Abstract
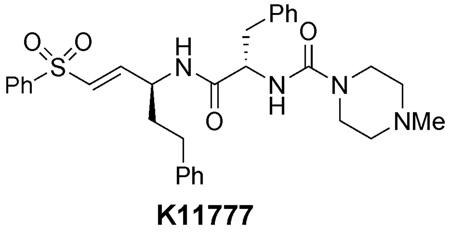
The importance of cysteine proteases in parasites, compounded with the lack of redundancy compared to their mammalian hosts makes proteases attractive targets for the development of new therapeutic agents. The binding mode of K11002 to cruzain, the major cysteine protease of Trypanosoma cruzi was used in the design of conformationally constrained inhibitors. Vinyl sulfone-containing macrocycles were synthesized via olefin ring-closing metathesis and evaluated against cruzain and the closely related cysteine protease, rhodesain.
Trypanosomes are parasitic protozoa responsible for several neglected diseases of global health importance including Chagas’ disease and sleeping sickness. Chagas’ disease, or American trypanosomiasis, is a chronic infection caused by the parasite, Trypanosoma cruzi, and is the leading cause of heart failure in many Latin American countries.1 T. cruzi is transmitted to humans through the bite of the triatomine bug or by transfusion of infected blood. The overall prevalence of human infection is estimated at 16 to 18 million cases with 13,000 deaths reported each year.2 Trypanosoma brucei gambiense and Trypanosoma brucei rhodesiense are the pathogenic agents of human African trypanosomiasis, or sleeping sickness. These parasites live extracellularly in blood and tissue fluids of the mammalian host and are transmitted by the bite of tsetse flies. The disease is endemic in certain regions of sub-Saharan Africa, covering about 50 million people in 36 countries. It is estimated that 50,000 to 70,000 people are currently infected; if left untreated, the disease in humans is fatal.3
Current drug therapy for trypanosomal diseases is not always effective and is often hampered by severe side effects.4 Thus, the identification of novel targets for trypanocidal agents is needed. One such target is the major cysteine protease of the parasitic organisms, which includes cruzain5 in T. cruzi and rhodesain6 in T. brucei rhodesiense. Both enzymes are clan CA proteases, share 70% similarity in primary structure, and are involved in critical roles in parasite survival, such as replication, penetration into host cells, nutrition at the expense of the host, and immunoevasion.7 Selective inhibitors of cruzain have been demonstrated to cure T. cruzi infection both in cell culture screens and in mouse models of Chagas’ disease. 8 In a recent report, a cruzain inhibitor was also found to be effective in treating Chagas’ disease in beagle dogs.9
