Abstract
Examination of the EST database of the light organ of the Hawaiian bobtail squid Euprymna scolopes revealed a sequence with similarity to complement C3. RACE yielded the full open reading frame of this protein. Analysis of the resultant sequence revealed that Es-C3 (E. scolopes-C3) has conserved residues and domains known to be critical for C3 function. The gene encoding C3 was expressed in all tissues tested, indicating that its expression is widely distributed throughout the animal’s body. Immunocytochemistry using an antibody against Es-C3 revealed that the protein is produced principally in the apical surfaces of epithelial cells. The finding of the gene encoding C3 in this mollusk extends the occurrence of this molecule to the lophotrochozoans, demonstrating that complement genes occur in all major branches of the animal kingdom.
Keywords: innate immunity, invertebrate immunity, Lophotrochozoa, mollusk, cephalopod, symbiosis, immunocytochemistry
1. Introduction
The complement system is a group of serum and membrane-associated proteins that plays an essential role in innate immune defenses. In vertebrates, the complement system not only serves as a bridge between the innate and adaptive responses, but several of its components also have been implicated in the enhancement and modulation of the adaptive immune response (reviewed in [1–3]). Among the known proteins of the complement system, component C3 is the central molecule where all known activation pathways (classical, lectin, and alternative) converge [4, 5]. Complement component C3 is a large, soluble polypeptide that belongs to the superfamily of thioester-containing proteins (TEPs) that also includes vertebrate complement components C4 and C5, the invertebrate and vertebrate alpha2-macroglobulins, and the thioester-containing proteins found in a variety of invertebrates (reviewed in [6]). Functional studies have shown that after the proteolytic activation of C3, the resulting C3a fragment is involved in inflammatory responses, while the C3b fragment serves as either an effective opsonin or as a recruiter of other proteins that will ultimately mediate the lysis of the foreign microorganism via perforin-like proteins or the formation of a membrane attack complex (MAC) [5, 7].
Since their discovery, complement molecules have been found and studied in a variety of organisms, principally vertebrates. This focus led to the previous misconception that complement only existed in the vertebrates and their chordate relatives. In 1998, a C3 ortholog was discovered in an echinoderm, the sea urchin Strongylocentrotus purpuratus [8]. This discovery demonstrated the presence of complement in invertebrates within the deuterostome clade, one of the three superphyla of bilaterian animals. More recently, complement C3-like proteins were identified in the horseshoe crab Carcinoscorpius rotundicauda, a member of the ecdysozoan superphylum [9], and in the cnidarian Swiftia exserta [10]. These findings demonstrated that the origin and evolution of the complement system dates back to the earliest radiations of the animal kingdom (for reviews see [11–15]. In all of these divergent groups, complement C3 proteins have several conserved structural and functional motifs, which are now considered characteristic of these molecules [5, 16]. Specifically, the C3 molecules are defined as those having: 1) an anaphylatoxin domain, which is released upon activation of C3 and is involved in the inflammatory response; 2) a thioester domain, responsible for the covalent attachment of the C3b fragment to its target; and finally, 3) a C345C domain located at the C-terminal region of the molecule [14]. One major group of organisms in which members of the complement system has not been identified is the third superphylum of bilaterians, the lophotrochozoans, which includes the mollusks (e.g., snails, clams, squid), annelids (e.g., earthworms, leeches), and flatworms.
Our laboratory focuses on the study of various aspects of the mutualistic relationship between the sepiolid squid Euprymna scolopes and the marine luminous bacterium Vibrio fischeri [17, 18]. In this beneficial symbiosis, the bacteria reside within a specialized structure called the light-organ, where they live in close contact with the host epithelia. The molecular mechanism(s) that the host E. scolopes uses to recognize, acquire, and maintain V. fischeri within a specific location, and without succumbing to pathogenic infection, are still largely unknown. However, studies of this system thus far suggest a well developed molecular communication between the host’s immune system and its symbionts including: the recognition of bacterial products [19, 20]; host production of reactive oxygen compounds [21–23]; host mucus secretion [24, 25]; and the migration and change in gene expression of squid hemocytes [26, 27].
In the present study, we identify and characterize a complement component C3 ortholog in E. scolopes. This report is the first to demonstrate the presence of a member of the complement family in the lophotrochozoans, thus extending the occurrence of complement throughout all branches of the animal kingdom.
2. Materials and methods
2.1 General procedures
Adult E. scolopes were collected from shallow sandy flats in Oahu, Hawaii, and breeding colonies were maintained in artificial seawater (ASW) in 450 L-recirculating aquaria. For studies with newly hatched juveniles, the animals were rinsed immediately upon hatching in filtered (0.22 μm) ASW and placed in individual glass vials containing 3 ml of ASW (nonsymbiotic) or 3 ml of ASW with 5 × 103 cells/ml of the wild-type V. fischeri strain ES114 (symbiotic) [28]. Colonization of the light organ of juvenile animals was monitored by measuring light output in a photometer (TD-20/20, Turner Designs, Inc., Sunnyvale, CA).
All chemicals used in this study were purchased from Sigma-Aldrich Co. (St. Louis, MO) unless otherwise stated.
2.2 RNA extraction
In preparation for RNA extraction, whole E. scolopes juveniles were anesthetized in 2% ethanol in ASW and stored in RNAlater (Ambion, Inc., Austin, TX) at −20° C until needed. Total RNA was extracted from squid juveniles (three animals per sample) by removing the RNAlater, transferring the samples to a microcentrifuge tube containing 1 ml of TRIzol reagent (Invitrogen, Corp., Carlsbad, CA), and homogenizing them with a Polytron PT-1200 homogenizer (Kinematica, Inc., Westbury, NY) following the manufacturer’s recommended protocol. The resulting total RNA (~20 mg) was DNase-treated and column purified with a combination of the RNase-Free DNase enzyme and RNeasy MinElute Cleanup Kit (Qiagen, Inc., Valencia, CA) to remove potential DNA contamination. The RNA concentration and purity of each sample was determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, LLC Subsidiary of Thermo Fisher Scientific, Inc., Waltham, MA) and its quality assessed by electrophoresis on a 1.5% agarose gel.
