Abstract
Purpose
Melanolipofuscin (MLF) is a complex granule, exhibiting properties of both melanosomes and lipofuscin (LF) granules, which accumulates in retinal pigment epithelial (RPE) cells and may contribute to the etiology of age-related macular degeneration (AMD). MLF accumulation has been reported by Feeney-Burns to more closely reflect the onset of AMD than the accumulation of lipofuscin. In an effort to assess the possible contribution MLF may have to the onset of AMD, we analyzed the phototoxicity and protein composition of MLF and compared those results to that of LF.
Methods
Specifically, we observed the accumulation of MLF in human RPE from different decades of life, and assessed the phototoxicity of these granules. We also employed fluorescence spectroscopy, atomic force microscopy, transmission and scanning electron microscopy and proteomic analysis to examine the composition of MLF granules in an effort to ascertain their origin.
Results
Our results show that MLF granules are phototoxic and their accumulation more closely reflects the onset of AMD than does LF accumulation. Our compositional analysis of MLF has shown that while these granules contain some similarities to LF granules, MLF is substantially different. Of significant interest is the finding that MLF, in contrast to LF, does not contain photoreceptor-specific proteins, suggesting that MLF may not originate from the phagocytosis of photoreceptor outer segments. Instead the presence of RPE- and melanosome-specific proteins would suggest that MLF accumulates as a result of the melanosomal autophagocytosis of RPE cells.
Conclusions
Our results provide significant insight into understanding the formation and toxicity of MLF and suggest a possible contribution to the etiology of retinal diseases.
Introduction
Several retinal diseases, including age-related macular degeneration (AMD), have been associated with the accumulation of autofluorescent granules in retinal pigment epithelial (RPE) cells. One such autofluorescent granule, lipofuscin (LF), may relate to the onset of these ocular diseases because it has been shown to generate reactive oxygen species via photosensitization with blue light [1-4]; which may cause damage and death of the RPE with subsequent death of the surrounding cells. However, as Feeney-Burns has reported [5], the accumulation of LF in human RPEs is not consistent with the onset of AMD. The most dramatic increase of LF in human RPEs, a 95% increase, occurs between young and middle-aged groups (defined as ages 1-20 and 21-60, respectively) while there is only a 21% increase between middle-aged and old-aged groups (ages 61-100) [5].
Another autofluorescent granule that accumulates in RPE cells and may contribute to the etiology of AMD is a complex granule exhibiting properties of both melanosomes and lipofuscin granules called melanolipofuscin (MLF). Although it is generally accepted that dermal melanin protects the skin from UV light damage, the biological function of RPE melanin is not completely understood. Melanin is known to absorb excess light passing through the eye, thereby reducing scatter and improving image resolution. It has also been suggested to play a photoprotective role in RPE cells [6,7] by scavenging reactive oxygen species (ROS) [8-10]. Evidence also exists for a phototoxic role for melanin in RPE cells, especially in aged cells, including measurable ROS photoproduction [6,9,11-13]. Melanosomes have been observed to undergo morphological and photophysical changes with age, possibly due to oxidation, which may result in diminished antioxidant potential. Studies have reported that aged human melanosomes are highly photoreactive and can result in RPE dysfunction, while young melanosomes appear to confer photoprotection [14-16]. With increasing age, a decrease in melanosomes in the RPE is observed along with an increase in melanolipofuscin (MLF) [17-19]. In contrast with the accumulation of LF in the RPE, MLF accumulation has been reported by Feeney-Burns to more closely reflect the onset of AMD. MLF exhibits a 15% increase between young and middle aged groups and a 50% increase between middle-aged and old-aged groups [5].
Oxidative stress has been suggested to be a major contributing factor for retinal degeneration in AMD. The retinas constant exposed to light and a relatively high oxygen pressure, which is close to that found in arterial blood, contributes to light-induced oxidative stress in the retina which may result in oxidative damage to biomolecules in these cells. RPE cells are post mitotic and therefore must respond to a life time of oxidative insult. While there are numerous mechanisms for preventing and combating oxidative injuries, by middle-age many of these anti-oxidative mechanisms have begun to break down, which can increase the susceptibility of RPE cells to accumulated damage. LF and MLF granules are thought to result from the accumulation of undegradable material in RPE cells. Modifications, including oxidation, may render the molecules in these granules undegradable by the cell, contributing to their accumulation.
While the identification of photoreceptor- and lysosomal-specific proteins in LF granules has provided evidence that LF originates from the accumulation of undigested material from the phagocytosis of photoreceptor disc in RPE lysosomes [20], little is known about the composition and origin of MLF granules. Two models for the origin of MLF have been suggested. The first model involves the autophagy of preexisting melansomes and their incorporation into accreting LF granules. This model is supported by the observation that phagosomes containing undegradable material fuse with melanosomes [7]. The second model is that melanin is synthesized de novo in lysosomes, which subsequently fuse with accreting LF granules. This model is supported by evidence that synthesis of melanin in depigmented RPE cells is seen in lysosomal compartments [19,21]. Knowledge of the composition of MLF could provide significant insight into the origin of these granules, and determining the phototoxicity of these granules could be useful for ascertaining MLF's role in the etiology of AMD and other retinal diseases.
In the present study we observed the accumulation of MLF in human RPE from different decades of life and assessed the phototoxicity of these granules. We also employed fluorescence spectroscopy, atomic force microscopy, transmission and scanning electron microscopy and proteomic analysis-using 1D gel electrophoresis coupled with ESI mass spectrometry-to examine the composition of MLF granules in an effort to ascertain their origin. Collectively these data provide significant insight into understanding the formation and toxicity of MLF and suggest a possible contribution to the etiology of retinal diseases. Specifically, these data do not provide direct support for either previously suggested hypothesis for the origin of MLF, but instead suggest that MLF accumulates as a result of the melanosomal autophagocytosis of RPE cells. To our knowledge this is the first report of the phototoxicity and biochemical analysis of retinal melanolipofuscin.
Methods
Lipofuscin and Melanolipofuscin Isolation and Fluorescence - Lipofuscin and melanolipofuscin granules were isolated as previously described [20,22], using a method that has been widely utilized for this process. Briefly, granules were isolated from human RPE from donor eyes, provided by Dr. Paul Bernstein of the Moran Eye Institute, University of Utah, Salt Lake City, UT. The time between donor death and enucleation was 1-4 h, after which the donor eyes were stored at 4 °C until dissection. Dissections were carried out by Dr. Bernstein's lab at the Moran 6-24 h after donor death in a dim light environment. RPE's were shipped to BYU on dry ice and stored at -75 °C until use. Lipofuscin granules were isolated from the band at the 0.3 M/1.0 M interface and melanolipofuscin granules were isolated from the bands at the 1.0 M/1.2 M and 1.2 M/1.4 M interface. The granules were removed from the gradients by inserting a needle through the side of the tube and extracting the bands sideways so as to minimize contamination of the bands. The material at the border of the LF and MLF bands was not removed from the gradients but was used as a buffer zone to keep the two samples separate from each other during the extraction process. Sucrose gradients were only briefly exposed to light while photographs of the gradients were taken. Fluorescence spectra of isolated lipofuscin and melanolipofuscin granules were acquired as described by Boulton et al. [22] using a Jobin Yvon Fluoromax-3 Spectrofluorometer (Edison, NJ).
