Abstract
Reactive oxygen species generated during oxidative stress can lead to unfavorable cellular consequences predominantly due to formation of 4-hydroxy-2-nonenal (HNE) during lipid peroxidation. Data-dependent and neutral loss (NL)-driven MS3 acquisition have been reported for the identification of HNE adducts by mass spectrometry-based proteomics. However, the limitation associated with this method is the ambiguity in correct assignment of HNE modification site when more than one candidate sites are present as MS3 is triggered on the neutral loss ion. We introduce NL-triggered electron capture dissociation tandem mass spectrometry (NL-ECD-MS/MS) for the characterization of HNE-modification sites in peptides. With this method performed using a hybrid linear ion trap-Fourier transform ion cyclotron resonance (FTICR) mass spectrometer, ECD in the FTICR unit of the instrument is initiated on precursor ions of peptides showing the neutral loss of 156 Da corresponding to an HNE molecule in the pre-scan acquired via collision-induced dissociation (CID) tandem mass spectrometry in the linear ion trap. Besides manifold advantages associated with ECD method of backbone fragmentation including extensive sequence fragments, ECD tends to retain the HNE group during MS/MS of precursor ion facilitating the correct localization of modification site. The results also suggest that predisposition of a peptide molecular ion to lose HNE during collision-induced dissociation-based fragmentation is independent of its charge state (2+ or 3+). In addition, we have demonstrated that coupling of solid-phase enrichment of HNE-modified peptides facilitates the detection of this posttranslational modification by NL-driven strategies for low abundance proteins that are susceptible to substoichiometric carbonylation during oxidative stress.
Oxidative stress typically results in peroxidation of polyunsaturated fatty acids present in lipids of the cell membrane bilayer and, as a consequence, various reactive aldehydes are generated. 4-Hydroxy-2-nonenal (HNE) is a highly reactiveα,β-unsaturated electrophilic aldehyde that has been shown to form Michael adducts with cysteine (Cys, C), histidine (His, H) and lysine (Lys, K) residues1 and have been reported to inactivate several enzymes.2 HNE–protein Michael adducts are one of the most useful biomarkers for the occurrence and/or extent of oxidative stress.3 HNE also forms Schiff- base with the ε-NH2 group of Lys.4 Identification of the sites of amino acid modification by this reactive end-product of lipid peroxidation will help to understand its impact on protein function and/or activity and, also, its effect on downstream targets or other interacting proteins.
Mass spectrometry-based approaches to characterize posttranslational HNE modification to proteins and peptides are promising as the peptide sequence and hence the protein identity and the position of HNE can be directly determined through tandem mass spectrometry (MS/MS). Collision-induced dissociation (CID) is the most widely used peptide fragmentation technique implemented in MS/MS. However, HNE-containing peptides may result in neutral loss of HNE (156 Da; 78 or 52 Da for doubly or triply charged peptides) from the precursor and/or product ions upon CID.5 Neutral loss-driven MS3 (NL-MS3) method for the characterization of HNE modified peptides has been reported recently.6,7 In this strategy, the neutral loss of HNE observed upon MS/MS of HNE-modified peptides triggers MS3 analysis of the neutral-loss product ion to reveal the sequence of the peptide. However, MS3 of the neutral loss ion provides no diagnostic mass tag that would allow for unambiguous HNE modification site identification, if more than one possible amino acid residues that could potentially react with HNE are present. Though neutral loss indicates the presence of modification by its characteristic signature tag, a significant challenge still persists in the determination of site(s) of HNE modification or of other post-translational modifications (PTMs) such as phosphorylation, sulfation, glycosylation and oxidation.8 As cleavage of the labile PTMs is favored over peptide backbone dissociation, neutral loss limits further fragmentation that would provide sequence information and, thus, may not afford identification of the modified peptide. Moreover, the difficulty to assign correctly the site of HNE modification is amplified when more than one possible candidate sites are present within the peptide, as the consensus sequence representing the chemical selectivity of HNE modification is lacking. A potential solution to these problems affecting the MS/MS characterization of HNE-modified peptides is to stabilize Michael adducts with sodium borohydride reduction, which prevents the neutral loss upon CID.9 However, this chemical conversion precludes the use of a subsequent and very attractive enrichment strategy applicable only to carbonylated peptides.7