We also extracted total RNA from adult squid tissues to study the occurrence of C3 mRNA in E. scolopes. Seven different tissues were dissected from adult animals that had been previously anesthetized in ASW with 2% ethanol. These tissues (light organ, light organ without central core, central core, white body, mantle, arm muscle, and gills) were stored in RNAlater (Ambion) and their total RNA was extracted as described above. To obtain hemocytes from the hemolymph of adult animals, blood was drawn from the cephalic artery using a syringe with 27-gauge needle. The hemolymph sample (200 μl; ~ 5000 hemocytes/μl) was centrifuged at 1000 × g and 4°C for 10 min to pellet the hemocytes. The serum then removed and total RNA from hemocytes was extracted using TRIzol as previously described before treatment with Turbo DNA-free reagents (Applied Byosystems/Ambion, Austin, TX) to remove potential genomic DNA contamination.
2.3 EST selection and Rapid Amplification of cDNA Ends (RACE)
Several E. scolopes EST sequences containing homology to human complement component C3 (GenBank Accession No. P01024) were identified from a previously constructed EST database derived from cDNA libraries of juvenile light organs [29]. One of these EST sequences (GenBank Accession No. DW284544) was used to obtain the full-length of the open reading frame encoding the putative C3 molecule in E. scolopes.
Purified total RNA samples from juvenile squid and specifically designed primers were used to amplify both 5′ and 3′ cDNA ends of the EST of interest using the GeneRacer Kit (Invitrogen) (Table 1). Resulting products from the RACE reactions were reamplified using nested primers, separated in agarose gels, extracted, and ligated into plasmids using the TOPO TA Cloning Kit for sequencing with ‘One Shot Top 10’ competent E. coli cells (Invitrogen) following the recommended protocols. Positively transformed cells were grown overnight at 37°C in LB (Luria Bertani) broth supplemented with 50 μg/ml kanamycin. Plasmids were then isolated and purified from bacteria using the Qiaprep Spin Mini Prep Kit (Qiagen), and 1 μg of each plasmid was used in sequencing reactions with M13 forward and reverse primers as well as BigDye Terminator v. 3.1 mix (Applied Biosystems, Foster City, CA).
Table 1.
Primers used for Es-C3 RACE and PCR protocols
| Primer Name | Sequence | Use |
|---|---|---|
| 1 Forward | 5′ ATTGTCCGCGTGTCAACGTGGGTGAAAT 3′ | Initial 5′ RACE |
| 1 Reverse | 5′ TCATGAGTCCCGAACTGTCAACCCAA 3′ | Initial 5′ RACE |
| 1 nForward | 5′ GGTGATGTCCAACAAGAGTTCGGA 3′ | Initial 5′ RACE (nested) |
| 1 nReverse | 5′ GTCCGAACTCTCTTGTTGGACATCAC 3′ | Initial 5′ RACE (nested) |
| 2 Forward | 5′ TGCTGTTCCGTTCTGTGAGCACTACT 3′ | Intial 5′ RACE |
| 2 Reverse | 5′ AGCAAGCAACACACTCTCTTTGAGCG 3′ | Initial 5′ RACE |
| 2 nForward | 5′ GCCAGCACCAATATACGTCTTCGT 3′ | Initial 5′ RACE (nested) |
| 2 nReverse | 5′ GTCTTGTCCGAATCACACAACAGG 3′ | initial 5′ RACE (nested) |
| Es-C3 Forward | 5′ TGGTTTTGTGTTGCACGATT 3′ | End-point PCR |
| Es-C3 Reverse | 5′ CGGAACAAAATGTTCGATGTT 3′ | End-point PCR |
| L21 Forward | 5′ GCCTTGGCTTGAGCCTTCAACTTT 3′ | End-point PCR |
| L21 Reverse | 5′ GGTGATCGTCAACAAACGCGTGAA 3′ | End-point PCR |
Sequencing was performed at the DNA Sequencing Laboratory, University of Wisconsin-Madison. Upon completing the first 5′-RACE reaction, the newly obtained sequence data confirmed the initial identification of the EST as a putative C3 homolog. Two more 5′-RACE reactions were necessary to reach the 5′ region of the molecule where a predicted start codon followed by an open reading frame could be identified. A 3′-RACE reaction confirmed the presence of a stop codon and subsequent poly-A tail at the 3′ end. Additional site-directed PCRs (polymerase chain reactions) were run in independent triplicates followed by sequencing reactions to fill-in gaps and verify the data sequence of this molecule.
2.4 Sequence analysis
All sequences were analyzed both at the nucleotide and amino acid levels using the Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information (NCBI; http://www.ncbi.mlm.nih.gov/blast/BLAST.cgi). Resulting RACE sequences were aligned and the contigs constructed using the sequence analysis software Vector NTI Advance Version 10.1.1 (Invitrogen). The predicted amino acid sequence of Es-C3 was compared to several other related polypeptides in the TEP (thioester-containing protein) superfamily. Comparison of the various TEP sequence alignments were used to construct an unrooted neighbor-joining tree (bootstrapped, 1000 iterations) based on the Kimura 2-parameter correction using CLUSTAL X 1.8 [30] with gapped positions excluded.
In addition, a rooted neighbor-joining tree (parameters as above) using the alpha-2-macroglobulin sequence from the Atlantic horseshoe crab was constructed to compare the placing of Es-C3 within the complement group of proteins. Maximum likelihood analyses were also performed on these data using the default parameters of the program proML in the PHYLIP (version 3.63) suite of programs [31]. Trees constructed by the neighbor-joining and maximum likelihood methods were compared using the Kishino-Hasegawa-Templeton test in proML [32].
2.5 Patterns of C3 gene expression
To determine the expression of Es-C3 in squid tissues, end-point PCR was carried out using specific primers (Table 1) in the 3′ end (5084–5411 nt) of the cDNA molecule. Eight adult squid samples were tested: whole light organ, light-organ central core (the epithelial tissue that houses the symbionts), light organ from which the central core had been removed, white body, gills, mantle, arm, and hemocytes. As positive and loading control for cDNA quality, primers for the ORF coding for the ribosomal protein L21 were also used for each tissue. The PCR reactions were performed using Paq5000 DNA polymerase from Stratagene (Agilent Technologies, Inc., Santa Clara CA) and the following thermocycler protocol: 2 min at 95°C; 35 cycles of 20 sec at 95°C, 20 sec at 55°C, and 30 sec at 72°C; with an extension of 4 min at 72°C. Products were separated in a 1.2% agarose gel, stained with ethidium bromide and visualized using a FluorChem 8400 imager (Alpha Innotech Corp., San Leandro, CA).