Lipofuscin and melanolipofuscin accumulation
To study the accumulation of LF and MLF over time, sucrose gradient centrifugation was employed using four groups of RPE, each consisting of 6 RPE and representing a different decade of life. The first group had an average age of 33±1.6 yrs; the second group had an average age of 43±0.9 yrs; the third group had an average age of 54.3±1.9 yrs; and the fourth group had an average age of 64±0.0 yrs. To compare the LF and MLF content in young and old eyes, sucrose gradients were run with 11 RPE from young eyes (average age of 21.2±5.9 yrs) and 14 RPE from old eyes (average age of 66.5±5.9 yrs). Pictures of the gradients were taken using an Olympus Camedia digital camera. Image J (National Institutes of Health) was used to measure the optical density of LF and MLF bands in the sucrose gradients.
All other experiments were performed using LF and MLF isolated from RPE's taken from a random donor population between 40 and 80 years old.
Cell culture
Human retinal pigment epithelial cells (ARPE-19; ATCC-CRL-2302) were grown in 24-well tissue culture plates in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS). Upon reaching confluency the fetal bovine serum (FBS) in the media was reduced to 1%. Cells were either maintained in RPMI 1640 media supplemented with 1% FBS or incubated in the same media which also contained about 300 melanolipofuscin or lipofuscin granules/cell for 24 h to allow for ingestion of the granules. After the 24 h incubation, the melanolipofuscin- or lipofuscin-fed RPE cells were transferred back to RPMI 1640 media supplemented with 1% FBS and maintained for 3 days before bioactivity assay.
Bioactivity assay
To investigate the bioactivity of MLF, ARPE-19 cells that were fed LF, MLF or neither (control cells) were either subjected to blue light (390-550 nm) for 48 h at an intensity of about 2.8 mW/cm2 or maintained in the dark. This intensity of light, or even higher intensities, have previously been used by investigators to determine the effect of blue light exposure on the retina [16,23]. Blue light was introduced into a 5% CO2 humidified cell incubator using a Mille Luce M1000 Fiber Optic Illuminator with a 150 W quartz halogen bulb, a 25 mm dichroic blue light filter, and a 48 inch fiber optic cable (Edmund Optics, Barrington, NJ). Photocytotoxicity of the lipofuscin and melanolipofuscin granules was assessed using Sulforhodamine B (JNCI 82, p1107) to measure cell viability. Briefly, cultures were fixed with trichloroacetic acid and stained with 0.4% sulforhodamine B in 1% acetic acid. The cultures were washed 4 times with 1% acetic acid to remove any unbound dye; protein-bound dye was extracted with 10 mM unbuffered Tris base and transferred to a 96 well culture plate. Absorbance was measured at 570 nm on a CERES UV900 HDi plate reading spectrophotometer (Bio-Tek Instruments, Winooski, VT).
Microscopy
MLF granules were prepared for scanning electron microscopy (SEM) analysis by drying the granules on a silicon wafer and sputter coating them with gold. The granules were analyzed on a Phillips XL30 ESEM FEG using a 5 kV accelerating potential. For transmission electron microscopy (TEM) analysis MLF granules were fixed in glutaraldehyde, postfixed in osmic acid, dehydrated and embedded in epoxy resin. 100 nm slices of the sample were imaged and photographed on a JOEL JEM 2000 FX.
MLF samples for atomic force microscopy (AFM) were prepared by drying the granules onto a mica slide. Images were taken with a Multimode IIIa AFM instrument with microfabricated Si cantilever tips (Nanoscience Instruments, Phoenix, AZ). Vibrational noise was dampened using an active isolation system (MOD1-M, Halcyonics, Goettingen, Germany). Typical imaging parameters were (a) tip resonance frequency, 55-65 kHz; (b) amplitude setpoint, 2.0-2.5 V; (c) scan rate, 2.0 Hz. Images were processed offline to remove the background slope using software bundled with the AFM instrument.
Flow cytometric analysis
To determine the size distribution and concentration (granules/unit volume), suspensions of MLF granules were diluted 1:100 and 1:1000 with PBS and 200,000 Flow Check High Intensity Green Alignment Beads (Polysciences, Inc., Warrington, PA), 5.726±0.375 μm in diameter, were added to each sample to serve as an internal standard. The samples were excited with an argon laser at 488 nm on a Beckman Coulter (Beckman, Fullerton, CA) EPICS-XL Flow Cytometer with EXPO 32 ADC software for flow cytometric analysis. The samples were analyzed for forward light scatter and autofluorescence by collection of data for 300 s, which allowed visualization of at least 10,000 beads and at least 95,000 MLF granules.
Total protein determination in melanolipofuscin
Melanolipofuscin granules were pelleted using centrifugation and lyophilized in an evaporative centrifugal concentrator. The granules were weighed using a Mettler UMT2 microbalance (Columbus, OH) to determine their total mass. After weighing the dried granules, the protein in these melanolipofuscin samples was quantified by solubilizing the granules in 1% SDS followed by the BCA Protein Assay (Pierce, Rockford, IL). Three independent measurements were used to calculate the percent protein and standard deviation.
1D gel electrophoresis and mass spectrometry
Melanlolipofuscin and lipofuscin granules containing 100 _g of protein were pelleted by centrifugation, solubilized in 4X Laemmli buffer (3% SDS, 0.17 M Tris pH 6.8, 35% glycerol, 3.5% 2-mercaptoethanol) and separated on a 10% SDS-PAGE gel (8.3x6.4x0.1 cm). The gel lanes were sliced into sections and the proteins were digested in-gel as described by Shevchenko et al. [24], injected onto a Jupiter C18 reversed-phase resin capillary column (150 μm ID, made in-house), and eluted using a gradient of 5-95% acetonitrile in 0.1% formic acid at a flow rate of 5 μl/min. On-line mass spectrometric analysis was performed on an Applied Biosystems QSTAR Pulsar i (Foster City, CA) using an API (atmospheric pressure ionization) source. Automated tandem mass spectrometry using information-dependent acquisition was run, collecting CID spectra for the three most intense ions from each survey scan excluding peaks chosen in the preceding 2 min. Fragmentation spectra were submitted to the Mascot (Matrix Science) website for peptide identification. Proteins in MLF granules from three independent preparations were examined. Each 1D gel lane containing MLF or LF proteins was cut into 24 gel slices for mass spectrometric analysis. Four gel slices (numbers 15, 16, 17, and 19) from two preps of LF granules were selected for analysis to provide a direct comparison of the differences in LF and MLF proteins. Relative quantization of proteins was estimated using the method of spectral counting [25].
Immunoblots
Human retinas were obtained from Dr. Paul Bernstein from the Moran Eye Institute to make a positive control for rhodopsin. A retina was gently triturated in 0.75 M sucrose, 0.68 mM calcium chloride, 20 mM tris and 1 mM DTT pH 7.4 to rupture the cells. The suspension was poured over 4 thicknesses of cheesecloth to remove debris. The sample was spun at 1475 xg for 20 min and the pellet was resuspended in 1% SDS. Total protein was determined using the BCA assay. An α-rhodopsin antibody (R4, polyclonal antibody, see Takemoto et al.) [26] was used at 1:1000 dilution in TBS-T.
Oxidized bovine serum albumin (BSA) samples were made by incubating BSA in hypochlorous acid at 37 °C for 30 min. LF, MLF and BSA samples were derivatized by incubating them in 5% sodium dodecyl sulfate and 10 mM 2,4-dinitrophenylhydrazine (DNPH) in 10% (v/v) trifluoroacetic acid for 30 min at room temperature. The solutions were neutralized by adding 2M Tris and Laemmli buffer (3% SDS, 0.17 M Tris pH 6.8, 35% glycerol, 3.5% 2-mercaptoethanol) and loaded directly onto a gel. Anti-DNP antibody from rabbit was purchased from Sigma.
