Abstract
Background
Plasmids are extrachromosomal elements that replicate autonomously, and many can be transmitted between bacterial cells through conjugation. Although the transcription pattern of genes on a plasmid can be altered by a change in host background, the expression range of plasmid genes that will result in phenotypic variation has not been quantitatively investigated.
Results
Using a microarray with evenly tiled probes at a density of 9 bp, we mapped and quantified the transcripts of the carbazole catabolic plasmid pCAR1 in its original host Pseudomonas resinovorans CA10 and the transconjugant P. putida KT2440(pCAR1) during growth on either carbazole or succinate as the sole carbon source. We identified the operons in pCAR1, which consisted of nearly identical transcription units despite the difference in host background during growth on the same carbon source. In accordance with previous studies, the catabolic operons for carbazole degradation were upregulated during growth on carbazole in both hosts. However, our tiling array results also showed that several operons flanking the transfer gene cluster were transcribed at significantly higher levels in the transconjugant than in the original host. The number of transcripts and the positions of the transcription start sites agreed with our quantitative RT-PCR and primer extension results.
Conclusion
Our tiling array results indicate that the levels of transcription for the operons on a plasmid can vary by host background. High-resolution mapping using an unbiased tiling array is a valuable tool for the simultaneous identification and quantification of prokaryotic transcriptomes including polycistronic operons and non-coding RNAs.
Background
Prokaryotic transcriptomes change not only in response to physiological parameters but also to genetic rearrangements mediated by mobile elements. Plasmids are extrachromosomal genetic elements that replicate autonomously, and many can be transmitted between different strains through conjugation. Plasmids provide benefits to their hosts, such as resistance to antibiotics or degradation of recalcitrant aromatic compounds [1]; however, in several cases, the carriage of a large plasmid results in changes in the transcriptome of the host chromosome [2-4]. Similar to the effects of plasmid carriage on the transcriptional network of the host chromosome, differences in host background can alter the transcription patterns of backbone and accessory genes on a plasmid. Many plasmid backbone genes essential for conjugative transfer, replication initiation, and active partitioning are regulated both autogenously and by host factors [5]. Additionally, a number of plasmid-encoded degradative accessory genes that constitute relatively large catabolic operons [6] are coordinately regulated by specific plasmid-encoded transcriptional regulators and chromosomal regulatory elements [7].
The 199,035-bp completely sequenced catabolic plasmid pCAR1, which was originally isolated from Pseudomonas resinovorans CA10, encodes the degradation pathway of carbazole, a nitrogen-containing recalcitrant aromatic compound [8-10]. pCAR1 carries the car and ant operons, both of which are induced by anthranilate, a carbazole intermediate, and which are under the control of the AraC/XylS family activator AntR [11,12]. pCAR1 has been characterized as a self-transmissible and narrow-host-range plasmid that belongs to incompatibility group P-7 (IncP-7); the conjugative transfer of pCAR1 enables the completely sequenced bacterium P. putida KT2440 [13] to grow on carbazole as the sole carbon source [14]. Using an expression microarray of the KT2440 chromosome coupled with pCAR1, we previously analyzed the differential expression of both pCAR1 and the KT2440 chromosome during growth on carbazole and succinate, and found that pCAR1 successfully functioned in the host and affected the chromosomal transcriptome [4]. However, we did not address whether and how the plasmid transcriptome in the original host differed from that in the transconjugant; thus, in this study, we focused on the transcriptomes of pCAR1 in the heterologous host bacteria.
Recent increases in microarray feature density have allowed the construction of tiling arrays, which contain overlapping probes that may be used to target any region of a sequenced genome. Transcriptome analyses using unbiased high-density tiling arrays have been used to detect individual exons of a spliced transcript and multifunctional and extensive transcription from both strands of human DNA [15]. As for prokaryotic transcriptomes, high-density tiling arrays aimed at the genome-wide determination of transcription start sites in E. coli and Caulobacter crescentus contain probes targeting intergenic regions [16,17]. In this study, we used an unbiased tiling array with evenly tiled probes at a 9-bp density to identify the entire operonic structure from transcription initiation to termination. The use of a high-resolution tiling array allowed us to visualize the transcriptome of the plasmid independent of the host background. A comparison of plasmid transcriptomes between the original host and transconjugant revealed similarities and differences in the transcription patterns of the plasmid.
Results and Discussion
Mapping of single-stranded cDNA using the pCAR1 tiling array
Two pCAR1-harboring strains, the original host P. resinovorans CA10 and the transconjugant P. putida KT2440(pCAR1), were grown to the exponential phase using carbazole or succinate as the sole source of carbon. Single-stranded cDNA was synthesized from total culture RNA using random primers. Although the Affymetrix RNA mapping protocol recommends synthesizing second-strand DNA from first-strand cDNA using a DNA polymerase, we hybridized the single-stranded cDNA to the tiling array in order to preserve the strand directionality of the prokaryotic transcriptome.
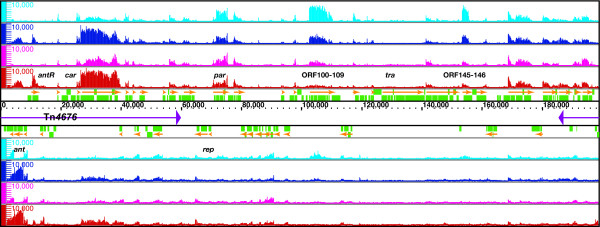
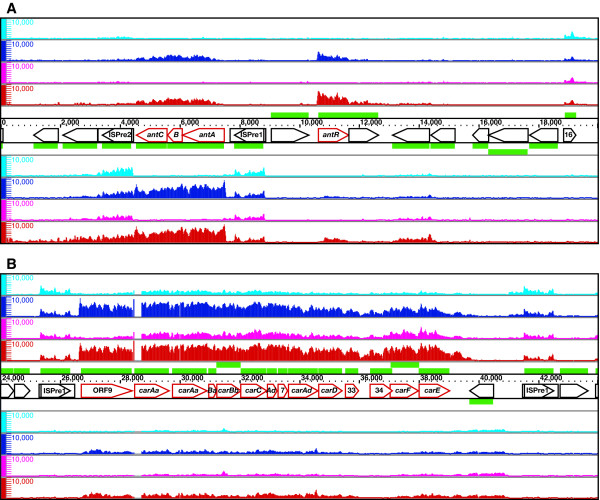
The resulting high-resolution map showed that the pCAR1 transcriptome consisted of identical transcription units regardless of the host background (Figure 1). Using the Integrated Genome Browser (IGB, Affymetrix), we identified a total of 49 transcription units that were transcribed continuously at a signal intensity above 100 (Table 1), including two overlapping operons that were transcribed from multiple initiation sites and three small transcripts identified as non-coding RNAs (ncRNAs). The average length of each transcription unit was 3.1 kb, and 72% (~144.2 kb) of the entire sequence of pCAR1 was transcribed.
