Abstract
Clinical trials of small interfering RNA (siRNA) targeting vascular endothelial growth factor-A (VEGFA) or its receptor VEGFR1 (also called FLT1), in patients with blinding choroidal neovascularization (CNV) from age-related macular degeneration, are premised on gene silencing by means of intracellular RNA interference (RNAi). We show instead that CNV inhibition is a siRNA-class effect: 21-nucleotide or longer siRNAs targeting non-mammalian genes, non-expressed genes, non-genomic sequences, pro- and anti-angiogenic genes, and RNAi-incompetent siRNAs all suppressed CNV in mice comparably to siRNAs targeting Vegfa or Vegfr1 without off-target RNAi or interferon-α/β activation. Non-targeted (against non-mammalian genes) and targeted (against Vegfa or Vegfr1) siRNA suppressed CNV via cell-surface toll-like receptor 3 (TLR3), its adaptor TRIF, and induction of interferon-γ and interleukin-12. Non-targeted siRNA suppressed dermal neovascularization in mice as effectively as Vegfa siRNA. siRNA-induced inhibition of neovascularization required a minimum length of 21 nucleotides, a bridging necessity in a modelled 2:1 TLR3–RNA complex. Choroidal endothelial cells from people expressing the TLR3 coding variant 412FF were refractory to extracellular siRNA-induced cytotoxicity, facilitating individualized pharmacogenetic therapy. Multiple human endothelial cell types expressed surface TLR3, indicating that generic siRNAs might treat angiogenic disorders that affect 8% of the world’s population, and that siRNAs might induce unanticipated vascular or immune effects.
Therapeutic application of long, double-stranded (ds)RNA-mediated RNAi and sequence-specific gene silencing through RNAi by short synthetic RNA duplexes is challenging because mammalian cells do not uptake ‘naked’ siRNA (whether chemically modified or not) without cell-permeating entities1–4. To minimize systemic exposure, initial clinical trials of siRNA were launched using intraocular injection in patients with CNV. CNV, wherein the retina is invaded by choroidal vessels beneath the retinal pigmented epithelium (RPE), is a late stage of age-related macular degeneration that afflicts 30–50 million people globally. The preclinical bases for trials of naked VEGFA siRNA (Bevasiranib) or VEGFR1 siRNA (AGN211745/siRNA-027) were single reports in mice5,6 that such siRNAs suppressed laser-injury-induced CNV, a model predictive of efficacy in humans7,8. These findings were interpreted as anomalous examples of local delivery surmounting the impediment to intracellular entry9–11. Instead, we show in two animal models that suppression of neovascularization is a generic property of siRNAs independent of sequence, target and internalization.
Sequence-independent angiogenesis suppression by siRNA
Numerous synthetic non-targeted 21-nucleotide duplex siRNAs from multiple vendors, when injected into the vitreous humour of wild-type mice, uniformly and dose-dependently suppressed CNV (Fig. 1a, b and Supplementary Fig. 1). siRNAs targeting jellyfish green fluorescent protein (Gfp) or firefly luciferase (Luc), non-ocular genes Bglap1 (bone-specific osteocalcin), Cdh16 (kidney-specific cadherin 16), or Sftpb (lung-specific surfactant protein B), or non-genomic random sequences (RS1–6), all suppressed CNV. This stereotypic effect, reproduced independently in the laboratories of J.A. and E.S., cannot be attributed to ‘off-target’ silencing due to sequence-specific mismatch tolerance12, nor is it an artefact of intraocular delivery because intraperitoneal administration of serum-stable 2′O-methyl-Luc-siRNA suppressed CNV (Supplementary Fig. 2). Furthermore, a siRNA with random sequence and containing proprietary chemical modifications precluding incorporation into RNA-induced silencing complex (RISC) also suppressed CNV (Fig. 1a), whereas a dsDNA analogue of Luc siRNA did not (Supplementary Fig. 3). Lipopolysaccharide did not reduce CNV, excluding endotoxin contamination as the source of angio-inhibition; however, nuclease digestion did abolish Luc-siRNA-induced CNV suppression. Collectively, these data indicate that angio-inhibition is a siRNA-class effect.
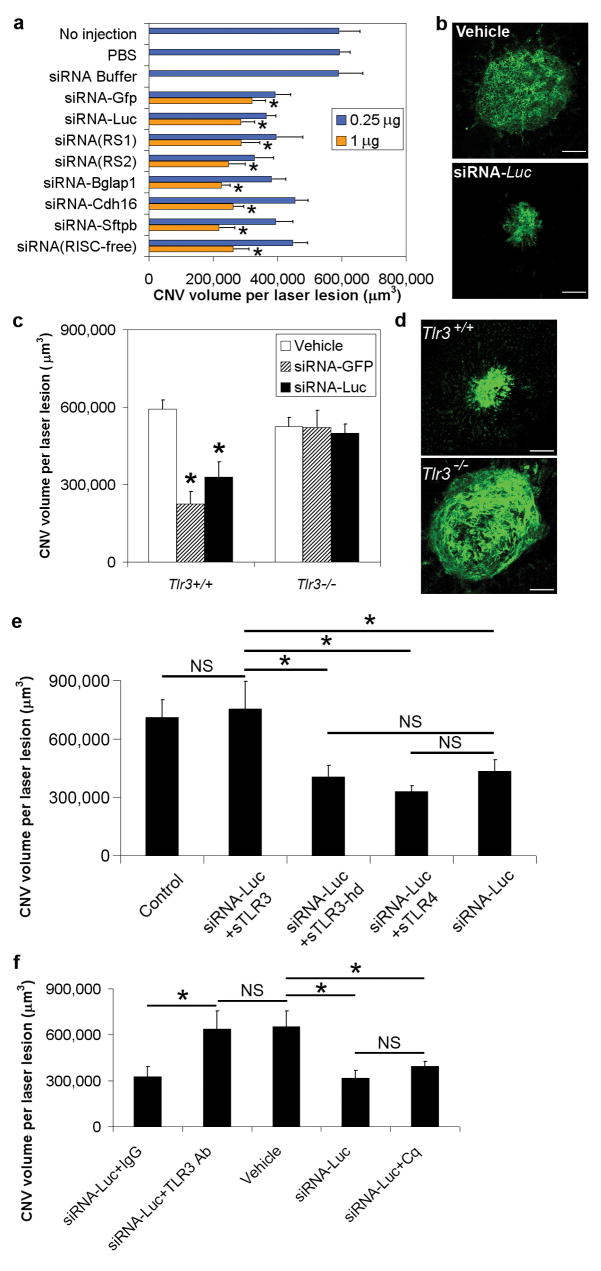
Figure 1. Sequence-independent CNV suppression by siRNA through TLR3.