A large number of cysteine protease inhibitors have been reported to date, several classes of which are potent, irreversible inhibitors. 10, 11 Based on the pioneering studies by Hanzlik12 and Palmer,13 our group has developed peptidyl vinyl sulfone inhibitors of parasitic cysteine proteases. 14, 15 The vinyl sulfones serve as Michael acceptors for the nucleophilic active site cysteine, and the peptidic backbone contains several hydrogen bond acceptors that interact with complementary residues in the active site. Several cruzain-inhibitor complexes have been solved by X-ray crystallography, which displayed the active site Cys25 of cruzain covalently bound to the vinyl sulfone unit of the inhibitor.16
With the aim to improve upon the lead compounds from previous studies and to develop an inhibitor with a broad spectrum of activity against a variety of parasitic hosts, we were interested in the design of conformationally constrained vinyl sulfones. Limiting conformational flexibility of the inhibitor or ligand is a well-established strategy to improve binding energies by decreasing the entropic barrier to binding of a particular conformation. Hence, in principle, by tethering distal segments of the inhibitors to form a rigid structure with a conformation favorable to binding, selectivity and/or potency of the inhibitor can be improved. This approach has been implemented in the design of various biologically active molecules such as aspartyl protease inhibitors,17 and Grb2 SH2 domain-binding ligands.18
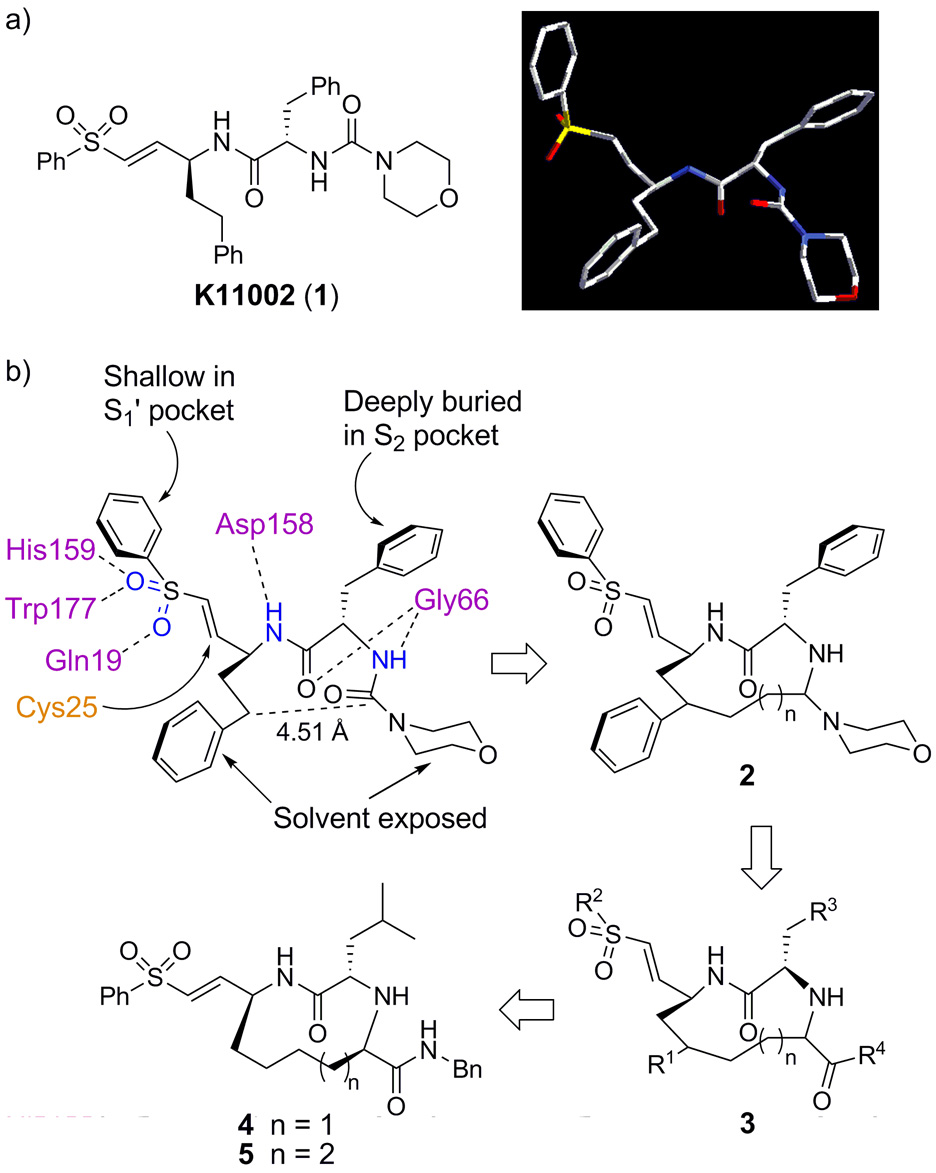
The crystal structures of cruzain with bound vinyl sulfonyl inhibitors are instrumental in elucidating the key factors that contribute to inhibitor binding. Examination of the crystal structures of cruzain with vinyl sulfones such as K11002 (1) reveal several highly conserved binding interactions.16 These include hydrogen bonding between the side chains of Gln19, His159, and Trp177 with the sulfonyl oxygen atoms, a hydrogen bond between the P1 nitrogen with the Asp158 peptide carbonyl, a hydrogen bond between the P2 carbonyl and the Gly66 amide, and a hydrogen bond between the P2 amide nitrogen with the Gly66 carbonyl (Figure 1b). 16 The S2 pocket is the primary recognition element for cruzain and all other enzymes in the papain class. The Phe side chain of K11002 and other inhibitors is deeply buried in the well-defined S2 pocket. However, the S1’, S1, and S3 pockets are very shallow and poorly defined, therefore the P1’, P1, and P3 groups are highly solvent exposed. In addition, the urea carbonyl does not participate in any interactions with the enzyme and thus was assumed to be non-essential to inhibitor binding.
Figure 1.
