Summary of Recent Advances
Two important classes of natural products are made by non-ribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs). With most biosynthetic intermediates covalently tethered during biogenesis, protein mass spectrometry (MS) has proven invaluable for their interrogation. New mass spectrometric assay formats (such as selective cofactor ejection and proteomics-style LC-MS) are showcased here in the context of functional insights into new breeds of NRPS/PKS enzymes, including the first characterization of an “iterative” PKS, the biosynthesis of the enediyne antitumor antibiotics, the study of a new strategy for PKS-initiation via a GNAT-like mechanism, and the analysis of branching strategies in so-called “AT-less” NRPS/PKS hybrid systems. The future of MS analysis of NRPS and PKS biosynthetic pathways lies in adoption and development of methods that continue bridging enzymology with proteomics as both fields continue their post-genomic acceleration.
Introduction
Many important bioactive and commercially valuable natural products are synthesized by non-ribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs). NRPS and PKS natural products represent an extreme amount of functional and structural diversity; this combined with the 10,000 – 700,000 MW polypeptides involved make precise characterization of their biosynthesis an arduous process. Protein mass spectrometry presents a growing set of new tools for the analysis of these biosynthetic systems, and continues to evolve as an invaluable platform for functional studies of NRPS and PKS characterization. Building on past reviews [1••,2], we focus here on the most recent developments in MS-based interrogation of NRPS and PKS systems with novel domains, domain organizations, and/or domain functions.
Non-ribosomal Peptide Synthetases and Polyketide Synthases
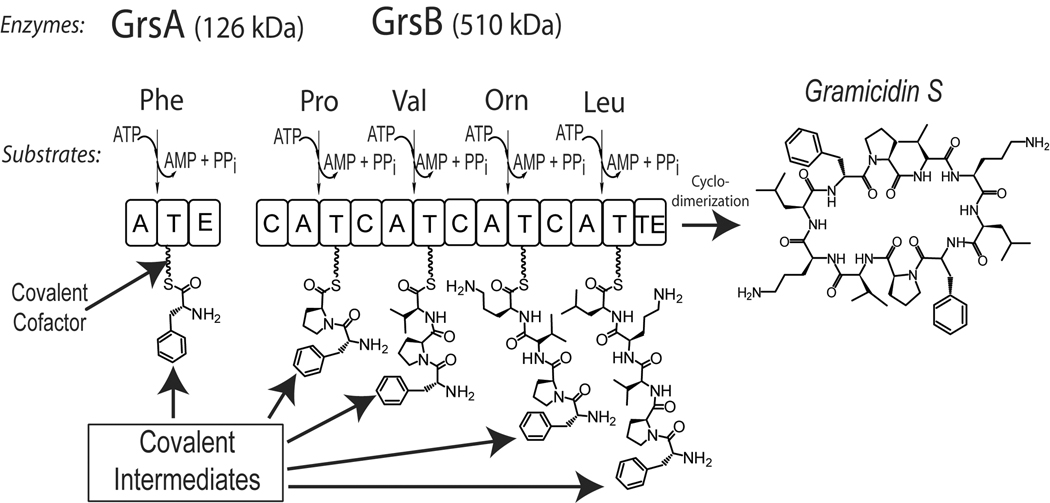
Biosynthetic processes carried out by NRPSs and PKSs have been extensively reviewed elsewhere [3••–5]. Briefly, NRPSs and PKSs are often >>100 kDa, multi-modular enzymes that contain a great variety of individual catalytic domains (e.g., the gramicidin S biosynthetic system shown in Figure 1). Each module is responsible for incorporation of one building block into the natural product in an assembly line fashion, with NRPs composed of amino acid monomer units and PKs built from malonyl monomer units. A module can be further broken down into domains, with each responsible for one biochemical transformation to the growing natural product. Domains responsible for activation of monomer units for incorporation into the natural product, transfer of monomer units to carrier proteins (thiolation (T) domains), and condensation of the growing natural product with a monomer unit control the core biosynthetic construction of NRPs and PKs. Additionally, tailoring domains can be distributed throughout NRPSs and PKSs, introducing diversity into NRP and PK structures.
Figure 1.
Non-ribosomal peptide synthesis exemplified by the prototypical biosynthetic system for the cyclic decapeptide gramicidin S in Bacillus brevis. Gramicidin S biosynthesis is catalyzed by two NRPSs, GrsA (126 kDa) and GrsB (510 kDa). Amino acid substrates are activated by adenylation domains (A) in an ATP-dependent fashion and loaded onto holo T domains at each of five phosphopantetheinyl covalent-binding sites. Sequential incorporation of five amino acids into a pentapeptide is followed by condensation of two pentapeptides and release/cyclization of the decapeptide catalyzed by the GrsB thioesterase domain (TE). Tailoring domain identification: E – epimerization domain that inverts the stereochemistry of phenylalanine from L to D.
A common feature of NRPSs and PKSs is the post-translational priming of inactive, apo T domains by a phosphopantetheinyl transferase [6] at a conserved serine residue with phosphopantetheine (Ppant) derived from coenzyme A (CoA) to generate active, holo enzyme (see “covalent cofactor” label in Figure 1). Addition of Ppant results in an available sulfhydryl group for covalent attachment of the biosynthetic intermediates, enabling the growing natural product to remain tethered to the synthase(s) during biogenesis. This covalent nature of biosynthesis makes NRPS and PKS biosynthetic processes ideal for analysis by large molecule mass spectrometry, as biosynthetic events can be measured down to the millidalton as a shift in mass to the apo T domain [7].