Electron capture dissociation (ECD) is an alternative fragmentation technique and usually implemented with FTICR mass analyzers.10 The partial neutralization of multiply charged peptides following capture of low-energy electrons results in inter-residue backbone cleavage.11 The typical fragmentation pathway of a peptide backbone in ECD consists of cleavage of the amine linkage (N-Cα) rather than at the amide bond characteristic to CID. ECD cleavage results in production of c- and z-type as opposed to that of b- and y-type fragment ions typically observed in CID.12 More recently, electron transfer dissociation (ETD) has been developed to induce fragmentation of the peptide backbone along pathways that are analogous to those observed in ECD. In ETD, a combination of gas-phase ion/ion chemistry and MS/MS is employed in which singly charged radical anions transfer an electron to multiply protonated peptides.13,14
The peptide bond dissociations upon ECD is “ergodic” in nature in which internal energy gained upon electron capture is randomized before dissociation.15–17 Nevertheless, this method of peptide fragmentation offers several advantages over CID. ECD allows labile PTMs (such as HNE) to remain attached during backbone fragmentation facilitating characterization of posttranslational modification, in contrast to CID where the labile modification group may be expelled first.18,19 Hence ECD permits the determination of the exact location of labile modifications with amino acid resolution.12
Mechanisms and fundamental features as well as benefits of ECD have been reviewed in detail.20 Several studies have demonstrated the efficiency of ECD in characterizing different types of posttranslational modifications such as phosphorylation,21,22 acylation,23 glycosylation,24 sulfation25 and other types of modifications in peptides and proteins. We also have shown previously the potential of ECD in retaining and identifying HNE modification sites in oxidized insulin B chain.19 The use of on-line separation of peptides prior to MS has been well known to decrease the complexity of spectra, reduce ion suppression in the spray and result in a preconcentration of the analytes.26 Implementation of electron-injection systems based on indirectly heated dispenser cathodes that provide higher electron fluxes, wider electron beams and better control of the electron energy12 has decreased electron irradiation time to millisecond range to afford ECD, which has enabled coupling of the technique with on-line liquid chromatography/FTICR-MS and made high-throughput data-dependent LC-ECD-MS/MS possible for characterization of proteins or analysis of peptides in tissue extracts.12,26–29 For the analysis of phosphopeptides, Sweet et al. have reported a novel method that exploits the neutral loss feature of CID and performs ECD on the precursor ion exhibiting a neutral loss of 98 Da (corresponding to H3PO4).30 This method, termed neutral loss triggered ECD (NL-ECD-MS/ MS), was applied for the identification of phosphorylation sites in α- and β-casein. While CID provided, along with the sequence of non-phosphorylated peptides, information on the occurrence of 98-Da neutral loss from the precursor, ECD was initiated to correctly determine the number and sites of phosphorylation in singly or multiply phosphorylated peptides.
To characterize HNE modifications, a NL-ECD-MS/MS approach would also be beneficial, since ECD will be triggered only on precursor ions that show HNE loss of 78 or 52 Th in CID spectra depending on the charge state of peptide (2+ or 3+), thereby reducing the duty cycle associated with full data-dependent ECD analysis. To our knowledge, the applicability of NL-ECD-MS/MS for the characterization of HNE-modified peptides has not been reported previously. The current work evaluates, in comparison with CID-based NL-MS3 technique,6 the performance of NL-ECD-MS/MS to characterize HNE modification.
EXPERIMENTAL SECTION
Materials
Human angiotensin (AGT) I peptide (DRVYIHPFHL) and AGT II octapeptide (DRVYIHPF) were purchased from Anaspec (San Jose, CA). Peptides LVLEVAQHLGESTVR and IVYGHLDDPANQEIER (which correspond to tryptic peptides of ATP synthase subunit beta and aconitate hydratase, respectively, found to be targets for HNE modification6) were custom synthesized by Peptide 2.0, Inc. (Chantilly, VA) and used without further purification. 4-Hydroxy-2-nonenal was obtained from Cayman Chemical (Ann Arbor, MI).
4-Hydroxy-2-Nonenal Modification of Peptides
HNE adducts of peptides (1 mg/ml) were prepared by reaction with 2 mM HNE in 0.1 M phosphate buffer, pH 7.4, at 37 °C for 2 h. Excess HNE was removed by extracting the solution three times with ethyl acetate. The resultant aqueous (stock) solutions of HNE-modified peptides were used without further purification in the experiments detailed below.