a, siRNAs targeting Gfp, Luc, random sequences (RS1, RS2), Bglap1, Cdh16, or Sftpb, and siRNA incapable of RNA-induced silencing (RISC-free), suppressed CNV in wild-type mice. n = 8–24; asterisk, P < 0.05 compared to no injection, phosphate buffered saline (PBS) and siRNA buffer. b, Representative examples of CNV in wild-type eye injected with vehicle (buffer) or Luc siRNA (1 μg). c, Gfp siRNA or Luc siRNA suppressed CNV in Tlr3+/+ but not Tlr3−/− mice. n = 16–18; asterisk, P < 0.05 compared to vehicle (buffer). d, Representative examples of CNV in Gfp-siRNA-injected (1 μg) Tlr3+/+ and Tlr3−/− eyes. e, CNV suppression in wild-type mice by Luc siRNA (0.25 μg) was abrogated by soluble TLR3 (sTLR3) but not soluble TLR4 or heat-denatured (hd) soluble TLR3 (all 2 μg). n = 8. f, CNV suppression in wild-type mice by Luc siRNA (1 μg) was abrogated by neutralizing anti-TLR3 antibodies (Ab; 0.2 μl) but not control IgG (0.2 μl) or chloroquine (Cq; 30 ng). n = 6–8; asterisk, P < 0.05. NS, not significant. Vehicle, buffer. All error bars indicate mean ± s.e.m. Scale bars in b, d are 100 μm.
Surface TLR3 mediates anti-angiogenic activity of siRNA
Consistent with reports that naked siRNAs are not internalized by mammalian cells1–3, fluorescein-conjugated Luc siRNA did not enter primary human choroidal endothelial cells (CECs) or mouse RPE and CECs (Supplementary Fig. 4). However, fluorescein–Luc-siRNA–chol, in which cholesterol is conjugated to the 3′ end to enable uptake3, was internalized by human and mouse cells, both of which express the mammalian homologue of Sid1, a protein involved in internalizing cholesterol-conjugated siRNAs13. We therefore sought a surface receptor mediating siRNA-induced angio-inhibition.
We proposed that siRNAs activate TLR3, a long double-stranded viral RNA sensor14, to suppress CNV. Indeed, non-targeted siRNAs did not suppress CNV in Tlr3−/− mice (Fig. 1c, d). The TLR3 activators polyinosinic:polycytidylic acid (poly(I:C))14 and in-vitro-transcribed long dsRNA15 suppressed CNV in wild-type mice, whereas the TLR3 non-activators polydeoxyinosinic:polydeoxycytidylic acid (poly(dI:dC))14 and 2-thiouridine (s2U)-modified in-vitro-transcribed dsRNA15 were ineffective. Poly(I:C) did not suppress CNV in Tlr3−/− mice (Supplementary Fig. 5).
Soluble TLR3, but not soluble TLR4 or heat-denatured soluble TLR3, abolished CNV suppression by Luc siRNA in wild-type mice (Fig. 1e), suggesting direct interaction of siRNA with TLR3. Fluorescein-conjugated Luc siRNA bound wild-type but not Tlr3−/− mouse eye sections in situ (Supplementary Fig. 6). Using flow cytometry to monitor binding of fluorescein–Luc siRNA to the surface of CD31+VEGFR2+ mouse choroidal endothelial cells, greater fluorescence was detected on wild-type than Tlr3−/− cells (Supplementary Fig. 7). Pre-incubation with soluble TLR3 or competition with poly(I:C) reduced fluorescein–Luc-siRNA binding to wild-type mouse CECs. These data extend cell-free system data that 20-nucleotide dsRNAs bind TLR3 (refs 16, 17) and support an in vivo siRNA–TLR3 interaction, although we cannot exclude accessory molecules enabling TLR3 activation. However, the only such facilitator reported so far, CD14 (ref. 18), was dispensable because Luc siRNA suppressed CNV in Cd14−/− mice (Supplementary Fig. 8).
CNV suppression by Luc siRNA was blocked by TLR3-neutralizing antibodies (Fig. 1f), suggesting that non-targeted siRNA signalled via surface TLR3. We confirmed in vivo surface TLR3 expression on mouse and human CECs by flow cytometry and immunofluorescence (Supplementary Fig. 9). To resolve the locus of TLR3 activation by siRNA, we used chloroquine, which inhibits endosomal TLR3 and TLR9. Chloroquine blocked the increase in CNV induced by CpG oligonucleotide, a TLR9 agonist, but did not prevent CNV suppression by Luc siRNA (Fig. 1f and Supplementary Fig. 10). Collectively, these data show that surface, not endosomal, TLR3 mediates extracellular siRNA-induced angio-inhibition.
Non-targeted siRNAs did not suppress CNV in TrifLps2 mice (Supplementary Fig. 11), which are deficient in signalling induced by TRIF (toll/interleukin (IL)-1-receptor-domain-containing adaptor-inducing interferon-β), the TLR3 adaptor protein19,20, confirming TLR3 indispensability. TLR3 signalling can diverge at the level of TRIF through a kinase cascade activating interferon regulatory factor (IRF)-3 or nuclear factor-κB (NF-κB)21. Non-targeted siRNAs suppressed CNV in Irf3−/− but not Nfkb1−/− mice (Supplementary Fig. 11), suggesting a TRIF–NF-κB cascade.
Duplex siRNAs containing immunostimulatory sequences such as UGUGU22 or GUCCUUCAA23 trigger off-target effects via TLR7, a single-stranded RNA sensor. However, none of our siRNAs contained these sequences (Supplementary Table). Also, non-targeted siRNAs suppressed CNV in Tlr7−/− mice (Supplementary Fig. 12). Furthermore, 2′O-methyl-Luc-siRNA suppressed CNV, excluding TLR7 involvement24.
Although it is improbable that cytosolic dsRNA sensors such as protein kinase R (PKR, encoded by Prkra), retinoic-acid-inducible gene 1 (RIG-I), and melanoma differentiation-associated gene 5 (MDA5)25–28 are involved in siRNA-mediated angio-inhibition because Luc siRNA did not enter cells, we excluded their involvement because siRNA activates PKR when transfected into cells29. Luc siRNA suppressed CNV in Prkra−/− just as in wild-type mice, suggesting that PKR has no involvement (Supplementary Fig. 13). Poly(I:C12U), an analogue of poly(I:C) that activates TLR3 but not MDA5 (ref. 30), suppressed CNV in wild-type mice as effectively as poly(I:C) (Supplementary Fig. 5). RIG-I participation is unlikely because in-vitro-transcribed dsRNA, which activates RIG-I, and poly(I:C), which does not25, suppressed CNV equally (Supplementary Fig. 5). Thus, TLR3 seems to be central to siRNA-induced angio-inhibition.