Results
Side by side comparison of the LF and MLF content in RPE from young (21.2±5.9 yr) and old (66.5±5.9 yr) individuals as seen in sucrose gradients is shown in Figure 1A. This figure confirms the presence of significant quantities of LF in RPEs from young individuals where very little if any MLF is present. In contrast RPEs from older individuals show significant quantities of MLF. This accumulation pattern is further evident when the optical density of the MLF and LF bands in sucrose density gradients is plotted versus the age of the RPE donor (Figure 1B).
Figure 1.
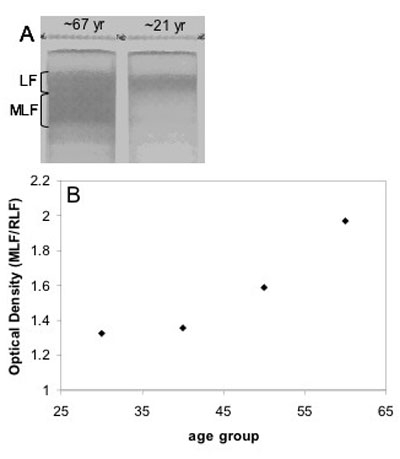
Melanolipofuscin accumulation. A: Comparison of Melanolipofuscin (MLF) granules from retinal pigment epithelium (RPE) of young (21.2±5.9 yrs) and old (66.5±5.9 yrs) human donors. B: Age of RPE donor versus optical density of MLF/LF plotted to show the accumulation of MLF throughout life. Lipofuscin (LF) granules are observed in human RPE from young individuals, whereas significant quantities of MLF do not appear to accumulate for decades afterward.
Analysis of the phototoxicity of MLF revealed that these granules cause a 58% decrease in cell viability in ARPE-19 cells fed with MLF and exposed to blue light for 48 h. This is compared to an 80% decrease in cell viability in ARPE-19 cells fed with the same number of granules of LF (Figure 2). Although this colorimetric assay provides an informative approximation of the phototoxicity of these granules, we are aware that the nature of ARPE-19 cells makes it difficult to precisely determine the phototoxicity of these granules. ARPE-19 cells migrate up the sides of tissue culture plates as they proliferate and phagocytose far fewer granules in this sideways position. These cells depress phototoxicity results because their inability to phagocytose granules inhibits them from undergoing light dependent cell death. Thus, these phototoxicity measurements are overly conservative. However, the relative comparison of LF and MLF phototoxicity is not affected.
Figure 2.
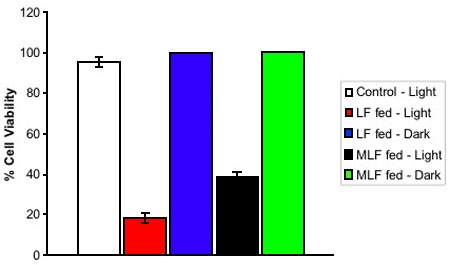
Bioactivity of lipofuscin and melanolipofuscin granules. Isolated lipofuscin (LF) and melanolipofuscin (MLF) granules were fed to ARPE-19 cells. Cells that were not fed LF or MLF were used as controls. Cells were then subjected to blue light irradiation (solid bars), or left in the dark (cross-hatched), for 48 h. Cell viability was determined using the Sulforhodamine B assay. Values represent mean of at least four independent measurements, error bars represent standard deviation. The phototoxicity of MLF granules in ARPE-19 cells is at least 72% as potent as that of LF granules, showing that MLF granules have the potential for deleterious affects on RPE cells in the retina.
Several physical measurements of MLF granules were made using fluorescence spectroscopy, electron microscopy, atomic force microscopy, and flow cytometry. Fluorescence of MLF and LF granules is shown in Figure 3. Both granules produce similar excitation spectra (data not shown), however, MLF granules have an emission maximum at 554 nm, whereas LF granules have an emission maximum at 578 nm. The similarity between the fluorescence spectra of these two granules is expected because of the A2E fluorophore present in both granules which dominates the spectra [27]. Apparently, melanin in MLF produces significantly less emission and appears to be negligible in comparison to the fluorescence of A2E (data not shown). The shoulder at about 470 nm in the fluorescence spectrum of MLF increased over time when exposed to light eventually becoming the maximum in the spectrum (data not shown). This change may result from the accumulation of damage on the proteins within MLF granules as a result of light exposure or from photo-isomerization of A2E or other lipids. This trend was also observed with LF (data not shown).
Figure 3.
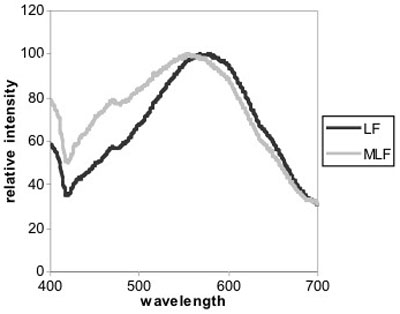
Fluorescence emission spectra of lipofuscin and melanolipofuscin. Fluorescence emission of lipofuscin (LF) and melanolipofuscin (MLF) monitored with excitation at 364 nm. Both granules produce similar excitation spectra (data not shown), however, MLF granules have an emission maximum at 554 nm, whereas LF granules have an emission maximum at 578 nm.
SEM and AFM analyses of MLF granules (Figure 4A-C) show nearly spherical granules with some surface features which suggest these granules are aggregates of smaller substructures. Transmission electron micrographs of MLF (Figure 4D) show these granules to contain inclusions of higher density, demonstrating that these granules are complex and not a mixed population of different granules.
Figure 4.
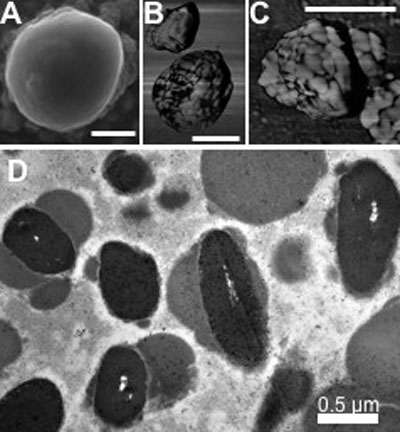
Microscopic structure of melanolipofuscin. A: Scanning electron micrograph of melanolipofuscin (MLF), showing nearly spherical granules with some surface features. B, C: Atomic force micrographs (phase images) showing MLF granules to be aggregates of about 200 nm and about 50 nm substructures. D: Transmission electron micrograph of MLF, shows these granules to contain inclusions of higher density, demonstrating that these granules are complexes of lipofuscin and melanin. Each bar represents 0.5 μm.
Flow cytometric (FC) analysis allowed a quantitative determination of granule size. Forward light scatter in FC instruments is directly proportional to the size of the objects passing through the beam. The MLF granules were found to have a mean diameter of 0.93 μm and a broad standard deviation of 0.60 μm. Flow cytometry also enabled a quantitative determination of the concentration of granules in our suspensions. Having a count of the MLF granules, we were able to determine their average weight, which proved to be 2.2±0.1 pg/granule. When compared to LF, MLF is about 35% larger but weighs about 69% more, again indicating the presence of a more dense substance.
To determine the percent protein composing MLF granules, the protein in a known quantity of MLF was solubilized in 1% SDS and quantified by BCA assay. MLF proved to consist of 60.7±6.4% total protein. Compared to LF, MLF contains more protein and therefore less extractable lipids (see Warburton 2005) [20].