Solid-Phase Enrichment
To obtain a complex tissue sample, 100 mg of mouse brain was incubated in 500 µl urea (8 M) for 30 min. The supernatant was aliquoted and reduced with 5 mM DTT at 65 °C for 30 min followed by alkylation with 20 mM iodoacetamide at room temperature in the dark for 30 min. The sample was diluted four fold with 50 mM ammonium bicarbonate and subsequently digested with trypsin (substrate/enzyme ratio of 100/1, wt/wt) at 37 °C for 18 hrs. One hundred microliters of the brain protein digest was acidified with acetic acid to pH 3.6 and mixed with 120 µl of reaction buffer (0.2% acetic acid, 10% acetonitrile pH 3.6) spiked with approximately 66, 88 and 20 picomoles of HNE-modified DRVYIHPF, LVLEVAQHLGESTVR and IVYGHLDDPANQEIER from their respective stock solutions. Five microliters of this mixture was used directly for LC-MS analysis to evaluate the mixture before enrichment.
For enrichment of HNE-modified peptides from the mouse brain tryptic digest, 4.2 mg of solid-phase hydrazine (SPH) reagent prepared as described by Roe et al.7 was added and the resulting mixture was rotated end-over-end overnight at room temperature. The SPH reagent was then pelleted and the supernatant was removed. The pellet was washed and the hydrazide-bound peptides were released by incubation with 200 µl of 10% formic acid for 30 min at 60 °C. This step was repeated once and the solutions containing the released peptides were combined. The eluate, containing the HNE-modified standard peptides was dried in an Eppendorf (Westbury, NY) Vacufuge concentrator and resuspended with 20 µl of 0.1% acetic acid. Five-microliter aliquot of the resuspended solution was used for LC-MS analysis by NL-ECD-MS/MS and NL-MS3.
ESI Mass Spectrometry
For analysis without LC separation, the modified peptides were first desalted on an octadecylsilica (C18) solid-phase (Ziptip pipette tips, Millipore, Billerica, MA). The bound peptides were washed with 0.1% acetic acid and, then, recovered by elution with water/methanol/acetic acid (49.5:49.5:1, v/v) and were electrosprayed directly through PicoTip (New Objective, Inc., Woburn MA) emitters.
LC–NL-MS3 and LC–NL- ECD-MS/MS
Neutral loss-driven MS3 and neutral loss-driven ECD tandem mass spectrometry were performed on a hybrid linear ion trap–FTICR (7-Tesla) mass spectrometer (LTQ-FT, Thermo Finnigan, San Jose, CA) equipped with an electrospray ionization source and operated with the Xcalibur (version 2.2) data acquisition software. Online high performance liquid chromatography was performed with an Eksigent nanoLC-2D system using a 15 cm × 75 µm PepMap C18 column (LC Packings) as the analytical column. Mobile phases consisted of solvent A (0.1% acetic acid and 99.9% water (v/v)) and B (0.1% acetic acid and 99.9% acetonitrile (v/v)). Five microliters of the HNE-modified peptide AGT-I and II, or peptide mixture of the HNE-modified LVLEVAQHLGESTVR and IVYGHLDDPANQEIER were automatically loaded onto the column and equilibrated for 5 min in 5% solvent B followed by a 90-min gradient to 40% solvent B at a constant flow rate of 250 nl/min. Analysis was performed using NL-initiated MS3 and NL-initiated ECD data-dependent acquisition mode.
NL-MS3 driven data-dependent acquisition was performed as described previously.6 If a neutral loss of 72 or 52 Th (from a doubly or triply charged HNE-modified peptide) is observed, the neutral loss fragment ion is then selected and subjected to another CID fragmentation (i.e., MS3) in the NL-MS3 method. Briefly, first an accurate m/z survey scan was performed in the FTICR cell followed by parallel MS/MS linear ion trap analysis of the top five most intense precursor ions selected from interim survey spectra (resolving power of <12,500 at m/z 400) obtained by fast Fourier transformation (FFT) about 0.15 s after starting the acquisition of the transients from the receiver plates of the FTICR unit. FTICR full-scan mass spectra were acquired at 100,000 mass resolving power (at m/z 400) from m/z 350 to 1500 using the automatic gain control mode of ion trapping (500000 target ion count). Collision-induced dissociation (CID) in the linear ion trap was performed using a 3.0 Th isolation width and 35% normalized collision energy with helium as the target gas. Isolation and subsequent CID fragmentation of ions exhibiting 78 or 52 Th difference (representing neutral loss of HNE from 2+ or 3+ precursor ions, respectively) from the precursor ion triggered, if the neutral loss fragment ions passed specified selection criteria (they were among the three most intense ions in the MS/MS spectra).
