have fragments of centrioles, which stain positively with an acetylated-tubulin antibody (Matsuura et al., 2004).
We compared POC1 localizations in mutants with doublet, singlet, or fragments of centrioles to its expression in wild-type Chlamydomonas (Figure 5). POC1 localizes to centrioles composed of only doublet or only singlet microtubules (Figure 5, B and C). The majority of bld10 cells had diffuse POC1 with no particular localization; however, whenever cells stained positively for acetylated tubulin, indicating the presence of centriole fragments, POC1 was precisely colocalized (Figure 5D). Fluorescence intensity quantification showed that wild-type basal bodies have a significantly higher amount of total POC1 than any of the three mutants (p < 0.005). This suggests that POC1 is incorporated into the core microtubule structure of basal bodies since it is reduced in mutants with either doublet or singlet microtubules. The mutant with the shortest centrioles, bld2, had the least POC1 recruited. This analysis demonstrates that POC1 can localize to centrioles lacking many of the structures found in mature centrioles, consistent with the above-mentioned data indicating early recruitment of POC1 to proBBs and procentrioles.
Figure 5.
POC1 localization in Chlamydomonas basal body mutants. (A) Wild-type Chlamydomonas cells stained with POC1 (red) and acetylated tubulin (green). Note staining at the base of the flagella (green). (B) uni3 cells stained with POC1 (red) and acetylated-tubulin (green), demonstrating that POC1 stains doublet microtubules. (C) bld2 cells stained with POC1 (red) and acetylated-tubulin (green), indicating that POC1 stains singlet microtubules. (D) bld10 cells stained with POC1 (red) and acetylated tubulin (green) showing that POC1 always colocalizes with acetylated tubulin positive centrioles. Merged images were stained with a nuclear stain (blue, DAPI). (E) Histogram of POC1 staining intensity indicating reduced POC1 recruitment levels in mutant basal bodies.
POC1 Localizes to Sites of Basal Body Fiber Attachment
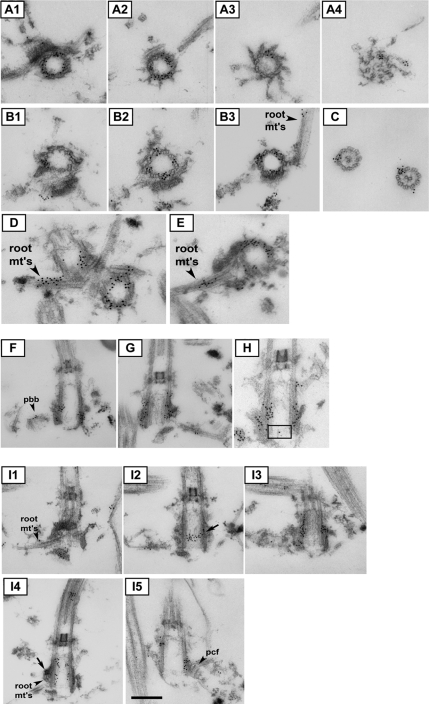
To get higher resolution information about POC1 localization, we used immuno-EM on isolated Chlamydomonas NFap preparations (Wright et al., 1985). POC1 protein was found to associate with the entire microtubule-based barrel structure that constitutes the basal body. Specifically, POC1 localizes to triplet microtubules (Figure 6, A and B), axonemal doublet microtubules (Figure 6C), and rootlet microtubules near the basal bodies (Figure 6, B, D, and E). Tangential sections confirm localization to the walls of the microtubule-based barrel (Figure 6, F–H). POC1 is absent from the centriole lumen, the transition zone, and the central pair microtubules in the axoneme (Figure 6, A, C, and G). Serial sections allowed us to examine where POC1 is found in detail throughout a centriole both in cross section and in tangentially cut sections. Serial sections indicate that POC1 is highly enriched in regions in which fibers attach to the basal bodies. In particular, POC1 localizes to both proximal and distal connecting fibers at the regions of attachment (Figure 6I). In fact, there is POC1 enrichment at the site of attachment of all fibers that are interacting with the basal body (Figure 6I). The basal body localization of POC1 to fiber attachment points demonstrates that the protein localizes in a highly asymmetric pattern on mature basal bodies. POC1 does not specifically localize to the cartwheel as was reported for Tetrahymena POC1 protein (Kilburn et al., 2007). This is seen for example in I2 of Figure 6 shown by gold particles located distal to the cartwheel, which is at the very base of the basal bodies (Ringo, 1967). In fact, POC1 localizes to both inner and outer walls of centrioles and is present on the entire length of the centriole but is completely absent from the centriole lumen (quantification of gold particle distribution given in Supplemental Figure S4). The pattern of POC1 localization implies that POC1 may be involved in establishing, maintaining, or stabilizing specialized microtubular structures. We suggest that POC1 is necessary for either formation or maintenance of doublet and triplet microtubules and is involved at the attachment site of fibers to the microtubule-based basal body structure.