Because of the possibility that the proteins in MLF granules are highly modified exhibiting highly heterogeneous populations and therefore unfocusable on 2D gels, we employed 1D SDS-PAGE coupled with automated LCMSMS to identify the protein constituents of MLF. Figure 5 shows representative 1D lanes of LF and MLF proteins and indicates the gel slices removed from the lanes. Proteomic analyses of the proteins in the 1D gel slices identified 110 proteins in MLF granules which are listed in Table 1. The proteome of MLF granules was compared to the proteome of other relevant organelles including RPE melanosomes, macrophage phagosomes, retinal LF, and melanocyte melanosomes. As indicated in Table 2, 23 proteins were previously identified in mature RPE melanosomes [28], 18 proteins were previously identified as part of the macrophage phagosome proteome [29], 14 proteins were previously identified in LF granules [20], and 7 proteins were identified in melanocyte melanosomes [30].
Figure 5.
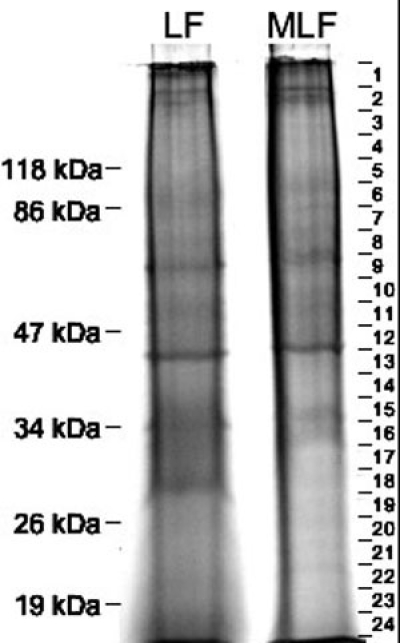
Electrophoresis of lipofuscin and melanolipofuscin proteins. Representative SDS-PAGE lanes of 50 μg of lipofuscin (LF) and melanolipofuscin (MLF) proteins. Mobility of molecular weight markers are indicated to the left. On the right, gel slices taken for subsequent in-gel digestion are shown. The lack of well-focused bands in the gel lane indicates microheterogeneous populations of the proteins, probably resulting from extensive modifications.
Table 1. Proteins identified in melanolipofuscin granules.
| Gel | Protein | Gel slice | Genbank Accession number | Synonyms | Subcellular location | Tissue specificity |
| 1,2 | acid ceramidaseIII | 13 | AAC73009 | putative heart protein | Lysosomal | Widely expressed |
| 1,2,3 | actin, betaIII | 11, 12, 13, 14, 15, 16 | AAH08633 | |||
| 2 | alpha actinin 4II | 6 | NP_004915 | F-actin cross linking protein | Nuclear and cytoplasmic | Widely expressed |
| 1 | alpha tubulin, ubiquitous | 1, 2, 4, 5, 6, 7, 9, 10, 13, 15, 17, 18, 19, 21 | NP_006073 | |||
| 3 | alpha tubulin 2 | 9, 10, 11, 24 | CAA25855 | |||
| 3 | alpha tubulin 4I | 10 | NP_079295 | |||
| 1,2,3 | alpha tubulin 6III | 1, 5, 10, 11, 12, 13, 14, 18, 20, 21, 23, 24 | AAH04949 | |||
| 3 | ankyrin 1IV | 3,4 | A35049 | Cytoplasmic surface of erythrocytic plasma membrane | Erythrocyte | |
| 1,2,3 | annexin A2I | 12, 13, 14, 15 | AAH23990 | |||
| 2 | annexin A5III | 15, 16 | AAH01429 | |||
| 2,3 | aspartate aminotransferase | 14 | NP_002071 | transaminase A, Glutamate oxaloacetate transaminase-2 | Mitochondrial matrix | |
| 3 | ATP Synthase subunit g | 24 | AAC61597 | |||
| 2,3 | ATP Synthase, H+ transporting, mitochondrial F0 complex, subunit 6 | 22 | AAQ88428 | |||
| 1,2,3 | ATP Synthase, H+ transporting, mitochondrial F0 complex, subunit b | 18 | AAH05960 | Mitochondrial | ||
| 2,3 | ATP Synthase, H+ transporting, mitochondrial F0 complex, subunit d | 22 | NP_006347 | |||
| 1,2,3 | ATP Synthase, H+ transporting, mitochondrial F0 complex, subunit f | 23 | AAH03678 | |||
| 1,2,3 | ATP Synthase, H+ transporting, mitochondrial F1 complex, alpha subunit | 2, 9,10 | AAP35873 | |||
| 1,2,3 | ATP Synthase, H+ transporting, mitochondrial F1 complex, beta subunitII,III | 10 | NP_001677 | Widely expressed | ||
| 2,3 | ATP Synthase, H+ transporting, mitochondrial F1 complex, gamma subunit | 16 | AAH16812 | Mitochondrial | ||
| 1,2,3 | ATP Synthase, H+ transporting, mitochondrial F1 complex, subunit o | 20 | AAV38639 | Mitochondrial matrix | ||
| 1,2,3 | beta tubulinII,III | 10, 11, 14, 15, 23 | AAH29529 | Ubiquitously expressed | ||
| 3 | beta tubulin polypeptide | 10, 11, 12, 13, 21 | AAH01938 | |||
| 1,2,3 | calnexinI,II,III | 6 | I53260 | MHC I antigen binding protein p88 | Type I membrane protein Endoplasmic reticulum | |
| 2,3 | cathepsin DI,II,III | 15, 16, 17 | AAP35556 | Lysosomal | ||
| 1,2 | cell death-regulatory protein GRIM19 | 23 | NP_057049 | NADH-ubiquinone oxidoreductase B16.6 subunit, Gene associated with retinoic-interferon-induced mortality 19 | Mitochondrial inner membrane protein | Widely expressed |
| 2,3 | cerebroside sulfate activator protein | 23 | AAA36594 | |||
| 2,3 | ceroid-lipofuscinosisI,II lysosomal pepstatin insensitive protease | 12, 13 | AAH14863 | tripeptidyl-peptidase I precursor | Lysosomal | All tissue |
| 1,2,3 | chromosome 10 open reading frame 58 | 19, 21 | AAQ89418 | SFLQ611 | ||
| 3 | chromosome 10 open reading frame 70 | 24 | AAH59168 | |||
| 3 | chromosome 20 open reading frame 3 | 12 | AAQ89435 | Type II membrane protein | ||
| 3 | chromosome 8 open reading frame 2 | 13 | AAQ88475 | Membrane-associated | Ubiquitous | |
| 2,3 | crystallin, alpha BII | 22 | AAP35416 | heat shock protein B5 | Lens as well as other tissues | |
| 2,3 | cytochrome c oxidase subunit III | 22 | CAA39187 | Integral membrane protein. Mitochondrial inner membrane | Ubiquitous | |
| 2 | cytochrome c oxidase subunit IV | 23 | AAV38628 | Mitochondrial inner membrane differentiated epidermis of palms and soles | ||
| 1 | cytokeratin 9IV | 1 | CAA82315 | Terminal | ||
| 1 | epidermal cytokeratin 2IV | 1 | AAC83410 | Epidermal tissue, Squamous metaplasias and carcinomas | ||
| 2 | erythryocyte membrane protein band 4.2IV | 7, 8 | NP_000110 | Membrane-associated and cytoplasmic | Erythrocyte | |
| 2 | gamma glutamyltransferase like activity 1 | 10, 11 | NP_004112 | Type II membrane protein | ||
| 2 | glucose regulated proteinIII protein, hsp A5 | 7 | CAB71335 | dnaK-type molecular chaperone, BiP | Endoplasmic reticulum lumen | |
| 1 | gp25L2 | 19 | CAA62380 | Type I membrane protein Endoplasmic reticulum | ||
| 2,3 | guanine nucleotide binding protein | 16 | AAA52584 | G(o) alpha subunit I | ||
| 1 | guanine-nucleotide binding protein, alpha transducing | 13 | NP_653082 | G protein | ||