Using Mass Spectrometry for NRPS/PKS Analysis
Many early mass spectrometric methods for NRPS and PKS analysis were recently extensively reviewed [1,2] and will be discussed only briefly here. Most often, the MS instrumentation of choice uses electrospray-ionization (ESI) with Fourier-Transform Mass Spectrometry (FTMS) [8] due to its ability to obtain high-resolution mass spectra of large peptide and protein species, detect small mass changes, and be coupled to a variety of tandem mass spectrometry (MS2) methods; however, several studies have seen success with other MS ionization and detection methods [9–11]. Both Top Down [12,13] and Bottom Up [14] MS strategies are applicable to NRPS and PKS investigations. Top Down MS analyzes intact proteins directly (no protease) and therefore has been most often applied to stand-alone T domains (only ~10–15 kDa) [15•,16••,17–19], although protein constructs 20–126 kDa have been studied directly [20,21]. The use of FTMS with >50,000 resolution for most recent studies has allowed detection of biosynthetic transformations involving small mass changes (e.g., from stable isotope labeling experiments or the mass increase of 1 Da resulting from amine transfer catalyzed by a novel tailoring domain in mycosubtilin biosynthesis [22]). This contrasts sharply with the resolving power of ~1,000 provided by ion trap instruments which have been used to track transformations involving large mass shifts (e.g., from cofactor or monomer unit loading).
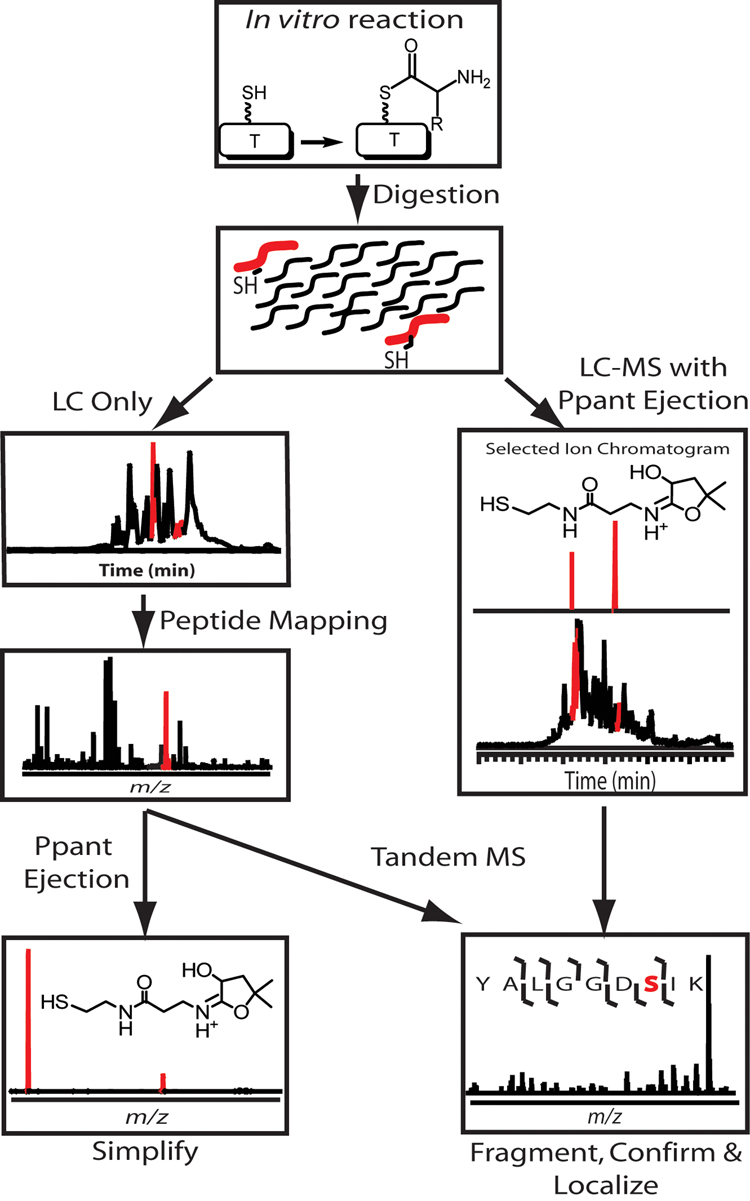
Briefly, in a typical Bottom Up FTMS experiment, in vitro NRPS and PKS reactions are conducted, followed by tryptic or cyanogen bromide digestion (Figure 2, top). The peptides are subjected to reverse phase liquid chromatography (RPLC), and eluent fractions are analyzed by “off-line” FTMS one-by-one for peptides of interest based on mass values of intact peptides (Figure 2, middle left). Peptide identities are confirmed by matching fragment ions and/or amino acid sequence information generated by MS2, and the Ppant modification localized to the active site serine residue (Figure 2, bottom). After the peptide is confirmed to contain the active site, loading, condensation and tailoring reactions involving mass changes are monitored [22–28•].
Figure 2.
General scheme for Bottom Up FTMS analysis of in vitro NRPS/PKS reactions. (Left) “Off-line” work flow. Peptides generated from digests of in vitro reactions are separated by off-line RPLC and each fraction analyzed individually by FTMS for active-site peptides, which are then characterized by MS2 and the Ppant ejection assay. (Right) “On-line” work flow. Peptide mixtures generated from digests of in vitro reactions are separated on-line by RPLC connected directly to the mass spectrometer, where MS2 and the Ppant ejection assay are conducted on the LC time scale. The on-line work flow greatly speeds up analysis time (total data collection time of ~1 h / experiment) and is more amenable to systems where less sample is available.