2.6 Immunocytochemistry
A 13-aa peptide corresponding to the 1054–1066 region of Es-C3 was synthesized (Peptide Synthesis Facility, Biotechnology Center, University of Wisconsin, Madison, WI) and conjugated to ovalbumin for purposes of rabbit immunization and polyclonal antibody production (Harlan Bioproducts for Science, Inc., Indianapolis, IN). The resulting antiserum was used to localize expression of Es-C3 by immunocytochemical staining of juvenile E. scolopes, collected shortly after hatching. All immunocytochemistry procedures were performed at 4°C unless specifications for a reagent recommended otherwise. After anesthetizing juvenile squids in 2% ethanol in ASW, the animals were fixed overnight in 4% paraformaldehyde in marine phosphate-buffered saline (mPBS: 50 mM sodium phosphate buffer with 0.45 M NaCl, pH 7.4). The animals were then rinsed 4 times (30 min each) with mPBS, after which the light organ was exposed by gently dissecting off the mantle and funnel, followed by permeabilization of the tissues by incubating for 2 days in mPBS with 1% Triton-X 100 (Promega Co., Madison, WI). The samples were then incubated overnight with a blocking solution (mPBS with 1% Triton-X 100, 0.5% bovine serum albumin, 1% goat serum) and then exposed to the Es-C3 antibody or, as a control, the preimmune serum at a 1:100 dilution in the blocking solution for 4 days. The samples were then washed 4 times (1 h each) with mPBs with 1% Triton-X 100, incubated with the blocking solution overnight and exposed to the secondary antibody (goat-anti rabbit conjugated with fluorescein isothyocyanate) for one day under dark conditions. Subsequently, the animals were washed with mPBS and treated, following the manufacturers’ protocols, with rhodamine phalloidin and TOTO-3 iodide (Molecular Probes, Invitrogen Co.) as counterstains. Finally, samples were mounted on glass slides with Vectashield (Vector Laboratories, Burlingame, CA) and visualized using a Zeiss LSM 510 confocal microscope (Carl Zeiss MicroImaging Inc., Thornwood, New York.
3. Results and discussion
3.1 Identification of Es-C3 as a complement component C3 homolog
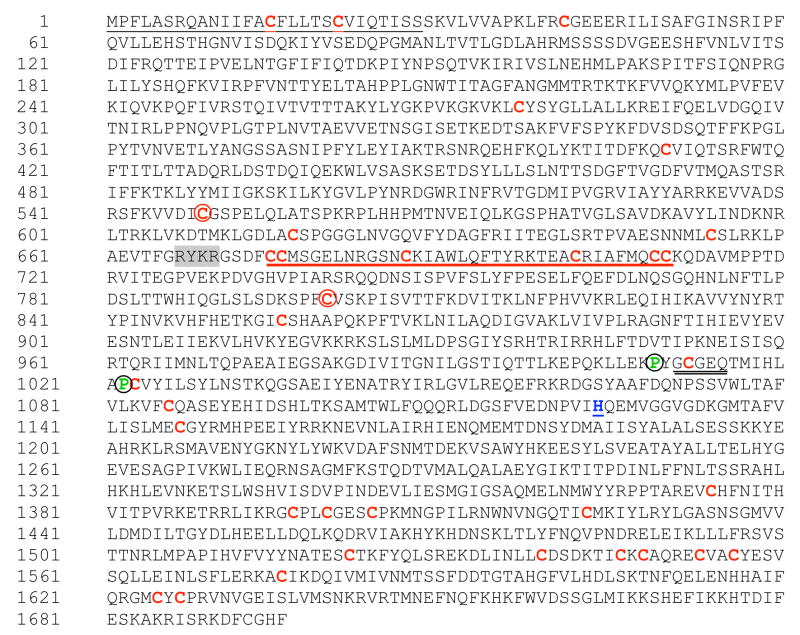
Characteristics of the derived amino acid sequence, as well as predicted domain structure, provided evidence that Es-C3 belongs to the C3 family of proteins. The 5′ and 3′ RACE of the selected E. scolopes EST (DW284544) yielded a cDNA of 5602 base pairs (bp) (GenBank Accession No. EU596375). The cDNA sequence predicted a start codon at nucleotide 291, a stop codon at nucleotide 5379, and a polyadenylation site (AATAAA) at nucleotide 5550. The deduced open reading frame (ORF) of 5088 bp encoded a putative protein of 1696 amino acid residues (Fig. 1). BLASTp analysis revealed that the deduced amino acid sequence of E. scolopes codes for an ortholog of the third component of the complement system (C3; Appendix A). The closest sequence homology was to amphioxus C3 (Accession No. BAB47146 [33]) with 46% similarity and 27% identity at the amino acid level. The two next closest sequences were the horseshoe crab C3 (Accession No. AAQ08323 [9]), with 45% amino acid similarity and 26% identity, and the coral C3 (Accession No. AAN86548 [10]) with 43% similarity and 23% identity.
Fig. 1.
The deduced amino-acid sequence of Es-C3. The ORF predicts a protein of 1696 amino acids with characteristic domains and motifs of other known complement C3 components. Highlighted are: the 28 amino-acid peptide of the secretion signal (black underline, N-terminus); the conserved cysteine residues (red), including the two cys (squared) that are thought to be part of the disulfide bond linking the α and β chains in the mature and activated C3b molecule; the ANATO domain (red underline); the thioester domain (double black underline); the neighboring proline residues (green, circled), known to be important for the thioester domain stability. The catalytic histidine residue (blue, underline); and, the potential processing site for the cleavage of the alpha and beta subunits (gray background).
Further analysis of the deduced polypeptide using the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/) predicted a secretion leader sequence from amino acids 1 to 28 (aa 1–22 in human C3). The squid C3-like molecule contains 34 cysteine residues compared to the 27 found in the human sequence. Seventeen of these 34 cysteine residues were located in the same position when Es-C3 was aligned with human C3, including the pair that is responsible for the covalent linkage of the alpha (a) and beta (b) chains in the mature C3 protein. In the human molecule, there are 9 additional pairs of cysteine residues creating disulfide bridges [34]; the Es-C3 sequence has eight of those pairs, suggesting a similar three-dimensional structure for the squid and human proteins.
The deduced Es-C3 sequence also had a putative cleavage site characteristic of C3 molecules. Studies of vertebrate C3 have suggested that it is synthesized as a single-chain pro-protein, with alpha (C-terminus) and beta (N-terminus) portions linked by a tetra arginine sequence. This RRRR sequence acts as a cleavage site and is removed post-translationally prior to secretion [5]. However, the sequence for this processing site varies among C3 molecules. For example, the putative cleavage sequence RKPR occurs in the Japanese lamprey C3 [33]; RRKR occurs in the C3 molecules of sea urchin, amphioxus, and a tunicate [8, 31, 36]; RKKR occurs in the horseshoe crab C3 [9]; and, RKRR occurs in the coral C3 [10]. Thus, alignment of the currently available C3 sequences suggests that RXXR is the consensus sequence for the beta-alpha cleavage site (X is frequently R or K). The most likely cleavage site in the Es-C3 molecule is RYKR, which occurs at amino acids 667–670 (Fig. 1).