| 1,2 | heat shock 70 kDa protein 9BIII 8, 9 AAH30634 mortalin-2 | |||||
| 1 | heat shock protein 27 responsive protein | 18, 19 | AAA62175 | estrogen-regulated protein, stress | Mitotic spindles in mitotic cells; nucleus during heat shock | |
| 2 | heat shock protein 60III | 10 | AAA36022 | chaperonin 60, mitochondrial matrix protein | Mitochondrial matrix | |
| 2,3 | heat shock protein gp96 regulated protein | 6 | AAK74072 | tumor rejection antigen, 94kD glucose | Endoplasmic Reticulum lumen | |
| 1,2,3 | hemoglobin beta chainIV | 23 | AAD19696 | Red blood cells | ||
| 3 | hemoglobin, alpha 2IV | 24 | AAN04486 | Red blood cells | ||
| 3 | hemoglobin, beta, mutantIV | 24 | AAD30656 | |||
| 2 | hydroxyacyl dehydrogenase, subunit A | 8 | NP_000173 | Long chain 3-hydroxyacyl-CoA dehydrogenase | Mitochondrial | |
| 2,3 | hydroxyacyl dehydrogenase, subunit B | 12 | NP_000174 | |||
| 1,2,3 | hypothetical protein MGC5508 | 20, 21 | NP_076997 | |||
| 1,2,3 | keratin 1IV | 11 | NP_006112.3 | |||
| 2,3 | keratin 10IV | 16 | KRHUO | |||
| 2 | keratin 2aIV | 6 | NP_000414 | |||
| 1 | keratin 6BIV | 1 | AAH34535 | Epithelial in oral mucosa, esophagus, papillae of tongue and hair follicle | ||
| 2,3 | microsomal epoxide hydrolase 1II | 12 | AAC41694 | Membrane-bound on microsomes | Liver | |
| 2 | microsomal glutathione S-transferase 3 | 23 | AAQ81301 | Integral membrane protein. | Widely expressed | |
| 2 | motor protein mitofilin | 6,7 | BAA04654 | Microsomal inner membrane protein | Mitochondrial inner mitochondrial; membrane | |
| 2 | myelin protein zeroI | 15, 19 | CAA83513 | |||
| 2,3 | myosin heavy chain 11II | 2 | NP_002465 | KIAA0866 | ||
| 2,3 | myosin heavy chain nonmuscle 10II | 2,4 | NP_005955 | |||
| 2,3 | myosin heavy chain nonmuscle form AIII | 2 | A61231 | MYH9 | ||
| 3 myosin light chain 3 | 24 AAA59853 | |||||
| 1,2,3 | Na+/K+ ATPase alpha chainI,II form AIII | 4, 5, 6 | A26641 | Integral membrane protein. | Skin and kidney | |
| 2 | NAD(P) transhydrogenase | 5 | CAA90428 | Outside the inner membrane on the Mitochondrial matrix side | ||
| 3 | NADH cytochrome b5 reductase | 14, 15 | CAA09006 | Diaphorase 1 | Membrane bound on ER and mitochondrial outer membrane | |
| 2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10 | 14 | NP_004535 | |||
| 2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 9 | 15, 16 | NP_004993 | |||
| 1,2,3 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | 19 | NP_066552 | |||
| 1,2 | NADH dehydrogenase (ubiquinone), Fe-S protein 1I | 6, 7, 8 | AAH30833 | NADH ubiquinone oxidoreductase | Matrix and cytoplasmic side of mitochondrial inner membrane | |
| 2,3 | NADH dehydrogenase (ubiquinone), Fe-S protein 2 | 12, 13 | CAH72148 | |||
| 1,2,3 | NADH dehydrogenase (ubiquinone), Fe-S protein 3 | 17, 18 | NP_004542 | NADH ubiquinone reductase | ||
| 1,2 | NADH dehydrogenase (ubiquinone), Fe-S protein 8 | 19, 20, 21 | NP_002487 | NADH-coenzyme Q reductase | ||
| 2 | NADH dehydrogenase precursor | 7, 8 | CAA43412 | Matrix and cytoplasmic side of mitochondrial inner membrane. | ||
| 3 | NADH ubiquinone oxidoreductase | 4 | AAH08146 | NDUFV1 protein | Matrix side of the mitochondrial inner membrane | |
| 2,3 | peptidylprolyl isomerase B | 22 | CSHUB | cyclophilin B | ||
| 3 | peroxiredoxin 1 | 20 | CAI13096 | Cytoplasmic | ||
| 1 | peroxiredoxin 2III | 21 | CAA80269 thioredoxin peroxidase, thio-specific antioxidant protein | Cytoplasmic | ||
| 2 | peroxiredoxin 3II | 19 | AAV38810 | Mitochondrial | ||
| 2,3 | predicted: similar to RIKEN cDNA 4732495G21 gene | 12, 15, 16 | NP_001017992 | |||
| 3 | prenylcysteine oxidase 1I 10 NP_057381 KIAA0908, PCL1 | Lysosomal | Ubiquitous | |||
| 3 | progesterone membrane binding protein | 18 | NP_006311 | |||
| 1,2,3 | progesterone receptor membrane component 1 | 19 | NP_006658 | Microsomal; membrane-bound | Widely expressed | |
| 1,2 | prohibitinI,III 17 AAS88903 | |||||
| 2,3 | protein disulfide isomeraseIII 60 precursor | 10 | S55507 | glucose regulated protein 58 kDa, ER | Endoplasmic reticulum lumen | |
| 2,3 | RAB11BII,III | 21 | NP_004209 | RAS oncogene family | ||
| 2,3 | RAP1BIII | 22 | AAH78173 | RAS oncogene family | ||
| 2,3 | retinal G protein coupled 17 receptor | 13, 15, | NP_002912 | peropsin, RGR | Retinal pigment epithelium | |
| 1,2,3 | retinal pigment epithelium specific proteinI,II 2, 6, 7, 8, 9, 10, 11, 12, 13, 15, 17, 24 | NP_000320 | RPE65 | Retinal pigment epithelium | ||
| 2,3 | retinol binding protein 3 | 4 | CAH74045 | |||
| 2 | retinol dehydrogenase 11 | 15 | AAH00112 | androgen-regulated short-chain dehydrogenase/reductase 1 | Type II membrane protein. Endoplasmic reticulum. | |
| 1,2,3 | retinol dehydrogenase 5 | 15, 16 | AAH28298 | 11-cis retinol dehydrogenase (11-cis and 9-cis)I,II | Membrane-associated | Retinal pigment epithelium |
| 3 | ribophorin II glycosyltransferase 48 kDa subunit 11 AAH02594 KIAA0115, dolichyl-diphosphooligosaccharide protein | Type I membrane protein | Endoplasmic reticulum | |||
| 3 | ribophorin II precursor glycosyltransferase 63 kDa subunit | 9 | B26168 | Dolichyl-diphosphooligosaccharide protein | Type I membrane protein Endoplasmic reticulum | Expressed in all tissues tested |
| 2 | serum albuminIV | 9 | CAA23754 | Secreted | Plasma | |
| 2,3 | solute carrier family 2, member 1II | 11 | NP_006507 | Glucose transporter type 1 | Integral membrane protein; primarily at the cell surface | |
| 2 | solute carrier family 25, member 1 | 17 | NP_005975 | |||
| 2 | solute carrier family 25, member 12 | 9 | NP_003696 | Integral membrane protein. Mitochondrial inner membrane. | Widely expressed | |
| 2 | solute carrier family 25, member 13 | 9 | NP_055066 | Integral membrane protein. Mitochondrial inner membrane. | Widely expressed | |
