Abstract
Leishmania major possesses, apparently uniquely, four families of ATG8-like genes, designated ATG8, ATG8A, ATG8B and ATG8C, and 25 genes in total. L. major ATG8 and examples from the ATG8A, ATG8B and ATG8C families are able to complement a Saccharomyces cerevisiae ATG8-deficient strain, indicating functional conservation. Whereas ATG8 has been shown to form putative autophagosomes during differentiation and starvation of L. major, ATG8A primarily form puncta in response to starvation - indicating a role for ATG8A in starvation-induced autophagy. Recombinant ATG8A was processed at the scissile glycine by recombinant ATG4.2 but not ATG4.1 cysteine peptidases of L. major and, consistent with this, ATG4.2-deficient L. major mutants were unable to process ATG8A and were less able to withstand starvation than wild type cells. GFP-ATG8-containing puncta were less abundant in ATG4.2 over-expression lines, in which unlipidated ATG8 predominated, which is consistent with ATG4.2 being an ATG8-deconjugating enzyme as well as an ATG8A-processing enzyme. In contrast, recombinant ATG8, ATG8B and ATG8C were all processed by ATG4.1, but not by ATG4.2. ATG8B and ATG8C both have a distinct subcellular location close to the flagellar pocket, but the occurrence of the GFP-labelled puncta suggest that they do not have a role in autophagy. L. major genes encoding possible ATG5, ATG10 and ATG12 homologues were found to complement their respective S. cerevisiae mutants, and ATG12 localised in part to ATG8-containing puncta, suggestive of a functional ATG5-ATG12 conjugation pathway in the parasite. L. major ATG12 is unusual as it requires C-terminal processing by an as yet unidentified peptidase.
Keywords: autophagy, Leishmania, protozoan parasite, ATG4, ATG8, ATG12
Introduction
Leishmania is a protozoan parasite that occurs as different morphological forms in its two hosts (mammals and sandflies) and greatly remodels its life cycle forms during differentiation. Within the alimentary tract of the sandfly vector Leishmania exists in two forms of promastigotes, the procyclic multiplicative form and the metacyclic, non-multiplicative mammal-infective form. Transformation to the smaller metacyclic form is known as metacyclogenesis. Within mammals Leishmania live in macrophages as small multiplicative, non-motile amastigotes (devoid of an external flagellum). Two parasites related to Leishmania, Trypanosoma brucei, the causative agent of sleeping sickness in man and nagana in cattle, and T. cruzi, responsible for Chagas disease, also have complicated life cycles involving a variety of morphological forms living in the mammalian and respective insect vectors; each undergoes extensive remodelling during differentiation to the subsequent form in the life cycle.
Autophagy is a conserved mechanism in eukaryotes whereby cytosolic proteins are transported in autophagosomes to the lysosomal network for degradation in response to nutrient deprivation. The amino acids generated are recycled and used for protein synthesis, thereby helping to maintain cellular homeostasis.1 In addition, autophagy plays roles in remodelling during cellular differentiation and development.2-4 Defects in autophagy in mammalian cells has been associated with a variety of disease states including cancer5-7 and neurodegenerative diseases,8,9 as well as susceptibility to bacterial and viral infections.10,11 Studies on trypanosomatids have shown that autophagy is crucial for differentiation between different life cycle forms and provision of nutrients under starvation conditions12-15 and it has been suggested that blocking autophagy could be a new strategy for fighting parasitic disease.12
The biogenesis of the dynamic membrane required for autophagosome formation in mammals and yeast requires the functioning of two conjugation pathways; those involving, respectively, ATG8 and ATG12/ATG5.16-20 ATG8 exists as multiple copies in mammals21-26 and plants,27 whereas yeast has just one; in all these cases, however, the ATG8s are of one type. ATG8 is synthesized as an inactive precursor, then it is activated via removal of a C-terminal polypeptide segment, by the clan CA, family C54 cysteine peptidase ATG4, to leave a C-terminal glycine.22,28,29 The exposed glycine is activated by the E1-like enzyme ATG7, transferred to the E2-like enzyme ATG3, and finally conjugated with the phospholipid, phosphatidylethanolamine (PE).30-33 PE anchors ATG8 to the autophagosomal membrane and adjacent ATG8 molecules tether together and thus mediate the formation of an autophagosome.34 The ATG8s are deconjugated from the PE before the autophagosomes enter the endosomal system in mammalian cells or the yeast vacuole, ATG4 also mediating this deconjugation.34-36 Thus ATG4 functions at two stages during autophagosome biogenesis and degradation. The absence of ATG8 does not negate the function of the core machinery proteins,37 and small autophagosome-like structures occur in yeast ATG8 null cells (designated atg8Δ).38 However, autophagosome size and the level of autophagy are regulated by the amount of Atg8 in yeast cells.36
Many ubiquitin-like modifier proteins similar to ATG8 have been identified, namely ubiquitin, neural precursor cell-expressed developmentally down-regulated 8 (Nedd8), small ubiquitin-related modifier (SUMO), Hub1, and ubiquitin-related modifier 1 (Urm1).39 They are similar to ATG8 in that they become conjugated to a primary amine to form an amide or isopeptide bond and subsequently are deconjugated to provide mechanisms that regulate cellular activities such as transcription, the cell cycle and autophagy. Of these, only ATG8 and ubiquitin have as yet been characterized in trypanosomatids 12,15,39 and the evidence is that ATG8 behaves similarly as in yeast and mammals.12,15 However, Leishmania is unusual in apparently having, based on predictions from genome mining, this type of ATG8 plus three others.15 One objective of this study was to experimentally test the hypothesis that all four groups indeed have the characteristics of ATG8s and to gain insights into their roles.
There is a multiplicity of ATG4s in higher eukaryotes. Mammals have four isoforms, autophagin-1, -2, -3 and -4, and their activities are thought to differ as only autophagin-1 and -3 can restore autophagy to Atg4Δ mutant yeast.40 Both ATG4s of Arabidopsis thaliana can cleave all its ATG8 copies and study of mutants defective in one or both of the AtATG4 genes (designated Δatg4a, Δatg4b or Δatg4a4b-1) suggest that redundancy exists between the AtAtg4s.41,42 Saccharomyces cerevisiae atg4Δ mutants are incapable of forming autophagosomes, and the expression in these mutants of ScAtg8 lacking residues beyond the scissile glycine residue resulted in autophagosome accumulation – indicating the role of ScAtg4 at the late stage of the autophagic pathway.35,43 Leishmania also has two ATG4s and, interestingly, mutants deficient in one of them (Δatg4.2) also accumulate putative autophagosomes.13 Thus a second aim of this study was to investigate whether the two ATG4s of Leishmania act differently towards the variety of Leishmania ATG8s and whether this may account for functional differences between them.
The second conjugation pathway necessary for autophagy in higher eukaryotes involves ATG12 and ATG5. ATG12 is activated by ATG7 and transferred to an E2-like enzyme, ATG10, whereupon it is conjugated to ATG5.44 The ATG12-ATG5 conjugate subsequently binds with ATG16 forming a multimeric complex of 350-800 kDa in size.45,46 The E3-like activity of the ATG12-ATG5 conjugate potentiates the formation of ATG8-PE; making the autophagic pathway analogous to ubiquitin-like pathways where an E3 conjugation ligase is required.47,48 The ATG12-ATG5 complex is absent from fully formed autophagosomes.
It has been reported, again based on genome mining, that these proteins are absent from trypanosomatids and so it was concluded that the ATG12/ATG5 pathway does not occur.12,49,50 The ATG12/ATG5 pathway is absent from organisms where microautophagy predominates51 and so its absence from T. brucei would correlate with evidence that the degradation of glycosomes in this parasites is via microautophagy.14 However, our studies have shown that macroautophagy occurs in Leishmania.13,15 Moreover, our in silico analyses led us to hypothesise that the proteins necessary for the ATG12/ATG5 pathway may indeed be encoded in the Leishmania genome, but they are significantly divergent from their yeast counterparts.15 Thus another aim of this study was to determine experimentally whether the proteins encoded by these putative genes of the ATG12/ATG5 pathway indeed function as postulated.
Results
The ATG4 and ATG8 proteins of L. major
Leishmania major has two ATG4 cysteine peptidases, designated ATG4.1 and ATG4.2. (see ref.13; http://www.genedb.org). The ORFs of the two ATG4 genes comprise 1185 bp and 1167 bp, encoding 394 and 388 amino acids with calculated molecular masses of 43.8 and 42.5 kDa, for ATG4.1 and ATG4.2, respectively. The predicted proteins are aligned with those of T. brucei, yeast, human and Arabidopsis in Figure S1A. The leishmanial proteins are typical of ATG4s in possessing an inhibitory loop over the active site pocket (Figure S1A; triple lines) and the conserved catalytic triad within the pocket (comprising C73, H241 and D239 in ATG4.1 and C92, H267 and D265 in ATG4.2; Fig. S1A, asterisks) typical of the proteins belonging to family C54 of Clan CA of cysteine peptidases.52,53 The residue adjacent to the catalytic triad that, based on evidence for the human enzyme HsAtg4B,52,53 is required for hydrolysis (Y54, marked with Δ in Figure S1A) is also conserved in both enzymes, whereas those implicated in recognition of the C-terminal region of the substrate ATG8 (W142, R229 and S316, marked with II in Fig. S1A) are present in ATG4.2, but are substituted with L159, V209, C296 in ATG4.1; suggesting perhaps some difference in substrate specificities.
Twenty five ORFs encode ATG8-like proteins in the L. major genome.15 The paralogue designated ATG8 (LmjF19.1630) has counterparts in the other sequenced genomes of Leishmania, L. infantum (LinJ19_V3.1660) and L. braziliensis (LbrM19_V2.1890). All are encoded by single copy genes with 35-46% identity with orthologues from yeast and higher eukaryotes. LmjF22.1300, another ATG8-like gene identified in L. major’s gene content, has the carboxyl-terminus scissile glycine that is exposed by the hydrolytic action of ATG4 prior to the activation and conjugation to PE by actions of E1-like and E2-like ATG proteins (* in Figure S2A(i)). Further, the LmjF22.1300 has amino acids C-terminal to the scissile glycine that is typical of ATG8. However, an insertion (of 58 amino acids) somewhat similar to that typical of ATG12s is also present in LmjF22.1300 (Figure S2A(ii). Thus based on sequence analyses alone LmjF22.1300 could be predicted to be either an ATG8 or an ATG12. Functional studies however, point to an ATG12 function for LmjF22.1300 (see below), so henceforth in this paper LmjF22.1300 is defined as ATG12.