(a) K11002 and its 3-D structure when bound to cruzain; (b) Design rationale for conformationally constrained inhibitors 4 and 5.
In designing a conformationally rigid inhibitor scaffold, we aimed to preserve the geometry of the peptide backbone and all of the critical hydrogen bonding interactions identified in Figure 1b. Since the P1 side chain and the urea carbonyl are not involved in specific binding interactions to the enzyme, they were selected as the sites to form a conformationally restricted linkage. The distance between the urea carbonyl carbon and the γ-carbon of homoPhe in 1 when bound to cruzain is 4.51 Å, therefore we reasoned that a 10- or 11-membered macrocycle would provide the optimal ring size to preserve the bioactive peptide backbone conformation. In addition, introduction of a linking unit between these two units was expected not to interfere with enzyme binding, since the P1 homoPhe residue is highly solvent exposed in the available crystal structures.16 We anticipated that 2 would not be chemically stable, due to the acid labile aminal moiety. This was addressed by replacing the aminal unit with an α-amino amide as shown in 3. Finally, we also elected to use leucine as the P2 residue as shown in 4 and 5 to facilitate the extension of this strategy to the synthesis of inhibitors that might be effective against other parasitic CA proteases that are much more specific at P2 than cruzain. We herein report the synthesis of two members of this class of compounds (4 and 5) and their inhibitory potency against cruzain and rhodesain.
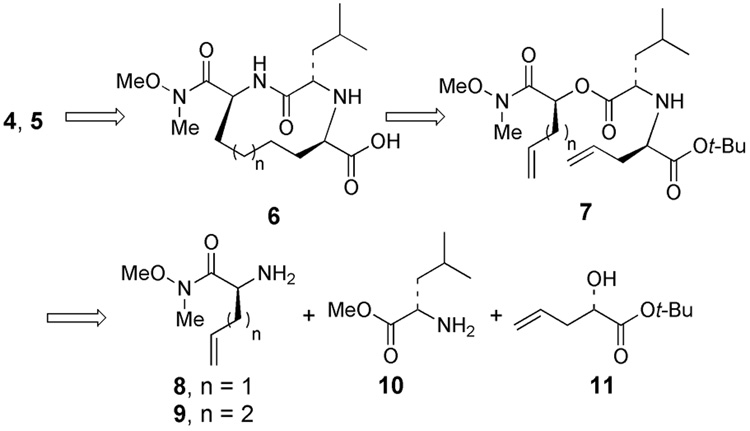
As illustrated in Figure 2, we sought to assemble inhibitors 4 and 5 from scaffold 6, which could be accessible from diene 7 via ring closing olefin metathesis. Diene 7, in turn, could be prepared from either allylglycine or homoallylglycine which serves as the “P1” fragment (8 or 9), leucine methyl ester (10), and α-hydroxyester 11.
Figure 2.
Disconnective analysis of inhibitors 4 and 5.
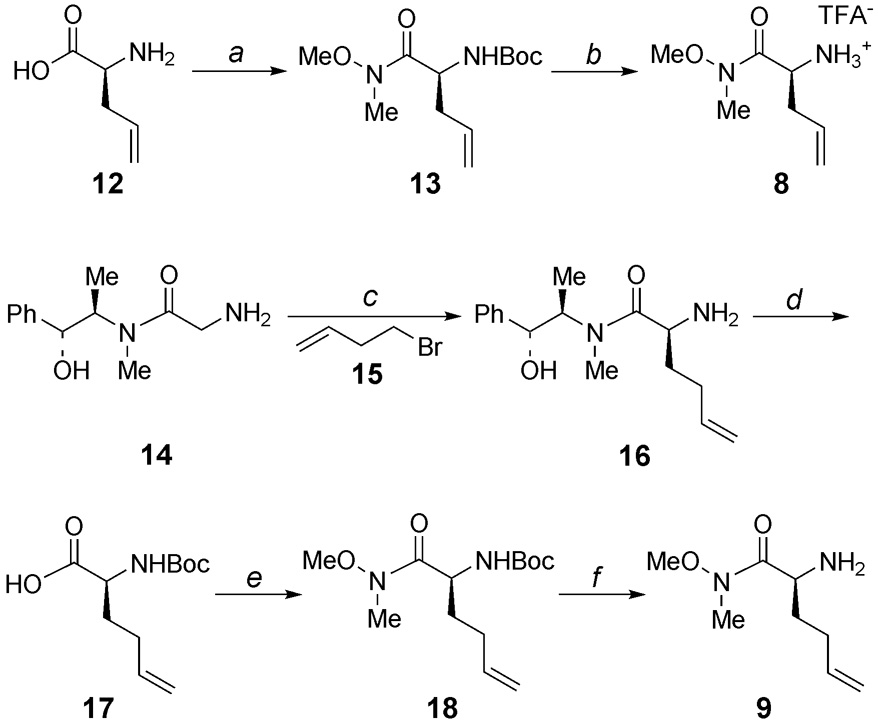
The synthesis of the “P1” fragments of 8 and 9 is illustrated in Scheme 1. Weinreb amide 13 was prepared by functionalization of allylglycine (12) as described by Borzilleri et al.19 Deprotection of the Boc group afforded amine 8 as a TFA salt. The synthesis of the Weinreb amide of homoallylglycine (9) began with asymmetric alkylation of pseudoephedrine glycinamide (14) 20 with 4-bromo-1-butene (15) to provide 16. Removal of the chiral auxiliary followed by Boc protection of the amine and coupling of the resulting acid with N,O-dimethylhydroxylamine21 gave Weinreb amide 18. Finally, deprotection of the Boc group provided homoallylglycine 9.