Figure 6.
Immuno-EM of Chlamydomonas POC1 reveals localization to triplet microtubules and sites of fiber attachment. (A1–A4) Immuno-EM of basal body serial sections showing POC1 localization throughout the length of the triplet microtubules. (B1–B3) Serial sections showing POC1 localization to triplet microtubules and rootlet microtubules (root mt's). (C) POC1 localizes to doublet microtubules of axonemes but is absent from central pair microtubules. (D and E) POC1 localizes to rootlet microtubules that are nearby and/or attached to centrioles. (F–H) Sections demonstrating that POC1 localizes to proBBs and occasionally at the cartwheel (cartwheel indicated by black box). (I1–I5) Serial sections through a longitudinally sectioned basal body. POC1 localizes to sites of rootlet microtubule attachment and sites where proximal and distal connecting fibers attach to the basal body (root mt's, rootlet microtubules; pcf proximal connecting fiber, arrow shows a high density of POC1 at rootlet microtubule connection and at proximal connecting fiber). All sections were labeled with anti-POC1 antibody and gold-conjugated secondary antibodies. Bar, 250 nm.
POC1 Localizes to Axonemal Doublet Microtubules
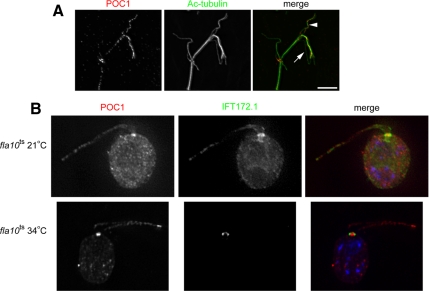
The localization of POC1 to microtubule triplets, which structurally resemble the axonemal microtubule doublets, led us to reexamine the localization of POC1 within the axoneme. We conducted a flagellar splay assay, which uses the ability of flagella to rip themselves apart (Johnson, 1998) to investigate whether POC1 localizes only to doublet microtubules. These experiments reveal that the doublet microtubules colocalize with POC1, whereas the singlet central pair microtubules fail to associate with POC1 (Figure 7A). This result, together with the complete lack of POC1 associated with cytoplasmic microtubules indicates that POC1 is not simply a general microtubule binding protein.
Figure 7.
Chlamydomonas POC1 associates with doublet microtubules in the axoneme and is not a component of the IFT machinery. (A) Flagellar splay assay demonstrates that POC1 (red) localizes specifically to the doublet microtubules (arrowhead) and is absent from central pair microtubules (arrow). Note basal body staining at the base of the splayed flagella with both POC1 and acetylated tubulin antibodies. Bar, 5 μm. (B) POC1 (red) localizes to flagella in fla10ts Chlamydomonas cells at both 21 and 34°C, unlike the IFT protein IFT172.1, which is absent at 34°C.
The punctate staining of POC1 in Chlamydomonas flagella resembles the staining pattern of many IFT proteins (Cole et al., 1998; Rosenbaum and Witman, 2002; Scholey, 2003). IFT proteins also localize in the vicinity of the basal body (Deane et al., 2001). Those similarities in localization raised the possibility that POC1 might be associated with IFT. To test whether POC1 is a structural component of the axoneme or a transitory IFT protein, we used a temperature-sensitive mutation in the axonemal-specific kinesin II motor subunit FLA10. This fla10 mutant is wild-type at 21°C but loses IFT protein in the axoneme and starts absorbing its flagella within 40 min at 34°C (Kozminski et al., 1995). We stained this temperature-sensitive mutant at 21°C and after 45 min at 34°C when all IFT proteins should be absent from flagella. Our results indicate that POC1 is in the flagella in both cases, unlike the IFT 172.1 protein, which disappears at 34°C, demonstrating that POC1 is a component of the axoneme rather than an IFT-associated protein (Figure 7B).