Anti-angiogenic innate immunity triggered by siRNA
siRNAs sometimes induce type I interferon (IFN)22,23,29,31–33. Although laser injury increased levels of IFN-α and IFN-β(IFN-α/β) in the RPE and choroid, they were not further induced by Luc siRNA (Supplementary Fig. 14). Luc siRNA suppressed CNV in Ifnar1−/− (IFN-α/βreceptor null) mice just as in wild-type mice (Fig. 2a). However, type II IFN-γ and IL-12, which are TLR3-inducible, were upregulated by Luc siRNA after laser injury (Fig. 2b). Recombinant IFN-γ and IL-12 suppressed CNV in wild-type mice (Fig. 2c), consistent with their anti-angiogenic properties34. Both were required for angiogenesis suppression because Luc siRNA did not suppress CNV in Ifng−/− or Il12a−/− mice (Fig. 2a).
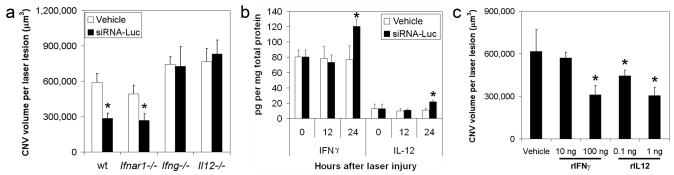
Figure 2. CNV suppression by siRNA is mediated by IFN-γ and IL-12.
a, Luc siRNA (1 μg) suppressed CNV in wild-type and Ifnar1−/− mice but not Ifng−/− or Il12−/− mice. n = 5–10. b, IFN-γ and IL-12 levels in RPE and choroid at 24 h after laser injury were higher in wild-type mouse eyes injected with Luc siRNA (1 μg) compared to vehicle-injected eyes. n = 12. c, Recombinant IFN-γ or IL-12 reduced CNV in wild-type mice. n = 8. Asterisk, P < 0.05 compared to vehicle (siRNA buffer). All error bars indicate mean ± s.e.m.
IFN-γ and IL-12 polarization is congruent with TRIF activation being biased towards NF-κB rather than IRF-3. Non-induction of IFN-α/βby synthetic siRNA is consistent with the absence of 5′-triphosphates33 and the dominant role of MDA5, which is not involved in siRNA-induced angio-inhibition (Supplementary Fig. 5), in mediating dsRNA-induced IFN-α/β(refs 25,28). Alternatively, laser injury might have saturated or inhibited IFN-α/βinduction.
Targeted siRNAs also suppress CNV via TLR3
We synthesized a siRNA targeting the same sequence as AGN211745 in Vegfr1 (Vegfr1 siRNA, complementary to mouse and human sequences). Vegfr1 siRNA suppressed CNV in wild-type mice, confirming an earlier report6, but not in Tlr3−/− mice (Fig. 3a), suggesting that it suppressed angiogenesis by means of the class effect, not RNAi. Supportive of this, Vegfr1 siRNA suppressed CNV as effectively as Luc siRNA in Vegfr1 tyrosine kinase−/− mice (Supplementary Fig. 15), which lack VEGFR1 signalling35. Also, Vegfr1 siRNA did not suppress Vegfr1 mRNA in the RPE and choroid of Tlr3−/− mice (Supplementary Fig. 15). Furthermore, Vegfr1-siRNA–chol did not suppress CNV in Tlr3−/− mice despite Vegfr1 knockdown (Fig. 3b and Supplementary Fig. 15), consonant with observations that VEGFR1 does not promote CNV36.
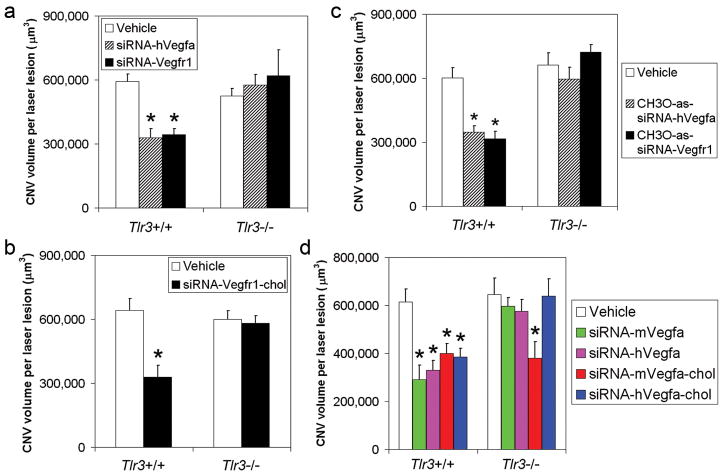
Figure 3. CNV suppression by targeted siRNAs is mediated by TLR3.
a, hVegfa siRNA and Vegfr1 siRNA suppressed CNV in Tlr3+/+ but not Tlr3−/− mice. b, Vegfr1-siRNA–chol suppressed CNV in Tlr3+/+ but not Tlr3−/− mice. n = 5–6. c, hVegfa siRNA and Vegfr1 siRNA, whose anti-sense (as) strands were modified with 5′-methoxy (CH3O) substitution, preventing incorporation into RISC, suppressed CNV in Tlr3+/+ but not Tlr3−/− mice. n = 9–14. d, siRNAs targeting mouse (m) or human (h) Vegfa reduced CNV in Tlr3+/+ but not in Tlr3−/− mice. mVegfa-siRNA–chol and hVegfa-siRNA–chol suppressed CNV in Tlr3+/+ mice; however, mVegfa-siRNA–chol but not hVegfa-siRNA–chol suppressed CNV in Tlr3−/− mice. n = 8–10. siRNAs were at 1 μg; asterisk, P < 0.05 compared to vehicle (siRNA buffer). All error bars indicate mean ± s.e.m.
CH3O-as-Vegfr1 siRNA (containing 5′-methoxy modification of the anti-sense strand, preventing phosphorylation of the 5′-hydroxyl end obligate for RISC incorporation and target cleavage37,38, thereby abolishing RNAi activity) suppressed CNV in wild-type mice as effectively as Vegfr1 siRNA and was ineffective in Tlr3−/− mice (Fig. 3c), confirming that Vegfr1 siRNA suppressed CNV through TLR3, not RNAi.