The reported NL-ECD-MS/MS approach for characterization of HNE modification in peptides was adapted from the method of mapping phosphorylation sites in tryptic peptides of β-casein and α-casein.30 In the NL-ECD-MS/MS method, the LTQ-FT performed a full mass scan (m/z 200–2000) during the first scan event followed by CID fragmentation of the most abundant peptides eluted at a particular chromatographic time point from the nano-LC column. Ultimately, if a neutral loss of 78 or 52 Th from a doubly or triply charged precursor ion was observed in the CID mass spectrum, then ECD fragmentation was initiated on the same precursor ion, as opposed to selection of the neutral loss peak in NL-MS3. Xcalibur 2.2 was used for data acquisition. For NL-ECD-MS/MS, FTICR acquisition parameters were adapted from Sweet et al.30 with slight modification. Briefly, full-scan mass spectra were acquired in the ICR cell with a resolving power of 50,000 at m/z 400. Precursor ions (selected from interim survey spectra obtained at resolving power of <12,500 at m/z 400) were isolated with an isolation width of 4 Th and subjected to CID in the linear ion trap with helium as the target gas. The dissociation of the precursor ion was induced using an activation time of 30 ms and at normalized collision energy of 35%. ECD of the precursor ion was triggered if a neutral loss of 156 Da (Δ of 78.0 or 52.0 Th from 2+ or 3+ charged peptide) was observed from one of the three most abundant fragment ions in the previous MS/MS scan. For ECD, precursor ions were isolated in the linear ion trap with an isolation width of 4 Th and transferred to the ICR cell. Ions were irradiated for 120 ms at 5 eV during ECD. The precursor ion that has been selected for ECD fragmentation is dynamically excluded from further MS/MS analysis for 180 s. The methods were evaluated using HNE-modified synthetic peptides.
RESULTS AND DISCUSSION
CID- and ECD-Based Fragmentation of HNE-Modified Peptides
Scheme 1 shows the steps incorporated in the NL-MS3 and NL-ECD-MS/MS experiments.
Scheme 1.
Flowcharts of NL-MS3 and NL-ECD-MS/MS acquisitions.
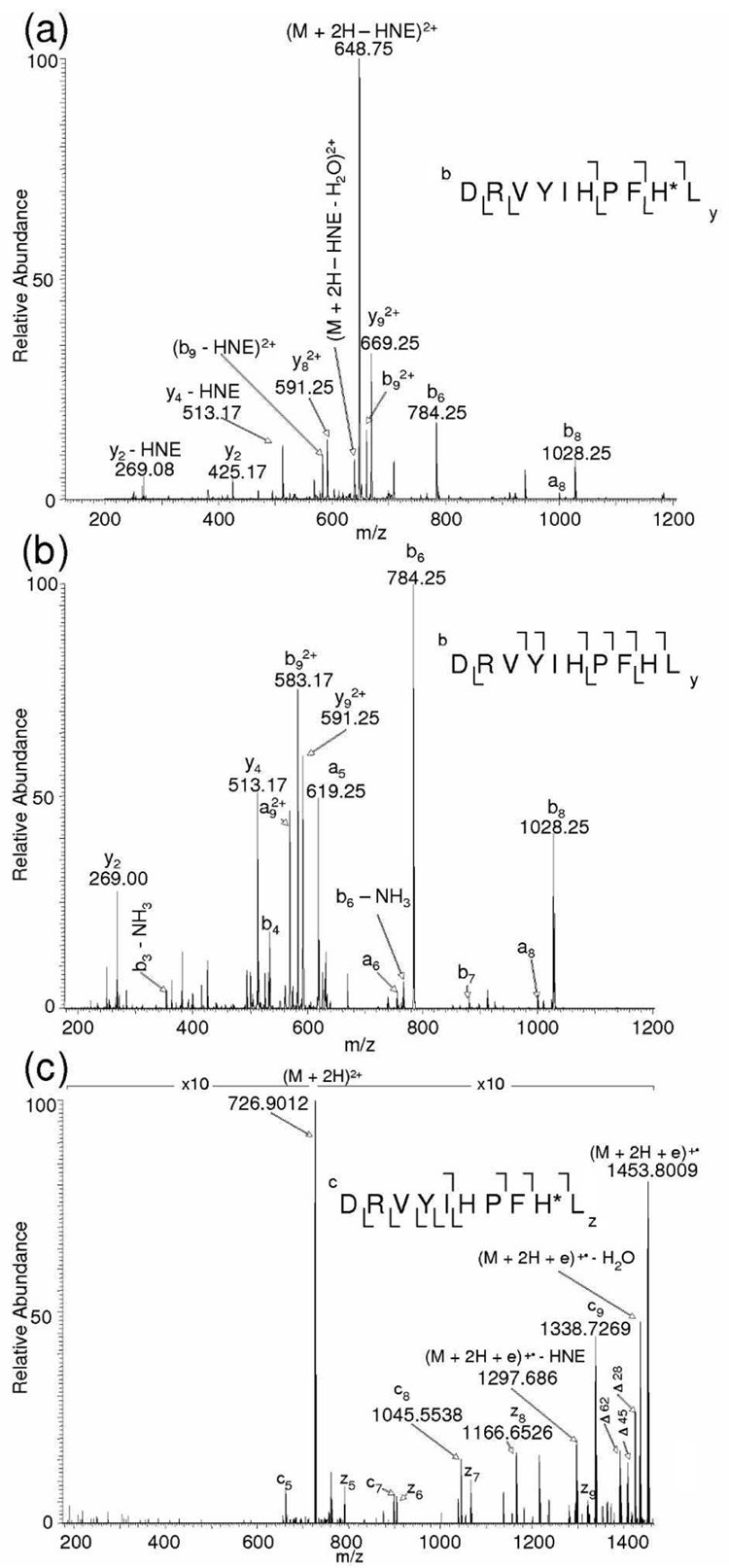
Figure 1a displays the CID MS/MS spectrum of [M + 2H]2+ of the HNE-modified AGT I peptide DRVYIHPFHL, obtained through ESI-MS without LC separation. This peptide has two potential HNE modification sites, His-6 and His-9, to form Michael adducts. The characteristic neutral loss peak observed at m/z 648.75 corresponds to the loss of HNE (156 Da) from the doubly charged precursor ion m/z 726.91. Also observed are the product ions b9 (m/z 583.17, 2+), y2 (m/z 269.08) and y4 (m/z 513.17) that are accompanied by the loss of HNE. The product ions b8 (m/z 1028.25) and b9 (m/z 661.25, 2+) show the presence