The proteins designated as the ATG8A, ATG8B and ATG8C families are encoded by gene arrays, those encoding ATG8A and ATG8B being interspersed, and appear to be unique to Leishmania. The scissile glycine is conserved in all the paralogues (Figure S2A, marked with *). Phylogenetic analysis suggests that the ATG8A, ATG8B and ATG8C families are distinct clades (Figure S2B). While in this analysis ATG8 clusters with orthologues from other organisms (Figure S2B), the L. major ATG12 occupies an intermediate position between the ATG8 and ATG12 clades.
L. major ATG8 proteins are selectively cleaved by the ATG4 cysteine peptidases
The specific cleavage of the C-terminus of Atg8 by Atg4 in S. cerevisiae involves the complex interaction of residues adjacent to the active site of the peptidase with the recognition site in ScAtg8 comprising the residues Tyr49, Leu50, Phe77 and Phe79.54 Three of these residues are conserved in Leishmania ATG8 (being Leu54, Phe81 and Phe83, with the Tyr being replaced by Phe53), whereas none appear to be conserved in the other protein groups in which these residues are substituted with Leu42, Ala43, Ala70 and Ala72; Leu44, Ala45, Thr72 and Ala74, and Ser42, Ala43, Ala71 and Thr73, respectively, in all ATG8A, ATG8B and ATG8C members (Figure S2A). Thus the functionality of these three groups of proteins as ATG8s was uncertain. To investigate if the four families of ATG8 are substrates for the ATG4s, a representative from each class (LmjF19.1630 for ATG8; LmjF19.0844 for ATG8A; LmjF19.0850 for ATG8B; LmjF09.0150 for ATG8C) were produced as recombinant proteins with His and HA tags at the N- and C-termini, respectively. His-ATG8-HA, His-ATG8A-HA, His-ATG8B-HA and His-ATG8C-HA encode proteins of molecular mass 17.2, 18.0, 15.8, and 17.0 kDa, respectively. The use of the HA tag allowed the parent proteins and the products of hydrolysis by Leishmania ATG4 to be distinguished. The study involved co-expression of individual His- and HA-tagged ATG8s and ATG4s in E. coli, with subsequent cell lysis and incubation to allow hydrolysis and then detection of the His-tagged proteins by Western analysis. Under these conditions only His-ATG8B-HA was susceptible to degradation or cleavage by E. coli peptidases such that a smaller protein was generated (Figure 1A, lane1). However, co-expression of the representative proteins of each family of ATG8 with either ATG4.1 or ATG4.2 followed by incubation of the lysate resulted in a variety of banding patterns (Figure 1A). The results indicate that ATG4.1 has proteolytic activity towards His-ATG8-HA (Figure 1A1, lane 2), His-ATG8B-HA (Figure 1A3, lane 2) and His-ATG8C-HA (Figure 1A4, lane 2), whereas ATG4.2 has proteolytic activity only towards His-ATG8A-HA (Figure 1A2, lane 3). With His-ATG8B-HA, the proteolytic cleavage by ATG4.1 was rapid and the cleavage product had a different molecular mass from the product generated by E. coli proteolysis alone (Figure 1A3, lane 1). Prolonged incubation of ATG4.2 with His-ATG8-HA (8-24 h) produced a limited hydrolysis, whereas ATG4.1 degraded all His-ATG8C-HA if the incubation period was greater than 1 h (data not shown). A similar assay carried out using His-ATG12-HA showed that the ATG4s have no hydrolytic action against this protein under the in vitro assay conditions (Figure 1A5).
Figure 1. Hydrolysis of L. major ATG8s by L. major ATG4s in vitro.
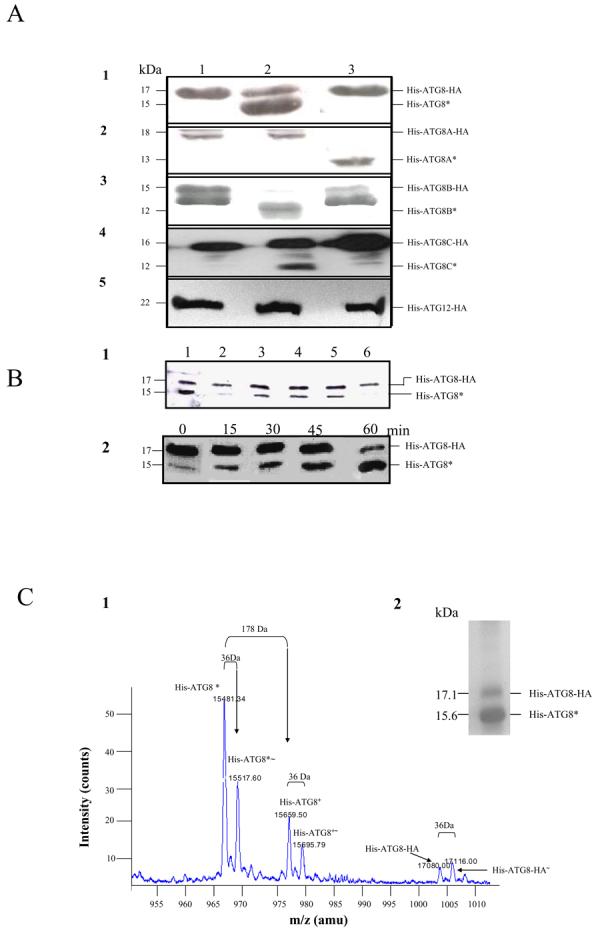
A. Soluble fractions of E. coli expressing His-ATG8-HA (row 1), His-ATG8A-HA (row 2), His-ATG8B-HA (row 3), His-ATG8C-HA (row 4) and His-ATG12-HA (row 5) alone (lane 1) or co-expressed with either ATG4.1 (lane 2) or ATG4.2 (lane 3) were incubated for 30 min at 30°C in 50 mM Tris-HCl, pH 8.0, 125 mM NaCl and 40 mM β-mercaptoethanol and then analysed by Western blot with α-His antibody. The cleaved ATG8 bands are labelled with an asterisk.
B. Hydrolysis of His-ATG8-HA by ATG4.1.
(B1) The hydrolysis was inhibited by 1 mM NEM (lane 2) and 1 mM iodoacetamide (lane 6) but unaffected by 1 mM PMSF (lane 3), 1 mM pepstatin (lane 4) and 10 mM 1,10-phenanthroline (lane 5). Lane 1 shows the control (no inhibitor added). Incubation was as described in A.
(B2) Hydrolysis of His-ATG8-HA by ATG4.1 was progressive over 60 min.
C. Analysis of the product of the cleavage of His-ATG8-HA by ATG4.1. Hydrolysis was over 30 min as described in A.
(C2) The product was purified by Ni+-affinity chromatography and analysed by SDS-PAGE, two main proteins were apparent.
(C1) The cleaved products were analysed by MALDI TOF. Six molecules were detected and their molecular masses are given.
Using His-ATG8-HA as a substrate, it was demonstrated that the proteolytic action by ATG4.1 was: (a) inhibited by 0.1 mM N-ethylmaleimide and 0.1 mM iodoacetamide (Figure 1B1, lanes 2 and 6, respectively) and unaffected by 1 mM E64, 10 μM pepstatin A, and 10 μM PMSF (Figure 1B1, lanes 3-5); and (b) time-dependent (Figure 1B2).
L. major His-ATG8-HA is cleaved by the ATG4 cysteine peptidases at the predicted scissile glycine residue
To determine the precise cleavage site of ATG4.1 on His-ATG8-HA, we carried out partial hydrolysis and purified the parent (His-ATG8-HA) and hydrolysed (His-ATG8*) proteins using affinity chromatography. Analysis of the eluant using SDS-PAGE showed the presence of just two detectable proteins, of sizes similar to those predicted for His-ATG8-HA and His-ATG8* (17189.79 and 15613.27 Da, respectively) (Figure 1C2). MALDI-TOF-MS analysis of this sample revealed three main peaks, each with a second peak of 36 Da greater mass (Figure 1C1). The three main peaks were designated His-ATG8*, His-ATG8+ and His-ATG8-HA, with the 36 Da partners marked with (∼). The mass of His-ATG8-HA was 131 Da lower than predicted, which is consistent with the removal of the initiator formyl t-RNA methionine during the translation of His-ATG8-HA (as reported previously.55,56 The mass difference between His-ATG8* and His-ATG8-HA is consistent with cleavage occurring between Gly121 and Gly122, towards the C-terminus of His-ATG8-HA. His-ATG8+ had 178 Da greater mass than His-ATG8*. This is likely to be due to N-gluconoylation of the exposed glycine on His-ATG8*, as previously reported to occur.57 The 36 Da partner peaks observed are consistent with chlorination of proteins and so the generation of second protein species, probably because the samples were maintained at pH 5.5 to prevent aggregation prior to analysis.58
Activity of L. major ATG4 towards the synthetic Fluorescence Resonance Energy Transfer (FRET) peptides Abz-peptidyl-Q-EDDnp
ATG4 peptidases cleave after a Gly residue, but what determines and is required for this specificity has not been analysed in detail. To investigate this we synthesised FRET peptides based on the amino acid residues surrounding the scissile Gly in the known and putative ATG8s (Figure S2A, underlined). In the case of LmjF19.0850 and LmjF19.0860, designated ATG8B.4 and ATG8B.5 respectively, which have three Gly residues in close proximity in the region of the likely cleavage, substrates including each of the glycine residues (Abz-A-M-G-G-Q-EDDnp and Abz-I-A-G-L-Q-EDDnp) were synthesised. We also synthesised analogues in which the putative scissile Gly was replaced by Ala. Using Abz-T-F-G-M-Q-EDDnp as substrate, it was shown that the activities were optimal at pH 7.5 and dependent upon reducing agents (10 mM DTT or 40 mM β-mercaptoethanol) (data not shown). Thus these conditions were used for the analyses.