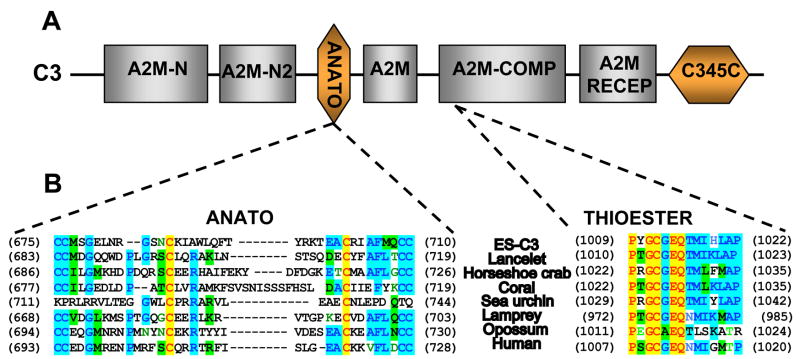
In addition to the presence of the two critical cysteine residues and the presence of a putative cleavage site, the presence of a putative anaphylatoxin (ANATO) domain (aa 675–710, Figs. 1 and 2) provided further support for the posttranslational maturation of the Es-C3 into alpha and beta subunits. The ANATO domain, a conserved character among complement molecules, is located at the N-terminus of the alpha chain. Sequence analysis of the putative Es-C3 ANATO domain revealed that it contains, in conserved locations, the six canonical cysteine residues known to be critical for its function (Fig. 2B) [33]. In addition, a putative cleavage site (IARS, aa 739–742) for the C3-convertase, which releases the ANATO domain and thereby creates C3b, was identified in a similar position as the LARS cleavage site of human C3. These sequence data suggest that the Es-C3 functions similarly to the C3 molecules of other animals.
Fig. 2.
Domain architecture of complement component. (A) The protein domains characteristic of complement component C3. The TEP superfamily domains (gray boxes): A2M-N, the alpha2-macroglobulin N terminus regions 1 and 2; A2M, the alpha2-macroglobulin C-terminus region; A2M-COMP, the alpha2-macroglobulin-like domain with the thioester domain; and, A2M RECEP, the alpha2-macroglobulin receptor domain. The domains of complement C3 molecule (orange boxes): ANATO, the anaphylatoxin domain; and, C345C, the complement C3/4/5 C-terminal region. (B) Alignment of the Es-C3 anaphylatoxin and thioester domains with their closest homologues and human C3. The anaphylatoxin (ANATO) domain of Es-C3 has the six canonical cysteine residues at conserved positions and the thioester motif (GCGEQ) with the surrounding amino acids, including the two flanking proline residues thought to be important in the formation, stability and function of this region [43]. Identical residues, red in yellow background; conserved residues, purple with blue background; similar residues, black with green background; weakly similar, green; and, non-similar, black.
Similar to other C3-like proteins, Es-C3 had the characteristic domains that occur in members of the C3/4/5 complement molecules (Fig. 2A). These domains included the five-residue thioester (TED) motif (GCGEQ) (Fig. 2B), the N- and C-terminal regions typical of A2Ms and TEPs, and the A2M receptor domain located at the C-terminus of the molecule. Because these characters are shared among most members of the TEP superfamily, we further analyzed the Es-C3 to obtain evidence that it contains the distinctive characteristics found only in complement molecules. The predicted Es-C3 protein has a similar sequence length to that of other known C3s. In addition, it possesses hallmark characteristics of the complement C3/4/5 family of proteins. In addition to the ANATO domain described above, which is specific to the C3/4/5 family, the Es-C3 has a histidine residue (H1124) located 109 amino acids downstream of the TED motif. This histidine residue, which is found 113 aa after the TED motif in human C3, is catalytic and functions in the formation of a covalent bond between C3b and its target molecules [37]. In addition, like human C3, the Es-C3 sequence also contains a glutamic acid (E1126) located two amino acids downstream from the catalytic histidine. This residue is thought to increase the nucleophilic strength and specificity of the TED to hydroxyl groups [38]. Finally, the Es-C3 has an extended (~200 residues) C-terminus region characteristic of C3/4/5 molecules, which is usually not found in A2Ms or invertebrate TEPs.
3.2 The phylogenetic position of Es-C3
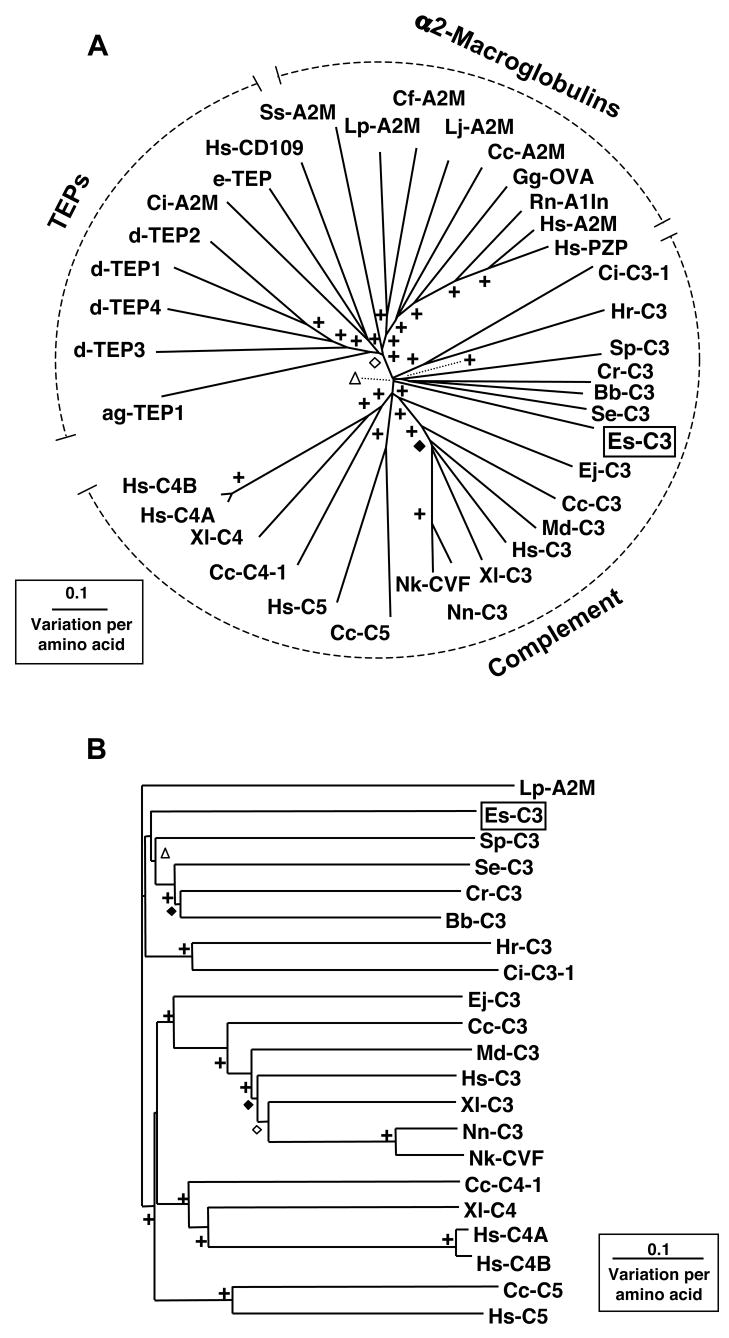
To determine how Es-C3 relates to other members of the TEP superfamily of proteins, a multiple alignment was performed using 36 additional sequences from the GenBank database. The results were used to construct an unrooted phylogenetic tree (Fig. 3A) using the neighbor-joining method. Comparisons within the complement group where performed using a rooted neighbor-joining tree (Fig. 3B). Trees constructed using the neighbor-joining and maximum likelihood methods were not found to be statistically different.