Figure 1.
High-resolution mapping of the pCAR1 transcriptome. Single-stranded cDNA from KT2440(pCAR1) grown on succinate (cyan) or carbazole (blue) and from CA10 grown on succinate (magenta) or carbazole (red) was mapped on both strands of pCAR1, which are transcribed from left to right (above) and from right to left (below). The identified transcripts and annotated ORFs are indicated by orange arrows and green rectangles, respectively. The scale bars indicate the signal intensities from 0 to 10,000 on a linear scale.
Table 1.
Transcription units of pCAR1 identified by tiling array analysis at 9-bp resolution.
| Unit | Direction | Transcription start site | Transcription termination site | Length | Genes or ORFs | KT_SUC sense | KT_SUC antisense | KT_CAR sense | KT_CAR antisense | CA_SUC sense | CA_SUC antisense | CA_CAR sense | CA_CAR antisense |
| #1 | - | 4440 | 3210 | 1230 | tnpA3 | 2164 | 428 | 1879 | 378 | 1386 | 236 | 2339 | 386 |
| #2 | - | 7530 | 4440 | 3090 | antA-antC | 86 | 86 | 4410 | 1399 | 136 | 78 | 4883 | 1810 |
| #3 | - | 8830 | 7440 | 1390 | tnpA2 | 1153 | 161 | 1266 | 201 | 929 | 124 | 851 | 100 |
| #4 | + | 10580 | 12310 | 1730 | antR | 54 | 45 | 1271 | 277 | 52 | 25 | 2902 | 430 |
| #5 | - | 14440 | 13310 | 1130 | ORF21 | 310 | 110 | 486 | 175 | 513 | 187 | 1325 | 491 |
| #6 | + | 18860 | 19730 | 870 | ORF16 | 678 | 124 | 395 | 141 | 465 | 84 | 405 | 72 |
| #7 | + | 25320 | 26350 | 1030 | tnpA1 | 1450 | 182 | 1375 | 219 | 1045 | 115 | 1001 | 104 |
| #8A | + | 26620 | 40170 | 13550 | ORF9-carE | 589 | 151 | 4068 | 646 | 1444 | 319 | 5481 | 946 |
| #8B | + | 27760 | 40170 | 12410 | carAa-carE | 645 | 159 | 3919 | 632 | 1552 | 358 | 5488 | 929 |
| #9 | - | 40980 | 39560 | 1420 | ORF37 | 576 | 142 | 658 | 252 | 936 | 182 | 875 | 186 |
| #10 | + | 41500 | 42530 | 1030 | tnpA4 | 1420 | 193 | 1320 | 217 | 1103 | 119 | 943 | 109 |
| #11 | + | 43880 | 44630 | 750 | ORF40 | 577 | 372 | 623 | 335 | 987 | 351 | 706 | 211 |
| #12 | - | 46080 | 44190 | 1890 | ORF42-41 | 898 | 300 | 903 | 460 | 653 | 234 | 637 | 173 |
| #13 | + | 46110 | 48370 | 2260 | tnpRaAa | 296 | 183 | 580 | 257 | 168 | 82 | 129 | 76 |
| #14 | - | 53780 | 50550 | 3230 | tnpS-ORF47 | 839 | 212 | 780 | 277 | 494 | 109 | 409 | 72 |
| #15 | + | 53890 | 55400 | 1510 | tnpT | 261 | 94 | 458 | 145 | 143 | 56 | 137 | 46 |
| #16 | + | 56180 | 58400 | 2220 | ORF53-54 | 719 | 216 | 582 | 222 | 1073 | 255 | 789 | 150 |
| #17 | + | 60210 | 64660 | 4450 | klaA-ORF59 | 356 | 120 | 269 | 105 | 353 | 130 | 368 | 119 |
| #18 | - | 68940 | 64440 | 4500 | ORF61-ORF60 | 217 | 101 | 214 | 93 | 246 | 128 | 267 | 101 |
| #19 | - | 70120 | 69120 | 1000 | repA | 347 | 67 | 235 | 65 | 362 | 82 | 372 | 85 |
| #20A | + | 70060 | 75580 | 5520 | parW-ORF67a | 2441 | 364 | 1316 | 175 | 1372 | 255 | 1257 | 202 |
| #20B | + | 71770 | 75580 | 3810 | parA-ORF67a | 3199 | 506 | 1713 | 231 | 1924 | 319 | 1562 | 221 |
| #21 | + | 77490 | 80400 | 2910 | pmr-ORF72 | 1163 | 269 | 769 | 184 | 752 | 198 | 728 | 181 |
| #22 | - | 81820 | 79480 | 2340 | ORF74-73 | 318 | 174 | 271 | 183 | 388 | 169 | 427 | 150 |
| #23 | - | 84010 | 82170 | 1840 | ORF77-75 | 401 | 88 | 301 | 76 | 294 | 78 | 411 | 87 |
| #24 | + | 85720 | 85870 | 150 | ncRNA1 | 1301 | 543 | 1168 | 470 | 403 | 150 | 1283 | 114 |
| #25 | - | 86510 | 84550 | 1960 | ORF81-79 | 249 | 61 | 291 | 72 | 196 | 48 | 262 | 60 |
| #26 | - | 90880 | 87680 | 3200 | ORF89-85 | 880 | 158 | 406 | 93 | 1051 | 211 | 620 | 128 |
| #27 | - | 92880 | 91500 | 1380 | ORF91 | 283 | 93 | 123 | 94 | 207 | 75 | 146 | 80 |