During tandem mass spectrometry using so-called “low-energy” ion activation methods (e.g., collision induced dissociation (CID) [29] or infrared multi-photon dissociation (IRMPD) [30]), the covalently bound Ppant arm is ejected from the peptide or protein, forming two specific marker ions at m/z values of 261.1267 and 359.1036 [31••] for a T domain in the holo state. These Ppant ejection products are also generated from T domains loaded with a natural product intermediate, and the ejected ions for covalent intermediates provide a simple way to identify the protein biosynthetic state. Ejection ions are typically less than 1 kDa and can be measured with sub-part-per-million (ppm) mass accuracy in high resolution instruments. Reduction of a >100 kDa protein ion to a <1 kDa Ppant ejection ion detected with low millidalton mass accuracy in FTMS instruments often results in unambiguous assignment of empirical formulas for covalent intermediates. Ppant ejection ions are detectable with ~0.2 Da mass accuracy in ion trap instruments, which is often sufficient for confirmation of an expected product. Additionally, MS3 of the ejected Ppant ion results in characteristic marker ions for use in confirming Ppant ejection [32]. The Ppant assay has proven entirely general for any thioester-bound species, and has greatly sped up the study of diverse systems [16,31,33–37] – even when used in a fraction-by-fraction search for active-site peptides (Figure 2, middle left).
In an effort to streamline analysis of complicated samples in Bottom Up analysis of NRPS and PKSs, an on-line RPLC-MS strategy has increasingly been used. Proteolytic digestions of in vitro reactions are loaded onto an RPLC column connected in-line with a mass spectrometer, and the LC eluent directed to the MS for fast, “on-line” analysis. Species entering the mass spectrometer can be subjected to a variety of MS and MS2 methods, including fragmentation in the ESI source (all ions) or after isolation of a single peptide’s ions for MS2 and the Ppant ejection assay for active-site peptide identification and characterization – even during analysis of highly complex mixtures of peptides (Figure 2, middle right). The observation of a Ppant ejection ion at a specific elution time alerts one to the presence of a peptide containing the T-domain active site, resulting in a significant reduction in the time required to identify the active-site containing peptide of interest.
The on-line LC-FTMS platform incorporating the Ppant ejection assay has been used to study activation and loading of a fatty acid chain in initiation of mycosubtilin biosynthesis [34], in development of a method for characterization of lipopeptide fatty acid tailoring enzymes [37•], and in study of the role of epimerization domains in intermodular peptide transfer [38]. Recently, the LC-Ppant assay was used to characterize an iterative PKS involved in aflatoxin B1 biosynthesis [39•]. In this study, LC-FTMS analysis was used to determine the relative abundances of seven acylated protein forms in the presence and absence of a product template (PT) domain, assisting in the biochemical characterization of ring formation in aflatoxin biosynthesis.
Recent Revelations: MS for Illuminating New Function
Application of FTMS to NRPS and PKS biosynthesis continues to increase as more biosynthetic systems are discovered and commercially-produced high-resolution MS instruments become available. These selected examples highlight not only the extreme mechanistic diversity of NRPSs and PKSs, but also the wide applicability of FTMS to characterization of covalent enzymology.