Scheme 1.
(a) (i) Boc2O, NaHCO3, H2O, THF, 0 to 23 °C, 12 h; (ii) CH3(CH3O)NH•HCl, HOBT, NMM, EDC, CH2Cl2, 0 to 23 °C, 12 h, 84% (two steps); (b) TFA, CH2Cl2, quant.; (c) LDA, LiCl, THF, 0 °C, 29 h, 62% (de > 20:1); (d) (i) NaOH, H2O, reflux, 3 h; (ii) Boc2O, NaOH, H2O, dioxane, 0 to 23 °C, 16 h, 82% (two steps); (e) CH3(CH3O)NH•HCl, HOBT, NMM, EDC, CHCl3, 0 to 23 °C, 14 h, 88%; (f) TFA, CH2Cl2, 0 23 °C, 1 h, quant.
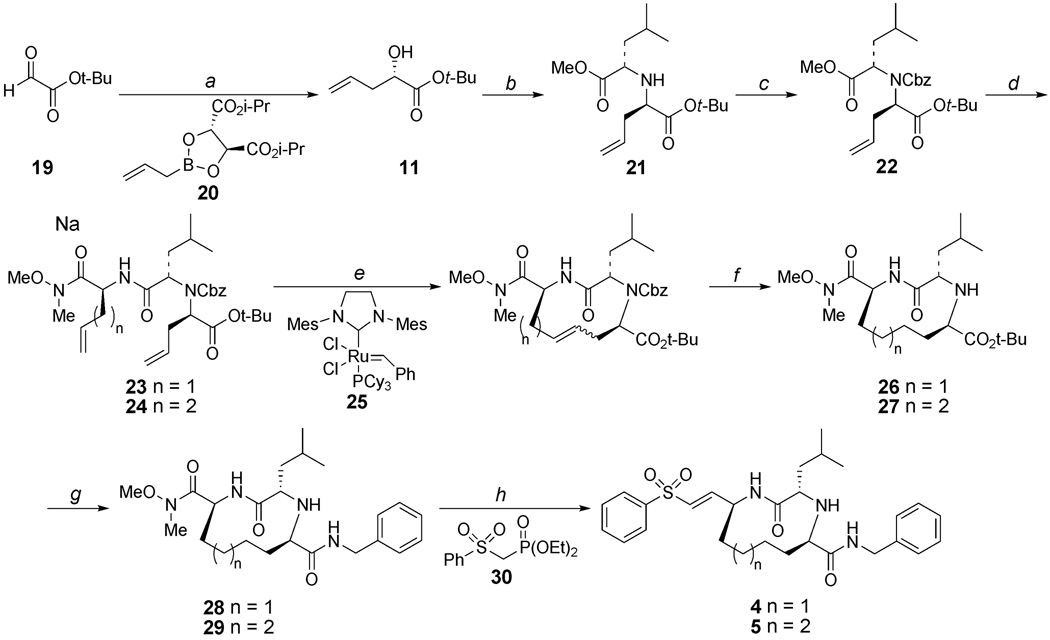
Allylboration of t-butyl glyoxylate 1922 with (S,S)-2023 gave α-hydroxyester 11 with 74% ee 24 (Scheme 2). This material was carried onto the next two steps without further enantiomeric enrichment. The alcohol was converted to the corresponding triflate and this was used to alkylate leucine methyl ester (10) which provided 21 as a ca. 5:1 diastereomeric mixture which was separated by silica column chromatography. Amine 21 was protected with a Cbz group using n-butyllithium as the base, since attempted ring-closing metathesis of the amine corresponding to 24 was unsuccessful presumably due to catalyst poisoning.25 The requirement for a strong base in this step is most likely due to the poor accessibility of the highly hindered amine proton. Selective hydrolysis of the methyl ester in compound 22, followed by coupling with Weinreb amide 8 or 9 gave dienes 23 and 24, respectively, which set the stage for macrocyclization via ring-closing olefin metathesis. 26 Macrocycle formation was achieved by treatment of either 23 or 24 with 20 mol % of second generation Grubbs’ catalyst 25 27 in 1,2-dichloroethane (0.8 mM substrate concentration). 28 Use of other solvents and higher reaction concentration led to substantial amounts of oligomers. The resulting alkene unit and Cbz group were removed by hydrogenation to yield 26 and 27. Cleavage of the t-butyl ester by treatment with TFA, followed by coupling of the resulting carboxylic acid with benzyl amine gave 28 and 29. Finally, the vinyl sulfone group was introduced by reduction of the Weinreb amide to the corresponding aldehyde, followed by Horner-Wadsworth-Emmons olefination with phosphonate 30.29 This yielded the targeted macrocyclic cysteine protease inhibitors 4 and 5 in 51 and 58% yield, respectively.
Scheme 2.