| #28 | + | 93360 | 94230 | 870 | ORF92 | 569 | 100 | 357 | 64 | 366 | 86 | 479 | 78 |
| #29 | - | 95580 | 94380 | 1200 | ORF93 | 541 | 187 | 404 | 149 | 482 | 152 | 559 | 169 |
| #30 | + | 97170 | 98100 | 930 | ORF95a | 848 | 146 | 627 | 121 | 607 | 110 | 587 | 107 |
| #31 | + | 102560 | 110380 | 7820 | ORF100-109 | 4564 | 554 | 1588 | 169 | 667 | 80 | 443 | 59 |
| #32 | - | 117040 | 113160 | 3880 | ORF115-111 | 264 | 76 | 129 | 58 | 302 | 93 | 165 | 70 |
| #33 | + | 117170 | 119180 | 2010 | ssb-recT | 719 | 103 | 249 | 33 | 208 | 34 | 114 | 19 |
| #34 | - | 119470 | 119330 | 140 | ncRNA2 | 806 | 725 | 656 | 545 | 242 | 254 | 237 | 168 |
| #35 | + | 121840 | 121950 | 110 | ncRNA3 | 1847 | 573 | 1562 | 472 | 581 | 125 | 694 | 129 |
| #36 | + | 123060 | 124110 | 1050 | ORF120 | 458 | 60 | 598 | 62 | 189 | 39 | 400 | 47 |
| #37 | + | 127040 | 140790 | 13750 | traG-ORF131 | 125 | 46 | 73 | 44 | 191 | 70 | 150 | 63 |
| #38 | + | 141370 | 152460 | 11090 | ORF134-143 | 252 | 150 | 204 | 188 | 235 | 120 | 187 | 118 |
| #39 | + | 153440 | 155620 | 2180 | ORF145-146 | 4464 | 645 | 2126 | 311 | 897 | 149 | 801 | 115 |
| #40 | + | 155840 | 161570 | 5730 | traF-trhG | 181 | 134 | 135 | 212 | 180 | 103 | 120 | 107 |
| #41 | - | 165130 | 161590 | 3540 | ORF154-150 | 193 | 60 | 180 | 102 | 217 | 63 | 222 | 47 |
| #42 | + | 168730 | 176860 | 8130 | ORF159-167 | 1295 | 263 | 1296 | 272 | 1082 | 214 | 1148 | 181 |
| #43 | - | 180180 | 179390 | 790 | tnpRb | 283 | 68 | 439 | 113 | 144 | 28 | 223 | 30 |
| #44 | + | 180340 | 185550 | 5210 | ORF170-175 | 717 | 165 | 899 | 218 | 492 | 106 | 765 | 153 |
| #45 | + | 185870 | 189340 | 3470 | tnpAcC | 310 | 307 | 1004 | 536 | 76 | 93 | 853 | 290 |
| #46 | + | 190420 | 192860 | 2440 | ORF179-181 | 746 | 489 | 640 | 349 | 664 | 379 | 608 | 308 |
| #47 | + | 193300 | 195590 | 2290 | ORF182-184 | 1451 | 227 | 1167 | 208 | 1942 | 330 | 1537 | 207 |
The transcription units of pCAR1 and their median signal intensities on both the sense and antisense strands are indicated; KT_SUC, KT2440(pCAR1) grown on succinate; KT_CAR, KT2440(pCAR1) grown on carbazole; CA_SUC, CA10 grown on succinate; CA_CAR, CA10 grown on carbazole. The positions of the transcription start and stop sites correspond to the pCAR1 nucleotide sequence [GenBank:AB088420].
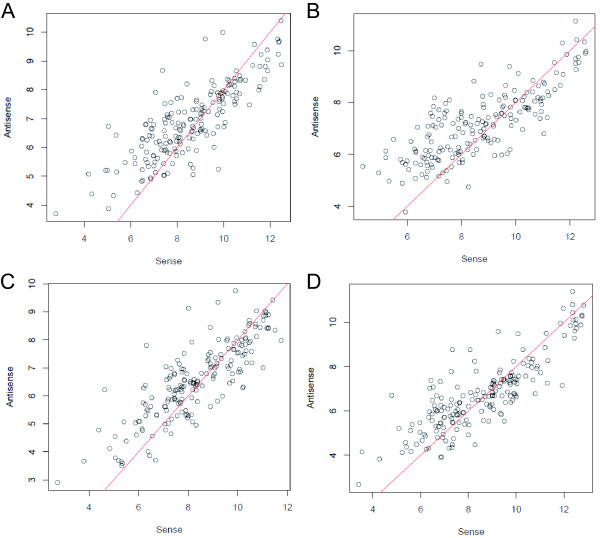
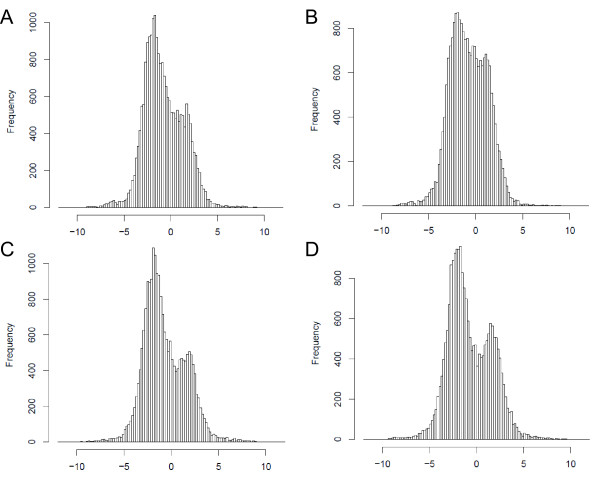
Unexpectedly, we observed numerous signals on the antisense strand in the coding regions. These signals likely represent experimental artifacts generated by secondary mispriming during reverse transcription using random primers [18,19] since no signal was detect on the antisense strand by hybridization of cDNA synthesized using strand-specific primers (data not shown). To estimate how much of the single-stranded cDNA synthesized using random primers contained these artifacts, the median signal intensities were calculated from probes located within the pCAR1 genes. The correlation coefficients between the sense and antisense values on a log2 scale were 0.79–0.84 (Figure 2). The plots were approximated to the line y = x-2, indicating that the intensity of the sense signals was approximately four times higher than that of the antisense signals. In addition, the log2 ratios of the signal intensities for each probe on one strand to those on the other strand were frequently 2 and -2 (Figure 3), suggesting that ~20% of the signals detected on the antisense strand in the coding regions represent artifacts. Therefore, we subsequently analyzed only the sense signal intensities.