Enediyne Biosynthesis
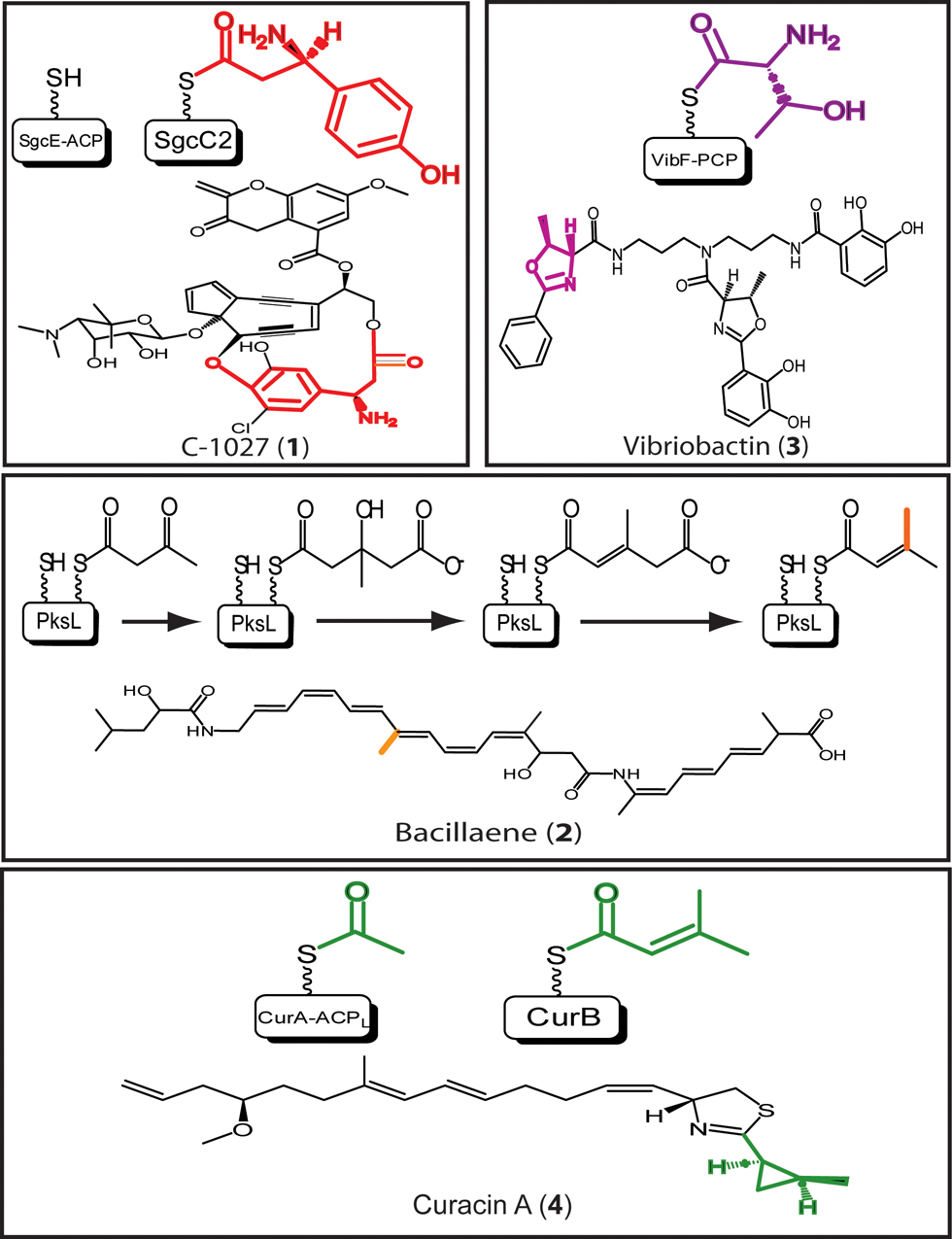
The enediyne antitumor antibiotic C-1027 (1) [40] is a member of a family of compounds characterized by their potent cytotoxicity and common biosynthetic pathways; eludication of these biosynthetic steps is crucial to exploiting the therapeutic potential of these compounds. Using FTMS, the substrate specificity of the amino-acid activating NRPS enzyme SgcC1 for β-tyrosine was confirmed, in addition to full kinetic characterization and loading efficiency of the enzyme (red in 1) [41]. The formation of an enzyme-bound intermediate during incorporation of the β-tyrosine functionality was confirmed by FTMS when the peptidyl carrier protein (an NRPS thiolation domain) SgcC2 was loaded with the β-tyrosinyl-adenylate formed by the aminomutase, SgcC4, and the adenylation domain, SgcC1 [42]. Bottom Up mass spectrometry was employed to characterize the self-phosphopantetheinylation of SgcE, a polyketide synthase with a domain architecture different from previously reported PKSs; SgcE contains a phosphopantetheinylating domain within the multi-domain PKS, as opposed to all previously reported PKSs which are serviced by a phosphopantetheinylating enzyme acting in trans [43••]. The biosynthesis of this complex natural product involves an extremely hybrid set of enzymes, ranging from homologues of enzymes involved in polyunsaturated fatty acid biosynthesis [43,44] to those involved in chorismate metabolism [45]. Novel enzymology is undoubtedly at play in this system, and it will be interesting to follow the role of FTMS in characterization of thiotemplate intermediates involved in iterative extension and tailoring.
Branching in NRPS/PKS Systems
Different biosynthetic systems for β-branch incorporation into natural products have been characterized by FTMS, including the incorporation of a methyl branch into bacillaene (orange in 2) [36••] and incorporation of two non-methyl β-branches into myxovirescin [35]. In the case of bacillaene, malonylation of the T domain AcpK by the trans-acting acyltransferase PksC was confirmed, followed by observation of the PksF-catalyzed decarboxylation of malonyl (Mal)-S-AcpK to acetyl (Ac)-S-AcpK; PksF shows homology to canonical ketosynthase domains in PKS biosynthesis and carries out the expected decarboxylation, yet lacks the active-site residue required for condensation reactions in natural product elongation. FTMS confirmed the 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase-like activity of PksG, catalyzing the transfer of the decarboxylated substrate on AcpK to polyketide intermediates on the two tandem T domains found in PksL(T2). Acetoacetyl (AcAc)-S-PksL(T2) was generated and observed as a shift of 84 and 168 Da from holo enzyme, and additional mass increases of 60 and 120 Da were observed when AcAc-S-PksL(T2) was incubated with PksG and Ac-S-AcpK, confirming transfer from AcpK to PksL(T2) and formation of HMG-S-PksL(T2). Characterization of the putative dehydratase PksH and decarboxylase PksI was complete when a loss of 18 Da per T domain in PksL was observed in the presence of PksH and HMG-S-PksL(T2) and an additional loss of 44 Da per T domain was observed with the inclusion of PksI. The elucidation of the entire β-branch incorporation by isoprenoid-like logic exemplifies the power of FTMS for biochemical characterization, and represents a virtual elimination of the need for other biochemical methods for a similar study.