| 2 | solute carrier family 25, member 24 | 12, 13 | NP_037518 | |||
| 2 | solute carrier family 25, member 3 | 16 | NP_005879 | |||
| 2 | solute carrier family 25, member 3 isoform b | 17 | NP_998776 | |||
| 1 | solute carrier family 25, member 4 | 15, 16, 17 | NP_001142 | ADP/ADT | Integral membrane protein. Mitochondrial inner membrane | |
| 2,3 | solute carrier family 25, member 4 | 15, 17, 18 | NP_001142 | |||
| 2 | solute carrier family 25, member 5II | 10, 14, 18 | NP_001143 | ADP/ATP carrier protein | ||
| 3 | solute carrier family 25, member 6 | 17 | AAA36750 | ADP.ATP translocase | ||
| 2,3 | solute carrier family 25, member A6 | 10, 16 | NP_001627 | |||
| 1,2,3 | solute carrier family 4, anion exchanger, member 1 | 2, 4, 18 | NP_000333 | |||
| 1,2,3 | spectrin, alphaII | 1, 2, 4 | NP_003118 | |||
| 1,2,3 | spectrin, betaII | 1, 2 | NP_003119.2 | |||
| 1 | succinate dehydrogenase comlex, subunit B, iron sulfur | 17 | NP_002991 | succinate-ubiquinone oxidoreductase iron sulfur subunit | Mitochondrial inner membrane | |
| 3 | succinate dehydrogenase complex, subunit AII | 9 | NP_004159 | Mitochondrial inner membrane | ||
| 1,2,3 | ubiquinol-cyctochrome c reductase, rieske iron-sulfur protein | 19, 20 | AAD38242 | Mitochondrial inner membrane | ||
| 2 | ubiquinol-cytochrome-c reductase | 18 | S00680 | |||
| 2 | ubiquinol-cytochrome-c reductase core protein III | 12, 13 | NP_003356 | Mitochondrial inner membrane | ||
| 3 | ubiquinol-cytochrome-c reductase core protein II | 12 | AAH00484 | UQCRC2 | Mitochondrial inner membrane; matrix side | |
| 1,2,3 | vimentinI,II,III | 13 | AAA61279 | beta tubulin, polypeptide | Highly expressed in fibroblasts, some expression in T and B lymphocytes | |
| 1,2,3 | voltage dependent anion channel 1III | 15, 16, 18 | NP_003365 | porin | Outer membrane of mitochondria and plasma muscle | Heart, liver and skeletal membrane |
| 1,2,3 | voltage dependent anion channel 2II | 15 | CAH73108 | porin | ||
| 1,2,3 | voltage dependent anion channel 3 | 16 | NP_005653 | Outer mitochondrial | membrane | Widely expressed |
MLF proteins were fractioned on a 1D gel. The gel lanes were sliced into sections and proteins were digested and analyzed using automated LC-MSMS and then identified using Mascot. Gel number refers to 3 separate preparations that were analyzed. I represents proteins that were previously identified as components of lipofuscin granules. II represents proteins that were previously identified as part of the melanosome proteome. III represents proteins that were previously identified as part of the macrophage phagosome proteome. IV represents proteins that are preparation contaminants.
Table 2. Melanolipofuscin proteome comparison with other organelles.
| Organelle | Total proteins | No. of common | Percent | Reference |
| Melanolipofuscin | 110 | - | - | this study |
| RPE melanosomes | 102 | 23 | 22.5 | [28] |
| Phagosomes | 140 | 18 | 12.9 | [29] |
| Lipofuscin | 36 | 14 | 38.9 | [20] |
| Melanocyte melanosomes | 68 | 7 | 10.3 | [30] |
The proteome of melanolipofuscin (MLF) was compared to the proteome of several relevant organelles. Organelles are listed in order of decreasing number of proteins in common with MLF.
Four gel slices from MLF and LF 1D gels were analyzed for a direct semi-quantitative comparison of RPE- and photoreceptor-specific proteins in these granules. Spectral counting of two photoreceptor-specific proteins, rhodopsin and peripherin, and two RPE-specific proteins, RGR and rpe65, from these four gel slices is show in Figure 6. The two photoreceptor-specific proteins were identified in LF granules but absent from MLF granules while RPE-specific proteins were more abundant in MLF granules. Although RGR was identified in LF granules, it appears to be about 58% less abundant than in MLF granules. A more comprehensive study of the proteins in LF granules was previously published by Warburton et al. [20].
Figure 6.
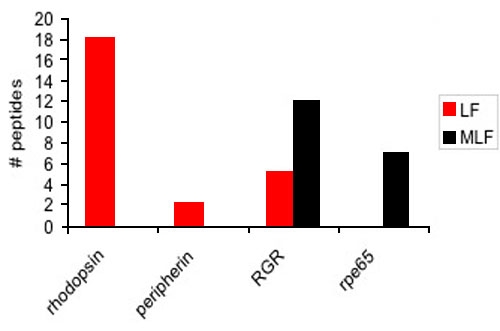
Semiquantitative analysis of photoreceptor- and retinal pigment epithelium-specific proteins in lipofuscin and melanolipofuscin granules. Spectral counting was performed on two photoreceptor-specific proteins, rhodopsin and peripherin, and two retinal pigment epithelium (RPE)-specific proteins, RGR and rpe65, in 4 gel slices from lipofuscin (LF) and melanolipofuscin (MLF) 1D gels. Photoreceptor-specific proteins were only identified in LF granules, while RPE-specific proteins were mainly identified in MLF granules. Although RGR was identified in LF granules it appeared to be about 58% less abundant than in MLF granules. This supports the hypothesis that LF granules originate from photoreceptors while MLF granules appear to originate from autophagy of RPE cells.
Rhodopsin was previously shown to be abundant in LF granules [20], however; this protein was only identified in a single gel slice from 1 of the 3 MLF preparations analyzed. In order to more quantitatively examine this apparent lack of rhodopsin in MLF granules, we performed an immunoblot of MLF proteins in which we used an α-RHO antibody to detect rhodopsin. Figure 7 shows that indeed no significant amount of rhodopsin was detected in MLF granules.
Figure 7.
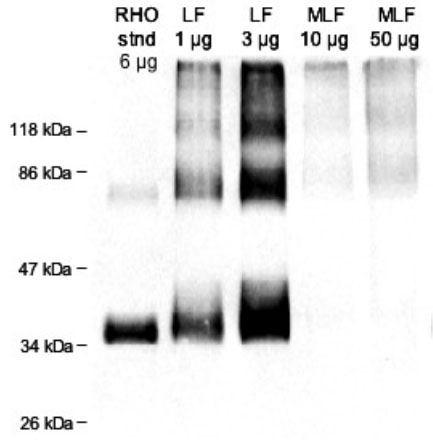
Rhodopsin immunoblot. Immunoblot following SDS-PAGE of 1 and 3 μg of total lipofuscin (LF) protein and 10 and 50 μg of total melanolipofuscin (MLF) protein. Shown for comparison is 6 μg of protein from photoreceptor cell membranes enriched from human retina. Rhodopsin runs on SDS-PAGE as a mixture of the monomer (about 30 kDa), dimer (about 60 kDa), and trimer (about 90 kDa). Although rhodopsin is seen to be present in abundance in LF granules, no significant quantity of rhodopsin is detected in MLF granules.