Fig. 3.
The phylogenetic position of Es-C3. (A) An Unrooted phylogenetic tree produced by the neighbor-joining method, using Kimura two-parameter distances between complement-like protein sequences of the organisms named on the tree. (B) Neighbor-joining phylogenetic tree of complement proteins rooted with the alpha-2-macroglobulin sequence from the Atlantic horseshoe crab (Lp-A2M). Gapped positions were excluded from the analysis. Symbols on branches represent percentage bootstrap values: +, 90 to 100%; ◇, 80 to 89%; ◆, 70 to 79%; △, 60 to 69%. Sequences used in analysis with their abbreviation and GenBank accession number are identified clockwise as they occur in the tree. Clustering with TEPs: ag-TEP1, Anopheles gambiae (mosquito) TEP1 (AAG00600); dTEP3, Drosophila melanogaster (fruit fly) TEP3 (CAB87809); d-TEP4, Drosophila melanogaster TEP4 (CAB87810); d-TEP1, Drosophila melanogaster TEP1 (CAB87807); d-TEP2, Drosophila melanogaster TEP2 (CAB87808); ci-A2M, Ciona intestinalis (sea squirt) A2M (CAD24311); e-TEP, Euphaedusa tau (snail) TEP (BAE44110); Hs-CD109, Homo sapiens (human) CD109 (AAN78483). Clustering with alpha2-macroglobulins: Ss-A2M, Scylla serrata (mud crab) A2M (ABD61456); Lp-A2M, Limulus polyphemus (Atlantic horseshoe crab) A2M (BAA19844); Cf-A2M, Chlamys farreri (scallop) A2M (AAR39412); Lj-A2M, Lethenteron japonicum (Japanese lamprey) A2M (BAA02762); Cc-A2M, Cyprinus carpio (carp) A2M (BAA85038); Gf-OVA, Gallus gallus (chicken) ovostatin (P20740); Rn-A1ln, Rattus norvegicus (rat) α1-inhibitor (P14046); Hs-A2M, Homo sapiens (human) A2M (P01023); Hs-PZP, Homo sapiens (human) pregnancy zone protein (CAA38255). Clustering with complement proteins - Ci-C3-1, Ciona intestinalis (sea squirt) C3-1 (Q8WPD8); Hr-C3, Halocynthia rorezi (ascidian) C3 (BAA75069); Sp-C3, Strongylocentrotus purpuratus (purple sea urchin) C3 (AAC14396); Cr-C3, Carcinoscorpius rotundicauda (Southeast Asian horseshoe crab) C3 (AAQ08323); Bb-C3, Branchiostoma belcheri (amphioxus) C3-like (BAB47146); Se-C3, Swiftia exserta (coral) C3-like (AAN86548); Es-C3, Euprymna scolopes (squid) C3 (EU596375); Ej-C3, Entosphenus japonicus (Japanese lamprey) C3 (Q00685); Cc-C3, Cyprinus carpio (carp) C3-H1 (BAA36618); Md-C3, Monodelphis domestica (opossum) C3 (XP_001378723); Hs-C3, Homo sapiens (human) C3 (P01024); Xl-C3, Xenopus laevis (clawed frog) C3 (AAB60608); Nn-C3, Naja naja (Indian cobra) C3 (Q01833); Nk-CVF, Naja kaouthia (monocled cobra) venom factor (I51018); Cc-C5, Cyprinus carpio (carp) C5 (BAC23058); Hs-C5, Homo sapiens (human) C5 (P01031); Cc-C4-1, Cyprinus carpio (carp) C4-1 (BAB03284); Xl-C4, Xenopus laevis (clawed frog) C4 (BAA11188); Hs-C4A, Homo sapiens (human) C4A (AAB59537); Hs-C4B, Homo sapiens (human) C4B (AAA99717).
Three major groups of proteins, the C3/4/5 components of the complement system, the A2Ms, and the invertebrate TEPs could be distinguished showing similar phylogenetic distribution to previously published trees of this protein family [9, 10, 37, 40]. The arrangement of these three groups shows that A2M and TEPs are more similar to one another than they are to complement. The complement group itself is further branched, with distinct C3, C4, and C5 groupings within the vertebrates, and a separate, although poorly bootstrapped, invertebrate C3 grouping. No sequences clustering with the C4 and C5 groups have been identified thus far in the invertebrates. These data suggest that vertebrate C4 and C5 may have evolved from a C3-like molecule as it has been suggested before (reviewed in Sahu A. and Lambris J. D.). Although the ascidians form a strongly bootstrapped group, there is no clear division between the deuterostomes and the non-deuterostomes within the invertebrate C3 grouping since the coral and horseshoe crab sequences group together with those of amphioxus and the sea urchin. These phylogenetic analyses support the initial BLAST results grouping Es-C3 with other complement C3 members. However, it stands alone in a separate, poorly bootstrap-supported branch among the invertebrate C3s. This finding reinforces the hypothesis that the protein is a complement C3 from an organism that is very different from those that have been sequenced so far, and perhaps when other C3s are identified in the lophotrochozoan superphylum, there will be other sequences that group with Es-C3.