Determining Inter- vs. Intra-Chain Flux for Dimeric Assembly Lines
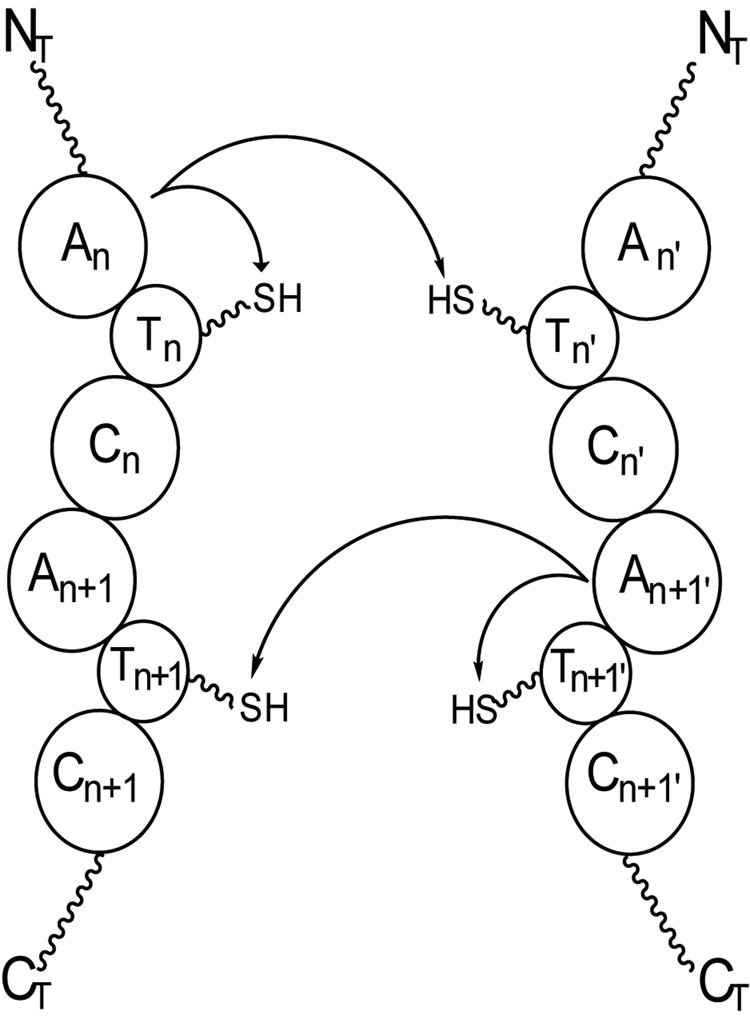
Vibriobactin (3) is a non-ribosomal peptide involved in iron acquisition in Vibrio cholerae [46], and a portion of the NRPS responsible for vibriobactin biosynthesis, VibF, exists as a protein dimer, with dimerization facilitated by a catalytically inactive condensation domain [47,48]. Mass spectrometry, in combination with generation of analytical mass mutant protein species (to distinguish between dimer partners), rapid quench methods, and kinetic analysis, was used in biophysical characterization of inter- and intra-chain acylation activities in the VibF dimer (purple in 3) [49•]. From the calculated abundances of acylated VibF T-domain active-site peptides detected by FTMS in protein combinations designed to promote inter- and/or intra-chain acyl transfer, a kinetic model of transfer was developed, revealing that while transfer of the acyl-intermediate within a single protein chain was more rapid than between dimer partners, there was no significant domination by either pathway. The method developed is easily transferred to other biosynthetic systems where similar processes involving transfer of intermediates in multimeric NRPS and PKS systems, thought to exist as megacomplexes in vivo [50], are under investigation (Figure 4).
Figure 4.
Representation of a possible interaction in a theoretical dimeric NRPS. Some NRPSs and many PKSs are known to exist as dimers in vitro, meaning that natural product biosynthesis can proceed by passing intermediates along the same polypeptide chain or between polypeptide chains in the complex. Here, an amino acid activated by An or An’ can be loaded onto either Tn or Tn’, with a similar scenario envisioned for the amino acid activated by An+1 and An+1’. This type of interaction can be monitored by FTMS, using methods similar to those utilized for the study of vibriobactin biosynthesis [49].
Biosynthesis of Curacin A
Recently, FTMS was used to elucidate biosynthetic steps involved in biosynthesis of the antiproliferative and cytotoxic mixed NRP/PK curacin A (4). A novel strategy for polyketide chain initiation was characterized, a strategy similar to that used by GCN5-related N-acetyltransferases (GNATs) [51]. Using Top Down and Bottom Up FTMS and the Ppant ejection assay, the GNAT-like domain of CurA was confirmed to decarboxylate malonyl-CoA and transfer the resulting acetyl substrate to an adjacent T domain. FTMS also assisted in characterization of two enoyl-CoA hydratase-like enzymes, CurE and the N-terminal domain of CurF [52,53••], putatively responsible for preliminary steps in generation of the curacin A cyclopropyl group (green in 4). The successive transformations catalyzed by the CurE/CurF enzyme pair were confirmed in vitro using full length CurE, a truncated form of CurF containing the N-terminal domain, and the stand-alone T domain CurB. HMG-loaded CurB (HMG-S-CurB) was generated, and incubation of HMG-S-CurB with the putative dehydratase CurE resulted in an observed loss of 18 Da from the loaded protein species; addition of CurF resulted in a total loss of 62 Da, confirming the decarboxylative action of CurF. The tandem activity of these enzymes in generation of a cyclopropyl precursor exemplifies another level of structural diversity encoded by a single enzyme family, similar to that of the branch incorporation described previously.
Conclusions: The Future of Mass Spectrometric Analysis of NRPSs and PKSs
As more MS-based analyses of NRPS and PKS systems shift to the streamlined LC-MS approaches, sample amounts required for analysis are reduced by at least one order-of-magnitude for small (~1 mm) inner diameter LC columns (from as much as 200 µg in off-line analysis to 10–50 µg for on-line LC-MS), and could be reduced at least two orders-of-magnitude for capillary-LC methods. Multiple T-domain active-site peptides are easily observable in a single LC-MS run, and are localized rapidly with the presence of multiple Ppant ejection ions. Additionally, the importance of purity of the protein sample is greatly reduced with the incorporation of the Ppant ejection assay with LC-MS platforms; this trend towards a more proteomics-like analysis system alleviates the requirement for large amounts of pure synthase enzyme.
From the few examples surveyed here, it is obvious that application of MS to the analysis of NRPS and PKS processes continues to expand and improve. The discovery rate of putative NRPS and PKS gene clusters continues to increase, and MS-based platforms are on trajectory to easily keep pace with studies of their biosynthetic machinery. As MS methods merge with those used in proteomics-based platforms [54], sample complexity will no longer be an issue. With several data acquisition and reduction tools already in place for proteomics, their application to thiotemplate systems opens the door to move beyond in vitro analysis for eventual study of endogenous biochemical processes ongoing in native producers of high-value natural products and their analogues.
Figure 3.
Structures of selected NRPS and PKS natural products discussed. Structural components of the natural products characterized by FTMS are colored. Thiolation domains and covalent intermediates detected are above the appropriate natural product structure.
Acknowledgments
This work was supported by an NIH Cell & Molecular Biology Training Grant to S.B.B. (T32 GM007283) and an NIH grant to N.L.K. (GM067725-05).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare no conflict of interest.