(a) 16, toluene, 4 Å mol. sieves, −78 °C, 3 h, 70% (74 % ee); (b) (i) Tf2O, pyridine, CH2Cl2, 0 to 23 °C, 30 min; (ii) 10, proton sponge, CHCl3, −78 to 23 °C, 18 h, 70% (two steps); (c) CbzCl, n-BuLi, Et2O, −78 to 23 °C, 72 h, 55%; (d) (i) NaOH, THF, MeOH, 0 °C; (ii) 8 or 9, EDC, HOBT, NMM, CH2Cl2, 0 to 23 °C, 14 h (two steps), 23 (77%), 24 (68%); (e) 25, 1,2-dichloroethane, 95 °C, 1 h; (f) H2, 10% Pd/C, EtOAc, EtOH, 36 h, 26 (65% from 23), 27 (67% from 24); (g) (i) TFA, CH2Cl2, 6 h; (ii) benzylamine, EDC, HOBT, NMM, CH2Cl2, 0 to 23 °C, 14 h (two steps), 28 (89%), 29 (73%); (h) (i), LiAlH4, THF, −10 °C 30 min; (ii) 30, NaH, THF, 0 to 23 °C; 4–8 h (two steps), 4 (58%), 5 (51%).
The activities of vinyl sulfones 4 and 5 as cysteine protease inhibitors were tested against cruzain and rhodesain. Recombinantly expressed cruzain or rhodesain was incubated with the inhibitor in sequential dilution followed by addition of Z-Phe-Arg-AMC as a fluorescent substrate. The increase in fluorescence produced by cleavage of the substrate allows determination of protease inhibition. IC50 values were determined in the liner portion of a plot of inhibition versus log of inhibitor concentration.
The results are summarized in Table 1. For comparison, K117778 was also assayed under similar conditions. The conformationally constrained compounds were substantially less active as inhibitors of cruzain and rhodesain compared to the acyclic vinyl sulfone. Owing to the very weak inhibitor potency, full kinetic analyses of these two inhibitors was not determined. Of the two macrocyclic inhibitors, 5 was the more effective, which suggests that the 11-membered macrocycle may have more flexibility to adopt a geometry that is more favorable for binding than the 10-membered ring scaffold of 4. Our current hypothesis is that P3 unit in 4 and 5 is sub-optimal compared to the P3 units in K11002 and other dipeptidyl vinylsulfonyl inhibitors.16 It is possible that the free amine in 4 or 5, which will be protonated under conditions of the enzyme assay, may negatively impact the binding of these inhibitors to the enzyme targets. It is also possible that the urea units of acyclic cysteine protease inhibitors (c.f., K11002) are much more important to inhibitor binding than initially assumed. Studies addressing these issues via the synthesis and evaluation of a second-generation series of conformationally constrained vinylsulfonyl cysteine protease inhibitors will be reported in due course.