We also synthesized a siRNA targeting the same sequence as Bevasiranib in human VEGFA (hVegfa siRNA, complementary to human VEGFA but mismatched to mouse Vegfa at position 11, critical for functional targeting39). Both hVegfa siRNA and CH3O-as-hVegfa-siRNA suppressed CNV in wild-type but not Tlr3−/− mice (Fig. 3a, c), demonstrating that hVegfa siRNA also suppresses CNV through TLR3, not RNAi. hVegfa-siRNA–chol did not suppress CNV in Tlr3−/− mice (Fig. 3d), indicating that this siRNA was incapable of functional targeting even after internalization due to critical nucleotide mismatch with mouse Vegfa. However, mVegfa-siRNA–chol (complementary to mouse Vegfa) suppressed CNV in Tlr3−/− mice. As further confirmation of the centrality of this nucleotide, mVegfa-siRNA–chol, but not hVegfa-siRNA–chol, suppressed Vegfa in the RPE and choroid of Tlr3−/− mice (Supplementary Fig. 16). Hence, siRNAs unaided by cell permeation cannot execute RNAi.
Luc siRNA suppressed CNV (51 ± 7%) in wild-type mice as effectively as mVegfa-siRNA–chol in Tlr3−/− mice (41 ± 11%; Fig. 3d) and anti-VEGFA neutralizing antibodies in wild-type mice (49 ± 7%; Supplementary Fig. 17), suggesting that TLR3 activation is as effective as VEGFA blockade, the current standard of care. Luc siRNA did not affect Vegfa, Vegfr1, or Vegfr2 mRNA levels in wild-type mice (Supplementary Fig. 18), so activity seems to be independent of VEGFA pathways. Despite the potential for mVegfa-siRNA–chol to activate TLR3 and knockdown Vegfa in wild-type mice, there was no angio-inhibitory synergy. This could be attributed to blockade of VEGFA-dependent signalling by IFN-γ (induced by siRNA) and potential crosstalk40. Indeed, CNV suppression by Luc siRNA or recombinant IFN-γ was not enhanced by co-administration of anti-VEGFA antibodies, consistent with convergence of anti-angiogenic mechanisms (Supplementary Fig. 18). Alternatively, incorporation of mVegfa-siRNA–chol into RISC might preclude TLR3 interaction.
Neither hVegfa siRNA nor Vegfr1 siRNA suppressed CNV in Ifng−/− or Il12a−/− mice (Supplementary Fig. 19), confirming that these targeted siRNAs suppressed CNV via the class effect, not RNAi. As an additional demonstration that siRNA suppresses CNV without target cleavage, Irf3 siRNA suppressed CNV in Irf3−/− mice as effectively as Luc siRNA (Supplementary Fig. 20), and Tlr3 siRNA suppressed CNV in wild-type mice without Tlr3 knockdown. This class effect was so robust that siRNA targeting Il12, which is anti-angiogenic in this model, also suppressed CNV in wild-type but not Tlr3−/− mice (Fig. 2c and Supplementary Fig. 21). These data demonstrate that non-targeted and targeted siRNA, without active intracellular delivery, suppressed CNV by means of extracellular TLR3 activation.
Minimum length of siRNA required to suppress CNV
The minimum length of dsRNA activating TLR3 in vivo is unclear. We found, unlike 21- and 23-nucleotide Luc siRNA, 7-, 13-, 16-, or 19-nucleotide versions did not suppress CNV (Fig. 4a). This, coupled with data that longer duplexes such as ~1,000-nucleotide dsRNA and poly(I:C) suppressed CNV (Supplementary Fig. 5), suggest that at least 21 nucleotides are required to activate TLR3. To understand the structural basis of the divergence between 19 and 21 nucleotides, we performed docking simulations between dimeric TLR3, which is required for signalling17,41,42, and dsRNA using crystal structures of TLR3 and 19-nucleotide dsRNA, the latter being shortened or extended as necessary.
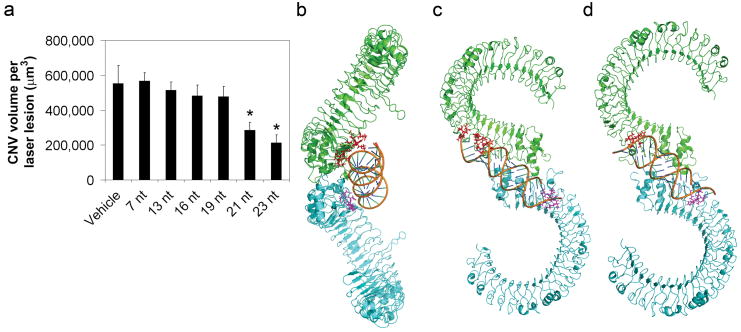
Figure 4. Minimum length for TLR3 activation.
a, 21-nucleotide or 23-nucleotide Luc siRNA but not truncated versions suppressed CNV in wild-type mice. n = 8–11; asterisk, P < 0.05 compared to vehicle (buffer); mean ± s.e.m. Equimolar amounts to 1 μg of 21-nucleotide Luc siRNA. nt, nucleotide. b, c, Orthogonal views of a model of TLR3 ectodomain dimer (green and cyan subunits shown as backbone ribbons) with computer-docked 21-nucleotide dsRNA. Protein modules interact via C-terminal domains to form a highly symmetrical dimer. The model incorporated potential interactions involving a larger set of TLR3 residues considered important in RNA binding (displayed as purple and red side chains on ectodomain subunits); these residues were within 4.5 Å of RNA. d, TLR3 dimer with docked 19-nucleotide dsRNA shows proximity to fewer putative RNA-binding residues. Binding of 19-nucleotide dsRNA was less favourable than 21-nucleotide dsRNA (ΔE = &minus308 kcal mol−1; 95% confidence interval: 261–355; n = 5; P = 0.008).
Docking of dsRNA of various lengths on a TLR3 ectodomain (ECD) monomer did not clarify the observed minimal length requirement for TLR3 activation because the ECD solenoid is approximately the same width as a single turn of an RNA helix. Therefore, a TLR3 monomer cannot discriminate between dsRNAs greater than about 10 nucleotides in length (data not shown), indicating that TLR3 dimerization is required for the observed length needed for activation. We developed a model of a 2:1 TLR3–dsRNA complex, constrained by these observations or interpretations of known data: (1) active TLR3 involves a specific ECD carboxy terminus dimer, formation of which is stabilized by dsRNA binding17,41,42; (2) TLR3 residues most critical for RNA binding and receptor activation, H539 and N541 (ref. 41), lie in a region 40–45 Å from the ECD C terminus; (3) dsRNA of 21-nucleotides or longer is required for receptor activation (Fig. 4a), suggesting that a dsRNA of approximately two full helical turns (~50 Å in length) is required to span clusters of RNA-binding residues on two TLR3 monomers.