References and Annotations
- 1.Dorrestein PC, Kelleher NL. Dissecting non-ribosomal and polyketide biosynthetic machineries using electrospray ionization Fourier-Transform mass spectrometry. Nat Prod Rep. 2006;23:893–918. doi: 10.1039/b511400b.This review provides detailed information on FTMS applications to the study of NRPS and PKS systems, including in-depth technical details, discussion of a variety of MS-specific assays, and summaries of the use of FTMS in the study of more than ten biosynthetic systems.
- 2.Kelleher NL, Hicks LM. Contemporary mass spectrometry for the direct detection of enzyme intermediates. Curr Opin Chem Biol. 2005;9:424–430. doi: 10.1016/j.cbpa.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 3.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097.This review provides detailed information on the mechanism and enzymology of NRPS and PKS biosynthesis, including discussion of catalytic domains, domain organization, and reaction mechanisms, using examples from numerous biosynthetic pathways.
- 4.Hill AM. The biosynthesis, molecular genetics and enzymology of the polyketide-derived metabolites. Nat Prod Rep. 2006;23:256–320. doi: 10.1039/b301028g. [DOI] [PubMed] [Google Scholar]
- 5.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides1. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 6.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 7.Stein T, Vater J, Kruft V, Otto A, Wittmann-Liebold B, Franke P, Panico M, McDowell R, Morris HR. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J Biol Chem. 1996;271:15428–15435. doi: 10.1074/jbc.271.26.15428. [DOI] [PubMed] [Google Scholar]
- 8.Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 9.Ma Y, Smith LH, Cox RJ, Beltran-Alvarez P, Arthur CJ, Simpson FRST. Catalytic relationships between type I and type II iterative polyketide synthases: The Aspergillus parasiticus norsolorinic acid synthase. Chembiochem. 2006;7:1951–1958. doi: 10.1002/cbic.200600341. [DOI] [PubMed] [Google Scholar]
- 10.Izumikawa M, Cheng Q, Moore BS. Priming type II polyketide synthases via a type II nonribosomal peptide synthetase mechanism. J Am Chem Soc. 2006;128:1428–1429. doi: 10.1021/ja0559707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arthur CJ, Szafranska AE, Long J, Mills J, Cox RJ, Findlow SC, Simpson TJ, Crump MP, Crosby J. The malonyl transferase activity of type II polyketide synthase acyl carrier proteins. Chem Biol. 2006;13:587–596. doi: 10.1016/j.chembiol.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 12.McLafferty FW, Breuker K, Jin M, Han X, Infusini G, Jiang H, Kong X, Begley TP. Top-down MS, a powerful complement to the high capabilities of proteolysis proteomics. Febs J. 2007;274:6256–6268. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 13.Reid GE, McLuckey SA. 'Top down' protein characterization via tandem mass spectrometry. J Mass Spectrom. 2002;37:663–675. doi: 10.1002/jms.346. [DOI] [PubMed] [Google Scholar]
- 14.Bogdanov B, Smith RD. Proteomics by FTICR mass spectrometry: top down and bottom up. Mass Spectrom Rev. 2005;24:168–200. doi: 10.1002/mas.20015. [DOI] [PubMed] [Google Scholar]
- 15.Dorrestein PC, Blackhall J, Straight PD, Fischbach MA, Garneau-Tsodikova S, Edwards DJ, McLaughlin S, Lin M, Gerwick WH, Kolter R, et al. Activity screening of carrier domains within nonribosomal peptide synthetases using complex substrate mixtures and large molecule mass spectrometry. Biochemistry. 2006;45:1537–1546. doi: 10.1021/bi052333k.The application of FTMS to screening of adenylation domain activity is discussed as the authors present a FTMS screen for determining substrates loaded onto thiolation domains using complex substrate mixtures.
- 16.Chan YA, Boyne MT, 2nd, Podevels AM, Klimowicz AK, Handelsman J, Kelleher NL, Thomas MG. Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc Natl Acad Sci U S A. 2006;103:14349–14354. doi: 10.1073/pnas.0603748103.This paper reports the incorporation of two new polyketide building blocks by enzymes normally restricted to incorporation of malonyl or methyl-malonyl units. The data also suggest use of a NRPS-like mechanism for PKS enzymology.