Recombinant ATG4.1 was active towards all the substrates tested, whilst ATG4.2 was active towards many of them (Table 1A). Notably, the two peptidases displayed different preferences for the various peptidyl substrates, as judged by specific activity. Most interestingly, activity was not dependent upon the presence of Gly; indeed peptidyl substrates in which Gly was replaced by Ala were hydrolysed at a greater rate than the parent substrate (Table 1A). Detailed analyses (Table 2) confirmed that Abz-A-M-A-A-EDDnp was better than Abz-A-M-G-G-EDDnp as a substrate for ATG4.1. These analyses revealed kcat/Km values of between 6 and 28 µM−1sec−1 with the various substrates using ATG4.1 (Table 2).
Table 1A.
Enzymatic activity of recombinant ATG4.1 and ATG4.2 towards fluorogenic Abz-X-X-X-X-Q-EDDnp substrates (25 μM). Values are the means ± SE.
| ATG8 homologue with sequence |
Substrates | Specific Activities (nmoles.min−1 mg protein−1) |
|
|---|---|---|---|
| ATG4.1 | ATG4.2 | ||
| Hs-MAP-LC3 | Abz-T-F-G-M-Q-EDDnp | 99.4 ± 9.2 | 59.0 ± 3.2 |
| ATG8 | Abz-T-Y-G-G-Q-EDDnp | 23.5 ± 7.7 | <5 |
| Abz-T-Y-A-G-Q-EDDnp | 111.6 ± 7.9 | <5 | |
| ATG8A | Abz-S-M-G-A-Q-EDDnp | 47.0 ± 9.2 | <5 |
| ATG8B | Abz-A-M-G-A-Q-EDDnp | 311. 6 ± 8.2 | 144.5 ± 3.4 |
| ATG8B | Abz-A-M-G-G-Q-EDDnp | 28.1 ± 3.4 | <5 |
| Abz-A-M-A-A-Q-EDDnp | 149.7 ± 3.8 | 22.3 ± 3.5 | |
| ATG8B | Abz-I-A-G-L-Q-EDDnp | 163.4 ± 4.2 | 83.7 ± 5.8 |
| Abz-I-A-A-L-Q-EDDnp | 152.0 ± 6.2 | 83.5 ± 7.3 | |
| ATG8C | Abz-C-M-G-A-Q-EDDnp | 128.9 ± 4.8 | 65.3 ± 8.4 |
Table 2.
Kinetics parameters of recombinant ATG4.1 activity towards Fluorescence Resonance Energy Transfer (FRET) peptides Abz-peptidyl-Q-EDDnp.
| Substrate | Km (μM) | kcat (sec−1) | kcat/Km (μM−1 sec−1) |
|---|---|---|---|
| Abz-T-F-G-M-Q-EDDnp | 0.6 ± 0.05 | 4.4 ±0.5 | 7.3 |
| Abz-T-Y-G-G-Q-EDDnp | 0.1 ± 0.05 | 1.1 ±0.4 | 10.7 |
| Abz-T-Y-A-G-Q-EDDnp | 0.6 ± 0.07 | 4.5 ±0.3 | 7.6 |
| Abz-S-M-G-M-A-EDDnp | 0.2 ± 0.05 | 1.6 ±0.5 | 7.8 |
| Abz-A-M-G-A-Q-EDDnp | 0.5 ± 0.05 | 8.8 ±0.0 | 17.7 |
| Abz-A-M-G-G-Q-EDDnp | 0.2 ± 0.06 | 1.4 ±0.2 | 6.6 |
| Abz-A-M-A-A-Q-EDDnp | 0.2 ± 0.07 | 5.7 ±0.3 | 28.4 |
| Abz-I-A-G-L-Q-EDDnp | 0.5 ± 0.04 | 4.9 ±0.3 | 9.9 |
| Abz-I-A-A-L-Q-EDDnp | 0.2 ± 0.09 | 5.4 ±0.2 | 26.8 |
| Abz-C-M-G-A-Q-EDDnp | 0.5 ± 0.08 | 6.2 ±0.3 | 12.3 |
The activities of both enzymes towards Abz-T-F-G-M-Q-EDDnp were inhibited almost completely by N-ethylmaleimide (0.1 mM) or iodoacetamide (1.0 mM), inhibited significantly by 1,10 phenanthroline (0.2 mM) or PMSF (1.0 mM) and unaffected by E64 (0.1 µM), pepstatin A (1.0 mM) or EDTA (0.5 mM) (Table 1B).
Table 1B.
Inhibitory activity of peptidase inhibitors against recombinant ATG4.1 and ATG4.2. ATG4 (0.5 μg) was incubated with the inhibitors at room temperature for 15 min, and residual activity against 25 μM Abz-T-F-G-M-Q-EDDnp analysed. Values are the means ± SE.
| Inhibitors | % Inhibition |
|
|---|---|---|
| ATG4.1 | ATG4.2 | |
| 0.1 mM N-Ethylmaleimide (NEM) | 99 ± 1.4 | 99 ± 1.9 |
| 1.0 mM Iodoacetamide | 99 ± 2.1 | 97 ± 3.2 |
| 0.2 mM 1,10 phenanthroline | 50 ± 9.3 | 43 ± 4.2 |
| 0.2 mM Phenylmethylsulfonyl fluoride (PMSF) | 22 ± 8.4 | 17 ± 5.7 |
| 1.0μM E64 | <1.0 ± 0.0 | < 0.1 ± 0.0 |
| 1.0 mM Pepstatin A | <1.0 ± 0.0 | <0.1 ± 0.0 |
| 0.5 mM EDTA | <1.0 ± 0.0 | <0.1 ± 0.0 |
L. major ATG8 genes complement ATG8-defective yeast strains
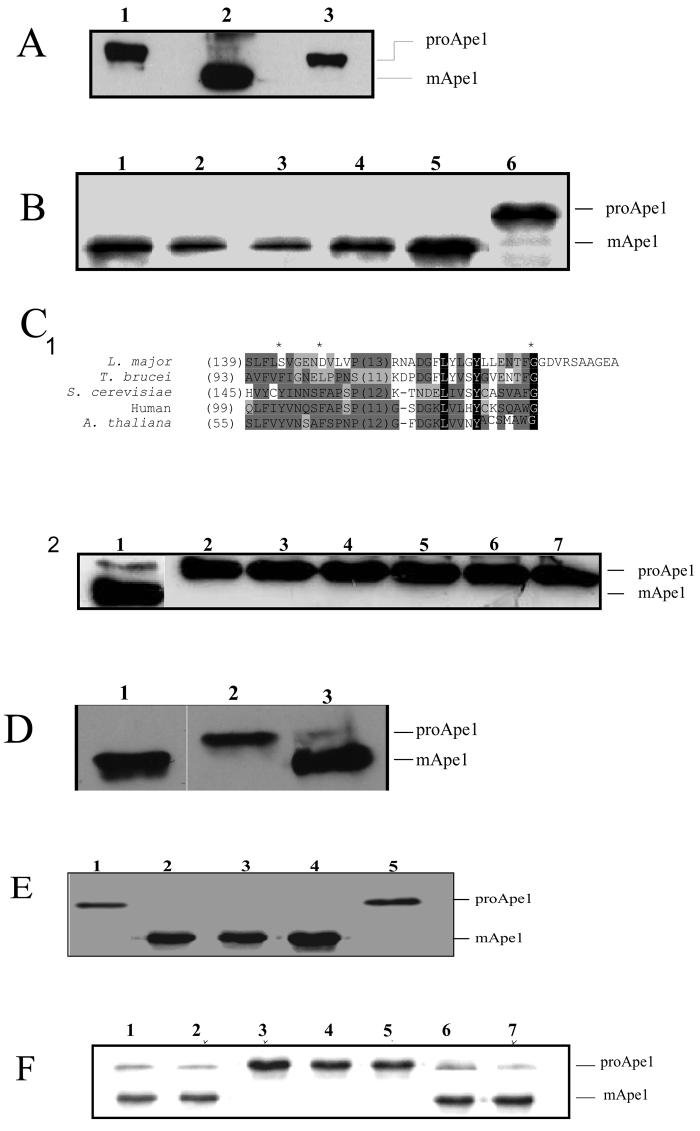
As the proteins designated ATG8A, ATG8B and ATG8C have only low identity with ATG8 orthologues from other organisms, we investigated if the respective genes are functional homologues by using yeast mutants deficient in the ATG8 gene homologue (the mutants are known as atg8Δ). The ORFs of ATG8, ATG8A.1, ATG8B.4, and ATG8C.1 were cloned into pCM185 plasmid, used to transform the atg8Δ null mutant of S. cerevisiae and the resulting cell lines assessed for their ability to process the vacuolar hydrolase aminopeptidase (Ape1) from its inactive precursor (PreApe1) to its mature form (mApe1), a process that is defective in the atg8Δ null mutant.40,59 The results using the control lines confirmed the validity of the assay (Figure 2A). The gene products of the representatives of all the predicted ATG8 families were able to complement the autophagic defect in the yeast atg8Δ null mutant (Figure 2B, lanes 3-6). These results provide evidence that all four of the predicted L. major ATG8 gene families can fulfil the ATG8 function, at least in yeast.
Figure 2. Complementation studies with L. major ATG genes in autophagy-defective Saccharomyces cerevisiae.
A. Western blot analysis of aminopeptidase I (Ape1) in yeast mutant atg8Δ (lane 1), wild type (lane 2) and yeast mutant atg12Δ (lane 3) S. cerevisiae. Cells were incubated under starvation conditions in SD(-N) medium for 16 h, lysed, and API processing was analyzed using rabbit α-Ape1 antibodies. ProApe1 and mature Ape1 (mApe1) are indicated.
B. Western blot analysis of Ape1 in atg8Δ yeast transformed with pCM185 containing L. major ATG8 homologues: ATG8 (lane 3), ATG8A (lane 4), ATG8B (lane 5), ATG8C (lane 2) or transformed with pCM185 only (lane 6). Wild type S. cerevisiae are shown in lane 1. The cells were cultured under starvation conditions in SD(-N) medium for 4 h and Ape1 processing was analyzed using rabbit α-API antibodies. ProApe1 and mature Ape1 (mApe1) are indicated.