Table 1.
Inhibition of cruzain and rhodesain by vinyl sulfone derivatives.
| IC50 (µM) |
||
|---|---|---|
| Compounds | Rhodesain | Cruzaina |
| 4 | 10 | >10 |
| 5 | 6 | 2 |
| K11777 | 0.1 | 0.004 |
Supplementary Material
Acknowledgements
This research was supported by a grant from the National Institutes of Allergy and Infectious Diseases (Program Project Grant AI 35707).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Lancet. 2006;368:619. doi: 10.1016/S0140-6736(06)69217-9. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. http://www.who.int/tdr/diseases/chagas/default.htm.
- 3.World Health Organization. http://www.cdc.gov/chagas/factsheet.html.
- 4.(a) Van den Bossche H. Nature. 1978;273:626. doi: 10.1038/273626a0. [DOI] [PubMed] [Google Scholar]; (b) Linares GEG, Ravaschino EL, Rodriguez JB. Curr. Med. Chem. 2006;13:335. doi: 10.2174/092986706775476043. [DOI] [PubMed] [Google Scholar]; (c) Castro JA, de Mecca MM, Bartel LC. Human & Experimental Toxicology. 2006;25:471. doi: 10.1191/0960327106het653oa. [DOI] [PubMed] [Google Scholar]
- 5.(a) Eakin AE, McGrath ME, McKerrow JH, Fletterick RJ, Craik CS. J. Biol. Chem. 1993;268:6115. [PubMed] [Google Scholar]; (b) McGrath ME, Eakin AE, Engel JC, McKerrow JH, Craik CS, Fletterick RJ. J. Mol. Biol. 1995;247:251. doi: 10.1006/jmbi.1994.0137. [DOI] [PubMed] [Google Scholar]
- 6.Caffrey CR, Hansell E, Lucas KD, Brinen LS, Alvarez-Hernandez A, Cheng J, Gwaltney SL, II, Roush WR, Stierhof Y-D, Bogyo M, Steverding D, McKerrow JH. Mol. Biochem. Parasitol. 2001;118:61. doi: 10.1016/s0166-6851(01)00368-1. [DOI] [PubMed] [Google Scholar]
- 7.McKerrow JH, Sun E, Rosenthal PJ, Bouvier J. Ann. Rev. of Microbiol. 1993;47:821. doi: 10.1146/annurev.mi.47.100193.004133. [DOI] [PubMed] [Google Scholar]
- 8.Engel JC, Doyle PS, Hsieh I, McKerrow JH. J. Exp. Med. 1998;188:725. doi: 10.1084/jem.188.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barr SC, Warner KL, Kornreic BG, Piscitelli, Wolfe A, Benet L, McKerrow JH. Antimicrob. Agents Chemother. 2005;49:5160. doi: 10.1128/AAC.49.12.5160-5161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powers JC, Asgian JL, Ekici OD, James KE. Chem. Rev. 2002;12:4639. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez-Hernandez A, Roush WR. Curr. Opin. Chem. Biol. 2002;6:459. doi: 10.1016/s1367-5931(02)00345-9. [DOI] [PubMed] [Google Scholar]
- 12.Liu S, Hanzlik RP. J. Med. Chem. 1992;35:1067. doi: 10.1021/jm00084a012. [DOI] [PubMed] [Google Scholar]
- 13.Palmer JT, Rasnick D, Klaus JL, Bromme D. J. Med. Chem. 1995;38:3193. doi: 10.1021/jm00017a002. [DOI] [PubMed] [Google Scholar]