RNAi of POC1 Reduces Centriole Duplication
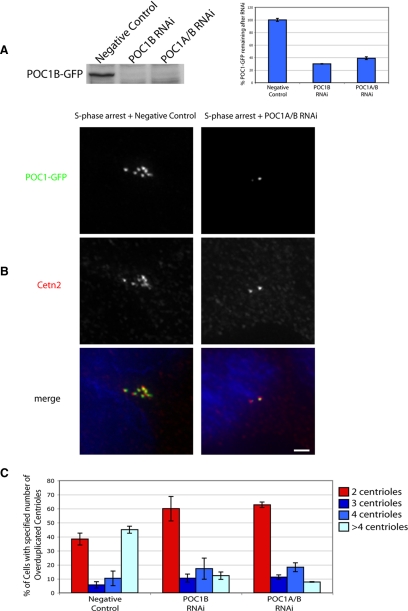
We took advantage of the fact that human U2OS cells overduplicate centrioles when S phase arrested, to examine the effect of POC1 depletion on centriole duplication (Habedanck et al., 2005). We were successfully able to knock down POC1 in a U2OS line stably expressing POC1-GFP, indicated by both fluorescence intensity quantification of GFP (data not shown) and by Western blot (Figure 8A). In untreated U2OS cells, S phase arrest caused cells to accumulate between two and 12 centrioles per cell (Figure 8B) so that a substantial fraction of cells have more than four centrioles, the maximum number seen in normal dividing cells. In contrast, the percentage of cells with more than four centrioles was significantly decreased (p < 0.05) in POC1 siRNA-treated cells in comparison with treatment with a negative control siRNA (Figure 8C). The POC1-depleted cells also had a significant increase in the percentage of cells that had only two centrioles. These data demonstrate that POC1 is necessary for centriole overduplication in U2OS cells and may suggest that POC1 plays a critical role in centriole duplication in cells that are not S phase arrested.
Figure 8.
POC1 depletion leads to a reduction in centriole overduplication. (A) Western blot of POC1-GFP in RNAi-treated U2OS cell lysate. Ponceau stain was used as a loading control (data not shown). Quantification of Western blot normalized to 100%, indicated ∼75 and 60% knockdown of POC1B-GFP by POC1B siRNA and POC1A/B siRNA, respectively. (B) S phase-arrested U2OS cells have overduplicated centrioles, but when POC1 is depleted this, overduplication is suppressed (green, POC1B; red, Cetn2). (C) The percent of cells with overduplicated centrioles is reduced in the presence of POC1 siRNA, whereas the number of cells with wild-type number of two centrioles increases. Bar, 5 μm.
Overexpression of POC1 Leads to Elongated Centriole-like Structures
To gain functional insight into the role of POC1 in centriole assembly, we overexpressed POC1-Cherry constructs to investigate the possibility of a dominant-negative effect. Overexpression of full-length POC1 or the C-terminal POC1 domain resulted in no measurable loss of POC1-GFP or centrin at the centrioles of stably expressing POC1-GFP HeLa or U2OS cells. Overexpression of POC1-WD40 resulted in a slight reduction (<30%) of POC1-GFP at the centriole (data not shown), but we failed to identify any associated phenotype with this slight loss of POC1-GFP at centrioles. However, we noticed that ∼5–10% of the full-length POC1-overexpressing cells showed a remarkable increase in centriole length indicated by both POC1-GFP and by centrin 2 staining (Figure 9A).
Figure 9.
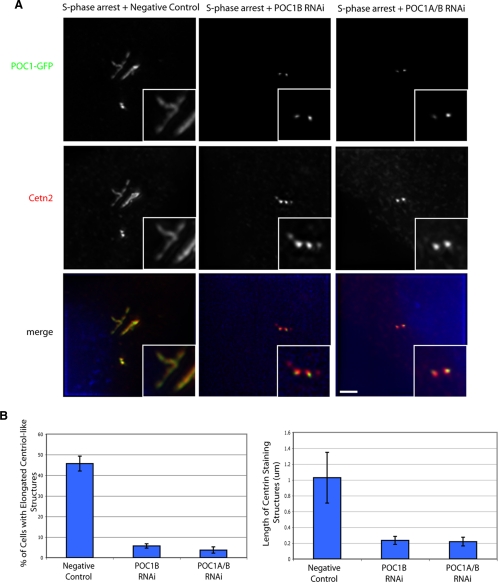
POC1 overexpression in human cells leads to elongated centriole-like structures, which is abolished when POC1 is knocked down by POC1 siRNA. (A) POC1B–GFP-expressing U2OS cells grown and treated with 3.2 μg/ml aphidicolin show a large percentage of cells with elongated centriole-like structures, which are POC1B-GFP positive and stain with Cetn2 (red). This overexpression phenotype is abolished in the presence of POC1 siRNA. (B) The percentage of cells with elongated centriole-like structures is dramatically reduced in the presence POC1 siRNA (left). The length of the elongated centriole-like structures was quantified and is also dramatically reduced in the presence of POC1 siRNA (right).
