Abstract
Human systemic lupus erythematosus (SLE) is an autoimmune disease characterized by autoantibodies to nuclear components with subsequent immune complex formation and deposition in multiple organs. A combination of genetic and environmental factors is required for disease development, but how the environment interacts with the immune system in genetically predisposed hosts to cause lupus is unclear. Recent evidence suggests that environmental agents may alter T cell chromatin structure and gene expression through effects on DNA methylation, a repressive epigenetic mechanism promoting chromatin inactivation, to cause lupus in people with the appropriate genetic background. DNA methylation is regulated by ERK pathway signaling, and abnormalities in ERK pathway signaling may contribute to immune dysfunction in lupus through epigenetic effects on gene expression. This article reviews current evidence for epigenetic abnormalities, and in particular DNA demethylation, in the pathogenesis of idiopathic and some forms of drug induced lupus, and how impaired ERK pathway signaling may contribute to the development of human lupus through effects on T cell DNA methylation.
Keywords: Lupus T cells, Epigenetics, DNA methylation, ERK pathway signaling, PKCδ
Take-Home messages.
Identical changes in T cell DNA methylation patterns, T cell gene expression, and T cell function are found in experimentally demethylated and lupus T cells.
Impaired ERK pathway signaling, due to impaired PKCδ phosphorylation has been proposed as the mechanism responsible for T cell demethylation.
A detailed analysis of PKCδ activation in hydralazine treated and lupus T cells may provide further insights into the pathogenesis of this puzzling disease.
Introduction
T cells do not play a direct role in lupus tissue damage. Rather, they promote the autoimmune response and are important for autoantibody production [1]. T cells from patients with active lupus have multiple biochemical abnormalities, resulting in functional aberrations that cause a lupus-like disease in animal models, and may contribute to the development of autoimmunity in humans. Recent evidence suggests that abnormal T cell ERK pathway (PKC → Ras→ Raf→ MEK→ ERK) signal transduction may be fundamental to some of these abnormalities through effects on DNA methylation [2]. This review summarizes current understanding of DNA methylation and gene expression, and how abnormal ERK pathway signaling can alter T cell DNA methylation, and consequently gene expression, to promote lupus development in genetically predisposed hosts.
DNA methylation and gene expression
DNA methylation refers to the post-synthetic methylation of deoxycytosine (dC) to form deoxymethylcytosine (dmC). dmC occurs primarily in CpG pairs. In general, methylation of regulatory elements suppresses gene expression, while regulatory elements of active genes are typically unmethylated. Methylation of regulatory elements tethers chromatin inactivation complexes that stabilize nearby chromatin in an inactive configuration [3]. DNA methylation patterns are established during development by the de novo DNA methyltransferases (Dnmt) 3a and 3b, then are replicated during mitosis by the maintenance methyltransferase Dnmt1 [3].
Mitotically inactive T cells express relatively low levels of Dnmt1 [4]. However, during mitosis Dnmt1 levels are increased in part by signals transmitted through the JNK and Ras-ERK pathways and interacting with AP-1 sites in an intronic promoter. Inhibiting either pathway inhibits DNA methylation in proliferating cells [5]. This suggests that environmental agents may affect the replication of DNA methylation patterns during mitosis by inhibiting these pathways, with the potential of altering gene expression.
DNA methlyation and lupus
Early studies demonstrated that inhibiting DNA methylation with 5-azacytidine (5-azaC) caused CD4+ T cell autoreactivity. The autoreactivity correlated with overexpression of the adhesion molecule LFA-1 (CD11a/CD18) [6] due to demethylation of sequences 5′ to the CD11a promoter [7], and overexpressing LFA-1 by transfection caused an identical autoreactivity [8]. Demethylated, autoreactive CD4+ T cells overstimulate antibody production by B cells and kill macrophages (Mø) [3], releasing apoptotic nuclear material that stimulates lupus-like autoantibodies [9]. Further, injecting experimentally demethylated CD4+ T cells into syngeneic mice causes anti-DNA antibodies and a lupus-like disease [8].