C1. Amino acid sequence alignment of the C-terminal end of L. major ATG12 with those of T. brucei, S. cerevisiae, human and A. thaliana. Identical and conserved amino acids are shaded in black and grey, respectively. Shade of grey depends upon number of residues conserved. The carboxyl-terminal glycine identified in A. thaliana ATG12 (Hanada et al., 2005) and the two hydrophobic residues (Y149 and F154) required for conjugation to ATG10 (Suzuki et al., 2006; Hanada et al., 2006) and essential for the formation of the ATG12-ATG5 conjugate are marked with an asterisk. L. major, LmjF22.1300; T. brucei, Tb927.7.3320; S. cerevisiae, P38316; human, AAH11033; A. thaliana, AAM70187. The numbers in parenthesis represent the amino acid number of the first residue in the alignment.
C2. Western blot analysis of Ape1 in atg12Δ S. cerevisiae transformed with pCM185 containing L. major ATG8 homologues: ATG8 (lane 3), ATG8A (lane 4), ATG8B (lane 5), ATG8C (lane 6), the putative L. major ATG12 (lane 7) or transformed with pCM185 only (lane 2). Wild type S. cerevisiae were used as the control (lane 1). The S. cerevisiae were cultured under starvation condition for 16 h and Ape1 processing was analyzed using a rabbit α-API antibodies. ProApe1 and mature Ape1 (mApe1) are indicated.
D. Western blot analysis of Ape1 in atg5Δ S. cerevisiae transformed with pCM185 containing L. major ATG5 (lane 3) and pCM185 only (lane 2) and treated as described in C2. Wild type S. cerevisiae was included as a positive control (lane 1).
E. Western blot analysis of Ape1 in atg10Δ S. cerevisiae transformed with pCM185 containing the L. major ATG10 homologue (lanes 2-3), or transformed with pCM185 only (lane 1) or ATG8 (lane 5) and treated as described in C2. Wild type S. cerevisiae was included as positive control (lane 4).
F. Western blot analysis of ApeI in atg8Δ S. cerevisiae (lanes 2-4) and atg12Δ S. cerevisiae (lanes 5-7) transformed with pCM185 containing ATG8g (lanes 2, 5), ATG12g (lanes 3, 6) and S. cerevisiae ATG12 (lanes 4, 7), respectively, and treated as described in C2. Wild type S. cerevisiae was included as positive control (lane 1).
L. major has functional homologues of proteins of the ATG12-ATG5 conjugation pathway
We have previously reported genes with some similarity to ATG5, ATG10 and ATG12 in the L. major genome, suggestive of the existence of an ATG12-ATG5 conjugation pathway in this organism.15,59 Based on evidence from other organisms, the ATG12-ATG5 pathway is likely to involve the interaction of the C-terminal Gly of a Leishmania ATG12 (Gly182 of LmjF22.1300, Figure 2C1, asterisk) with Lys143 on ATG5 via an isopeptide bond, a reaction mediated by a Cys residue of ATG7 and ATG10.60-62 To investigate the functionality of the putative ATG5 (LmjF30.0980), ATG10 (LmjF31.3105) and ATG12 (LmjF22.1300), we performed yeast complementation assays, with constructs comprising the ORFs cloned into the pCM185 plasmid, in their corresponding yeast null mutants. The predicted ATG5 was able to rescue the defective autophagic pathway in the atg5Δ yeast mutant (Figure 2D, lane 3) as effectively as the S. cerevisiae Atg5 (Figure 2D, lane 1), similarly the predicted ATG10 complemented the atg10Δ yeast mutant (Figure 2E, lanes 2-3). L. major ATG8 was unable to complement the yeast atg12Δ mutant (Figure 2E, lane 5).
Full length ATG12, however, also did not complement the autophagic defect in atg12Δ yeast mutant (Figure 2C2, lane 7). Representative genes from the ATG8, ATG8A, ATG8B and ATG8A gene families also failed to complement the autophagy defect of the atg12Δ yeast mutant (Figure 2C2, lanes 3-6). ATG12 from a wide variety of organisms characterized to date 63-66 all terminate at an exposed glycine (Figure 2C1), the residue that interacts with other ATG5 and ATG16. We thus hypothesised that as ATG12 of L. major encodes amino acids C-terminal to the scissile glycine, it would need to be processed prior to being functional in the ATG5-ATG12 conjugation pathway and the lack of functionality of ATG12 in the yeast atg12Δ yeast mutant was due to the yeast lacking the appropriate peptidase to expose the glycine residue so that it was available for conjugation. We tested this hypothesis using mutated genes for ATG12 and ATG8 that terminated at the key glycine (designated ATG12g and ATG8g, respectively). ATG12g did not complement an autophagic defect in the atg8Δ yeast mutant (Figure 2F, lane 3) while ATG8g did (Figure 2F, lane 2). However, ATG12g did complement the atg12Δ yeast mutant (Figure 2F, lane 6), similarly to ScATG12 itself (Figure 2F, lane 7). ATG8g did not complement an autophagic defect in the atg12Δ yeast mutant (Figure 2F, lane 5). These results suggest that LmjF22.1300 is a functional ATG12, but unlike homologues reported in other species to date it needs to be cleaved prior to conjugation to ATG5 and ATG16.
ATG12 forms punctate structures in L. major promastigotes
It is known that ATG8 N-terminally tagged with GFP becomes associated with punctate structures (putative autophagosomes) in Leishmania promastigotes, especially under the starvation conditions and during differentiation between its life cycle forms.13 To discern the functional similarity and differences between ATG8 and ATG12, we compared the localisation of ATG12 N-terminally tagged to a red fluorescent protein (and so designated RFP-ATG12) and GFP-ATG8-containing puncta in the cytoplasm of L. major promastigotes. In nutrient-rich medium, RFP-ATG12 had a diffuse cytosolic distribution (data not shown). Under starvation conditions, most cells redistributed RFP-ATG12 into a punctate structure ∼500 nm wide (Figure 3A1). Promastigotes co-expressing both RFP-ATG12 and GFP-ATG8 produced, under starvation, some punctate structures displaying red and green fluorescence. This is consistent with both proteins being involved in autophagosome formation (Figure 3A1). As RFP-ATG12 produced structures that overlapped with GFP-ATG8-containing puncta, we investigated whether each protein became lipidated. Two GFP-ATG8 proteins were detected in starved promastigotes, the non-lipidated GFP-ATG8* (∼ 40 kDa) and the rapidly migrating lipidated GFP-ATG8-PE (Figure 3B1). However, for ATG12 only one major protein was detected at 50 kDa, which is consistent with the predicted mass of the fusion protein (Figure 3B2). On longer exposure a trace of higher molecular mass GFP-ATG12 could be detected (not shown), which could be an ATG12-ATG5 conjugate. These findings are consistent with ATG12 having a role in autophagosome biogenesis, and one that is different from that of ATG8.
Figure 3. Localization of ATG12 in L. major promastigotes.
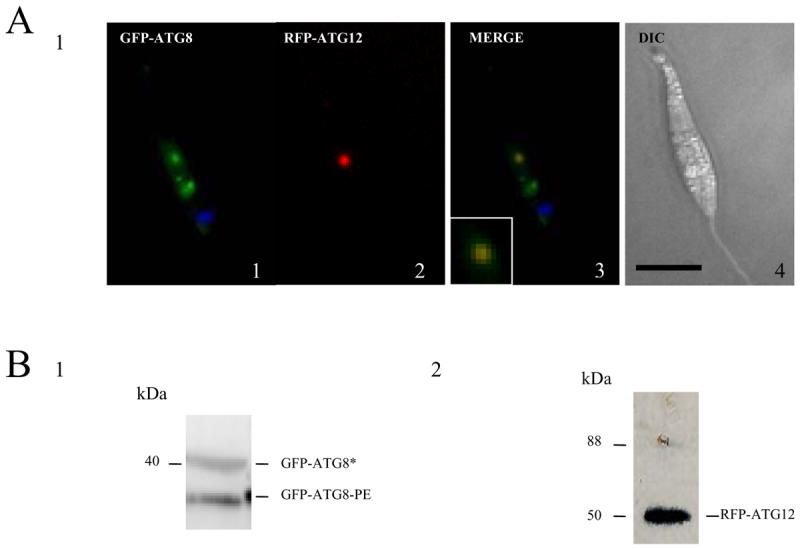
A. Occurrence of RFP-ATG12-containing structures (A2) and GFP-ATG8-containing putative autophagosomes (A1) in wild type L. major promastigotes expressing both proteins. The position of the kinetoplast is shown (blue) using DAPI staining. L. major procyclic promastigotes were suspended in nutrient-deprived medium for 120 min and observed by fluorescence microscopy. The merge of the RFP-ATG12 and GFP-ATG8 images, the co-labelled structure is enlarged at the bottom left, and the DIC image of the promastigote are shown in A3 and A4, respectively. Scale bar, 10 μm.
B. Western blot analysis using α-GFP (B1) and α-RFP (B2) antibodies on L. major procyclic promastigote cell extracts (5-8 × 106 cells ml−1) transfected with GFP-ATG8 and RFP-ATG12. The α-GFP antibody detected a faster migrating lipidated ATG8 (labelled GFP-ATG8-PE) and a higher molecular mass unlipidated ATG8 (labelled GFP-ATG8*), resolved in a urea SDS-PAGE gel (B1). The anti-RFP antibody showed just a single protein (labelled RFP-ATG12) consistent with the predicted size of the fusion protein (B2).