On S phase arrest in POC1–GFP-overexpressing U2OS cells, we found that a large proportion of cells (>45%) that had elongated centriole-like structures that were positive for POC1-GFP but also stained with centrin (Figure 9A). These elongated centriole-like structures also stain with antibodies against acetylated tubulin and polyglutamylated tubulin, both which mark the more stable microtubules that constitute the centriole (Supplemental Figure S6; (Piperno et al., 1987; Kann et al., 2003). Additionally, γ-tubulin, which can associate specifically with centrioles and is a component of the pericentriolar material, but its not found within cilia (Fuller et al., 1995; Dibbayawan et al., 1995), colocalizes with these POC1–GFP-positive elongated centriole-like structures. The distal appendage marker ODF2 (Ishikawa et al., 2005) fails to colocalize along the length of these elongated structures and rather localizes to only the mother centriole (Supplemental Figure S6). The average length of these elongated centriole-like structures was >1 μm and in some instances extended several micrometers in length (Figure 9B). These structures are very similar to the phenotype seen by depletion of either Cep97 or CP110 (Spektor et al., 2007), although those authors interpreted the structures as primary cilia. Due to the presence in these structures of centrin, which localizes to centrioles and only in the transition zone of axonemes (Laoukili et al., 2000), and of γ-tubulin, we argue that these elongated centriole-like structures are not primary cilia but rather elongated centrioles.
To confirm that the presence of elongated centriole-like structures was dependent on POC1 levels, we transfected cells with siRNA targeting either POC1B alone or simultaneous knockdown of POC1A and POC1B. Depletion of either POC1B or simultaneous depletion of both POC1A and POC1B caused a significant reduction in percent of cells with elongated centriole structures (<10%) in comparison with negative control siRNA (>45%; p < 0.05) (Figure 9B). Additionally the average length of centrin-staining structures within cells was reduced from >1 μm to ∼0.2 μm in both POC1 siRNA treatments (Figure 9B). Wild-type centriole length, measured by both POC1-GFP and centrin fluorescence, is ∼0.2 μm, which is similar to the POC1-depleted cells (data not shown). These data together suggest that overexpression of POC1 leads to an elongation of centrioles. This phenotype seems to be dependent specifically on POC1 because reduced expression of POC1 restores centrioles to their normal length. We therefore suggest that POC1 is involved in both centriole duplication and in centriole length control.
DISCUSSION
Human Disease Proteins Are Highly Represented in the Centriole Proteome
Here, we document an expansion of the Chlamydomonas centriole proteome based on version three of the Chlamydomonas genome (Merchant et al., 2007). The total expanded proteome includes 12 potential human disease proteins (summarized in Table 1), confirming the importance of a thorough investigation of centrioles. In fact, known human disease genes encode >17% of the cross-validated Chlamydomonas centriole proteins (Supplemental Table 1).
Table 1.
Human diseases represented by the centriole proteome
| Proteins involved in cystic kidney diseases | Proteins involved in cone-rod dystrophies | Proteins involved in other human diseases |
|---|---|---|
| POC3 (CEP290/NPHP6) | POC3 (CEP290/NPHP6) | POC9 (Rib72-like protein)-juvenile myoclonic epilepsy |
| POC10 (NPHP4) | POC10 (NPHP4) | DIP13-Sjogrens antigen |
| POC12 (MKS1) | BUG28 (RPGR1) | BUG30 (Ro/SSA) |
| BUG29 (AHI1/Jouberin) | BUG29 (AHI1/Jouberin) | BUG11 (OFD1) |
| BUG11 (OFD1) | POC7 (UNC119) | BUG21 (PACRG)-male infertility |
Due to the ubiquitous distribution of cilia in the human body, mutations in genes encoding for proteins localized in cilia and basal bodies can result in systemic diseases that involve a wide range of symptoms, including but not limited to polydactyly, polycystic kidney disease, and retinal degeneration. In particular, two classes of ciliary disease genes are highly represented among the basal body proteome (Table 1): cystic kidney disease syndromes and cone-rod dystrophy syndrome genes. Mutations in POC3 (CEP290/NPHP6), POC10 (NPHP4), POC12 (MKS1), BUG29 (AHI1/Jouberin), and BUG11 (OFD1) cause a broad spectrum of phenotypes, with cystic kidney disease being among the most common. The diseases that are caused by mutations in these genes (Nephronopththsis, Meckel syndrome, Joubert's syndrome, and oral-facial-digital syndrome) are all known ciliopathies (King et al., 1984; Feather et al., 1997; den Hollander et al., 2006; Ferrante et al., 2006; Kyttala et al., 2006; Parisi et al., 2006; Valente et al., 2006; Tory et al., 2007; Jauregui et al., 2008), and we hypothesize that these diseases arise from defects in centriole structure, function, or both.