We proposed that C terminus dimerization of TLR3 ECDs creates a scaffold for RNA binding in which clusters of functional RNA-binding residues are separated by approximately 50 Å. This is possible with significant overlap of the C terminus domains, as would be required for effective dimerization.
Using these constraints, computer docking identified a highly symmetrical C terminus dimer of TLR3 ECD with clusters of RNA-binding residues on the same side of the complex and spaced appropriately (N541 to N541 distance of 50 Å). This dimer was used to dock either 21-nucleotide or 19-nucleotide dsRNA, under the constraint that docked complexes place H539 and N541 on both TLR3 monomers within 5 Å of RNA and be as symmetrical as possible43. The resulting 2:1 complexes (Fig. 4b–d) were subjected to short molecular dynamics simulations, from which five time points were sampled and energy minimized, indicating that the 21-nucleotide complex was approximately 300 kcal mol−1 energetically more favourable than the 19-nucleotide complex. This is consistent with experimental results showing a loss in anti-angiogenic activity for dsRNA with fewer than 21-nucleotides (Fig. 4a). Furthermore, 19-nucleotide dsRNA was unable to make contact with as many of the putative RNA-binding residues as 21-nucleotide dsRNA (Fig. 4c, d and Supplementary Figs 22 and 23). The model also allows for binding of longer dsRNAs such as poly(I:C).
We considered possible interactions between dsRNA and an extended set of putative RNA-binding residues including, in addition to H539 and N541, E442, N466, K467, R489, N515, N516, N517, N540 and N572 (refs 17, 44). Compared to 19-nucleotide dsRNA, 21-nucleotide dsRNA made contact (within 4.5 Å) with 80% more side chains. Thus, this model accords with the possibility that these TLR3 residues may contribute in varying degrees to RNA binding, if not actual receptor activation.
This model predicts that although siRNAs shorter than 21 nucleotides cannot bridge functional residues on TLR3 dimers, they can bind just as well as longer siRNAs to TLR3 monomers (Fig. 4d and Supplementary Fig. 23). Indeed, excess 19-nucleotide Luc siRNA prevented CNV suppression by 21-nucleotide Luc siRNA (Supplementary Fig. 24) in a dose-dependent manner, suggesting that the shorter siRNA can be an inactive competitor by saturating binding sites on TLR3 monomers and interfere with the ability of longer siRNAs to bridge two monomers to form the active dimeric signalling complex. The specificity of this competition was demonstrated by the inability of a dsDNA analogue of 19-nucleotide Luc siRNA to interfere with CNV suppression by 21-nucleotide Luc siRNA.
Clinical implications
Of the known non-synonymous single-nucleotide polymorphisms (SNPs) in TLR3, two (N284I, rs5743316 and L412F, rs3775291) are hypomorphic (associated with a reduction in cell surface expression and functional activation of receptor)45. Variance for rs3775291 is prevalent in many populations (minor allele frequency ranges from 0.26 to 0.40 in Asian and European samples) whereas variance for rs5743316 is rare (minor allele frequency <0.01). Therefore we sought to determine whether non-targeted siRNA therapy would have differential effects on patients with different homozygous genotypes at rs3775291 (we did not identify any patients with variance at rs5743316). We scanned a library of primary human CECs isolated from single donors for cells homozygous at rs3775291. To determine the fate of these cells when exposed to extracellular non-targeted siRNA, we assayed their survival in serum-supplemented culture in the presence of serum-stable 2′O-methyl-Luc-siRNA without transfection reagents. Non-targeted siRNA induced a 38% reduction in survival of 412LL human CECs (consistent with known cytotoxic effects of TLR3 activation), but a 3% increase in that of 412FF human CECs (Supplementary Fig. 25), suggesting that the TLR3 412 genotype is an important determinant of the cell survival response to siRNA and consistent with TLR3-mediated angio-inhibition. However, further confirmation is required to assign differential survival solely to this SNP and to support a role for pharmacogenetic testing of patients before initiating siRNA therapeutics. Apart from CECs, pericytes and leukocytes have critical roles in angiogenesis; defining their contributions awaits the development of tissue-specific TLR3 knockouts.
To determine the potential for siRNA-induced angio-inhibition in other organs, we interrogated human endothelial cells from aorta, dermis, lung and umbilical vein for TLR3 expression. Surface TLR3 was detected on all of these cells (Supplementary Fig. 26). Serum-stable 21-nucleotide 2′O-methyl-Luc-siRNA, but not a 7-nucleotide version, suppressed dermal angiogenesis induced by excisional wounding in wild-type mice as effectively as 21-nucleotide 2′O-methyl-mVegfa-siRNA (Supplementary Fig. 27), recapitulating the length-dependent anti-angiogenic class effect of siRNA in CNV. The broad angio-inhibitory effect of siRNA and widespread surface expression of TLR3 on endothelial cells suggest that people with the 412LL genotype might be suitable candidates for exploring the anti-angiogenic utility of non-targeted siRNA, whereas targeted cell-permeating siRNA therapies would be better suited for 412FF individuals. Analysing SNPs of genes downstream from TLR3 might yield refined genomic profiles predictive of response to targeted and non-targeted siRNAs.
Discussion
Our results demonstrate that 21-nucleotide or longer dsRNAs are anti-angiogenic. We show that two investigational siRNAs in clinical trials owe their anti-angiogenic effect in mice not to target knockdown but to TLR3 activation. Our findings support investigations of non-targeted dsRNAs as generic anti-CNV agents as effective as anti-VEGFA antibodies, the current standard of care8, while avoiding potential neurotoxicity resulting from chronic administration of the latter46.
Our docking model, although not a high-resolution structure, confirms a potential interaction between 21-nucleotide siRNA and TLR3, and structurally explains TLR3’s remarkable length-based discrimination of dsRNAs. Although duplexes shorter than 21 nucleotides did not suppress CNV, our model suggests that they could interact with TLR3 with a free energy of binding below the threshold for dimer stabilization and receptor activation. Such discrimination between interaction and activation, if it exists, might also be attributed to unknown accessory proteins that chaperone siRNA to TLR3. An outstanding question is whether differential binding to monomeric versus active signalling dimeric TLR3 underlies the divergence between 21-nucleotide and shorter siRNAs.