- 17.Garneau S, Dorrestein PC, Kelleher NL, Walsh CT. Characterization of the formation of the pyrrole moiety during clorobiocin and coumermycin A1 biosynthesis. Biochemistry. 2005;44:2770–2780. doi: 10.1021/bi0476329. [DOI] [PubMed] [Google Scholar]
- 18.Dorrestein PC, Yeh E, Garneau-Tsodikova S, Kelleher NL, Walsh CT. Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. Proc Natl Acad Sci U S A. 2005;102:13843–13848. doi: 10.1073/pnas.0506964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garneau-Tsodikova S, Dorrestein PC, Kelleher NL, Walsh CT. Protein assembly line components in prodigiosin biosynthesis: characterization of PigA,G,H,I,J. J Am Chem Soc. 2006;128:12600–12601. doi: 10.1021/ja063611l. [DOI] [PubMed] [Google Scholar]
- 20.Hicks LM, Mazur MT, Miller LM, Dorrestein PC, Schnarr NA, Khosla C, Kelleher NL. Investigating nonribosomal peptide and polyketide biosynthesis by direct detection of intermediates on >70 kDa polypeptides by using Fourier-transform mass spectrometry. Chembiochem. 2006;7:904–907. doi: 10.1002/cbic.200500416. [DOI] [PubMed] [Google Scholar]
- 21.Dorrestein PC, Van Lanen SG, Li W, Zhao C, Deng Z, Shen B, Kelleher NL. The bifunctional glyceryl transferase/phosphatase OzmB belonging to the HAD superfamily that diverts 1,3-bisphosphoglycerate into polyketide biosynthesis. J Am Chem Soc. 2006;128:10386–10387. doi: 10.1021/ja0639362. [DOI] [PubMed] [Google Scholar]
- 22.Aron ZD, Dorrestein PC, Blackhall JR, Kelleher NL, Walsh CT. Characterization of a new tailoring domain in polyketide biogenesis: the amine transferase domain of MycA in the mycosubtilin gene cluster. J Am Chem Soc. 2005;127:14986–14987. doi: 10.1021/ja055247g. [DOI] [PubMed] [Google Scholar]
- 23.Hicks LM, Moffitt MC, Beer LL, Moore BS, Kelleher NL. Structural characterization of in vitro and in vivo intermediates on the loading module of microcystin synthetase. ACS Chem Biol. 2006;1:93–102. doi: 10.1021/cb500007v. [DOI] [PubMed] [Google Scholar]
- 24.Hicks LM, O'Connor SE, Mazur MT, Walsh CT, Kelleher NL. Mass spectrometric interrogation of thioester-bound intermediates in the initial stages of epothilone biosynthesis. Chem Biol. 2004;11:327–335. doi: 10.1016/j.chembiol.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Gatto GJ, Jr, McLoughlin SM, Kelleher NL, Walsh CT. Elucidating the substrate specificity and condensation domain activity of FkbP, the FK520 pipecolate-incorporating enzyme. Biochemistry. 2005;44:5993–6002. doi: 10.1021/bi050230w. [DOI] [PubMed] [Google Scholar]
- 26.Miller LM, Mazur MT, McLoughlin SM, Kelleher NL. Parallel interrogation of covalent intermediates in the biosynthesis of gramicidin S using high-resolution mass spectrometry. Protein Sci. 2005;14:2702–2712. doi: 10.1110/ps.051553705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLoughlin SM, Kelleher NL. Monitoring multiple active sites on thiotemplate enzymes in parallel: a molecular movie of yersiniabactin bioassembly. J Am Chem Soc. 2005;127:14984–14985. doi: 10.1021/ja0555264. [DOI] [PubMed] [Google Scholar]
- 28.Schnarr NA, Chen AY, Cane DE, Khosla C. Analysis of covalently bound polyketide intermediates on 6-deoxyerythronolide B synthase by tandem proteolysis-mass spectrometry. Biochemistry. 2005;44:11836–11842. doi: 10.1021/bi0510781.A Bottom Up MS assay using low-resolution instrumentation is presented as applied to the study of a prototypical PKS system, including the use of MS for competition and kinetic analysis of differential selection of a variety of intermediates by a ketosynthase domain. Active-site peptides from multiple domains with covalently-tethered intermediates were identified.
- 29.Wells JM, McLuckey SA. Collision-induced dissociation (CID) of peptides and proteins. Methods Enzymol. 2005;402:148–185. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 30.Little DP, Speir JP, Senko MW, O'Connor PB, McLafferty FW. Infrared multiphoton dissociation of large multiply charged ions for biomolecule sequencing. Anal Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 31.Dorrestein PC, Bumpus SB, Calderone CT, Garneau-Tsodikova S, Aron ZD, Straight PD, Kolter R, Walsh CT, Kelleher NL. Facile detection of acyl and peptidyl intermediates on thiotemplate carrier domains via phosphopantetheinyl elimination reactions during tandem mass spectrometry. Biochemistry. 2006;45:12756–12766. doi: 10.1021/bi061169d.The development and implementation of the Ppant ejection assay for NRPS and PKS analysis is discussed in detail, in addition to presentation of numerous examples of its applications in a variety of systems.
- 32.Meluzzi D, Zheng WH, Hensler M, Nizet V, Dorrestein PC. Top-down mass spectrometry on low-resolution instruments: Characterization of phosphopantetheinylated carrier domains in polyketide and non-ribosomal biosynthetic pathways. Bioorg Med Chem Lett. 2007 doi: 10.1016/j.bmcl.2007.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly WL, Boyne MT, 2nd, Yeh E, Vosburg DA, Galonic DP, Kelleher NL, Walsh CT. Characterization of the aminocarboxycyclopropane-forming enzyme CmaC. Biochemistry. 2007;46:359–368. doi: 10.1021/bi061930j. [DOI] [PubMed] [Google Scholar]
- 34.Hansen DB, Bumpus SB, Aron ZD, Kelleher NL, Walsh CT. The loading module of mycosubtilin: an adenylation domain with fatty acid selectivity. J Am Chem Soc. 2007;129:6366–6367. doi: 10.1021/ja070890j. [DOI] [PubMed] [Google Scholar]
- 35.Calderone CT, Iwig DF, Dorrestein PC, Kelleher NL, Walsh CT. Incorporation of nonmethyl branches by isoprenoid-like logic: multiple beta-alkylation events in the biosynthesis of myxovirescin A1. Chem Biol. 2007;14:835–846. doi: 10.1016/j.chembiol.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Convergence of isoprene and polyketide biosynthetic machinery: isoprenyl-S-carrier proteins in the pksX pathway of Bacillus subtilis. Proc Natl Acad Sci U S A. 2006;103:8977–8982. doi: 10.1073/pnas.0603148103.This paper discusses the use of FTMS for eludication of the mechanism of β-branch incorporation by isoprenoid-like logic in the biosynthesis of bacillaene, highlighting the ability of FTMS to confirm the entire reaction sequence. A similar mechanism is reported for a different system in [35].