In addition to gene products implicated in cystic kidney disease syndromes, our basal body proteome also includes products of genes connected with retinal degeneration. Mutations in BUG28 (RPGR1) and POC7 (UNC119) are associated with cone-rod dystrophy, a name given to a wide range of eye conditions causing deterioration of the cones and rods in the retina, which often leads to blindness (Kobayashi et al., 2000; Gerber et al., 2001; Hameed et al., 2003; Koenekoop, 2005; Adams et al., 2008). All human diseases represented by the centriole proteome are summarized in Table 1. Due to the heterogeneity of ciliopathies, many proteins are associated with more than one human disease.
One possibility is that the disease genes found in the centriole proteome encode for structurally conserved core centriole proteins that are necessary for establishing or maintaining the integrity of the complex triplet microtubules structure. Another possibility is that these proteins are involved in ciliogenesis-related functions such as IFT docking. In the future, it will be important to not only examine the composition of centrioles but also to understand their precise function in regard to the cell cycle and ciliogenesis.
POC1 Is Involved in Centriole Duplication and Centriole Length Control
The Vienna Drosophila RNAi Center reports that a line expressing an RNA interference construct encoding for POC1 is lethal (Dietzl et al., 2007), suggesting POC1 may be essential. Consistent with this, we have not been able to obtain sustained knockdown of POC1 in Chlamydomonas or in human cells by stable expression of RNA interference constructs. Thus, to investigate the function of POC1, we turned to methods such as overexpression and transient siRNA knockdown in human cells. We have demonstrated that POC1 is necessary for the formation of newly duplicated daughter centrioles. Using the standard U2OS centriole-overduplication system, we found that depletion of POC1 strongly reduces the amount of newly formed daughter centrioles. This is exciting because there are only a handful of candidate centriole proteins known to be involved in centriole duplication (Pelletier et al., 2006; Dobbelaere et al., 2008). That cells depleted for both POC1A and POC1B show the absence of centrin-staining procentrioles, together with the localization of POC1 protein to procentrioles that we have demonstrated, suggests that POC1 may be involved in an early stage of centriole assembly.
Overexpression of POC1 in S phase-arrested cells causes a large increase in the percentage of cells with elongated centriole-like structures. These elongated centrioles are unique for a number of reasons. First, they stain with centrin and γ-tubulin, which are both specific for centrioles, indicating they are not simply abnormal cilia or microtubule bundles. Second, these fibers, despite having an average length of ∼1 μm can extend over several micrometers, far longer than normal centrioles can ever become. Third, the elongated centriole-like structures are caused directly by the overexpression of POC1 because subsequent depletion of the protein almost eliminates these fibers. Mechanisms have been identified for controlling the size of a number of cellular structures (Marshall, 2004), but there is currently little information about how centriole length may be regulated. Identification of a protein involved in centriole length control is significant in that it will provide a starting point to identify the mechanisms that control centriole size.
Because POC1 is an early recruited protein to the centriole that is subsequently found along the whole length of the centriole barrel, it is possible that it is intimately involved in determining and influencing centriole length. The total amount of POC1 that becomes incorporated into a centriole may be directly proportional to the length of centrioles, which could explain why overexpression of POC1 causes such a drastic increase in centriole length. Future ultrastructural analysis of these elongated structures by using electron microscopy will be required to further define the nature of the elongation defect.
Relation of POC1 to Ciliary Motility and Centriole Length or Complexity
It is interesting to note that the POC1 gene is apparently absent from the C. elegans genome. This is intriguing because C. elegans centrioles are extremely short and composed of singlet rather than doublet microtubules (Inglis et al., 2007). Worms have lost other conserved centriole proteins and structures, such as centrin and ε- and δ-tubulin along with the cartwheel structure, indicating that nematodes are highly divergent when it comes to both centrioles and cilia (Pelletier et al., 2006; Azimzadeh and Bornens, 2007). C. elegans also lacks motile cilia, possibly suggesting that POC1 might be associated with motility. Although is formally possible that other proteins in C. elegans play the function of POC1, the simplest interpretation is that the short, reduced centrioles of nematodes do not require POC1 protein, which could be consistent with a role for POC1 either in ciliary motility or in centriole elongation.
Drosophila lack motile cilia in all cell types except sperm. Moreover, the centrioles in all cells other than sperm are extremely short. Thus, if POC1 is involved in either centriole elongation or ciliary motility, we would expect the POC1 gene to be most highly expressed in testis where the sperm are forming. Indeed, Drosophila POC1 mRNA is highly up-regulated (>4 times higher expression than in any other adult tissue) in testes (Chintapalli et al., 2007). In vertebrates, many cell types form nonmotile primary cilia. We have shown that POC1 localizes to the basal bodies of human nonmotile cilia (Supplementary Figure S5), indicating that POC1 is not strictly limited to basal bodies of motile cilia.