Because of the extensive modifications on proteins in LF granules that have been previously reported [20], we used immunoblot techniques to detect oxidative modifications on proteins in MLF granules. Dinitrophenylhydrazine (DNPH) was used to derivatize protein carbonyls, a common product of protein oxidation, and was detected using an α-DNP antibody. Figure 8 shows that the degree of oxidative modifications on proteins in MLF and LF granules is both extensive and comparable, though not identical.
Figure 8.
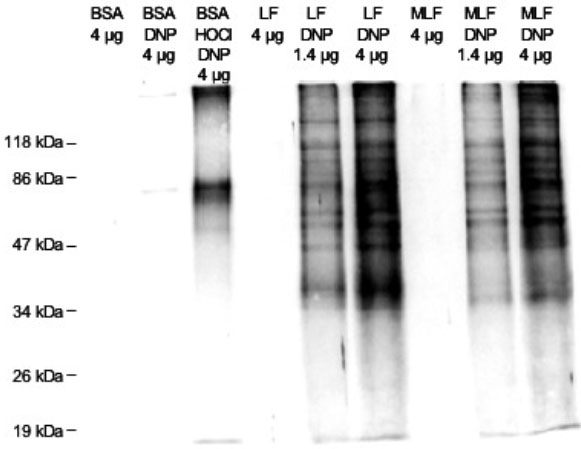
Dinitrophenyl immunoblot. Lipofuscin (LF) and melanolipofuscin (MLF) proteins, 1.4 and 4 μg, that had been derivatized with dinitrophenylhydrazine (DNPH) or not (control) were run on SDS-PAGE, transferred to nitrocellulose, and probed with an α-DNP antibody to show the derivatized protein carbonyls, a common product of protein oxidative damage. Shown for comparison and to demonstrate specificity are lanes of bovine serum albumin (BSA), BSA treated with DNPH, and BSA oxidized with hypochlorite then treated with DNPH. Oxidative damage on proteins in LF and MLF granules is both extensive and comparable, though not identical.
Discussion
Sucrose density gradients of human RPE from different decades of life illustrated that MLF is virtually non-existent in the RPE of younger individuals even though LF granules appear to be abundant in these RPE and has been detected in RPE as young as 18 years of age (data not shown). Consistent with Feeney-Burns results [5], MLF does not begin to accumulate significantly until midlife. This accumulation of MLF later in life is consistent with the onset of AMD which affects 2% of individuals over 50 and 30% of individuals over 75 [31]. This correlation may suggest that MLF contributes to the etiology of AMD.
Of significant interest is the fact the MLF is biologically active, showing a light-dependent decrease in cell viability in ARPE-19 cells fed MLF and placed in blue light for 48 h. To our knowledge this is the first report of the phototoxicity of MLF. The phototoxicity of MLF granules in ARPE-19 cells is at least 72% as potent as that of LF granules. These data show that MLF granules have the potential for deleterious affects on RPE cells in the retina.
The physical characteristics of MLF granules support the description of MLF as a complex granule with characteristics of both melanosomes and LF. The most compelling characteristic of MLF is the protein complement identified in the granules. Of the 110 proteins identified as components of MLF, 23 were previously identified in mature RPE melanosomes [28], 18 were previously identified as part of the macrophage phagosome proteome [29], 14 were previously identified in LF granules [20], and 7 were identified in melanocyte melanosomes [30]. As expected, MLF granules appear to be considerably more similar to RPE melanosomes than to melanocyte melanosomes. While LF and MLF granules contain a significant number of similar proteins, these proteins appear to be related to the lysosomal processes which these granules both participate in. However, the lack of similar cell specific proteins would suggest different origins of the material being degraded.
Of interest is the presence of RPE65, which we previously identified in LF granules where it appeared to be far less abundant than we observe in MLF granules. RPE65 was previously identified in 3 of 15 gel slices from a 1D lane of LF proteins and in 12 of 24 gel slices from a 1D lane of MLF proteins. RPE65 plays a key role in the isomerization of retinol as part of the visual cycle in the RPE and is therefore crucial to proper visual acuity. In contrast to RPE65, rhodopsin-which is abundant in LF is practically absent from MLF.
Of significant interest is the finding that MLF, in contrast to LF, does not contain photoreceptor-specific proteins, suggesting that MLF does not originate from the phagocytosis of photoreceptor outer segments as does LF, or by the fusion of melanosomes and lipofuscin. Instead, the presence of RPE- and melanosome-specific proteins would suggest that MLF accumulates as a result of the melanosomal autophagocytosis of RPE cells. Our results appear to support neither of the two previously proposed hypotheses for the origin of MLF, because both hypotheses suggest the fusion of LF granules with additional material to form MLF. Our results instead suggest a new hypothesis for the origin of MLF which excludes the involvement of previously existing LF granules. This new hypothesis for the formation of MLF granules is supported by recent evidence that melanosomes function as specialized lysosomes. Evidence for this specialized function includes the related biogenesis of melanosomes and lysosomes [32,33], the observed fusion between phagosomes and melanosomes [7], and the presence of lysosomal enzymes in melanosomes [28].
The proteins in MLF granules were shown to be extensively modified by oxidative damage. The degree of oxidative damage is comparable to that found on LF proteins. The prevalence of oxidative damage on these proteins may render them undegradable by the cell and therefore lead to their accumulation in MLF granules.
Collectively these data provide significant insight into understanding the formation and toxicity of retinal MLF and suggest a new theory for its formation as well as support a possible contribution to the etiology of retinal diseases. Our findings suggest that MLF might result from the accumulation of undegradable material perhaps undegradable due to oxidative damage in the autophagocytic melanosomes of RPE cells. Furthermore, MLF granules might pose serious risk of photosensitization of these cells allowing blue light to produce cell death by liberation of reactive oxygen species, perhaps contributing to AMD.
Acknowledgements
We gratefully thank and acknowledge: Dr. Paul S. Bernstein for providing human RPE and retinal samples, Dr. Barry M. Willardson for providing the R4 antibody, Dr. Paul B. Savage for the use of the fluorimeter, Dr. Nolan F. Mangelson for the use of the microbalance, and the staff of the BYU Microscopy Lab for their assistance. This research was supported by the Brigham Young University College of Physical and Mathematical Sciences and the Department of Chemistry and Biochemistry.