Tlr3+/+ and Tlr3−/− mice exhibited similar CNV responses in the absence of exogenous dsRNAs, suggesting that TLR3 was dispensable to basal angiogenesis and that endogenous dsRNAs (for example, microRNAs) do not activate TLR3 or influence angiogenesis in this model. Elucidating the discriminants among potential TLR3 ligands will advance understanding of self/non-self immune recognition.
It is tempting to speculate that TLR3 activation and anti-angiogenic dampening by viral transcripts abundant in the human retina–choroid interface (G. S. Hageman, personal communication) might explain why few patients with early age-related macular degeneration undergo the angiogenic switch despite the onslaught of inflammatory stimuli that promote CNV47. Similarly, investigations of SNPs in TLR3 and downstream genes might prove fruitful in revealing new risk-conferring and protective alleles for CNV and atrophic age-related macular degeneration, and predict efficacy of angiogenesis inhibition and susceptibility to adverse vascular effects.
Further studies will determine whether siRNAs, apart from inducing (un)desirable effects on the vasculature, also influence TLR3-mediated antimicrobial immune responses and non-canonical adverse effects of TLR3 activation on pregnancy48, immune privilege49 and neuronal growth50, especially when delivered systemically. Judicious approaches to enhance therapeutic specificity include modifications abrogating TLR3 activation and reducing lengths below 21 nucleotides. Conversely, genomic fingerprinting of patients could hasten angio-inhibitory applications of generic siRNA while avoiding hazards of specific gene targeting.
METHODS SUMMARY
Laser photocoagulation was performed to induce CNV in mice as previously described36,47. CNV volumes were measured by scanning laser confocal microscopy using fluorescein-isothiocyanate-conjugated Griffonia simplicifolia isolectin B4 (Vector Laboratories), and compared by hierarchical logistic regression using repeated measures analysis. Chemical siRNA sequences for treatments were purchased from Dharmacon or Ambion, Inc. Sense and anti-sense strands of siRNA were annealed per the manufacturer’s instructions, formulated in either sterile siRNA buffer (Dharmacon) or nuclease-free PBS, and injected into the vitreous cavity in a total volume of 1 μl with a 33-gauge Exmire microsyringe (Ito Corporation). The docked complex structures were obtained through rigid-body docking using a modified version of the program FTDock with the X-ray structures of TLR3 ligand-binding domain (Protein Data Bank (PDB) entry 2A0Z) and a 19-nucleotide RNA duplex (PDB entry 1QC0) as the initial structures, the latter being shortened or extended as necessary to generate various lengths of dsRNA with 2-nucleotide overhangs at each end. Final docked 2:1 TLR3–dsRNA complexes were subjected to a short minimization and an implicit solvent molecular dynamics simulation to refine the structures, which were then used for energy calculations. Application of biological information as described above greatly improved the docking results by constraining the sampling space and generating models consistent with the experimental binding data.
METHODS
Animals
C57BL/6J, Ifng−/−, Il12a−/−, Nfkb1−/−, Tlr3−/−, Tlr3+/+ and TrifLps2 mice were purchased from The Jackson Laboratory. Vegfr1 tyrosine kinase−/−, Ifnar1−/− (gift from H. W. Virgin), Irf3−/− (gift from T. Taniguchi via M. David), Prkra−/− (gift from R. H. Silverman via G. Luo) and Tlr7−/−(obtained from S. Akira via D. A. Golenbock) mice have been previously described35,51–53.
CNV
Laser photocoagulation (OcuLight GL, IRIDEX) was performed on both eyes of each 6–8-week-old mice to induce CNV as previously described36,47. CNV volumes were measured by scanning laser confocal microscopy (TCS SP, Leica) as reported36,47 with 0.5% FITC-conjugated Isolectin B4 (Vector). Results are expressed as mean ± s.e.m. with P < 0.05 considered statistically significant.
Drug treatments
siRNAs formulated in siRNA buffer (Dharmacon) or PBS (Sigma-Aldrich), chloroquine (Invivogen), poly(I:C) (Invivogen), poly(dI:dC) (Invivogen), poly(I:C12U) (Ampligen; Bioclones), and recombinant soluble mouse TLR3 (R&D Systems), mouse soluble TLR4 (R&D Systems), neutralizing rat antibodies against mouse TLR3 (eBioscience; ref. 48), immunostimulatory CpG oligodeoxynucleotide (ODN), GpC ODN (Invivogen), recombinant IFN-γ (Peprotech), recombinant IL-12 (eBioscience), neutralizing rat antibodies against mouse IL-12 (R&D Systems) and isotype control IgGs (R&D Systems or eBioscience as appropriate) were dissolved in PBS and injected into the vitreous cavity in a total volume of 1 μl. In some experiments, siRNA or vehicle was injected intraperitoneally.
siRNA
Chemical siRNA sequences and modifications are listed in the Supplementary Table. All siRNAs except RS3–RS6 (Ambion) were purchased from Dharmacon.
Protein expression
ELISAs were used to quantify IFN-α/β/γ (PBL) with normalization to total protein (Bio-Rad).
Gene expression
Real-time reverse transcriptase PCR (RT–PCR) was performed (Realplex, Eppendorf) on total mouse RPE and choroid RNA (RNAqueous, Ambion). Primers and Taqman probes for Vegfa were described in ref. 54. SYBR green was used for all other assays. Primers for TLR3 were purchased (Superarray), and were custom designed for Vegfr1 and Vegfr2 (Supplementary Methods).
Flow cytometry
For surface TLR3 staining, human CECs, isolated as previously reported55, HAECs (human aortic endothelial cells; gift from E. Reed), HPAECs (human pulmonary artery endothelial cells; Clonetics), HDMECs (human dermal microvascular endothelial cells; Clonetics) and HUVECs (human umbilical vein endothelial cells; Clonetics) cultivated in MCDB-131 media (Gibco) with EGM-2MV (omitting gentamicin, hydrocortisone, Clonetics) were blocked and incubated with PE-conjugated anti-human TLR3 (20 μg ml−1, Imgenex) and FITC-conjugated CD31 (BD Biosciences). For intracellular TLR3 staining, FITC–CD31-labelled cells were fixed and permeabilized with Leucoperm (Serotec) and stained for TLR3 in the presence of 10% mouse serum. PE-conjugated mouse IgGκ1 isotype served as control (BD Biosciences). Samples were analysed on a FACSCalibur (Becton Dickinson) with a minimum of 10,000 events using Kolmogorov–Smirnov statistics.