The overall conclusion from these phylogenetic considerations is that POC1 seems to correlate with larger, more complex centrioles, that are capable of acting as basal bodies for motile cilia. Basal bodies, which have the appearance of a symmetrical cylinder, also have an inherent asymmetry due to the asymmetric attachment of various fibers and appendages. Proteins such as VFL1 and Centrin are known to localize on mature basal bodies in an asymmetric manner and have been hypothesized to confer orientational information to the two adjacent mature basal bodies and to the newly duplicating probasal bodies (Salisbury et al., 1998; Silflow et al., 2001; Geimer and Melkonian, 2005). The distribution of POC1 on these fibrous attachment points confers a rotational asymmetry that may be important for either setting up basal body orientation that is essential for subsequent ciliary beating or for establishing the cytoplasmic location of probasal body formation, which is also determined by the inherent asymmetry of the mature basal body. POC1 thus may play a role in establishing and maintaining the connections between basal bodies, which has implications for ciliary beat patterns and planar cell polarity.
CONCLUSIONS
We have shown that POC1 is likely to play a key role in determining many aspects of centriole biology. The localization of POC1 to Chlamydomonas centrioles at sites of fibrous attachments suggest that it may play a central role in establishing centriole orientation within the context of a cell and/or organism. Additionally, POC1 localizes to and seems to be essential for the emergence of newly formed daughter centrioles during centriole duplication. POC1, which localizes along the entire triplet microtubule structure of mature centrioles is also directly involved in determining centriole length. Future studies focusing on POC1 interacting proteins and how it becomes incorporated into centrioles to determine length control will help provide insights into the complex triplet microtubules structure that epitomizes a centriole.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Marshall laboratory for invaluable discussion and critical review of the manuscript; J. Salisbury, M. Hirono, T. Ikeda, R. Kamiya, M. Bornens, J. Azimzadeh, and D. Cole for reagents; and E. Harris and the Chlamydomonas Genetics Center for providing strains. A special thanks to C. Carroll for help with several experimental details. L.C.K. is supported by an American Heart Association predoctoral fellowship. W.F.M. acknowledges support from National Institutes of Health grant R01 GM-077004, a W. M. Keck Foundation Distinguished Young Scholar in Medical Research award, and the Searle Scholars Program.
Abbreviations used:
- BB
basal body
- BUG
basal body protein with up-regulated gene
- IFT
intraflagellar transport
- NFAp
nucleoflagellar apparatus
- POC
proteome of the centriole protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-06-0619) on December 24, 2008.
REFERENCES
- Adams M., Smith U. M., Logan C. V., Johnson C. A. Recent advances in the molecular pathology, cell biology and genetics of ciliopathies. J. Med. Genet. 2008;45:257–267. doi: 10.1136/jmg.2007.054999. [DOI] [PubMed] [Google Scholar]
- Afzelius B. A. Cilia-related diseases. J. Pathol. 2004;204:470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Ansley S. J., et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Bornens M. Structure and duplication of the centrosome. J. Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M. K., Carmo N., Balloux F., Callaini G., Glover D. M. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Broadhead R., et al. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander A. I., et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., Muller-Reichert T., Pelletier L., Habermann B., Desai A., Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Dawe H. R., Farr H., Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- Deane J. A., Cole D. G., Seeley E. S., Diener D. R., Rosenbaum J. L. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 2001;11:1586–1590. doi: 10.1016/s0960-9822(01)00484-5. [DOI] [PubMed] [Google Scholar]
- Delattre M., Canard C., Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Dibbayawan T. P., Harper J.D.I., Elliott J. E., Gunning B.E.S., Marc J. A γ-tubulin that associated specifically with centrioles in HeLa cells and the basal body complex in Chlamydomonas. Cell Biol. Int. 1995;19:559–567. doi: 10.1006/cbir.1995.1102. [DOI] [PubMed] [Google Scholar]