Figure 2.
Scatterplots of the median signal intensities (log2) within a sense region and its antisense region. (A) KT2440(pCAR1) grown on succinate; r = 0.82 (n = 190). (B) KT2440(pCAR1) grown on carbazole; r = 0.79 (n = 190). (C) CA10 grown on succinate; r = 0.83 (n = 190). (D) CA10 grown on carbazole; r = 0.84 (n = 190). Red lines, y = x-2.
Figure 3.
Histograms showing the log2 ratio of the signal intensities for the sense and antisense strands. The frequency of the signal intensity ratios for a total of 21,875 probe pairs was plotted. All four histograms have two peaks (at 2 and -2), indicating that most of the ratios were 4 or 0.25. (A) KT2440(pCAR1) grown on succinate. (B) KT2440(pCAR1) grown on carbazole. (C) CA10 grown on succinate. (D) CA10 grown on carbazole.
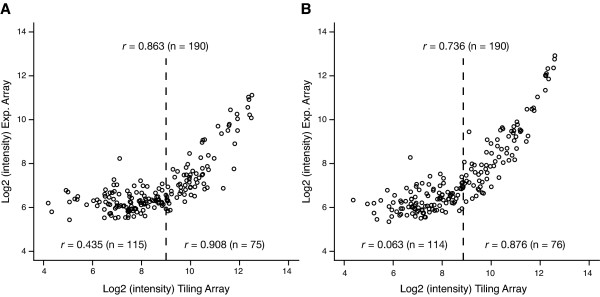
To estimate the sensitivity of the tiling array, the median signal intensities of the pCAR1 genes in KT2440(pCAR1) were compared to those calculated as part of our previous expression array analysis [4]. Overall, the signal intensities of the pCAR1 genes, especially the frequently transcribed genes, exhibited a strong correlation between the two microarray platforms (Figure 4). However, a significant decrease in the Pearson correlation coefficient was detected in the low-signal-intensity plots, which were distributed more widely in the tiling array analysis than in the expression array analysis, indicating that tiling arrays are superior in terms of sensitivity to traditional gene-centered microarrays.
Figure 4.
Comparison of the dynamic range between the tiling array and the expression array platforms. The plots represent the median signal intensities (log2) of 190 pCAR1 genes calculated from the tiling and the expression arrays. The correlation coefficients are for the signal intensities of the overall genes, highly transcribed genes (> 29 in the tiling array), and weakly transcribed genes (< 29).
Transcription pattern of the pCAR1 accessory genes
pCAR1 carries a 72.8-kb class II transposon, Tn4676 (1–59, 548 and 185, 774–199, 035 on pCAR1), which confers the ability to degrade carbazole [20]. Within Tn4676, the ant operon is transcribed from the inducible promoter Pant [11], whereas the car operon is transcribed from the inducible promoter Pant upstream of open reading frame (ORF) 9 and additionally from the constitutive promoter PcarAa within the coding region of ORF9 [12]. The AraC/XylS family transcriptional regulator AntR, which is encoded on Tn4676, activates the Pant promoter in response to anthranilate, an intermediate of the carbazole degradation pathway. The transcription of antR originates from an RpoN-dependent promoter and is induced during growth on carbazole [4]. As expected, our tiling array results indicated that growth on carbazole strongly induced these catabolic operons in both hosts compared to growth on succinate (Figure 5A and 5B). The 3.1- and 1.7-kb transcripts of antABC and antR were significantly induced during growth on carbazole (Figure 5A). The ~13-kb transcript of the car operon was also detected; it was induced during growth on carbazole and constitutively produced during growth on succinate (Figure 5B). Intriguingly, the level of transcription of the lower car operon, which is composed of carFE, was apparently higher in CA10 than in KT2440(pCAR1) regardless of the carbon source. The transcription start sites of antA, antR, and ORF9 indicated by our tiling array (Figure 5A and 5B) agreed with those identified through primer extension analysis [4,11]. Although our previous data showed constitutive transcription of the car operon from 385-bp upstream of carAa [12], our tiling array analysis detected only slightly constitutive transcription in both strains during growth on succinate and could not identify the transcription start site (Figure 5B).
Figure 5.
High-resolution mapping of the catabolic operons induced during growth on carbazole, the ant operon and antR (A), and the car operon (B). Single-stranded cDNA from KT2440(pCAR1) grown on succinate (cyan) or carbazole (blue) and from CA10 grown on succinate (magenta) or carbazole (red) was mapped on both strands of pCAR1, which are transcribed from left to right (above) and from right to left (below). Pentagons represent the pCAR1 genes and their transcriptional directions; the gene names or ORF numbers are indicated therein. The scale bars indicate the signal intensities from 0 to 10,000 on a linear scale. pCAR1 carries duplicate carAa genes [9]; several probes within the first copy of carAa were deleted by the Affymetrix Custom Array Design Program.
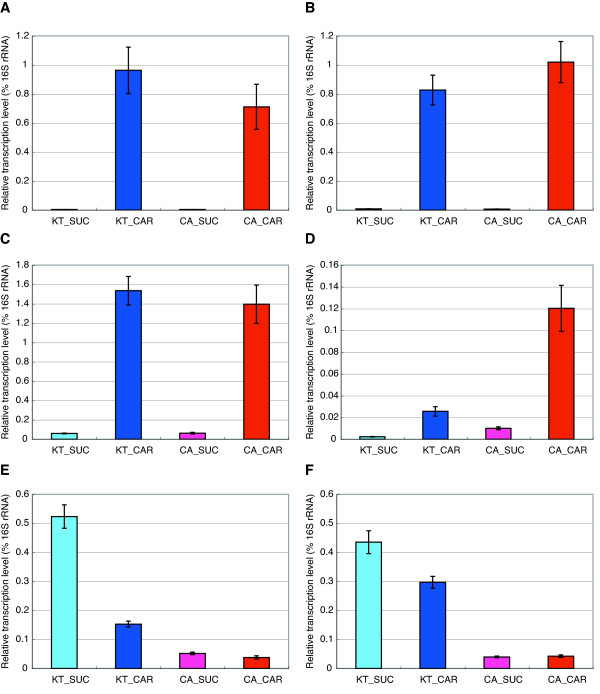
To verify the transcription levels estimated by our tiling array analysis, we performed quantitative RT-PCR and calculated the relative abundance of the pCAR1 mRNAs normalized by 16S rRNA. During growth on carbazole, the transcription of antA and antR was significantly increased by ~200- and ~100-fold to the same level in both hosts (Figure 6A and 6B). The transcription of the upper car operon, which originates from the dual promoters, was also upregulated by ~20-fold with a negligible difference between the hosts (Figure 6C). In contrast, the transcription of the lower car operon was significantly higher in CA10 than in KT2440(pCAR1) (Figure 6D). These results suggest that the car operon possesses additional promoters that are preferentially activated in the original host.