3.3 mRNA expression of Es-C3 in E. scolopes
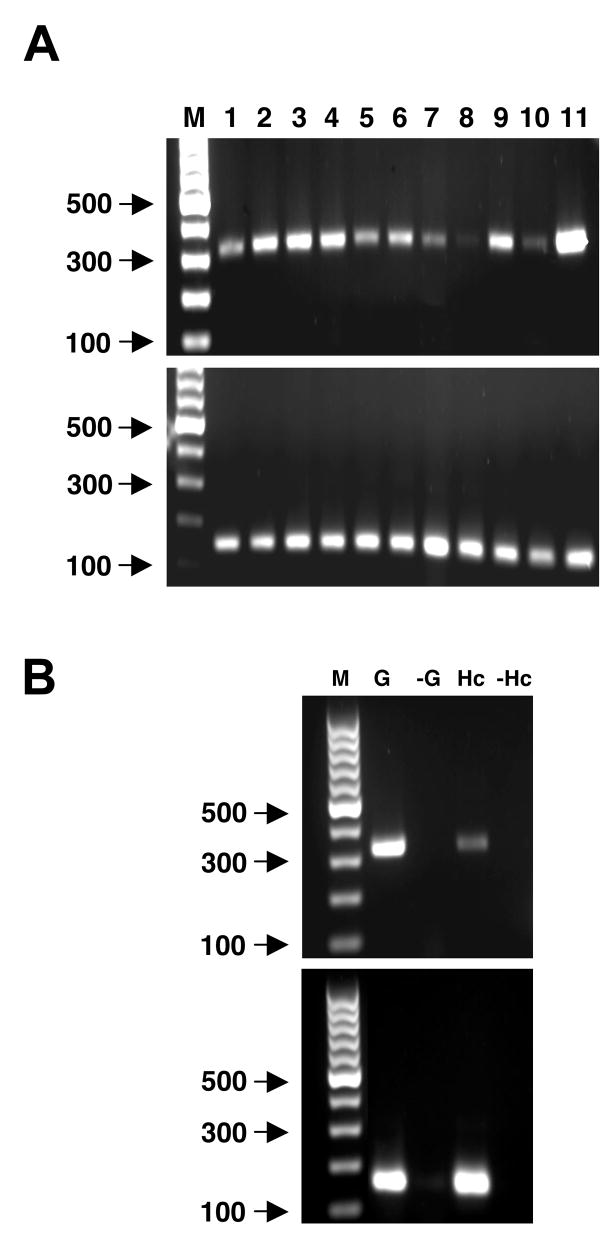
To determine patterns of expression of the Es-C3 transcript, we analyzed total RNA samples from whole juveniles or adult tissues for the amplification of a PCR product using primers specific for a region located in the 3′ end of the nucleotide sequence (Fig. 4A, top). A positive control to test for quality of the RNA samples was also performed using primers designed for a ribosomal protein (Fig 4A, bottom). Our results revealed the presence of transcripts in all juvenile samples (hatchling, nonsymbiotic and symbiotic), suggesting that C3 is constitutively expressed in juvenile animals. In the adult, all tissues tested also produced products confirming the presence of Es-C3 mRNA (Fig 4A, lanes 3–9). Although these studies did not use quantitative PCR, they suggest lower levels of Es-C3 transcript in some tissues, such as the hemocytes and arm tissue. Because in other invertebrates, hemocytes are the primary or sole site of C3 synthesis [8, 34, 41], we performed PCR on hemocytes with higher levels of mRNA. These experiments confirmed the presence of Es-C3 transcript in these cells (Fig. 4C). More studies, such as in-situ hybridization, need to be done to determine the specific source of C3 expression and synthesis in E. scolopes. Because vascularized tissues such as the gills appeared to have higher levels of message than isolated hemocytes, it is unlikely that all Es-C3 message in such tissues is due to hemocyte mRNA. It is more likely that more than one cell type produces C3 in E. scolopes. In vertebrates, complement C3 expression has been reported in many other cell types such as hepatocytes, fibroblasts, endothelial and epithelial cells, and adipocytes among others [42].
Fig. 4.
Relative expression of Es-C3 in Euprymna scolopes. (A) Top: the relative expression of the complement C3 transcript in cDNA pools of whole juveniles and adult squid tissues; bottom: the corresponding loading controls using the ribosomal protein L21. Lanes: M = standard bp markers; juvenile squid, 1–3: 1 = whole hatchling, 2 = whole 48-hour nonsymbiotic animal, 3 = whole 48-hour symbiotic animal; adult tissue samples, 4–11: 4 = whole light organ, 5 = light organ without symbiont-containing central core, 6 = symbiont-containing central core, 7 = white body, 8 = hemocytes, 9 = mantle, 10 = arm, and 11 = gills. (B) Confirmation of Es-C3 transcript in hemocytes. Gill cDNA was used as a positive control for Es-C3 and L21. G and Hc, gill and hemocyte cDNA, respectively; -G and -HC, control reactions run with no reverse transcriptase to assess genomic DNA contamination in gill and hemocyte cDNA, respectively. Results shown are representative of a minimum of three independent and separate RNA extractions and PCR procedures for each sample in the figure.