L. major ATG8 paralogues form puncta in promastigotes
We recently demonstrated that GFP-ATG8 can be used as a marker for tracking putative autophagosomes in L. major.13, 15 As the four families of ATG8 in L. major can complement a yeast ATG8 gene-deletion mutant and are substrates for ATG4s, we predicted that they would associate with the putative autophagosomes in L. major in vivo. GFP tagged ATG8A, ATG8B and ATG8C were expressed in L. major promastigotes (designated WT(pN-ATG8A), WT(pN-ATG8B), WT(pN-ATG8C)) and characterised as described previously.13 As predicted, GFP-ATG8A, GFP-ATG8B and GFP-ATG8C did form punctate structures under certain conditions (Figure 4A). Interestingly, the single punctum in WT(pN-ATG8B) and WT(pN-ATG8C) was always located close to the flagellar pocket (Figure 4A3 and 4A4), the only site of endocytosis and exocytosis in the cell. Most early log phase promastigotes of WT(pN-ATG8A), WT(pN-ATG8B) and WT(pN-ATG8C) in nutrient-rich medium had the GFP-tagged proteins evenly distributed throughout the cytoplasm (similar to the diffuse pattern described previously for GFP-ATG8 transgenic cell lines at logarithmic phase of growth,13 although 2-10% had a single GFP-labelled punctum in the cytosol (Figure 4B, 0 h). However, in contrast to the pattern described previously for GFP-ATG8, no peak in punctate structures was observed for GFP-ATG8A, GFP-ATG8B or GFP-ATG8C during metacyclogenesis, and the proportion of cells containing puncta remained consistently low during growth in nutrient rich medium (data not shown). Suspension of the promastigotes of WT(pN-ATG8A), WT(pN-ATG8B and WT(pN-ATG8C) in nutrient-deprived medium (PBS) resulted in the production of puncta in most WT(pN-ATG8A) cells by the end of the 4 h incubation (Figure 4A.1 and 4B), with there being 5-8 puncta per cell (Figure 4A1). In contrast, there was no significant increase in the occurrence of puncta in WT(pN-ATG8B) and WT(pN-ATG8C) upon starvation (Figure 4B). These data suggest that ATG8 and ATG8A are associated with starvation-induced puncta (putative autophagosomes), but that ATG8B and ATG8C are associated with other structures in the cell.
Figure 4. Localisation of ATG8 family members in L. major promastigotes.

A. The occurrence of punctate structures in live L. major promastigotes containing GFP-ATG8 (row 1), GFP-ATG8A (row 2), GFP-ATG8B (row 3) and GFP-ATG8C (row 4). Promastigotes expressing GFP-ATG8A were visualised after 4 h incubation in nutrient-deprived medium. Arrowheads indicate punctate structures. Scale bar, 10 μm.
B. Quantification of punctate structure formation in promastigotes after incubation for 4 h in nutrient-deprived medium. The number of promastigotes containing at least one GFP-ATG8 punctum is expressed as a percentage of all GFP-expressing cells. The data are means ± SE from three series of measurements from three independent experiments. *, data for GFP-ATG8 and GFP-ATG8A cell lines after starvation differed significantly from those for cells maintained in nutrient-rich medium (p<0.01).
C. The appearance of live Δatg4.2 promastigotes containing GFP-ATG8A (C1) or GFP-ATG8 (C2) after 4 h starvation in nutrient-deprived medium. Arrowheads indicate puncta. Scale bar, 10 μm.
To analyse the possible involvement of Leishmania ATG4.2 in the formation of these puncta in vivo, we investigated the formation of structures containing GFP-ATG8A, GFP-ATG8B and GFP-ATG8C in a L. major mutant lacking ATG4.2 (Δatg4.2, 13). Δatg4.2 promastigotes transfected with the GFP-ATG8A, GFP-ATG8B and GPF-ATG8C constructs (designated Δatg4.2[pN-ATG8A], Δatg4.2[pN-ATG8B], Δatg4.2[pN-ATG8C]) were assessed for the formation of puncta formation under starvation conditions. Fluorescence microscopic analysis of the distribution of these ATG8 homologues in the Δatg4.2 cell line in nutrient-rich medium prior to their transfer to starvation medium was similar to that of the wild type cell lines transfected with the same construct, in that they were diffusely distributed throughout the cytosol with just a small proportion of cells containing a single punctum (data not shown). This is in contrast to the greater number of ATG8-containing structures that were present in Δatg4.2[pN-ATG8] than in WT(pN-ATG8) under similar conditions (Besteiro et al., 2006). As with WT(pN-ATG8B) and WT(pN-ATG8C), suspension of Δatg4.2[pN-ATG8B] and Δatg4.2[pN-ATG8C] promastigotes in nutrient-deprived medium did not result in a significant increase in puncta. Interestingly, however, starvation of Δatg4.2[pN-ATG8A] did not result in the increased occurrence of puncta as occurred with WT[pN-ATG8A] (Figure 4C1 and comparison with Figure 4A2). Δatg4.2[pN-ATG8] cells on the other hand retained the enhanced number of puncta also under these conditions (Figure 4C2 and comparison with Figure 4A1). These results are consistent with and support the biochemical data reported above that ATG4.2 is able to hydrolyse ATG8A rapidly and suggest that this processing could be involved in autophagosome formation in vivo.
ATG4.2 has a post-lipidation role in L. major autophagic pathway involving ATG8
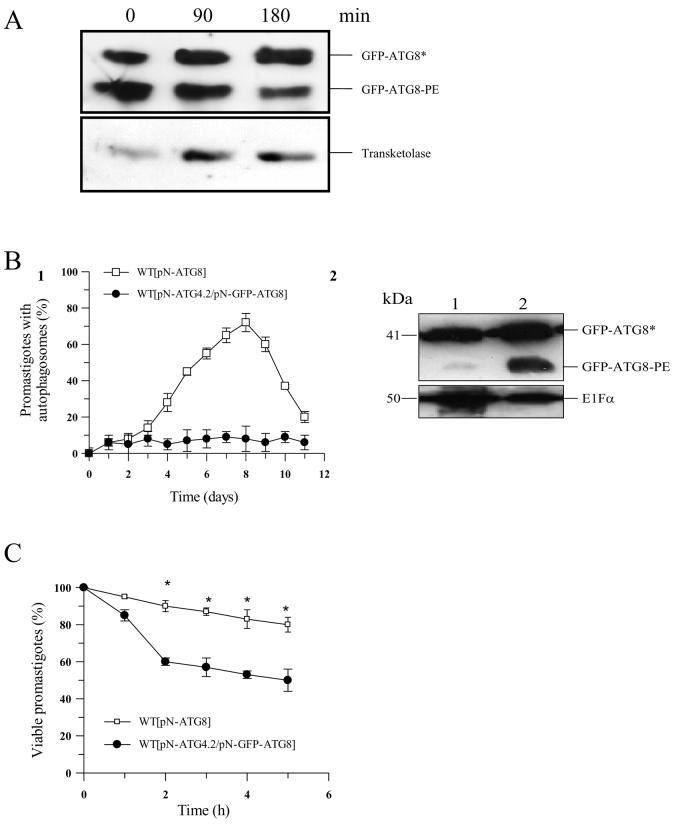
The enhanced number of GFP-ATG8-containing puncta in Δatg4.2 relative to wild type cells (see ref. 13 and Figure 4C2) led us to hypothesise that ATG4.2 is a deconjugating enzyme for lipidated ATG8. We have previously shown by Western blot analysis that extracts from the Δatg4.2 cell line over-expressing GFP-ATG8 contained relatively more GFP-ATG8 conjugated to PE than wild type cells similarly expressing GFP-ATG8.13 To investigate this further, we developed an in vitro deconjugation assay that involved the incubation of purified recombinant ATG4.2 with lysate from Δatg4.2[pN-ATG8] for 3 h. Western blot analysis carried out on aliquots collected at 90 min intervals showed a reduction in GFP-ATG8-PE (Figure 5A). Next, ATG4.2 was over-expressed in WT [pN-GFP-ATG8] to produce the WT [pN-ATG4.2/pN-GFP-ATG8] line and assessed for its ability to form putative autophagosomes. Microscopic analysis of the cell line revealed that GFP-ATG8-containing puncta did not become more abundant as the line progressed from log to early stationary phase of growth (Figure 5B1). Western blot analysis of cell extracts from early stationary phase promastigotes from both cell lines showed that the WT [pN-ATG4.2/pN-GFP-ATG8] had predominantly the unlipidated form of GFP-ATG8 (Figure 5B2, lane 1), consistent with the rapid cleavage of ATG8 from PE on the nascent structures (putative autophagosomes) preventing them becoming fully formed. One predicted consequence of this is that WT [pN-ATG4.2/pN-GFP-ATG8] promastigotes would be less able to withstand starvation, a process that requires a functional autophagic pathway. This was found to be the case (Figure 5C). Our studies have shown that the enzyme has only low activity towards ATG8 itself (Figure 1A1), but presumably it has better activity towards ATG8 bound to PE. These data together suggest that in L. major ATG4.2 is a deconjugating enzyme for ATG8 and that the increased amount of ATG4.2 in the transgenic line results in the deconjugation of PE from ATG8 before the putative autophagosomes can be properly formed.
Figure 5. A role for ATG4.2 in processing lipidated ATG8 in L. major.
A. Western blot analysis using α-GFP antibody on Δatg4.2 [pN-ATG8] promastigote cell extracts (5-8 × 106 cells ml−1) show an abundance of lipidated ATG8 (labelled GFP-ATG8-PE) (lane 0). Incubation of the cell extract with recombinant ATG4.2 for 90 and 180 min at 30°C resulted in a decrease in GFP-ATG8-PE (lanes 2 and 3). Transketolase was used as an internal loading control.
B1. WT[pN-ATG8] (open squares) and WT[pN-ATG4.2/ pN-GFP-ATG8] (closed circles) promastigotes were compared for the occurrence of putative autophagosomes during growth in vitro in normal medium. Data are means ± SD from 3 independent experiments.
B2. Western blot analysis of extracts from early stationary phase WT[pN-ATG4.2/pN-GFP-ATG8] (lane 1) and WT[pN-GFP-ATG8] (lane 2) promastigotes separated by SDS-PAGE containing 6 M urea and detected with α-GFP antibody. Cleaved ATG8 (GFP-ATG8*) and lipidated ATG8 (GFP-ATG8-PE) are indicated. EF1α was used as an internal loading control.
C. Sensitivity to starvation of WT[pN-ATG8] (open squares) and WT[pN-ATG4.2/pN-GFPATG8] (closed circles) L. major promastigotes. Cells were incubated in PBS and their viability assessed by the MTT assay. Data are means ± SD from four replicates. *: data for WT[pN-ATG4.2/pN-GFP-ATG8] and WT[pN-GFP-ATG8] promastigotes differed significantly (P < 0.05).