Figure 6.
Quantification of antA (A), antR (B), ORF9 (C), carF (D), ORF100 (E), and ORF145 (F) transcription. Quantitative RT-PCR was performed using single-stranded cDNA from KT2440(pCAR1) grown on succinate (cyan) or carbazole (blue) and from CA10 grown on succinate (magenta) or carbazole (red). The relative amount of each pCAR1 transcript to 16S rRNA (%) is indicated.
Tn4676 encodes the transposase TnpAc and its repressor TnpC at one end and the cointegrate resolvase TnpST at the other. The transcription of tnpAc was induced during growth on carbazole in both hosts, consistent with previous microarray results [4], but the basal level of transcription for tnpAc was higher in KT2440(pCAR1) than in CA10 during growth on succinate (Table 1). The levels of the divergent tnpS and tnpT transcripts were unaffected by the growth conditions and were slightly higher in KT2440(pCAR1) (Table 1). It is noteworthy that Tn4676 contains three identical ISPre1 insertion sequences, two of which are located on either end of the car operon, with the third located upstream of the ant operon [10]. Our tiling array data revealed nearly identical transcription patterns within these insertion sequences (Figure 5A and 5B). This indicates the equivalent detection sensitivities of the contiguous probes, because transcripts of ISPre1 were detected equally on both strands using different probes shifted by several nucleotides.
Transcription pattern of IncP-7 plasmid backbone
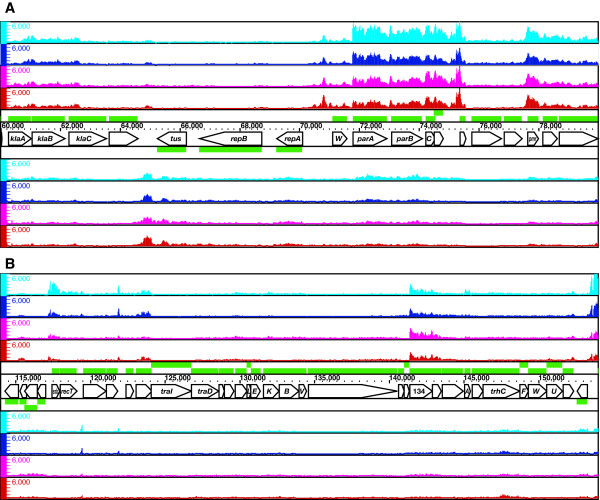
The pCAR1 plasmid contains divergent repA and parWABC genes for its replication and active partitioning, which are conserved among IncP-7 plasmids [10,21,22]. Our tiling array analysis showed that repA was transcribed at a low level and that the level of transcription was unaffected by the host background or carbon source (Figure 7A and Table 1). The transcription of the par gene cluster originated from parW and parA, although the promoter activity of parA was much higher than that of parW (Figure 7A).
Figure 7.
High-resolution mapping of the IncP-7 plasmid backbone, rep and par (A) and tra (B). Single-stranded cDNA from KT2440(pCAR1) grown on succinate (cyan) or carbazole (blue) and from CA10 grown on succinate (magenta) or carbazole (red) was mapped on both strands of pCAR1, which are transcribed from left to right (above) and from right to left (below). Pentagons represent the pCAR1 genes and their transcriptional directions; the gene names or ORF numbers are indicated therein. The scale bars indicate the signal intensities from 0 to 6,000 on a linear scale.
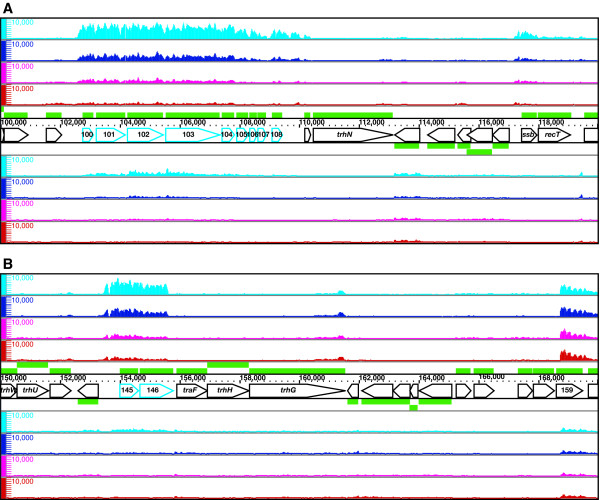
A large gene cluster encoding the conjugative transfer apparatus of IncP-7 plasmids has been reported only for the self-transmissible plasmid pCAR1 [12]. The host range and conjugative transfer frequency of pCAR1 depends on the recipient and donor species; pCAR1 efficiently transfers between species with the same genetic background [23]. High-resolution mapping revealed that the overall transfer gene cluster was transcribed at a low level (Figure 7B); however, two operons flanking the transfer gene cluster (from ORF100 to ORF109 and from ORF145 to ORF146, respectively) were transcribed at significantly higher levels in the transconjugant strain KT2440(pCAR1) than in the original host strain CA10 (Figure 8A and 8B).
Figure 8.
High-resolution mapping of ORF100-108 (A) and ORF145-146 (B), differentially transcribed operons between KT2440(pCAR1) and CA10. Single-stranded cDNA from KT2440(pCAR1) grown on succinate (cyan) or carbazole (blue) and from CA10 grown on succinate (magenta) or carbazole (red) was mapped on both strands of pCAR1, which are transcribed from left to right (above) and from right to left (below). Pentagons represent the pCAR1 genes and their transcriptional directions; the gene names or ORF numbers are indicated therein. The scale bars indicate the signal intensities from 0 to 10,000 on a linear scale.