- Dietzl G., et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Josue F., Suijkerbuijk S., Baum B., Tapon N., Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. K., Morrissette N. S., Preble A. M., Rackley C., Stanga J. Epsilon-tubulin is an essential component of the centriole. Mol. Biol. Cell. 2002;13:3859–3869. doi: 10.1091/mbc.E02-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. K., Trabuco E. C. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell. 1998;9:1293–1308. doi: 10.1091/mbc.9.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feather S. A., Winyard P.J.D., Dodd S., Woolf A. S. Oral-facial-digital syndrome type 1 is another dominant polycystic kidney disease: clinical, radiological and histopathological features of a new kindred. Nephrol. Dial. Transplant. 1997;12:1354–1361. doi: 10.1093/ndt/12.7.1354. [DOI] [PubMed] [Google Scholar]
- Ferrante M. I., Zullo A., Barra A., Bimonte S., Messaddeq N., Studer M., Dolle P., Franco B. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat. Cell Biol. 2006;38:112–117. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- Fuller S. D., Gowen B. E., Reinsch S, Sawyer A, Buendia B., Wepf R., Karsenti E. The core of the mammalian centriole contains gamma-tubulin. Curr. Biol. 1995;5:1384–1393. doi: 10.1016/s0960-9822(95)00276-4. [DOI] [PubMed] [Google Scholar]
- Geimer S., Melkonian M. Centrin scaffold in Chlamydomonas reinhardtii revealed by immunoelectron microscopy. Eukoryot. Cell. 2005;4:1253–1263. doi: 10.1128/EC.4.7.1253-1263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber S., et al. Complete exon-intron structure of the RPGR-interacting protein (RPGRIP1) gene allows the identification of mutations underlying Leber congenital amaurosis. Eur. J. Hum. Genet. 2001;9:561–571. doi: 10.1038/sj.ejhg.5200689. [DOI] [PubMed] [Google Scholar]
- Goodenough U. W., StClair H. S. BALD-2, a mutation affecting the formation of doublet and triplet sets of microtubules in Chlamydomonas reinhardtii. J. Cell Biol. 1975;66:480–491. doi: 10.1083/jcb.66.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y. D., Wilkinson C. J., Nigg E. A. The polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hameed A., Abid A., Aziz A., Ismail M., Mehdi S. Q., Khaliq S. Evidence of RPGRIP1 gene mutations associated with recessive cone-rod dystrophy. J. Med. Genet. 2003;40:616–619. doi: 10.1136/jmg.40.8.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H. The Chlamydomonas Sourebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego, CA: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E. H., Linck R. W. Two proteins isolated from sea urchin sperm flagella: structural components common to the stable microtubules of axonemes and centrioles. J. Cell Sci. 1998;111:585–595. doi: 10.1242/jcs.111.5.585. [DOI] [PubMed] [Google Scholar]
- Hiraki M., Nakazawa Y., Kamiya R., Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr. Biol. 2007;17:1–6. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Ikeda T., Morikawa K., Kamiya R. Axonemal localizations of Chlamydomonas PACRG, a homologue of the human Parkin-coregulated gene product. Cell Motil. Cytoskeleton. 2007;64:814–821. doi: 10.1002/cm.20225. [DOI] [PubMed] [Google Scholar]
- Inglis P. N., Ou G., Leroux M. R., Scholey J. M. The sensory cilia of. Caenorhabditis elegans. WormBook. 2007;8:1–22. doi: 10.1895/wormbook.1.126.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Kubo A., Tsukita S., Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- Jauregui A. R., Nguyen K.C.Q., Hall D. H., Barr M. M. The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J. Cell Biol. 2008;180:973–988. doi: 10.1083/jcb.200707090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. The axonemal microtubules of the Chlamydomonas flagellum differ in tubulin isoform content. J. Cell Sci. 1998;111:313–320. doi: 10.1242/jcs.111.3.313. [DOI] [PubMed] [Google Scholar]
- Kann M., Soues S., Levilliers N., Fouquet J. Glutamylated tubulin: diversity of expression and distribution of isoforms. Cell Motil. Cytoskeleton. 2003;55:14–25. doi: 10.1002/cm.10107. [DOI] [PubMed] [Google Scholar]
- Keller L. C., Romijn E. P., Zamora I., Yates J. R., III, Marshall W. F. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Kilburn C. L., Pearson C. G., Romijn E. P., Meehl J. B., Giddings T. H., Culver B. P., Yates J. R., III, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. D., Dudgeon J., Stephenson J. B. Joubert's syndrome with retinal dysplasia: neonatal tachypnea as the clue to a genetic brain-eye malformation. Arch. Dis. Child. 1984;59:709–718. doi: 10.1136/adc.59.8.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Higashide T., Hamasaki D., Kubota S., Sakuma H., An W., Fujimaki T., McLaren M. J., Weleber R. G., Inana G. HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest. Ophthalmol. Vis. Sci. 2000;41:3268–3277. [PubMed] [Google Scholar]
- Koenekoop R. K. RPGRIP1 is mutated in Leber congenital amaurosis: a mini-review. Ophthalmic. Genet. 2005;26:175–179. doi: 10.1080/13816810500374441. [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Beech P., Rosenbaum J. L. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J. Cell Biol. 1995;131:1517–1527. [Google Scholar]