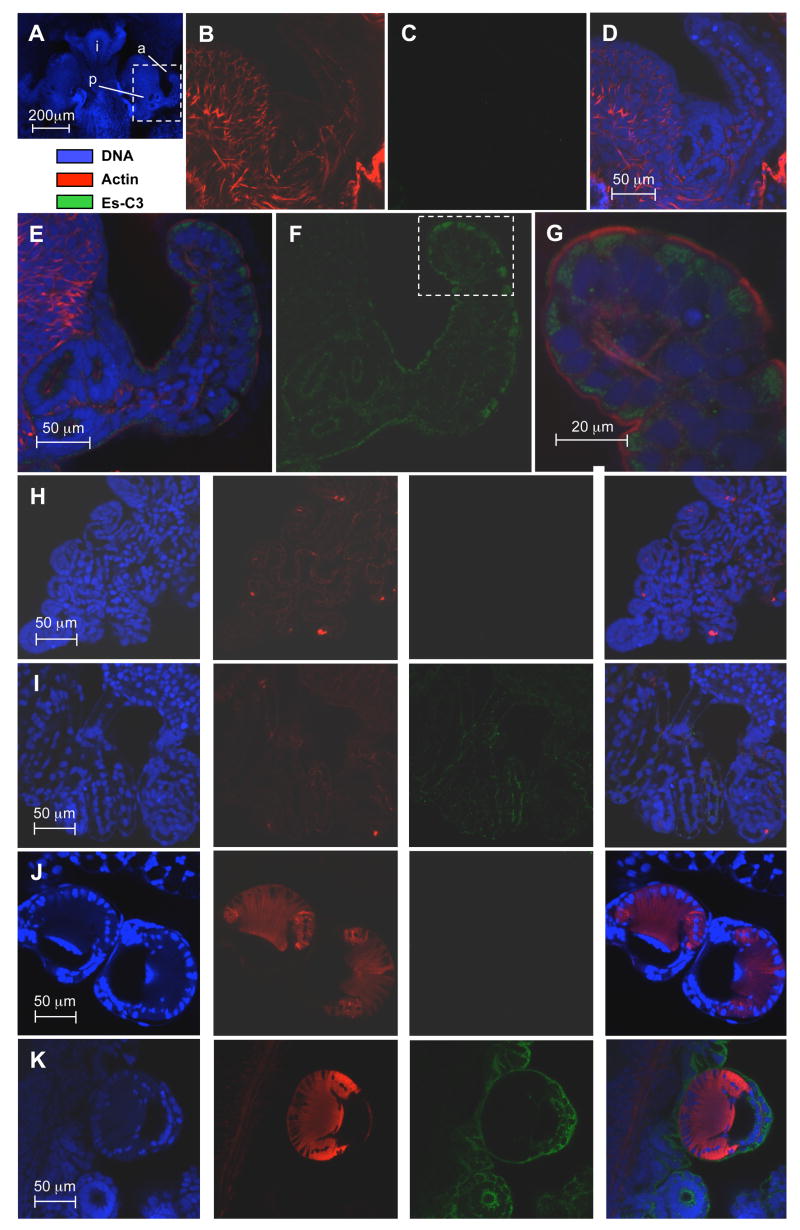
To characterize the protein production of the putative C3 homologue in E. scolopes, we incubated fixed juvenile animals with a rabbit polyclonal antibody made against a region in the α-chain of the polypeptide following previously described immunocytochemical protocols and observed the samples by confocal microscopy (Fig. 5). The rabbit pre-immune serum showed no cross reactivity above background in any tissue tested. In contrast, in the animals treated with anti-Es-C3 antibody, cross reactivity occurred in the apical surfaces of epithelial cells of all tissues analyzed, including those of the light organ surface. The antibody bound to discrete regions within the cytoplasm of cells in a punctate distribution suggesting that this protein is stored in vesicles. Unlike other animals, where C3 is present in the blood [15], we observed no cross reactivity in blood vessels.
Fig 5.
Localization of Es-C3 in juvenile E. scolopes by confocal microscopy. (A) For orientation, the juvenile light organ, and adjacent tissues, labeled solely with the nuclear DNA stain TOTO-3 iodide (intestine, i). The area enclosed in the white box, a portion of one side of the organ including the anterior appendage (a) of the superficial ciliated field and the pore region (p) where symbionts enter, is magnified in panels B–G. (B–D) A representative preimmune control, demonstrating low cross reactivity against anti-Es-C3. (E–G) Light organ treated with anti-Es-C3 antibody conjugated with FITC. Antibody cross reactivity localized to the apical surface of the light organ epithelial cells. Panels and G are composite images of all three labels; panel F shows only the signal received from the fluorescein dye, revealing that Es-C3 appears to be present in discrete areas within the cells. (H, I) Labeling in gill epithelial; H panels, preimmune controls; I panels, anti-Es-C3. (J, K) Labeling of the arm suckers. J panels, preimmune controls; K panels, anti-Es-C3. In panels H–K, right most images are the composite of all three labels. In these other tissues, Es-C3 is also found in the cytoplasm of cells in a similar distribution to that found in the light organ epithelia. All images are representative of at least three separate replicate treatments.
4. Conclusion
This study provides evidence for the presence of a complement C3 ortholog in the Hawaiian bobtail squid Euprymna scolopes. The discovery of Es-C3 extends the occurrence of complement components to all three superphyla of bilaterian animals, the Deuterostomia, the Ecdysozoan, and the Lopothochozoa. This finding, coupled with the report of complement molecules in cnidarians, renders the complement system one of the most diverse and ancient immune recognition and defense mechanisms known to date.
Further studies on the functional characteristics of Es-C3 in E. scolopes should provide further insight into the conserved characters of the molecule critical for function. In addition, because the study of complement proteins has largely been restricted to their function in pathogenesis, their study in the E. scolopes-V. fischeri symbiosis offers the opportunity to discover possible roles for complement proteins in the acquisition and maintenance of beneficial animal-microbe relationships.
Supplementary Material
Supplementary data associated with this article can be found in the online version at (site provided by publisher).
Acknowledgments
We thank N. Dinguirard and A. Wier for helpful comments on the manuscript. This work was funded by NIH grants RR R01-12294 to EG Ruby and R01-AI50661 to MMN, and NSF IOS 0517007 to MM-N and EG Ruby.