Suspensions of C57BL/6J RPE and choroid cells (106) were isolated 1 day after laser injury and incubated with APC anti-mouse CD31 antibody (20 μg ml−1, BD Biosciences) and PE anti-mouse VEGFR2 antibody (20 μg ml−1; BD Biosciences). For surface TLR3 staining, cells were incubated with anti-mouse TLR3 (10 μg ml−1, R&D Systems) that was pre-conjugated to Alexa Fluor 488 (Invitrogen). For intracellular TLR3 staining, CD31/VEGFR2-labelled cells were subjected to fixation and permeabilization followed by incubation with anti-mouse TLR3 (5 μg ml−1). FITC-conjugated rat IgG2a isotype (BD Biosciences) served as a control.
Fluorescein–siRNA interaction assays
CD31/VEGFR2-labelled RPE and choroid cells (106) isolated from C57BL/6J or Tlr3−/− mice 1 day after laser injury were incubated with fluorescein–Luc-siRNA (100 μg ml−1) for 30 min on ice. To determine the specificity Luc siRNA binding to TLR3, fluorescein–Luc-siRNA was pre-incubated with either soluble TLR3, TLR4, poly(I:C), or poly(dI:dC).
Fluorescein–siRNA uptake in mouse RPE and choroid cells
C57BL/6J RPE and choroid cells (106) were harvested immediately after laser injury, incubated with 21-nucleotide fluorescein–Luc-siRNA, 21-nucleotide fluorescein–Luc-siRNA–chol (both 100 μg ml−1) or PBS vehicle in phenol-free M199 media (Gibco) plus 10% FBS for 1 h at 37 °C and treated with 150 μl of 0.05% trypsin (Gibco) for 3 min at 37 °C to remove extracellular siRNA.
Immunofluorescent microscopy
Human CECs (105) were incubated with 21-nucleotide fluorescein–Luc-siRNA or 21-nucleotide fluorescein–Luc-siRNA–chol (both 100 μg ml−1) for 1 h at 37 °C.
Fluorescein–siRNA binding in mouse RPE and choroid tissue
Frozen sections of C57BL/6J or Tlr3−/− mice prepared 3 days after laser injury and intravitreous injection of Luc siRNA (1 μg) were blocked with 3% BSA and incubated with 21-nucleotide fluorescein–Luc-siRNA (10 μg ml−1) for 1 h at 37 °C. Slides were fixed with 4% PFA, blocked in 10% normal rabbit serum, incubated with rabbit anti-FITC antibody (1:100, Zymed) overnight at 4 °C followed by anti-rabbit Alexa Fluor 488 (1:200, Invitrogen).
TLR3 localization
C57BL/6J mouse eye sections (10 μm) were fixed in ice-cold acetone for 10 min at 4 °C, blocked, and incubated with rabbit anti-mouse TLR3 (1:50, Imgenex) overnight at 4 °C followed by Alexa Fluor 594 goat anti-rabbit IgG (1:200, Invitrogen) For co-labelling of CECs, sections were incubated with rat anti-mouse CD31 (1:50, BD Biosciences) followed by goat anti-rat IgG conjugated to Alexa Fluor 488 (1:200). Human CECs, isolated from donated eyes using anti-CD31 antibody-coated magnetic beads (BD Biosciences and Invitrogen, respectively) as previously described55, were grown in chamber slides (Lab-Tek). For surface staining, cells were blocked with PBS-1% BSA and incubated with mouse anti-human TLR3 (20 μg ml−1, eBioscience). For surface membrane co-localization, cells were stained with rabbit anti-CD46 (1:50, Santa Cruz). Goat anti-mouse Alexa Fluor 488 and donkey anti-rabbit Cy3 (both 1:400) were used as secondary antibodies. Mouse IgGκ1 isotype antibody (20 μg ml−1, eBioscience) served as a control.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank collaborators for gifts of knockout mouse strains; R. King, L. Xu and K. Emerson for technical assistance; R. Mohan, S. Bondada, R. A. Brekken, M. W. Fannon, T. S. Khurana, B. J. Raisler, P. A. Pearson, J. E. Springer, J. G. Woodward, A. M. Rao, G. S. Rao and K. Ambati for discussions; and C. Liu and R. J. Kryscio for statistical guidance. J.A. was supported by NEI/NIH, Burroughs Wellcome Fund Clinical Scientist Award in Translational Research, Macula Vision Research Foundation (MVRF), E. Matilda Ziegler Foundation for the Blind, Dr. E. Vernon Smith and Eloise C. Smith Macular Degeneration Endowed Chair, Lew R. Wassermann Merit (LRWM) and Physician Scientist Awards (Research to Prevent Blindness (RPB)), American Health Assistance Foundation, University of Kentucky University Research Professorship, and departmental challenge grant from RPB; A.T. by Japan Society for the Promotion of Science for Young Scientists; R.J.C.A by RPB Medical Student Fellowship; K.Z. by NEI/NIH, RPB LRWM award, MVRF, and VA Merit Award; B.A. by Clayton Foundation for Research; J.R.S by NEI/NIH and RPB Career Development Award; B.K.A. by NEI/NIH, VA Merit Award, and Department of Defense; and E.W.T. by NC Biotechnology Center.