- Kyttala M., Talila J., Salonen R., Kopra O., Kohischmidt N., Paavola-Sakki P., Peltonen L., Kestila M. MSK1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat. Genet. 2006;38:155–157. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- Laoukili J., Perret E., Middendorp S., Houcine O., Guennou C., Marano F., Bornens M., Tournier F. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J. Cell Sci. 2000;113:1355–1364. doi: 10.1242/jcs.113.8.1355. [DOI] [PubMed] [Google Scholar]
- Liu Q., Tan G., Levenkova N., Li T., Pugh E. N., Jr., Rux J. J., Speicher D. W., Pierce E. A. The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F. Cellular length control systems. Annu. Rev. Cell Dev. Biol. 2004;20:677–693. doi: 10.1146/annurev.cellbio.20.012103.094437. [DOI] [PubMed] [Google Scholar]
- Marshall W. F. The cell biological basis of ciliary disease. J. Cell Biol. 2008;180:17–21. doi: 10.1083/jcb.200710085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K., Lefebvre P. A., Kamiya R., Hirono M. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J. Cell Biol. 2004;165:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S. S., et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- O'Toole E. T., Giddings T. H., McIntosh J. R., Dutcher S. K. Three-dimensional organization of basal bodies from wild-type and delta-tubulin strains of Chlamydomonas reinhardtii. Mol. Biol. Cell. 2003;14:2999–3012. doi: 10.1091/mbc.E02-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M. A., et al. AHI1 mutations cause both retinal dystrophy and renal cystic disease in Joubert syndrome. J. Med. Genet. 2006;43:334–339. doi: 10.1136/jmg.2005.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Rosenbaum J. L. Intraflagellar transport and cilia-dependent diseases. Trends Cell Biol. 2002;12:551–555. doi: 10.1016/s0962-8924(02)02410-8. [DOI] [PubMed] [Google Scholar]
- Pelletier L., O'Toole E., Schwager A., Hyman A. A., Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Piperno G., LeDizet M., Chang X. Microtubules containing acetylated α-tubulin in mammalian cells in culture. J. Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringo D. L. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J. Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J. L., Witman G. B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Salisbury J. L. Roots. J. Eukaryot. Microbiol. 1998;45:28–32. doi: 10.1111/j.1550-7408.1998.tb05064.x. [DOI] [PubMed] [Google Scholar]
- Scholey J. M. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Shu X., et al. RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Hum. Mol. Genet. 2005;14:1183–1197. doi: 10.1093/hmg/ddi129. [DOI] [PubMed] [Google Scholar]
- Silflow C. D., LaVoie M., Tam L., Tousey S., Sanders M., Wu W., Borodovsky M., Lefebvre P. A. The Vfl1 protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 2001;153:63–74. doi: 10.1083/jcb.153.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. C., Northey J.G.B., Gary J., Pearlman R. E., Siu K. W. Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J. Proteome Res. 2005;4:909–919. doi: 10.1021/pr050013h. [DOI] [PubMed] [Google Scholar]
- Snell W. J., Dentler W. L., Haimo L. T., Binder L. I., Rosenbaum J. L. Assembly of chick brain tubulin onto isolated basal bodies of Chlamydomonas reinhardtii. Science. 1974;26:357–360. doi: 10.1126/science.185.4148.357. [DOI] [PubMed] [Google Scholar]
- Sorokin S. P. Reconstruction of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- Spektor A., Tsang W. Y., Khoo D., Dynlacht B. D. Cep197 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Tory K., et al. High NPHP1 and NPHP6 mutation rate in patients with Joubert syndrome and nephronophthisis: potential epistatic effect of NPHP6 and AHI1 mutations in patients with NPHP1 mutations. Clin. J. Am. Soc. Nephrol. 2007;18:1566–1575. doi: 10.1681/ASN.2006101164. [DOI] [PubMed] [Google Scholar]
- Valente E. M., et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- Vorobjev I. A., Chentsov Y. S. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 1982;98:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M. P., Wolters D., Yates J. R., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wright R. L., Salisbury J., Jarvik J. W. A nucleus-basal body connector in Chlamydomonas reinhardtii that may function in basal body localization or segregation. J. Cell Biol. 1985;101:1903–1912. doi: 10.1083/jcb.101.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder B. K. Role of primary cilia in the pathogenesis of polycystic kidney disease. J. Am. Soc. Nephrol. 2007;18:1381–1388. doi: 10.1681/ASN.2006111215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.