References
- 1.Beatty S, Koh H, Phil M, Henson D, Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2000;45:115–34. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 2.Winkler BS, Boulton ME, Gottsch JD, Sternberg P. Oxidative damage and age-related macular degeneration. Mol Vis. 1999;5:32. http://www.molvis.org/molvis/v5/a32/ [PMC free article] [PubMed] [Google Scholar]
- 3.Wassell J, Davies S, Bardsley W, Boulton M. The photoreactivity of the retinal age pigment lipofuscin. J Biol Chem. 1999;274:23828–32. doi: 10.1074/jbc.274.34.23828. [DOI] [PubMed] [Google Scholar]
- 4.Rozanowska M, Jarvis-Evans J, Korytowski W, Boulton ME, Burke JM, Sarna T. Blue light-induced reactivity of retinal age pigment. In vitro generation of oxygen-reactive species. J Biol Chem. 1995;270:18825–30. doi: 10.1074/jbc.270.32.18825. [DOI] [PubMed] [Google Scholar]
- 5.Feeney-Burns L, Hilderbrand ES, Eldridge S. Aging human RPE: morphometric analysis of macular, equatorial, and peripheral cells. Invest Ophthalmol Vis Sci. 1984;25:195–200. [PubMed] [Google Scholar]
- 6.Sarna T. Properties and function of the ocular melanin--a photobiophysical view. J Photochem Photobiol B. 1992;12:215–58. doi: 10.1016/1011-1344(92)85027-r. [DOI] [PubMed] [Google Scholar]
- 7.Schraermeyer U, Peters S, Thumann G, Kociok N, Heimann K. Melanin granules of retinal pigment epithelium are connected with the lysosomal degradation pathway. Exp Eye Res. 1999;68:237–45. doi: 10.1006/exer.1998.0596. [DOI] [PubMed] [Google Scholar]
- 8.Dunford R, Land EJ, Rozanowska M, Sarna T, Truscott TG. Interaction of melanin with carbon- and oxygen-centered radicals from methanol and ethanol. Free Radic Biol Med. 1995;19:735–40. doi: 10.1016/0891-5849(95)00059-7. [DOI] [PubMed] [Google Scholar]
- 9.Rozanowska M, Sarna T, Land EJ, Truscott TG. Free radical scavenging properties of melanin interaction of eu- and pheo-melanin models with reducing and oxidising radicals. Free Radic Biol Med. 1999;26:518–25. doi: 10.1016/s0891-5849(98)00234-2. [DOI] [PubMed] [Google Scholar]
- 10.Sarna T, Menon IA, Sealy RC. Photosensitization of melanins: a comparative study. Photochem Photobiol. 1985;42:529–32. doi: 10.1111/j.1751-1097.1985.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 11.Boulton M, Rozanowska M, Rozanowski B. Retinal photodamage. J Photochem Photobiol B. 2001;64:144–61. doi: 10.1016/s1011-1344(01)00227-5. [DOI] [PubMed] [Google Scholar]
- 12.Korytowski W, Kalyanaraman B, Menon IA, Sarna T, Sealy RC. Reaction of superoxide anions with melanins: electron spin resonance and spin trapping studies. Biochim Biophys Acta. 1986;882:145–53. doi: 10.1016/0304-4165(86)90149-2. [DOI] [PubMed] [Google Scholar]
- 13.Sarna T, Burke JM, Korytowski W, Rozanowska M, Skumatz CM, Zareba A, Zareba M. Loss of melanin from human RPE with aging: possible role of melanin photooxidation. Exp Eye Res. 2003;76:89–98. doi: 10.1016/s0014-4835(02)00247-6. [DOI] [PubMed] [Google Scholar]
- 14.Dayhaw-Barker P, Davies S, Shamsi FA, Rozanowska M, Rozanowska B, Boulton M. The phototoxicity of aged RPE melanosomes. Invest Ophthalmol Vis Sci. 2001;42:S755. [Google Scholar]
- 15.Gaillard E, Hill C, Griffiths D. UVC and visible light damage to re-pigmented RPE cells. ARVO Annual Meeting; 2003 May 4-9; Fort Lauderdale (FL). [Google Scholar]
- 16.Rozanowska M, Korytowski W, Rozanowski B, Skumatz C, Boulton ME, Burke JM, Sarna T. Photoreactivity of aged human RPE melanosomes: a comparison with lipofuscin. Invest Ophthalmol Vis Sci. 2002;43:2088–96. [PubMed] [Google Scholar]
- 17.Boulton M. Ageing of the retinal pigment epithelium. In: Osborne NN, Chader G, editors. Progress in retinal research. Vol 11. Oxford: Pergamon; 1991. p. 125-51. [Google Scholar]
- 18.Feeney L. Lipofuscin and melanin of human retinal pigment epithelium. Fluorescence, enzyme cytochemical, and ultrastructural studies. Invest Ophthalmol Vis Sci. 1978;17:583–600. [PubMed] [Google Scholar]
- 19.Schraermeyer U, Stieve H. A newly discovered pathway of melanin formation in cultured retinal pigment epithelium of cattle. Cell Tissue Res. 1994;276:273–9. doi: 10.1007/BF00306113. [DOI] [PubMed] [Google Scholar]
- 20.Warburton S, Southwick K, Hardman RM, Secrest AM, Grow RK, Xin H, Woolley AT, Burton GF, Thulin CD. Examining the proteins of functional retinal lipofuscin using proteomic analysis as a guide for understanding its origin. Mol Vis. 2005;11:1122–34. http://www.molvis.org/molvis/v11/a130/ [PubMed] [Google Scholar]
- 21.Dorey CK, Torres X, Swart T. Evidence of melanogenesis in porcine retinal pigment epithelial cells in vitro. Exp Eye Res. 1990;50:1–10. doi: 10.1016/0014-4835(90)90003-d. [DOI] [PubMed] [Google Scholar]
- 22.Boulton M, Docchio F, Dayhaw-Barker P, Ramponi R, Cubeddu R. Age-related changes in the morphology, absorption and fluorescence of melanosomes and lipofuscin granules of the retinal pigment epithelium. Vision Res. 1990;30:1291–303. doi: 10.1016/0042-6989(90)90003-4. [DOI] [PubMed] [Google Scholar]
- 23.Sparrow JR, Nakanishi K, Parish CA. The lipofuscin fluorophore A2E mediates blue light-induced damage to retinal pigmented epithelial cells. Invest Ophthalmol Vis Sci. 2000;41:1981–9. [PubMed] [Google Scholar]
- 24.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 25.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Takemoto DJ, Spooner B, Takemoto LJ. Antisera to synthetic peptides of bovine rhodopsin: use as site-specific probes of disc membrane changes in retinal dystrophic dogs. Biochem Biophys Res Commun. 1985;132:438–44. doi: 10.1016/0006-291x(85)91041-1. [DOI] [PubMed] [Google Scholar]
- 27.Vollmer-Snarr HR, Pew MR, Alvarez ML, Cameron DJ, Chen Z, Walker GL, Price JL, Swallow JL. Amino-retinoid compounds in the human retinal pigment epithelium. In: Retinal Degenerative Diseases. Hollyfield JG, Anderson RE, LaVail MM, eds. New York, Springer; 2006. p. 69-74. [DOI] [PubMed] [Google Scholar]
- 28.Azarian SM, McLeod I, Lillo C, Gibbs D, Yates JR, Williams DS. Proteomic analysis of mature melanosomes from the retinal pigmented epithelium. J Proteome Res. 2006;5:521–9. doi: 10.1021/pr0502323. [DOI] [PubMed] [Google Scholar]
- 29.Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. The phagosome proteome: insight into phagosome functions. J Cell Biol. 2001;152:165–80. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basrur V, Yang F, Kushimoto T, Higashimoto Y, Yasumoto K, Valencia J, Muller J, Vieira WD, Watabe H, Shabanowitz J, Hearing VJ, Hunt DF, Appella E. Proteomic analysis of early melanosomes: identification of novel melanosomal proteins. J Proteome Res. 2003;2:69–79. doi: 10.1021/pr025562r. [DOI] [PubMed] [Google Scholar]
- 31.Eye Disease Case-Control Study Group Antioxidant status and neovascular age-related macular degeneration. Arch Ophthalmol. 1993;111:104–9. doi: 10.1001/archopht.1993.01090010108035. Errata in: Arch Ophthalmol 1993; 111:1228. Arch Ophthalmol 1993; 111:1366. Arch Ophthalmol 1993; 111:1499. [DOI] [PubMed] [Google Scholar]
- 32.Orlow SJ. Melanosomes are specialized members of the lysosomal lineage of organelles. J Invest Dermatol. 1995;105:3–7. doi: 10.1111/1523-1747.ep12312291. [DOI] [PubMed] [Google Scholar]
- 33.Raposo G, Marks MS. The dark side of lysosome-related organelles: specialization of the endocytic pathway for melanosome biogenesis. Traffic. 2002;3:237–48. doi: 10.1034/j.1600-0854.2002.030401.x. [DOI] [PubMed] [Google Scholar]