The obsevation that a DNA methylation inhibitor can cause a lupus-like disease suggests that drugs which cause a lupus-like disease might be DNA methylation inhibitors. Hydralazine and procainamide cause anti-nuclear antibodies in a majority of people receiving these drugs, and a lupus-like disease in a subset [10]. Both were found to inhibit T cell DNA methylation and cause LFA-1 overexpression and autoreactivity [11], and the demethylated T cells caused a lupus-like disease in mice like 5-azaC [8]. Procainamide is a competitive Dnmt1 inhibitor [12] while hydralazine decreases Dnmt1 levels by inhibiting ERK pathway signaling [13].
T cells from lupus patients similarly have decreased Dnmt1 and dmC levels (14). Lupus T cells also overexpress LFA-1 [6], due to demethylation of the same sequences affected by 5-azaC [7], and kill Mø [6], like the demethylation model. Identical demethylation and overexpression of perforin, CD70 and CD40L is also seen in experimentally demethylated and lupus T cells [3].
Patients with active lupus have impaired T cell ERK1/2 phosphorylation, similar to hydralazine treated T cells. The degree of impairment is proportional to disease activity [15]. Further, inhibiting T cell ERK1/2 phosphorylation with MEK inhibitors decreases Dnmt1 expression and dmC content, causes LFA-1 overexpression, makes T cells autoreactive, and the treated T cells induce anti-DNA antibodies in animal models [3]. CD4+ T cells treated with ERK pathway and Dnmt inhibitors also demethylate and overexpress TNFSF7, encoding the B cell costimulatory molecule CD70, and identical demethylation and overexpression of CD70 is found in lupus T cells [16, 17]. These latter studies demonstrate identical effects of Dnmt inhibitors, ERK pathway inhibitors, and lupus, on TNFSF7 promoter methylation and gene expression.
Mapping the lupus ERK pathway signaling defect
Since T cell ERK pathway signaling is decreased in lupus [15], hydralazine inhibits ERK pathway signaling [13], and ERK pathway signaling regulates Dnmt1 levels [15], decreased ERK pathway signaling may be fundamental to human SLE through effects on DNA methylation. The ERK pathway defect was isolated by stimulating lupus or hydralazine treated T cells with PMA and comparing PKC, ERK, MEK, and Raf phosphorylation. Identical phosphorylation decreases were observed in ERK, MEK and Raf. No effects were seen on PKCα or PKCθ. However, PKCδ phosphorylation was diminished in lupus and hydralazine treated T cells. Further, PMA stimulated, primary human CD4+ T cells treated with Rottlerin, a selective PKCδ inhibitor, and CD4+ T cells transfected with a dominant negative PKCδ, demonstrated identical decreases in ERK phosphorylation with concomitant TNFSF7 promoter demethylation and CD70 overexpression, similar to lupus and hydralazine treated-T cells.
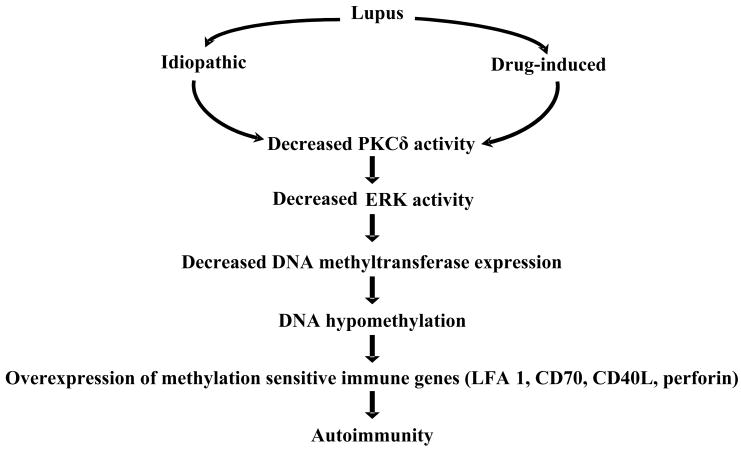
This supports a direct link between PKCδ and ERK [18] and suggests that abnormalities in the PKC-ERK pathway, due to impaired PKCδ activation, may contribute to human lupus through effects on DNA methylation in T cells and perhaps other cells (Fig 1). This is supported by reports that mice genetically deficient in PKCδ develop a lupus-like disease [19]. The mechanisms causing impaired PKCδ activation at present are unknown.
Figure 1.