- 37.Kopp F, Linne U, Oberthur M, Marahiel MA. Harnessing the chemical activation inherent to carrier protein-bound thioesters for the characterization of lipopeptide fatty acid tailoring enzymes. J Am Chem Soc. 2008;130:2656–2666. doi: 10.1021/ja078081n.This article discusses the use of on-line LC-MS and the Ppant ejection assay in characterization of three enzymes involved in the biosynthesis of calcium-dependent antibiotic; FTMS and Ppant ejection were applied to confirm phosphopantetheinylation of a putative thiolation domain and characterize an oxidase with intrinsic epoxidase activity that acts at covalently-bound intermediates.
- 38.Stein DB, Linne U, Hahn M, Marahiel MA. Impact of epimerization domains on the intermodular transfer of enzyme-bound intermediates in nonribosomal peptide synthesis. Chembiochem. 2006;7:1807–1814. doi: 10.1002/cbic.200600192. [DOI] [PubMed] [Google Scholar]
- 39.Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711.Application of on-line LC-FTMS and the Ppant ejection assay to characterization of an iterative polyketide synthase is presented, including observation of multiple intermediates on a single thiolation domain; discussed is a product template domain responsible for mediation of a cyclization cascade of the iteratively-incorporated malonyl units.
- 40.Liu W, Christenson SD, Standage S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 41.Van Lanen SG, Lin S, Dorrestein PC, Kelleher NL, Shen B. Substrate specificity of the adenylation enzyme SgcC1 involved in the biosynthesis of the enediyne antitumor antibiotic C-1027. J Biol Chem. 2006;281:29633–29640. doi: 10.1074/jbc.M605887200. [DOI] [PubMed] [Google Scholar]
- 42.Van Lanen SG, Dorrestein PC, Christenson SD, Liu W, Ju J, Kelleher NL, Shen B. Biosynthesis of the beta-amino acid moiety of the enediyne antitumor antibiotic C-1027 featuring beta-amino acyl-S-carrier protein intermediates. J Am Chem Soc. 2005;127:11594–11595. doi: 10.1021/ja052871k. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Van Lanen SG, Ju J, Liu W, Dorrestein PC, Li W, Kelleher NL, Shen B. A phosphopantetheinylating polyketide synthase producing a linear polyene to initiate enediyne antitumor antibiotic biosynthesis. Proc Natl Acad Sci U S A. 2008;105:1460–1465. doi: 10.1073/pnas.0711625105.These three manuscripts [41–43] discuss the application of FTMS to study of a variety of steps in the biosynthesis of the enediyne compound C-1027, specifically characterization of a PKS with novel domain structure and determination of SgcC1 substrate specificity for β-tyrosine.
- 44.Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, Valentine R, Lardizabal K, Domergue F, Yamada A, et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. 2001;293:290–293. doi: 10.1126/science.1059593. [DOI] [PubMed] [Google Scholar]
- 45.Van Lanen SG, Lin S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027 involves a new branching point in chorismate metabolism. Proc Natl Acad Sci U S A. 2008;105:494–499. doi: 10.1073/pnas.0708750105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffiths GL, Sigel SP, Payne SM, Neilands JB. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem. 1984;259:383–385. [PubMed] [Google Scholar]
- 47.Hillson NJ, Balibar CJ, Walsh CT. Catalytically inactive condensation domain C1 is responsible for the dimerization of the VibF subunit of vibriobactin synthetase. Biochemistry. 2004;43:11344–11351. doi: 10.1021/bi0489199. [DOI] [PubMed] [Google Scholar]
- 48.Hillson NJ, Walsh CT. Dimeric structure of the six-domain VibF subunit of vibriobactin synthetase: mutant domain activity regain and ultracentrifugation studies. Biochemistry. 2003;42:766–775. doi: 10.1021/bi026903h. [DOI] [PubMed] [Google Scholar]
- 49.Hicks LM, Balibar CJ, Walsh CT, Kelleher NL, Hillson NJ. Probing intra- versus interchain kinetic preferences of L-Thr acylation on dimeric VibF with mass spectrometry. Biophys J. 2006;91:2609–2619. doi: 10.1529/biophysj.106.084848.FTMS is used in combination with a variety of analytical techniques in a biophysical study of the dimeric protein VibF; the authors determined kinetic parameters for inter- and intra-chain acylation activities using MS measurements.
- 50.Straight PD, Fishbach MA, Walsh CT, Rudner DZ, Kolter R. A singular enzymatic megacomplex from Bacillus subtilis. PNAS. 2007;104:305–310. doi: 10.1073/pnas.0609073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu L, Geders TW, Wang B, Gerwick WH, Hakansson K, Smith JL, Sherman DH. GNAT-like strategy for polyketide chain initiation. Science. 2007;318:970–974. doi: 10.1126/science.1148790. [DOI] [PubMed] [Google Scholar]
- 52.Gu L, Jia J, Liu H, Hakansson K, Gerwick WH, Sherman DH. Metabolic coupling of dehydration and decarboxylation in the curacin A pathway: functional identification of a mechanistically diverse enzyme pair. J Am Chem Soc. 2006;128:9014–9015. doi: 10.1021/ja0626382. [DOI] [PubMed] [Google Scholar]
- 53.Geders TW, Gu L, Mowers JC, Liu H, Gerwick WH, Hakansson K, Sherman DH, Smith JL. Crystal structure of the ECH2 catalytic domain of CurF from Lyngbya majuscula. Insights into a decarboxylase involved in polyketide chain beta-branching. J Biol Chem. 2007;282:35954–35963. doi: 10.1074/jbc.M703921200.These three manuscripts [51–23] discuss two biosynthetic components of the curacin A biosynthetic pathway for which FTMS was used in characterization. Of particular interest is the report of a novel loading mechanism for PKS initiation using a GNAT-like strategy, an example of the extreme mechanistic diversity in these systems.
- 54.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]