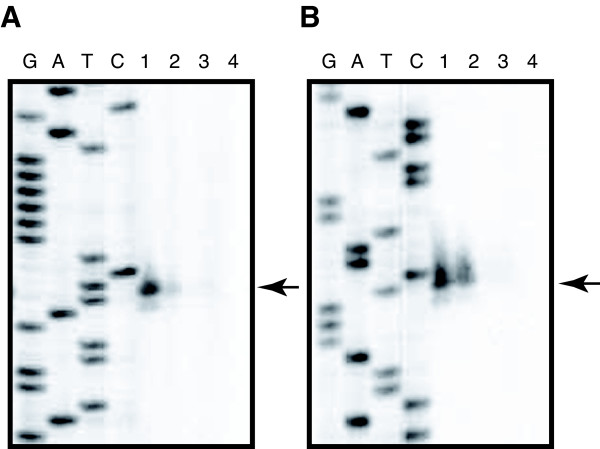
Quantitative RT-PCR verified that the transcription levels of ORF100 and ORF145 were significantly higher in KT2440(pCAR1) than in CA10 and during growth on succinate than on carbazole (Figure 6E and 6F). To characterize the promoters of ORF100 and ORF145, we performed a primer extension analysis using the same total RNA used in our tiling array analysis. The transcription start sites were mapped to 141-nt and 522-nt upstream of the translation start sites of ORF100 and ORF145, respectively (Figure 9A and 9B), in agreement with our tiling array data. The signal intensities of the primer extension products also corresponded to our quantitative RT-PCR results. However, we were unable to identify a consensus motif between the promoter regions of ORF100 and ORF145. These results suggest that the two operons are regulated by different mechanisms in the two hosts.
Figure 9.
Mapping of the transcription start sites for ORF100 (A) and ORF145 (B). Primer extension was performed using equal amounts of total RNA from KT2440(pCAR1) grown on succinate (lane 1) or carbazole (lane 2) and from CA10 grown on succinate (lane 3) or carbazole (lane 4). Lanes G, A, T, and C indicate the sequence ladder obtained using the same primer.
A search of the ACLAME database http://aclame.ulb.ac.be[24] indicated that the genes within ORF100-103 are conserved in the IncH plasmid R478 [25], the IncT plasmid Rts1 [26], and the integrating conjugative elements SXT [27] and R391 [28]. ORF101 encodes a MoxR-like AAA+ ATPase with a C-terminal CbbQ domain that is associated with various cellular activities [29]. Many genes in the AAA+ family have been found in close proximity to genes that encode proteins with von Willebrand factor type A (VWA) domains [29], and we found that ORF103 actually encodes a VWA protein. The VWA domain is a well-studied domain associated with cell adhesion, extracellular matrix proteins, and integrin receptors [30]. ORF145 and ORF146, which encode a putative nickase and primase, respectively, are located upstream of the traF-trhH-trhG gene cluster, which is involved in pilus assembly [23]; however, they were transcribed separately. Our high-resolution map showed a significant decrease in the transcription of ORF146 at approximately nucleotide position 155,630 (Figure 8B), raising the possibility that ORF146 is dysfunctional or at least rarely transcribed. Since the conjugative host range and transfer frequency of pCAR1 are thought to depend on the donor strain [23], these findings suggest that ORF100-109 and ORF145-146 are involved in the conjugative transfer of pCAR1 and that the transcription levels of these operons in donor cells may affect the behavior of pCAR1.
Conclusion
Using a tiling array, we identified the complete transcriptome of pCAR1 and demonstrated that the level of transcription of several operons was host-dependent. Many of the genes carried on bacterial plasmids have not been adequately annotated or functionally characterized; thus, functional genomic approaches are necessary to identify novel genes with important roles in the relationship between plasmids and their hosts. This study demonstrates that the plasmid transcriptome is affected by the host background, while recent studies have indicated that the carriage of a large plasmid results in a change in the transcriptome of the host chromosome [2-4]. Therefore, we conclude that conjugative transfer results in bidirectional alterations in the transcriptional networks of the plasmid and host chromosome.
The unbiased nature of tiling arrays allows the simultaneous identification and quantification of the prokaryotic transcriptome including polycistronic operons and non-coding RNAs. The transcription start sites identified by tiling array analysis may be verified by primer extension analysis. The number of transcripts indicated by quantitative RT-PCR is comparable among different growth conditions and genetic backgrounds. Tiling array technology is a powerful tool for the analysis of prokaryotic transcriptomes, and it may be used to determine the complete operonic structure of a bacterial genome.
Methods
Bacterial strains and growth conditions
Pseudomonas resinovorans CA10 [8] and P. putida KT2440(pCAR1) [14] were grown in Luria broth and nitrogen-plus mineral medium-4 (NMM-4) as described previously [4]. For RNA extraction, 100 ml of NMM-4 supplemented with 1.0 mg/ml of carbazole or sodium succinate were inoculated with cells from an overnight culture in Luria broth to obtain an initial optical density at 600 nm (OD600) of 0.05. The cells were grown at 30°C on a rotary shaker set at 120 rpm with monitoring of the OD600 or the number of colony-forming units (CFU)/ml to the exponential phase.
Tiling array design
A high-density oligonucleotide microarray covering all 199,035-bp of the pCAR1 plasmid [GenBank:AB088420] was designed using the CustomExpress Array Program (Affymetrix, Santa Clara, CA). The array contained a total of 88,460 probes consisting of pairs of 25-mer perfect match (PM) probes overlapping at 9-nt offsets, and corresponding mismatch (MM) probes with a one-base mismatch at the thirteenth nucleotide.
RNA preparation, labeling and hybridization
Two biological replicates were independently prepared. Approximately 1 × 109 cells of each strain at the exponential phase were mixed with RNA Protect Bacteria Reagent (Qiagen, Valencia, CA) as recommended by the manufacturer. Total RNA was extracted using NucleoSpin RNA II (Macherey-Nagel GmbH & Co. KG, Düren, Germany). The eluted RNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI) at 37°C for 30 min. Following DNase inactivation by the addition of the provided stop reagent and subsequent incubation at 65°C for 10 min, the total RNA was repurified using NucleoSpin RNA Clean-Up (Macherey-Nagel).