- 14.(a) Roush WR, Gwaltney SL, II, Cheng J, Scheidt KA, McKerrow JH, Hansell E. J. Am. Chem. Soc. 1998;120:10994. [Google Scholar]; (b) Roush WR, Cheng J, Knapp-Reed B, Alvarez-Hernandez A, McKerrow JH, Hansell E, Engel JC. Bioorg. Med. Chem. Lett. 2001;11:2759. doi: 10.1016/s0960-894x(01)00566-2. [DOI] [PubMed] [Google Scholar]; (c) Shenai BR, Lee BJ, Alvarez-Hernandez A, Chong PY, Emal CD, Neitz RJ, Roush WR, Rosenthal PJ. Antimicrob. Agents Chemother. 2003;47:154. doi: 10.1128/AAC.47.1.154-160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Emal CD, Hansell E, Alvarez-Hernandez A, Chong PY, Knapp-Reed B, McKerrow JH, Roush WR. doi: 10.1016/s0960-894x(01)00566-2. to be submitted. [DOI] [PubMed] [Google Scholar]; (e) Emal CD, Alvarez-Hernandez A, Truong Y, Miller MW, Nora PG, Hansell E, Doyle PS, McKerrow JH, Roush WR. to be submitted. [Google Scholar]
- 15.Jaishankar P, Hansell E, Zhao D-M, Doyle PS, McKerrow JH, Renslo AR. Bioorg. Med. Chem. Lett. 2008;18:624. doi: 10.1016/j.bmcl.2007.11.070. [DOI] [PubMed] [Google Scholar]
- 16.Brinen LS, Hansell E, Cheng J, Roush WR, McKerrow JH, Fletterick RJ. Structure. 2000;8:831. doi: 10.1016/s0969-2126(00)00173-8. [DOI] [PubMed] [Google Scholar]
- 17.Hanessian S, Yang G, Rondeau J-M, Neumann U, Betschart C, Tintelnot-Blomley M. J. Med. Chem. 2006;49:4544. doi: 10.1021/jm060154a. [DOI] [PubMed] [Google Scholar]
- 18.Gao Y, Voigt J, Wu JX, Yang D, Burke TR., Jr Bioorg. Med. Chem. Lett. 2001;11:1889. doi: 10.1016/s0960-894x(01)00316-x. [DOI] [PubMed] [Google Scholar]
- 19.Borzilleri RM, Zheng X, Schmidt RJ, Johnson JA, Kim S-H, DiMarco JD, Fairchild CR, Gougoutas JZ, Lee FYF, Long BH, Vite GD. J. Am. Chem. Soc. 2000;122:8890. [Google Scholar]
- 20.Myers AG, Gleason JL, Yoon T, Kung DW. J. Am. Chem. Soc. 1997;119:656. [Google Scholar]
- 21.Nahm S, Weinreb SM. Tetrahedron Lett. 1981;22:3815. [Google Scholar]
- 22.Subasinghe N, Schulte M, Chan MY-M, Roon RJ, Koerner JF, Johnson RL. J. Med. Chem. 1990;33:2734. doi: 10.1021/jm00172a009. [DOI] [PubMed] [Google Scholar]
- 23.(a) Roush WR, Walts AE, Hoong LK. J. Am. Chem. Soc. 1985;107:8186. [Google Scholar]; (b) Roush WR, Hoong LK, Palmer MAJ, Park JC. J. Org. Chem. 1990;55:4109. [Google Scholar]; (c) Roush WR, Hoong LK, Palmer MAJ, Straub JA, Palkowitz AD. J. Org. Chem. 1990;55:4117. [Google Scholar]
- 24.Enantiomeric excess was determined by Mosher ester analysis ( Dale JA, Mosher HS. J. Am. Chem. Soc. 1973;95:512.). Absolute stereochemistry was confirmed by reduction of the ester to the corresponding diol and comparing with published data ( Enders D, Lenzen A, Müller M. Synthesis. 2004;9:1486.).
- 25.Shon Y-S, Lee TR. Tetrahedron Lett. 1997;38:1283. [Google Scholar]
- 26.Grubbs RH, Chang S. Tetrahedron. 1998;54:4413. [Google Scholar]
- 27.Scholl M, Ding S, Lee CW, Grubbs RH. Org. Lett. 1999;1:953. doi: 10.1021/ol990909q. [DOI] [PubMed] [Google Scholar]
- 28.Dekker FJ, de Mol NJ, Fisher MJE, Kemmink J, Liskamp RM. Org. Biomol. Chem. 2003;1:3297. doi: 10.1039/b306681a. [DOI] [PubMed] [Google Scholar]
- 29.Enders D, von Berg S, Jandeleit B. Org. Synth. 2000;78:169. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.