Discussion
The occurrence of autophagy in a wide variety of organisms ranging from protozoa to humans is indicative of its indispensable contribution to the life of eukaryotes.7,67 Much is known of the molecular mechanisms of autophagy in mammals and yeast,67 but far less about the process in lower eukaryotes such as Leishmania. We have previously shown that autophagy has a role in the transformation of Leishmania, 13,15 but predictions based on bioinformatic analyses suggested differences in the autophagic machinery from that underpinning autophagy in mammals; most notably the occurrence of multiple ATG8-like genes and the uncertainty about the existence of the two conjugation pathways described for higher eukaryotes.15,49,50,68 Thus, the major aim of this study was to verify experimentally whether the multiple ATG8s encoded in the Leishmania genome function as ATG8s and whether or not they have different roles and also to verify whether the genes identified as potential components of the ATG12-ATG5 conjugation pathway in the L. major genome15 are indeed functional homologues.
The yeast complementation approach has provided compelling evidence that L. major ATG8, ATG8A, ATG8B and ATG8C can indeed carry out the function of the yeast Atg8 (Figure 2). Moreover, examples of the three families of ATG8-like proteins were found to form GFP-labelled puncta in promastigotes transfected with GFP-tagged ATG8s (Figure 4). However, the puncta occurrence, location and number differed between the four families. GFP-ATG8B and GFP-ATG8C occur as punctate structures in only a low proportion of cells during growth in normal medium and, unlike GFP-ATG8,13 no peak in puncta formation was observed during metacyclogenesis or in response to starvation. In addition, ATG8B and ATG8C do not co-localise with ATG8-labelled puncta (data not shown). These data suggest that while ATG8B and ATG8C are likely to have ATG8-like role, this appears to be related not to the putative autophagosomes but to another as yet unidentified vesicle. Interestingly, GFP-ATG8B and ATG8C-containing puncta were always located close to the flagellar pocket, perhaps pointing to a potential role in endocytic trafficking or recycling. Thus these data suggest that ATG8B and ATG8C do not have a role in macroautophagy which is in contrast to the mammalian situation where the three ATG8 homologues (GATE-16, GABARAP and LC3) are more similar (all are in the ATG8 clade, Figure S2B) and all are involved in autophagy.32,33,69
ATG8A of L. major differs from each of the other families of ATG8s. It rarely was observed in punctate structures during growth in nutrient rich medium, but a strong induction of ATG8A labelled puncta was observed in response to nutrient deprivation - indicating a potential role in starvation-induced autophagy. Its role in autophagy was reinforced by the finding that it often co-localised with ATG8-labelled puncta (Figure 4). However, its distribution, even when in puncta, and occurrence, the puncta did not increase during metacyclogenesis, indicate clearly that ATG8A differs in its detailed role from that of ATG8 itself.
One common feature between yeast and higher eukaryotes is the existence of just one type of ATG8, albeit there are multiple copies in the latter but just one copy in yeast.27,69-72 Thus the multiple families of ATG8s in Leishmania is most unusual, and indeed it has no known parallel (not occurring even in other trypanosomatid genera such as Trypanosoma). The ATG8 families are found in different Leishmania species that cause diverse diseases (for example L. infantum, L. braziliensis and L. mexicana), so they are likely to have conserved functions in the parasite. We have now shown that they are all ATG8-like in action but that there are clear differences. Discovering the parts that each plays is the next goal, but as ATG8A, ATG8B and ATG8C occur as multicopy gene families it will be problematic to investigate this through generation of ATG8 family-specific knockout lines.
L. major also has two homologues of ATG4 (ATG4.1 and ATG4.2)13 and we hypothesised that part of the selective functioning of the ATG8s could be mediated through differential cleavage by the differing ATG4s. We investigated the specificity of the two ATG4s towards ATG8 and the 3 families of ATG8-like proteins using an in vitro cleavage assay. The results (Figure 1) indicate clear specificity of cleavage between different ATG4.1 and ATG4.2 and the four ATG8 families. The mechanism determining this specificity remains uncertain, however, the two ATG4s showed some similarity in their ability to cleave short peptide fluorogenic substrates (designed on the basis of the cleavage sequence in each class of ATG8; Table 2) and so the specificity of the ATG4s towards the ATG8s must be controlled by other residues than the amino acids close to the cleavage site. Interestingly, the results of this approach using short peptide fluorogenic substrates show that the scissile glycine residue is not a requirement for hydrolysis; previously it has also been shown that in the action of HsAtg4b on a 9-mer the scissile glycine plays no role in the hydrolysis of ATG8.53 Thus residues other than this glycine and the adjacent amino acids must be important in determining the substrate binding of the enzyme, as previously postulated.35,73
Our investigation has provided firm evidence for the existence in L. major of functional equivalents of proteins of the ATG12-ATG5 pathway. LmjF22.1300 has moderate levels of identity to both ATG8 and ATG12 and it failed to reconstitute the autophagic defect in atg12Δ yeast mutant lines. However, the gene encoding a truncated form not encoding residues beyond the scissile glycine fully restored autophagy to atg12Δ yeast (Figure 2F). In addition, LmjF22.1300 was unlike ATG8 in that under starvation stress it located to a single large punctum in most cells with as few as 13% having multiple structures and we could find no evidence that it was lipidated. Thus we have tentatively designated this as ATG12. Nevertheless it is an unusual ATG12 in that it requires cleavage prior to conjugation to ATG5, a situation that does not occur in yeast or mammalian cells. In vitro assays would suggest that neither ATG4.1 nor ATG4.2 can process ATG12, so it must be that a different peptidase cleaves ATG12 in vivo. A syntenic homologue to LmjF22.1300 is present in other trypanosomatids; T. cruzi (designated TcATG8.2) and T. brucei (Tb927.7.3320). Our findings with L. major ATG12 are similar to the reported findings for TcATG8.2 in that the protein forms punctate structures in T. cruzi, but less so than TcATG8.1, and this apparently does not involve conjugation to PE.12 It is possible, therefore, that TcATG8.2 is also a functional ATG12. The yeast functional assays used also provided support for the predicted gene identification for LmjF30.0980 (ATG5) and LmjF31.3105 (ATG10), as they could restore autophagy to yeast mutants lacking the ATG5 and ATG10 yeast genes. LmjF30.0980 and LmjF31.3105 also have syntenic homologues in T. cruzi and T. brucei, so our results suggest that L. major, T. brucei and T. cruzi are similar to other eukaryotes studied in having two conjugation pathways involved in autophagosome biogenesis.
Findings with mammalian cells and yeast are all in accordance with ATG4s having a deconjugation role. A down-regulation of HsAtg4b in HEK293 cells can increase endogenous LC3-PE and GABARAP-PE levels.33 A mutant ATG8 construct lacking residues beyond the scissile glycine expressed in atg4Δ S. cerevisiae, which is incapable of forming autophagosomes, resulted in a build up of autophagosomes that cannot be delivered to the vacuole – indicating the importance of the role of ATG4 at the late stage of the autophagic pathway.36,74 Our previous study suggested that L. major ATG4.2 is involved in deconjugating ATG8 from putative autophagosomes.13 We have now shown that L. major ATG4.2 has delipidation activity and can directly delipidate ATG8-PE to ATG8 (Figure 5A). Moreover, over-expression of ATG4.2 greatly reduced GFP-ATG8 puncta in wild type lines (Figure 5B). This is consistent with a greatly increased rate of deconjugation of ATG8 from the surface of the putative autophagosomes before they are fully formed and thus their enhanced clearance from the cytosol, as has been described in other systems.33,34,43 Thus our findings on the effects of the absence or over-expression of ATG4.2 on the location and form of GFP-ATG8 in Leishmania accords well with the roles of ATG4s in mammals. The enhanced susceptibility of these L. major lines to the stress of starvation (see ref. 13 and Figure 5C) is also indicative that both lines lack fully functional autophagic machinery. Thus although our data show that ATG4.1 and ATG4.2 in Leishmania have differences in specificity towards substrates and are of different importance in certain roles, there appears to be redundancy between the two ATG4s. Further evidence for this is provided by the finding that ATG4.2 is capable of processing ATG8 during prolonged incubation in vitro (this study) and that Δatg4.1-deficient lines expressing GFP-ATG8 form putative autophagosomes in vivo (Williams, Mottram and Coombs, unpublished).
Materials and Methods
Identification, cloning, expression and purification of genes involved in autophagy
Genes potentially involved in autophagy in L. major have been reported.15 The open reading frames (ORFs) of ATG4.1 (LmjF32.3890) and ATG4.2 (LmjF30.0270), ATG8 (LmjF19.1630), ATG8A (LmjF19.0840), ATG8B (LmjF19.0850) and ATG8C (LmjF09.0150) were amplified by PCR experiments using the Expand High Fidelity PCR system (Roche Molecular Biochemicals) from L. major genomic DNA, isolated as described by (75), using gene-specific primers modified with appropriate restriction sites and epitope tags (to facilitate cloning into their respective expression vectors and resolution of cleavage products, respectively) as detailed in Table S1. All PCR assays were carried out in a GeneAmp 9600 PCR system (PerkinElmer Life Sciences) for 30 cycles of denaturation (94°C, 15 s), annealing (65°C, 15 s) and extension (72°C, 2 min). Each ORF was verified by nucleotide sequencing (MBSU, University of Glasgow) and cloned into the pET expression vectors (Invitrogen) pre-digested with appropriate restriction enzymes to produce the plasmids detailed in Table S1(i). BL21(DE3) (Invitrogen) were transformed with plasmids to provide the strains listed in Table S2 (ii). Protein expression was carried out overnight at 15°C after induction with 1- 2 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Recombinant proteins were purified using an affinity chromatography column (bioCAD® 700E work station) and eluants of the ATG4s and ATG8s were dialysed against 50 mM Tris-HCl, pH 7.9, 125 mM NaCl, 10% glycerol at 4°C overnight and stored at 4°C with 1 mM DDT for ATG4 and 1% urea for ATG8. In some instances, storage buffer for ATG8 was acidified to pH 5.5 with 1 mM HCl to prevent aggregation.