Single-stranded cDNA was synthesized in 60 μl of 1× First Strand Buffer (Invitrogen, Carlsbad, CA) containing 12 μg of total RNA, 750 ng of random primers (Invitrogen), 1,500 U of SuperScript II (Invitrogen), 60 U of RNaseOUT (Invitrogen), 10 mM DTT (Invitrogen), 0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dGTP, 0.4 mM dTTP, and 0.1 mM dUTP (Roche Applied Science, Mannheim, Germany). After denaturation of the RNA and random primers at 70°C for 10 min and annealing at 25°C for 10 min, the remaining reagents were added, and the reaction mixture was incubated at 25°C for 10 min, 37°C for 60 min, 42°C for 60 min, and 70°C for 10 min. Following cDNA synthesis, the template RNA was degraded with one-third volume of 1N NaOH at 65°C for 30 min; one-third volume of 1N HCl was added to neutralize the reaction mixture prior to cleanup. The cDNA was purified using a QIAquick PCR Purification Kit (Qiagen).
The cDNA was fragmented and labeled using a GeneChip WT Double-Stranded DNA Terminal Labeling Kit (Affymetrix). The purified single-stranded cDNA (~5 μg) was fragmented in 48 μl of 1× cDNA Fragmentation Buffer containing 15 U of Uracil DNA Glycosylase (UDG) and 225 U of Apurinic/Apyrimidinic Endonuclease 1 (APE1) at 37°C for 60 min followed by incubation at 93°C for 2 min; the majority of the fragmented cDNA was 50–200 bp in length. The fragmented cDNA was labeled in 60 μl of 1× TdT Buffer containing 60 U of Terminal Deoxynucleotidyl Transferase (TdT) and 0.083 mM GeneChip DNA Labeling Reagent at 37°C for 60 min followed by incubation at 70°C for 2 min.
The labeled cDNA was mixed with 50 pM control oligonucleotide B2 (Affymetrix), 1× Hybridization Mix (Affymetrix), and 7% DMSO in a total volume of 200 μl and denatured at 95°C for 5 min. For each array, 130 μl of the hybridization cocktail were hybridized at 45°C for 16 h with a rotation rate of 60 rpm using a GeneChip Hybridization Oven 640 (Affymetrix). The chips were then washed and stained using a Hybridization, Wash, and Stain Kit (Affymetrix) according to the FlexFS450-0002 protocol for GeneChip Fluidics station 450 (Affymetrix). Signals were detected using GeneChip Scanner 3000 7G (Affymetrix).
Tiling array data analysis
The signal intensity for each probe was computed using Affymetrix Tiling Analysis Software v1.1 (TAS), which uses non-parametric quantile normalization and a Hodges-Lehmann estimator for fold-enrichment (Affymetrix Tiling Array Software v1.1 User's Guide). The intensities were linearly scaled so that the median was 100. The PM and MM intensity pairs were mapped to the plasmid genome. For each position to which a probe pair was mapped, a dataset was generated consisting of all pairs mapping within a window of ± 30 bp, which defines the number of bases extending from the position being analyzed so that every probe in the 61-bp region was included in the signal and p-value analysis. The pseudomedian was calculated using a sliding window across the genome as an estimate of the signal intensity per probe position. The significance of the signal intensity was calculated with p < 10-3 taken as the threshold.
The intensities from two independent CEL files were analyzed by the MVA plot of TAS; the correlation coefficients between the biological replicates were > 0.95. Two CEL files from independent replicates were converted into BAR files using two separate BPMAP files for the forward and reverse strands. The data were visualized using the IGB package (Affymetrix). We used the median signal intensities of the interior probes as an indicator of the expression level of each gene.
Quantitative RT-PCR
Quantitative RT-PCR was performed using the ABI 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA) as described previously [4]. The primers used for quantitative RT-PCR (Additional file 1) were designed using the Primer3 program [31]. All products were between 100 and 150 bp in length. For normalization, 16S rRNA was used as an internal standard. The univ16S-F and univ16S-R primer set used to measure the transcription of 16S rRNA was designed based on the 16S rRNA sequences from P. putida KT2440 [GenBank:AE015451] and P. resinovorans CA10 [GenBank:AB047273]. All reactions were performed a minimum of three times, and the data were normalized using the average of the internal standard.
Primer extension
Primer extension analysis was performed using a Li-Cor model 4200L-2 auto-DNA sequencer as described previously [4]. We used the IRD800-labeled primer ORF100-R2 5'-AGGATTCGAGTTCACGGGTA-3' or ORF145-R3 5'-ACGCGGTCAGCACTTTAACT-3' (Aloka, Ltd., Tokyo, Japan), which anneals to the region from +57 to +76 of ORF100 or +353 to +372 of ORF145 relative to the annotated translation start site. A sequence ladder was obtained using the same primer and the appropriate cover clone of pCAR1 [10] as template.
Accession number
The data were deposited in NCBI's Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE10862.
Authors' contributions
MM conceived the study, carried out the experiments, and drafted the manuscript. MM, HNi, and MS analyzed the data. HNi, MS, HY, and HNo assisted in the preparation of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Primers used for quantitative RT-PCR. Nucleotide sequences of primers used for quantitative RT-PCR.
Acknowledgments
Acknowledgements
This work was supported by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and a Grant-in-Aid (Hazardous Chemicals) from the Ministry of Agriculture, Forestry, and Fisheries of Japan (HC-07-2325-3) to H. Nojiri. MM was supported by Research fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Contributor Information
Masatoshi Miyakoshi, Email: mmiyakoshi@ige.tohoku.ac.jp.
Hiromi Nishida, Email: hnishida@iu.a.u-tokyo.ac.jp.
Masaki Shintani, Email: smasaki@os.rim.or.jp.
Hisakazu Yamane, Email: ayamane@mail.ecc.u-tokyo.ac.jp.
Hideaki Nojiri, Email: anojiri@mail.ecc.u-tokyo.ac.jp.