Proposed mechanism by which epigenetic modifications in T cells may induce idiopathic and drug-induced lupus. Decreased ERK pathway signaling due to impaired PKCδ decreases DNA methylation what modifies gene expression making T cells autoreactive.
Summary
Current evidence indicates that lupus is an epigenetic disease caused by inhibiting T cell DNA methylation in a genetically predisposed host. The methylation defect traces to impaired PKCδ activation, causing decreased ERK pathway signaling and a failure to upregulate Dnmt1 during mitosis. Better understanding of the PKCδ defect may suggest ways to prevent lupus in predisposed hosts.
Acknowledgments
This work was supported by PHS grants AR42525, AG25877 and ES015214, and a Merit grant from the Dept. of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kotzin BL. Systemic lupus erythematosus. Cell. 1996;85:303–306. doi: 10.1016/s0092-8674(00)81108-3. [DOI] [PubMed] [Google Scholar]
- 2.Kammer GM, Perl A, Richardson BC, Tsokos GC. Abnormal T cell signal transduction in systemic lupus erythematosus. Arthritis Rheum. 2002;46:1139–1154. doi: 10.1002/art.10192. [DOI] [PubMed] [Google Scholar]
- 3.Richardson B. Primer: epigenetics of autoimmunity. Nat Clin Pract Rheumatol. 2007;3:521–527. doi: 10.1038/ncprheum0573. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, Deng C, Hemati N, Hanash SM, Richardson BC. Effect of mitogenic stimulation and DNA methylation on human T cell DNA methyltransferase expression and activity. J Immunol. 1997;159:1303–1309. [PubMed] [Google Scholar]
- 5.MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327–11337. doi: 10.1074/jbc.270.19.11327. [DOI] [PubMed] [Google Scholar]
- 6.Richardson BC, Strahler JR, Pivirotto TS, Quddus J, Bayliss GE, Gross LA, O’Rourke KS, Powers D, Hanash SM, Johnson MA. Phenotypic and functional similarities between 5-azacytidine-treated T cells and a T cell subset in patients with active systemic lupus erythematosus. Arthritis Rheum. 1992;35:647–662. doi: 10.1002/art.1780350608. [DOI] [PubMed] [Google Scholar]
- 7.Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, Richardson B. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 2002;46:1282–1291. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 8.Yung R, Powers D, Johnson K, Amento E, Carr D, Laing T, Yang J, Chang S, Hemati N, Richardson B. Mechanisms of drug-induced lupus. II. T cells overexpressing lymphocyte function-associated antigen 1 become autoreactive and cause a lupus-like disease in syngeneic mice. J Clin Invest. 1996;97:2866–2871. doi: 10.1172/JCI118743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny MF, Chandaroy P, Killen PD, Caricchio R, Lewis EE, Richardson BC, Lee KD, Gavalchin J, Kaplan MJ. Accelerated macrophage apoptosis induces autoantibody formation and organ damage in systemic lupus erythematosus. J Immunol. 2006;176:2095–2104. doi: 10.4049/jimmunol.176.4.2095. [DOI] [PubMed] [Google Scholar]
- 10.Yung RL, Richardson BC. Drug-induced lupus. Rheum Dis Clin North Am. 1994;20:61–86. [PubMed] [Google Scholar]
- 11.Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. 1988;140:2197–2200. [PubMed] [Google Scholar]
- 12.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005;280:40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 14.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 15.Deng C, Kaplan MJ, Yang J, Ray D, Zhang Z, McCune WJ, Hanash SM, Richardson BC. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 2001;44:397–407. doi: 10.1002/1529-0131(200102)44:2<397::AID-ANR59>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 16.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. 2005;174:6212–6219. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 17.Oelke K, Lu Q, Richardson D, Wu A, Deng C, Hanash S, Richardson B. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004;50:1850–1860. doi: 10.1002/art.20255. [DOI] [PubMed] [Google Scholar]
- 18.Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol. 2007;179:5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto A, Nakayama K, Imaki H, Hirose S, Jiang Y, Abe M, Tsukiyama T, Nagahama H, Ohno S, Hatakeyama S, et al. Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cdelta. Nature. 2002;416:865–869. doi: 10.1038/416865a. [DOI] [PubMed] [Google Scholar]