Footnotes
Author Contributions M.E.K., K.Y., A.T., M.N., J.Z.B., R.J.C.A., S.I., M.I. and E.S. performed CNV experiments. T.A.W. and L.A.D. performed dermal experiments. V.C. and E.W.T. performed modelling simulations. Y.P, B.A. and J.R.S. performed cytotoxicity experiments. D.G., Z.Y. and K.Z. performed genotyping. K.K. provided reagents. J.A. conceived and directed the project, and, with assistance from E.W.T., B.K.A., K.K., B.A. and J.R.S., wrote the paper. All authors had the opportunity to discuss the results and comment on the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Chiu YL, Ali A, Chu CY, Cao H, Rana TM. Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chem Biol. 2004;11:1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Saleh MC, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nature Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 4.Song E, et al. Antibody mediated in vivo delivery of small interfering RNAs via cellsurface receptors. Nature Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 5.Reich SJ, et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis. 2003;9:210–216. [PubMed] [Google Scholar]
- 6.Shen J, et al. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 2006;13:225–234. doi: 10.1038/sj.gt.3302641. [DOI] [PubMed] [Google Scholar]
- 7.Krzystolik MG, et al. Prevention of experimental choroidal neovascularization with intravitreal anti-vascular endothelial growth factor antibody fragment. Arch Ophthalmol. 2002;120:338–346. doi: 10.1001/archopht.120.3.338. [DOI] [PubMed] [Google Scholar]
- 8.Rosenfeld PJ, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1419–1431. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 9.Corey DR. RNA learns from antisense. Nature Chem Biol. 2007;3:8–11. doi: 10.1038/nchembio0107-8. [DOI] [PubMed] [Google Scholar]
- 10.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nature Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nature Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 12.Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 13.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nature Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 14.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of doublestranded RNA and activation of NF-kB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 15.Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Bell JK, et al. The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc Natl Acad Sci USA. 2005;102:10976–10980. doi: 10.1073/pnas.0505077102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranjith-Kumar CT, et al. Biochemical and functional analyses of the human Tolllike receptor 3 ectodomain. J Biol Chem. 2007;282:7668–7678. doi: 10.1074/jbc.M610946200. [DOI] [PubMed] [Google Scholar]
- 18.Lee HK, Dunzendorfer S, Soldau K, Tobias PS. Double-stranded RNAmediated TLR3 activation is enhanced by CD14. Immunity. 2006;24:153–163. doi: 10.1016/j.immuni.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 20.Hoebe K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Z, Mak TW, Sen G, Li X. Toll-like receptor 3-mediated activation of NF-kB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-b. Proc Natl Acad Sci USA. 2004;101:3533–3538. doi: 10.1073/pnas.0308496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Judge AD, et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 23.Hornung V, et al. Sequence-specific potent induction of IFN-a by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 24.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 26.Meurs E, et al. Molecular cloning and characterization of the human doublestranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 27.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in doublestranded RNA-induced innate antiviral responses. Nature Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 28.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nature Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 30.Gowen BB, et al. TLR3 is essential for the induction of protective immunity against Punta Toro Virus infection by the double-stranded RNA (dsRNA), poly(I:C12U), but not Poly(I:C): differential recognition of synthetic dsRNA molecules. J Immunol. 2007;178:5200–5208. doi: 10.4049/jimmunol.178.8.5200. [DOI] [PubMed] [Google Scholar]
- 31.Heidel JD, Hu S, Liu XF, Triche TJ, Davis ME. Lack of interferon response in animals to naked siRNAs. Nature Biotechnol. 2004;22:1579–1582. doi: 10.1038/nbt1038. [DOI] [PubMed] [Google Scholar]
- 32.Kariko K, Bhuyan P, Capodici J, Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J Immunol. 2004;172:6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- 33.Kim DH, et al. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nature Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 34.Voest EE, et al. Inhibition of angiogenesis in vivo by interleukin 12. J Natl Cancer Inst. 1995;87:581–586. doi: 10.1093/jnci/87.8.581. [DOI] [PubMed] [Google Scholar]
- 35.Hiratsuka S, Minowa O, Kuno J, Noda T, Shibuya M. Flt-1 lacking the tyrosine kinase domain is sufficient for normal development and angiogenesis in mice. Proc Natl Acad Sci USA. 1998;95:9349–9354. doi: 10.1073/pnas.95.16.9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nozaki M, et al. Loss of SPARC-mediated VEGFR-1 suppression after injury reveals a novel antiangiogenic activity of VEGF-A. J Clin Invest. 2006;116:422–429. doi: 10.1172/JCI26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 38.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Singlestranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 39.Birmingham A, et al. 39 UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nature Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 40.Battle TE, Lynch RA, Frank DA. Signal transducer and activator of transcription 1 activation in endothelial cells is a negative regulator of angiogenesis. Cancer Res. 2006;66:3649–3657. doi: 10.1158/0008-5472.CAN-05-3612. [DOI] [PubMed] [Google Scholar]
- 41.Bell JK, Askins J, Hall PR, Davies DR, Segal DM. The dsRNA binding site of human Toll-like receptor 3. Proc Natl Acad Sci USA. 2006;103:8792–8797. doi: 10.1073/pnas.0603245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 43.Gay NJ, Gangloff M, Weber AN. Toll-like receptors as molecular switches. Nature Rev Immunol. 2006;6:693–698. doi: 10.1038/nri1916. [DOI] [PubMed] [Google Scholar]
- 44.Takada E, et al. C-terminal LRRs of human Toll-like receptor 3 control receptor dimerization and signal transmission. Mol Immunol. 2007;44:3633–3640. doi: 10.1016/j.molimm.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Ranjith-Kumar CT, et al. Effects of single nucleotide polymorphisms on Toll-like Receptor 3 activity and expression in cultured cells. J Biol Chem. 2007;282:17696–17705. doi: 10.1074/jbc.M700209200. [DOI] [PubMed] [Google Scholar]
- 46.Nishijima K, et al. Vascular endothelial growth factor-A is a survival factor for retinal neurons and a critical neuroprotectant during the adaptive response to ischemic injury. Am J Pathol. 2007;171:53–67. doi: 10.2353/ajpath.2007.061237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nozaki M, et al. Drusen complement components C3a and C5a promote choroidal neovascularization. Proc Natl Acad Sci USA. 2006;103:2328–2333. doi: 10.1073/pnas.0408835103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin Y, Liang Z, Chen Y, Zeng Y. TLR3-involved modulation of pregnancy tolerance in double-stranded RNA-stimulated NOD/SCID mice. J Immunol. 2006;176:4147–4154. doi: 10.4049/jimmunol.176.7.4147. [DOI] [PubMed] [Google Scholar]
- 49.Lang KS, et al. Immunoprivileged status of the liver is controlled by Toll-like receptor 3 signaling. J Clin Invest. 2006;116:2456–2463. doi: 10.1172/JCI28349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cameron JS, et al. Toll-like receptor 3 is a potent negative regulator of axonal growth in mammals. J Neurosci. 2007;27:13033–13041. doi: 10.1523/JNEUROSCI.4290-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller U, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 52.Sato M, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-a/b gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 53.Hemmi H, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nature Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, et al. Different effects of glucose starvation on expression and stability of VEGF mRNA isoforms in murine ovarian cancer cells. Biochem Biophys Res Commun. 2002;292:860–868. doi: 10.1006/bbrc.2002.6710. [DOI] [PubMed] [Google Scholar]
- 55.Smith JR, et al. Unique gene expression profiles of donor-matched human retinal and choroidal vascular endothelial cells. Invest Ophthalmol Vis Sci. 2007;48:2676–2684. doi: 10.1167/iovs.06-0598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.