Assay of cleavage of ATG8s in vitro
Pellets of E. coli co-expressing each ATG8 homologue with either ATG4.1 or ATG4.2 (Table S2(ii)) were lysed in reaction buffer (containing 25 mM Tris-HCl, pH 7.4, 50 mM KCl, 10 mM β-mercaptoethanol). The supernatant protein obtained after centrifugation at 13,000 × g for 30 min at 4°C was adjusted to 5 mg ml−1 and incubated at 30°C for a fixed time (specified by the particular experiment). The action of different peptidase inhibitors on the hydrolytic action of ATG4 on ATG8 was determined by adding the inhibitors prior to disruption of the cell membrane. In the deconjugation assay, Δatg4.2 promastigotes expressing GFP-ATG8-PE were lysed in reaction buffer and the supernatant was incubated with recombinant ATG4.2 for up to 180 min. Reactions were stopped with SDS-PAGE loading buffer (1 mM Tris-HCl pH 6.8, 20% glycerol, 10% β-mercaptoethanol and 40% bromophenol blue). The hydrolytic actions of the recombinant ATG4.1 and ATG4.2 on His-ATG8-HA, His-ATG8A-HA, His-ATG8B-HA and His-ATG8C-HA and recombinant ATG4.2 on GFP-ATG8-PE were determined by Western blot analysis with α-His-tag and α-GFP antibodies (Pierce). Supernatants containing each ATG8 homologue and GFP-ATG8-PE but lacking ATG4 were used as controls.
MALDI-TOF MS analysis
Electronspray ionization (ESI)–MS/MS was carried out on a hybrid quadrupole time-of-flight mass spectrometer, Q.STAR (Applied Biosystems). Solution containing partially-cleaved ATG8 treated with 1% formic acid was loaded into a borosilicate nanoflow (micromass), and sent into the ESI source. MS/MS data were processed by Analyst QS software, which is capable of deconvoluting the peaks of varying charge state, to obtain protein masses.
Activity analyses of ATG4s
Enzymatic activities of purified recombinant ATG4.1 and ATG4.2 towards Fluorescence Resonance Energy Transfer (FRET) peptides (Abz-X-X-X-X-Q-EDDnp) as substrates, synthesized by LJ, were analysed in an assay (1 ml) comprising substrate and 10 ng of recombinant protein in 50 mM Tris/HCl, pH 7.9, 125 mM NaCl, 10 mM dithiothreitol. Peptide hydrolysis was measured using the LS 50-B PerkinElmer spectrofluorometer (λexcitation, 320 nm; λemission, 420 nm) at 30°C for 15 min after initiation with the enzyme. Specific activities were determined using 2.5 mM substrate. Kinetics parameters (Km, Vmax and kcat/Km) were determined using substrate concentrations ranging from 0.5-10 mM and calculated using the Grafit software. For inhibition experiments, the reaction mixture was pre-incubated for 30 min at 20°C with inhibitors and residual activities determined.
Yeast complementation studies
The ORFs of ATG5 (LmjF32.3890), ATG12 (LmjF22.1300) and ATG10 (LmjF19.1630) were amplified by PCR experiments using the Expand High Fidelity PCR system (Roche Molecular Biochemicals) from L. major genomic DNA isolated as described by Medina-Acosta et al 75 using gene-specific primers modified with appropriate restriction sites (to facilitate cloning into their respective expression vector) as detailed in Table S1. All PCR assays were carried out in a GeneAmp 9600 PCR system (PerkinElmer Life Sciences) for 30 cycles of denaturation (94°C, 15 s), annealing (65°C, 15 s) and extension (72°C, 2 min). Each ORF was verified by nucleotide sequencing (MBSU, University of Glasgow) and cloned into the pCM185 expression vectors (EUROSCARF) pre-digested with appropriate restriction enzymes to produce the plasmids detailed in Table S1(iii). Each plasmid was subsequently used to transform their respective mutant strain of S. cerevisiae (EUROSCARF) by the lithium acetate method.76 The resultant transformed lines and wild type lines (Table S2(iv) and (iii)) were grown in YNB medium containing 0.5% ammonium sulphate and amino acids but lacking uracil until the optical density at 600 nm (OD600) was 1.0, whereupon they were split into two aliquots. One aliquot was washed twice with SD(-N) and incubated in the same medium for up to 16 h at 30°C to induce starvation. The second aliquot was washed and resuspended in fresh growth medium and incubated under the same conditions as a control. The cells were then harvested by centrifugation (3,000 × g, 4°C, 5 min), suspended in Laemmli’s sample buffer and the cells walls disrupted with lysis buffer (containing 0.1% lyticase). Yeast proteins were resolved by SDS-PAGE, transferred to a nitrocellulose membrane and analysed by Western blotting with α-(pro)aminopeptidase I primary antibodies (a gift from Prof D. Klionsky) and horseradish peroxidise-conjugated anti-rabbit secondary antibodies. The bands were visualized with the ECLTM detection system (Amersham Biosciences).
Occurrence in L. major transgenic lines of puncta associated with ATG proteins
L. major (MHOM/JL/80/Friedlin) promastigotes were used throughout this study. The Δatg4.2 null mutant (in which both alleles of the gene encoding the cysteine peptidase ATG4.2 had been deleted) was generated as described previously.77 Generation of the GFP-ATG8 fusion construct is also described in Besteiro et al.13 To generate GFP-ATG8A, GFP-ATG8B, GFP-ATG8C and RFP-ATG12, their respective ORFs amplified with primers modified with the BgIII/XhoI restriction sites (Table S1(ii)) were sub-cloned into GFP-containing pNUS-GFPnH and RFP-containing pNUS-RFPnH vectors, respectively,78 yielding the plasmids detailed in Table S1(ii). These constructs were used to transfect L. major wild type and Δatg4.2 lines as previously described.79 All lines generated (Table S2(i)) were grown in complete medium (modified Eagle’s Medium [designated HOMEM] with 10% (v/v) heat-inactivated foetal calf serum (HiFCS)) at 25 °C as described in Williams et al.80 In vitro procyclic promastigote-metacyclic promastigote differentiation was monitored by growing promastigotes in complete medium to stationary phase of growth (1-3 × 107 cells ml−1) whereupon the proportion of metacyclic promastigotes was determined as described.81,82 Autophagy was induced in promastigotes expressing ATG8 homologues by incubation in PBS at 2 × 107 cells ml−1 for up to 5 h as previously described.15 Puncta biogenesis was monitored in these starved promastigotes by fluorescence microscopy using Zeiss Axioplan2 imaging and Axiophot2 microscope equipped with FITC or Rhodamine filter sets, respectively; images were captured at intervals and along the Z-axis and processed using VELOCITY (Improvision). The presence and number of GFP-ATG8s- or RFP-ATG12-containing vesicles within these cells was assessed by observing at least 100 cells from 3 independent experiments. Unless otherwise stated, early log and late log/early stationary phase cells of all cell lines corresponds to 1-5 ×106 and 5-9 ×106 parasites ml−1, respectively.
Parasite viability
Viability of the L. major wild type and Δatg4.2 lines subjected to starvation conditions was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay as described previously.15 Briefly, 4 × 106 stationary phase promastigotes were further incubated for 45 min at 37°C with MTT to a final concentration of 1 mg ml−1, whereupon absorbance at 620 nm was measured using a microtitre plate reader.
Antibodies and immunoblotting
Western blot analysis was performed on lysates from L. major or E. coli lysates separated on standard 12.5% Laemmli separating gels (SDS-PAGE) containing 6 M urea (Urea-SDS-PAGE) at pH 9.2. The primary α-GFP antibody (Abcam Ltd) was used at 1:2000 dilution, whereas α-His antibodies (Pierce), α-(pro)aminopeptidase 1 antibodies (a gift from Prof D. Klionsky) and the secondary anti-rabbit IgG antibody were used at a 1:5000 dilution. The West-Pico chemiluminescence detection system (Pierce) was used to visualize the bands of antigens.
Supplementary Material
Acknowledgements
We thank Richard J. Burchmore of the Sir Henry Wellcome Functional Genomics Facility, Faculty of Biomedical and Life Sciences, University of Glasgow for the mass spectrometry analyses. This study was funded by the Medical Research Council [grant numbers G9722968, G0000508, G0700127]. We thank the European Saccharomyces cerevisiae Archive for Functional analysis (EUROSCARF), Frankfurt, Germany for the yeast mutant lines and yeast expression plasmids and Prof. D. J. Klionsky, University of Michigan, USA, for the α-(pro)aminopeptidase antibodies.
Abbreviations
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- Ape
vacuolar hydrolase aminopeptidase
- WT
wild type
- GFP
green fluorescence protein
- RFP
red fluorescence protein
- ORF
open reading frame
- DTT
Dithiothreitol
- PBS
phosphate-buffered saline
- PE
phosphotidylethanolamine
- MALDI-TOF
Matrix Assisted Laser Desorption Ionization-Time Of Flight
- Abz
2-aminobenzoyl
- EDDnp
ethylenediamine-2,4-dinitrophenyl
- kDa
kilodaltons
- LC3
microtubule associate protein light chain-3
- GATE-16
Golgi-associated ATPase enhancer of 16 kDa
- GABARAP
GABA receptor-associated protein.