References
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Harr B, Schlötterer C. Gene expression analysis indicates extensive genotype-specific crosstalk between the conjugative F-plasmid and the E. coli chromosome. BMC Microbiol. 2006;6:80. doi: 10.1186/1471-2180-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M, Fookes M, Ivens A, Mangen MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- Miyakoshi M, Shintani M, Terabayashi T, Kai S, Yamane H, Nojiri H. Transcriptome analysis of Pseudomonas putida KT2440 harboring the completely sequenced IncP-7 plasmid pCAR1. J Bacteriol. 2007;189:6849–6860. doi: 10.1128/JB.00684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM. Transcription regulatory circuits in bacterial plasmids. Biochem Soc Trans. 2006;34:1072–1074. doi: 10.1042/BST0341072. [DOI] [PubMed] [Google Scholar]
- Williams PA, Jones RM, Zylstra G. Genomics of catabolic plasmid. In: Ramos JL, editor. Pseudomonas. Vol. 1. Kluwer Academic/Plenum Publishers; 2004. pp. 165–195. [Google Scholar]
- Ramos JL, Marqués S, Timmis KN. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu Rev Microbiol. 1997;51:341–372. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- Ouchiyama N, Zhang Y, Omori T, Kodama T. Biodegradation of carbazole by Pseudomonas spp. CA06 and CA10. Biosci Biotechnol Biochem. 1993;57:455–460. [Google Scholar]
- Nojiri H, Sekiguchi H, Maeda K, Urata M, Nakai S, Yoshida T, Habe H, Omori T. Genetic characterization and evolutionary implications of a car gene cluster in the carbazole degrader Pseudomonas sp. strain CA10. J Bacteriol. 2001;183:3663–3679. doi: 10.1128/JB.183.12.3663-3679.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Nojiri H, Shintani M, Yoshida T, Habe H, Omori T. Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J Mol Biol. 2003;326:21–33. doi: 10.1016/S0022-2836(02)01400-6. [DOI] [PubMed] [Google Scholar]
- Urata M, Miyakoshi M, Kai S, Maeda K, Habe H, Omori T, Yamane H, Nojiri H. Transcriptional regulation of the ant operon, encoding two-component anthranilate 1,2-dioxygenase, on the carbazole-degradative plasmid pCAR1 of Pseudomonas resinovorans strain CA10. J Bacteriol. 2004;186:6815–6823. doi: 10.1128/JB.186.20.6815-6823.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakoshi M, Urata M, Habe H, Omori T, Yamane H, Nojiri H. Differentiation of carbazole catabolic operons by replacement of the regulated promoter via transposition of an insertion sequence. J Biol Chem. 2006;281:8450–8457. doi: 10.1074/jbc.M600234200. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VAP, Fouts DE, Gill SR, Pop M, Holmes M, Brinkac L, Beanan M, DeBoy RT, Daugherty S, Kolonay J, Madupu R, Nelson W, White O, Peterson J, Khouri H, Hance I, Chris Lee P, Holtzapple E, Scanlan D, Tran K, Moazzez A, Utterback T, Rizzo M, Lee K, Kosack D, Moestl D, Wedler H, Lauber J, Stjepandic D, Hoheisel J, Straetz M, Heim S, Kiewitz C, Eisen J, Timmis KN, Düsterhöft A, Tümmler B, Fraser CM. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- Shintani M, Yano H, Habe H, Omori T, Yamane H, Tsuda M, Nojiri H. Characterization of the replication, maintenance, and transfer features of the IncP-7 plasmid pCAR1, which carries genes involved in carbazole and dioxin degradation. Appl Environ Microbiol. 2006;72:3206–3216. doi: 10.1128/AEM.72.5.3206-3216.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Selinger DW, Cheung KJ, Mei R, Johansson EM, Richmond CS, Blattner FR, Lockhart DJ, Church GM. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, Tan MH, Hillson NJ, Hu P, Shapiro L, McAdams HH. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, Tammana H, Gingeras TR. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosome 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Shintani M, Yoshida T, Habe H, Omori T, Nojiri H. Large plasmid pCAR2 and class II transposon Tn4676 are functional mobile genetic elements to distribute the carbazole/dioxin-degradative car gene cluster in different bacteria. Appl Microbiol Biotechnol. 2005;67:370–382. doi: 10.1007/s00253-004-1778-0. [DOI] [PubMed] [Google Scholar]
- Li W, Shi J, Wang X, Han Y, Tong W, Ma L, Liu B, Cai B. Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene. 2004;336:231–240. doi: 10.1016/j.gene.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Yano H, Garruto CE, Sota M, Ohtsubo Y, Nagata Y, Zylstra GJ, Williams PA, Tsuda M. Complete sequence determination combined with analysis of transposition/site-specific recombination events to explain genetic organization of IncP-7 TOL plasmid pWW53 and related mobile genetic elements. J Mol Biol. 2007;369:11–26. doi: 10.1016/j.jmb.2007.02.098. [DOI] [PubMed] [Google Scholar]
- Shintani M, Habe H, Tsuda M, Omori T, Yamane T, Nojiri H. Recipient range of IncP-7 conjugative plasmid pCAR2 from Pseudomonas putida HS01 is broader than from other Pseudomonas strains. Biotechnol Lett. 2005;27:1847–1853. doi: 10.1007/s10529-005-3892-1. [DOI] [PubMed] [Google Scholar]
- Leplae R, Hebrant A, Wodak SJ, Toussaint A. ACLAME: a classification of mobile genetic elements. Nucleic Acid Res. 2004;32:D45–49. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour MW, Thompson NR, Sanders M, Parkhill J, Taylor DE. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid. 2004;52:182–202. doi: 10.1016/j.plasmid.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Murata T, Ohnishi M, Ara T, Kaneko J, Han C-G, Li YF, Takashima K, Nojima H, Nakayama K, Kaji A, Kamio Y, Miki T, Mori H, Ohtsubo E, Terawaki Y, Hayashi T. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J Bacteriol. 2002;184:3194–3202. doi: 10.1128/JB.184.12.3194-3202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol. 2002;184:4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böltner D, MacMahon C, Pembroke T, Strike P, Osborn AM. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J Bacteriol. 2002;184:5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider J, Houry WA. MoxR AAA+ ATPases: a novel family of molecular chaperones? J Struct Biol. 2006;156:200–209. doi: 10.1016/j.jsb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. 2002;13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editor. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for quantitative RT-PCR. Nucleotide sequences of primers used for quantitative RT-PCR.