Abbreviations and symbols used
- aa
amino acid(s)
- ANATO
Anaphylatoxin
- ASW
artificial seawater
- A2M
alpha2-macroglobulin
- bp
base pairs
- ORF
open reading frame
- PCR
polymerase chain reaction
- RACE
rapid amplification of cDNA ends
- TED
thioester domain
- TEPs
thioester-containing proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carroll MC. The complement system in regulation of adaptive immunity. Nature Immunol. 2004;5:981–6. doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- 2.Morgan BP, Marchbank KJ, Longhi MP, Harris CL, Gallimore AM. Complement: central to innate immunity and bridging to adaptive responses. Immunol Lett. 2005;97:171–9. doi: 10.1016/j.imlet.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen CH, Leslie RG. Complement’s participation in acquired immunity. J Leuk Biol. 2002;72:249–61. [PubMed] [Google Scholar]
- 4.Volanakis J. Overview of the complement system. In: Volanakis J, Frank M, editors. The Human Complement System in Health and Disease. Marcel Dekker; New York: 1998. pp. 9–32. [Google Scholar]
- 5.Sahu A, Lambris JD. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immun Rev. 2001;180:35–48. doi: 10.1034/j.1600-065x.2001.1800103.x. [DOI] [PubMed] [Google Scholar]
- 6.Blandin S, Levashina EA. Thioester-containing proteins and insect immunity. Mol Immunol. 2004;40:903–8. doi: 10.1016/j.molimm.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Nair SV, Del Valle H, Gross PS, Terwilliger DP, Smith LC. Macroarray analysis of coelomocyte gene expression in response to LPS in the sea urchin. Identification of unexpected immune diversity in an invertebrate. Physiol Gen. 2005;22:33–47. doi: 10.1152/physiolgenomics.00052.2005. [DOI] [PubMed] [Google Scholar]
- 8.Al-Sharif WZ, Sunyer JO, Lambris JD, Smith LC. Sea urchin coelomocytes specifically express a homologue of the complement component C3. J Immunol. 1998;160:2983–97. [PubMed] [Google Scholar]
- 9.Zhu Y, Thangamani S, Ho B, Ding JL. The ancient origin of the complement system. EMBO J. 2005;24:382–94. doi: 10.1038/sj.emboj.7600533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dishaw LJ, Smith SL, Bigger CH. Characterization of a C3-like cDNA in a coral: phylogenetic implications. Immunogenetics. 2005;57:535–48. doi: 10.1007/s00251-005-0005-1. [DOI] [PubMed] [Google Scholar]
- 11.Fujita T, Matsushita M, Endo Y. The lectin-complement pathway--its role in innate immunity and evolution. Immun Rev. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- 12.Nonaka M, Yoshizaki F. Primitive complement system of invertebrates. Immun Rev. 2004;198:203–15. doi: 10.1111/j.0105-2896.2004.00118.x. [DOI] [PubMed] [Google Scholar]
- 13.Nonaka M, Yoshizaki F. Evolution of the complement system. Mol Immunol. 2004;40:897–902. doi: 10.1016/j.molimm.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Nonaka M, Kimura A. Genomic view of the evolution of the complement system. Immunogenetics. 2006;58:701–13. doi: 10.1007/s00251-006-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinto MR, Melillo D, Giacomelli S, Sfyroera G, Lambris JD. Ancient origin of the complement system: emerging invertebrate models. Adv Exp Med Biol. 2007;598:372–88. doi: 10.1007/978-0-387-71767-8_26. [DOI] [PubMed] [Google Scholar]
- 16.Janssen BJ, Huizinga EG, Raaijmakers HC, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–11. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 17.Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–42. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 18.Visick KL, Ruby EG. Vibrio fischeri and its host: it takes two to tango. Curr Opin Microbiol. 2006;9:632–8. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Foster JS, Apicella MA, McFall-Ngai MJ. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol. 2000;226:242–54. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- 20.Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–8. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 21.Weis VM, Small AL, McFall-Ngai MJ. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc Natl Acad Sci USA. 1996;93:13683–8. doi: 10.1073/pnas.93.24.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Small AL, McFall-Ngai MJ. A halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J Cellul Biochem. 1999;72:445–57. [PubMed] [Google Scholar]
- 23.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–51. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- 24.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci USA. 2000;97:10231–5. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol. 2002;68:5113–22. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyholm SV, McFall-Ngai MJ. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- 27.Koropatnick TA, Kimbell JR, McFall-Ngai MJ. Responses of host hemocytes during the initiation of the squid-Vibrio symbiosis. Biol Bull. 2007;212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- 28.Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–6. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chun CK, Scheetz TE, Bonaldo M, de F, Brown B, Clemens A, Crookes-Goodson WJ, Crouch K, DeMartini T, Eyestone M, Goodson MS, Janssens B, Kimbell JL, Koropatnick TA, Kucaba T, Smith C, Stewart JJ, Tong D, Troll JV, Webster S, Winhall-Rice J, Yap C, Casavant TL, McFall-Ngai MJ, Soares MB. An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics. 2006;7:154–63. doi: 10.1186/1471-2164-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felsenstein J. PHYLIP: phylogeny inference package (version 3.2) Cladistics. 1989;5:164–6. [Google Scholar]
- 32.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol. 1989;29:170–9. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki MM, Satoh N, Nonaka M. C6-like and C3-like molecules from the cephalochordate, amphioxus, suggest a cytolytic complement system in invertebrates. J Mol Evol. 2002;54:671–9. doi: 10.1007/s00239-001-0068-z. [DOI] [PubMed] [Google Scholar]
- 34.Dolmer K, Sottrup-Jensen L. Disulfide bridges in human complement component C3b. FEBS Lett. 1993;315:85–90. doi: 10.1016/0014-5793(93)81139-q. [DOI] [PubMed] [Google Scholar]
- 35.Nonaka M, Takahashi M. Complete complementary DNA sequence of the third component of complement of lamprey. Implication for the evolution of thioester containing proteins. J Immunol. 1992;148:3290–5. [PubMed] [Google Scholar]
- 36.Nonaka M, Azumi K. Opsonic complement system of the solitary ascidian, Halocynthia roretzi. Dev Comp Immunol. 1999;23:421–7. doi: 10.1016/s0145-305x(99)00021-x. [DOI] [PubMed] [Google Scholar]
- 37.Law SK, Dodds AW. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 1997;6:263–74. doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM. X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2. Science. 1998;280:1277–81. doi: 10.1126/science.280.5367.1277. [DOI] [PubMed] [Google Scholar]
- 39.Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell. 2001;104:709–18. doi: 10.1016/s0092-8674(01)00267-7. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Song L, Li C, Zhao J, Wang H, Gao Q, Xu W. Molecular cloning and characterization of a thioester-containing protein from Zhikong scallop Chlamys farreri. Mol Immunol. 2007;44:3492–500. doi: 10.1016/j.molimm.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 41.Marino R, Kimura Y, De Santis R, Lambris JD, Pinto MR. Complement in urochordates: cloning and characterization of two C3-like genes in the ascidian Ciona intestinalis. Immunogenetics. 2002;53:1055–64. doi: 10.1007/s00251-001-0421-9. [DOI] [PubMed] [Google Scholar]
- 42.Botto M. Part 3, C3 family. In: Morley BJ, Walport MJ, editors. The complement facts book. San Diego, CA: Academic Press; 2000. pp. 87–94. [Google Scholar]
- 43.Isaac L, Isenman DE. Structural requirements for thioester bond formation in human complement component C3. Reassessment of the role of thioester bond integrity on the conformation of C3. J Biol Chem. 1992;267:10062–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data associated with this article can be found in the online version at (site provided by publisher).