References
- 1.Yoshimori T. New insights into molecular mechanisms and roles of autophagy. Cell Struct and Funct. 2004;29(supplement):13. [Google Scholar]
- 2.Boland B, Nixon RA. Neuronal macroautophagy: from development to degeneration. Mol Aspects Med. 2006;27:503–19. doi: 10.1016/j.mam.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 4.Penaloza C, Orlanski S, Ye Y, Entezari-Zaher T, Javdan M, Zakeri Z. Cell death in mammalian development. Current Pharmaceutical Design. 2008;14:184–96. doi: 10.2174/138161208783378789. [DOI] [PubMed] [Google Scholar]
- 5.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 6.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 9.Rajawat YS, Bossis I. Autophagy in aging and in neurodegenerative disorders. Hormones (Athens) 2008;7:46–61. doi: 10.14310/horm.2002.1111037. [DOI] [PubMed] [Google Scholar]
- 10.Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease) Am J Pathol. 2005;167:1713–28. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HK, Iwasaki A. Autophagy and antiviral immunity. Curr Opin Immunol. 2008;20:23–9. doi: 10.1016/j.coi.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alvarez VE, Kosec G, Sant'Anna C, Turk V, Cazzulo JJ, Turk B. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J Biol Chem. 2008;283:3454–64. doi: 10.1074/jbc.M708474200. [DOI] [PubMed] [Google Scholar]
- 13.Besteiro S, Williams RAM, Morrison LS, Coombs GH, Mottram JC. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J Biol Chem. 2006;281:11384–96. doi: 10.1074/jbc.M512307200. [DOI] [PubMed] [Google Scholar]
- 14.Herman M, Perez-Morga D, Schtickzelle N, Michels PA. Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy. 2008;4:294–308. doi: 10.4161/auto.5443. [DOI] [PubMed] [Google Scholar]
- 15.Williams RA, Tetley L, Mottram JC, Coombs GH. Cysteine peptidases CPA and CPB are vital for autophagy and differentiation in Leishmania mexicana. Mol Microbiol. 2006;61:655–74. doi: 10.1111/j.1365-2958.2006.05274.x. [DOI] [PubMed] [Google Scholar]
- 16.Dunn WA., Jr. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–43. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 17.Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 18.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–88. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–80. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 20.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 21.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278:51841–50. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 22.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994;269:11492–7. [PubMed] [Google Scholar]
- 24.Mann SS, Hammarback JA. Gene localization and developmental expression of light chain 3: a common subunit of microtubule-associated protein 1A(MAP1A) and MAP1B. J Neurosci Res. 1996;43:535–44. doi: 10.1002/(SICI)1097-4547(19960301)43:5<535::AID-JNR3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 25.Sagiv Y, Legesse-Miller A, Porat A, Elazar Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 2000;19:1494–504. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- 27.Ketelaar T, Voss C, Dimmock SA, Thumm M, Hussey PJ. Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett. 2004;567:302–6. doi: 10.1016/j.febslet.2004.04.088. [DOI] [PubMed] [Google Scholar]
- 28.Scherz-Shouval R, Sagiv Y, Shorer H, Elazar Z. The COOH terminus of GATE-16, an intra-Golgi transport modulator, is cleaved by the human cysteine protease HsApg4A. J Biol Chem. 2003;278:14053–8. doi: 10.1074/jbc.M212108200. [DOI] [PubMed] [Google Scholar]
- 29.Tanida I, Komatsu M, Ueno T, Kominami E. GATE-16 and GABARAP are authentic modifiers mediated by Apg7 and Apg3. Biochem Biophys Res Commun. 2003;300:637–44. doi: 10.1016/s0006-291x(02)02907-8. [DOI] [PubMed] [Google Scholar]
- 30.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 31.Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004;279:40584–92. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- 32.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem. 2006;281:3017–24. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 33.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–18. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 35.Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem. 2004;279:47704–10. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- 36.Xie Z, Nair U, Klionsky DJ. Atg8 Controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-12-1292. Epub May 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–61. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 38.Abeliovich H, Dunn WA, Jr., Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–34. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ponder EL, Bogyo M. Ubiquitin-like modifiers and their deconjugating enzymes in medically important parasitic protozoa. Eukaryot Cell. 2007;6:1943–52. doi: 10.1128/EC.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–8. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 41.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–83. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimoto K. Recent advances in plant autophagy research: plant atg mutants. Tanpakushitsu Kakusan Koso. 2006;51:1532–6. [PubMed] [Google Scholar]
- 43.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujioka Y, Noda NN, Fujii K, Yoshimoto K, Ohsumi Y, Inagaki F. In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J Biol Chem. 2008;283:1921–8. doi: 10.1074/jbc.M706214200. [DOI] [PubMed] [Google Scholar]
- 45.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–25. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 46.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 47.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 48.Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, Roussel MF, et al. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat Struct Mol Biol. 2004;11:927–35. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herman M, Gillies S, Michels PA, Rigden DL. Autophagy and related processes in trypanosomatids - Insights from genomic and bioinformatic analyses. Autophagy. 2006;2:107–18. doi: 10.4161/auto.2.2.2369. [DOI] [PubMed] [Google Scholar]
- 50.Rigden DJ, Herman M, Gillies S, Michels PA. Implications of a genomic search for autophagy-related genes in trypanosomatids. Biochem Soc Trans. 2005;33:972–4. doi: 10.1042/BST20050972. [DOI] [PubMed] [Google Scholar]
- 51.Mukaiyama H, Baba M, Osumi M, Aoyagi S, Kato N, Ohsumi Y, et al. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol Biol Cell. 2004;15:58–70. doi: 10.1091/mbc.E03-05-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumanomidou T, Mizushima T, Komatsu M, Suzuki A, Tanida I, Sou YS, et al. The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J Mol Biol. 2006;355:612–8. doi: 10.1016/j.jmb.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 53.Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J Biol Chem. 2005;280:40058–65. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- 54.Fass E, Amar N, Elazar Z. Identification of essential residues for the C-terminal cleavage of the mammalian LC3: a lesson from yeast Atg8. Autophagy. 2007;3:48–50. doi: 10.4161/auto.3417. [DOI] [PubMed] [Google Scholar]
- 55.Mazel D, Pochet S, Marliere P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 1994;13:914–23. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang J, Hernandez G, LeMaster DM. Increased peptide deformylase activity for N-formylmethionine processing of proteins overexpressed in Escherichia coli: application to homogeneous rubredoxin production. Protein Expr Purif. 2004;36:100–5. doi: 10.1016/j.pep.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 57.Geoghegan KF, Dixon HB, Rosner PJ, Hoth LR, Lanzetti AJ, Borzilleri KA, et al. Spontaneous alpha-N-6-phosphogluconoylation of a “His tag” in Escherichia coli: the cause of extra mass of 258 or 178 Da in fusion proteins. Anal Biochem. 1999;267:169–84. doi: 10.1006/abio.1998.2990. [DOI] [PubMed] [Google Scholar]
- 58.Chae YK, Im H, Zhao Q, Doelling JH, Vierstra RD, Markley JL. Prevention of aggregation after refolding by balanced stabilization-destabilization: production of the Arabidopsis thaliana protein APG8a (At4g21980) for NMR structure determination. Protein Expr Purif. 2004;34:280–3. doi: 10.1016/j.pep.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 59.Ohsumi Y. Molecular mechanism of autophagy in yeast, Saccharomyces cerevisiae. Philos Trans R Soc Lond B Biol Sci. 1999;354:1577–80. doi: 10.1098/rstb.1999.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–79. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–41. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–92. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 63.Hanada T, Ohsumi Y. Structure-function relationship of Atg12, a ubiquitin-like modifier essential for autophagy. Autophagy. 2005;1:110–8. doi: 10.4161/auto.1.2.1858. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy. 2005;1:119–26. doi: 10.4161/auto.1.2.1859. [DOI] [PubMed] [Google Scholar]
- 65.Umemiya R, Matsuo T, Hatta T, Sakakibara S, Boldbaatar D, Fujisaki K. Cloning and characterization of an autophagy-related gene, ATG12, from the three-host tick Haemaphysalis longicornis. Insect Biochem Mol Biol. 2007;37:975–84. doi: 10.1016/j.ibmb.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 66.Umemiya R, Matsuo T, Hatta T, Sakakibara S, Boldbaatar D, Fujisaki K. Autophagy-related genes from a tick, Haemaphysalis longicornis. Autophagy. 2008;4:79–81. doi: 10.4161/auto.5143. [DOI] [PubMed] [Google Scholar]
- 67.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 68.Muthuvijayan V, Marten MR. In silico reconstruction of nutrient-sensing signal transduction pathways in Aspergillus nidulans. In Silico Biol. 2004;4:605–31. [PubMed] [Google Scholar]
- 69.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 70.Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–93. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanida I, Wakabayashi M, Kanematsu T, Minematsu-Ikeguchi N, Sou YS, Hirata M, et al. Lysosomal turnover of GABARAP-phospholipid conjugate is activated during differentiation of C2C12 cells to myotubes without inactivation of the mTor kinase-signaling pathway. Autophagy. 2006;2:264–71. doi: 10.4161/auto.2871. [DOI] [PubMed] [Google Scholar]
- 72.Wu J, Dang Y, Su W, Liu C, Ma H, Shan Y, et al. Molecular cloning and characterization of rat LC3A and LC3B--two novel markers of autophagosome. Biochem Biophys Res Commun. 2006;339:437–42. doi: 10.1016/j.bbrc.2005.10.211. [DOI] [PubMed] [Google Scholar]
- 73.He H, Dang Y, Dai F, Guo Z, Wu J, She X, et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem. 2003;278:29278–87. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- 74.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Medina-Acosta E, Cross GA. Rapid isolation of DNA from trypanosomatid protozoa using a simple 'mini-prep' procedure. Mol Biochem Parasitol. 1993;59:327–9. doi: 10.1016/0166-6851(93)90231-l. [DOI] [PubMed] [Google Scholar]
- 76.Ausubel F, Moore DD, Seidman JG, Struhe K. Current Protocols in Molecular Biology. John Wiley & Sons Inc.; New York: 1989. [Google Scholar]
- 77.Mottram JC, Souza AE, Hutchison JE, Carter R, Frame MJ, Coombs GH. Evidence from disruption of the lmcpb gene array of Leishmania mexicana that cysteine proteinases are virulence factors. Proc Natl Acad Sci USA. 1996;93:6008–13. doi: 10.1073/pnas.93.12.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tetaud E, Lecuix I, Sheldrake T, Baltz T, Fairlamb AH. A new expression vector for Crithidia fasciculata and Leishmania. Mol Biochem Parasitol. 2002;120:195–204. doi: 10.1016/s0166-6851(02)00002-6. [DOI] [PubMed] [Google Scholar]
- 79.Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol. 2003;128:217–228. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 80.Williams RAM, Kelly SM, Mottram JC, Coombs GH. 3-mercaptopyruvate sulfurtransferase of Leishmania contains an unusual C-terminal extension and is involved in thioredoxin and antioxidant metabolism. J Biol Chem. 2003;278:1480–6. doi: 10.1074/jbc.M209395200. [DOI] [PubMed] [Google Scholar]
- 81.Leon LL, Temporal RM, Soares MJ, Grimaldi JG. Proteinase activities during temperature-induced stage differentiation of species complexes of Leishmania. Acta Trop. 1994;56:289–98. doi: 10.1016/0001-706x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 82.Bates PA, Tetley L. Leishmania mexicana: induction of metacyclogenesis by cultivation of promastigotes at acidic pH. Exp Parasitol. 1993;76:412–23. doi: 10.1006/expr.1993.1050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.