of HNE at His-9 and the overall peptide fragmentation efficiency was 56%. MS3 of the neutral loss ion m/z 648.75 was performed manually in the linear ion trap (LTQ) that produced the fragment ions (Figure 1b) necessary to identify the peptide sequence. Since a neutral loss of HNE was observed during CID analysis of the modified AGT I peptide, this peptide was used to evaluate the performance of NL-ECD-MS/MS for the analysis of HNE modification. In the NL-ECD-MS-MS method, the observed neutral loss of 78 Th from the precursor ion m/z 726.9012 (m/zcalc 726.9035; 3.2 ppm error in mass accuracy) during the CID-MS/MS scan triggered ECD of the same precursor ion in the subsequent MS/MS scan (Figure 1c). In this ECD spectrum, the backbone fragmentation resulted in 8 out of the total 8 inter-residue bonds (the cleavage of the N-terminal side of proline is not considered due to its resistance to ECD because of the cyclic structure of this residue) being cleaved, and HNE was retained on the peptide as shown by the ECD product ions c8 (m/z 1045.5538) and c9 (m/z 13–38.7269). This result led to the unequivocal mapping of the HNE modification site to His-9 in the AGT-I peptide. A loss of HNE moiety from the charge reduced precursor ion was observed; however, it did not interfere in the correct identification of the site of modification. Generally, the backbone fragments of ECD tend to retain post-translational modifications, such as phosphorylation,21,22 acylation23, glycosylation,24 and sulfation25. Also, there are no fragment ions consistent with the modification of His-6. These results corroborate the fact that neutral loss of HNE (m/z 78 for the doubly charged peptide) in CID-based fragmentation serves as a signature tag that shows the presence of HNE-modified peptides in complex mixtures. The extensive backbone fragmentation by ECD, while retaining the labile HNE group, allows for direct localization of the HNE-modified residues. In addition to c, z product ions, the losses of 28, 45 and 62 Da from charge reduced precursor ions are also observed in the ECD spectrum.31
Figure 1.
(a) ESI-MS/MS spectrum of the HNE-modified AGT I peptide, DRVYIHPFH*L, from its (M + 2H)2+ precursor ion at m/z 726.91, obtained through direct infusion; (b) MS/MS/MS (MS3) spectrum showing another round of CID performed manually on the neutral loss ion, m/z 648.75, observed in the MS/MS spectrum; (c) ECD-FTICR MS/MS spectrum of the (M + 2H)2+ ion m/z 726.9012 of DRVYIHPFH*L. The mass difference between fragment ions c8 (m/z 1045.5538) and c9 (m/z 1338.7269) is 156 Da higher compared to the ECD mass spectrum of the unmodified peptide (not shown). An asterisk after the one-letter code of an amino acid residue denotes its HNE modification.
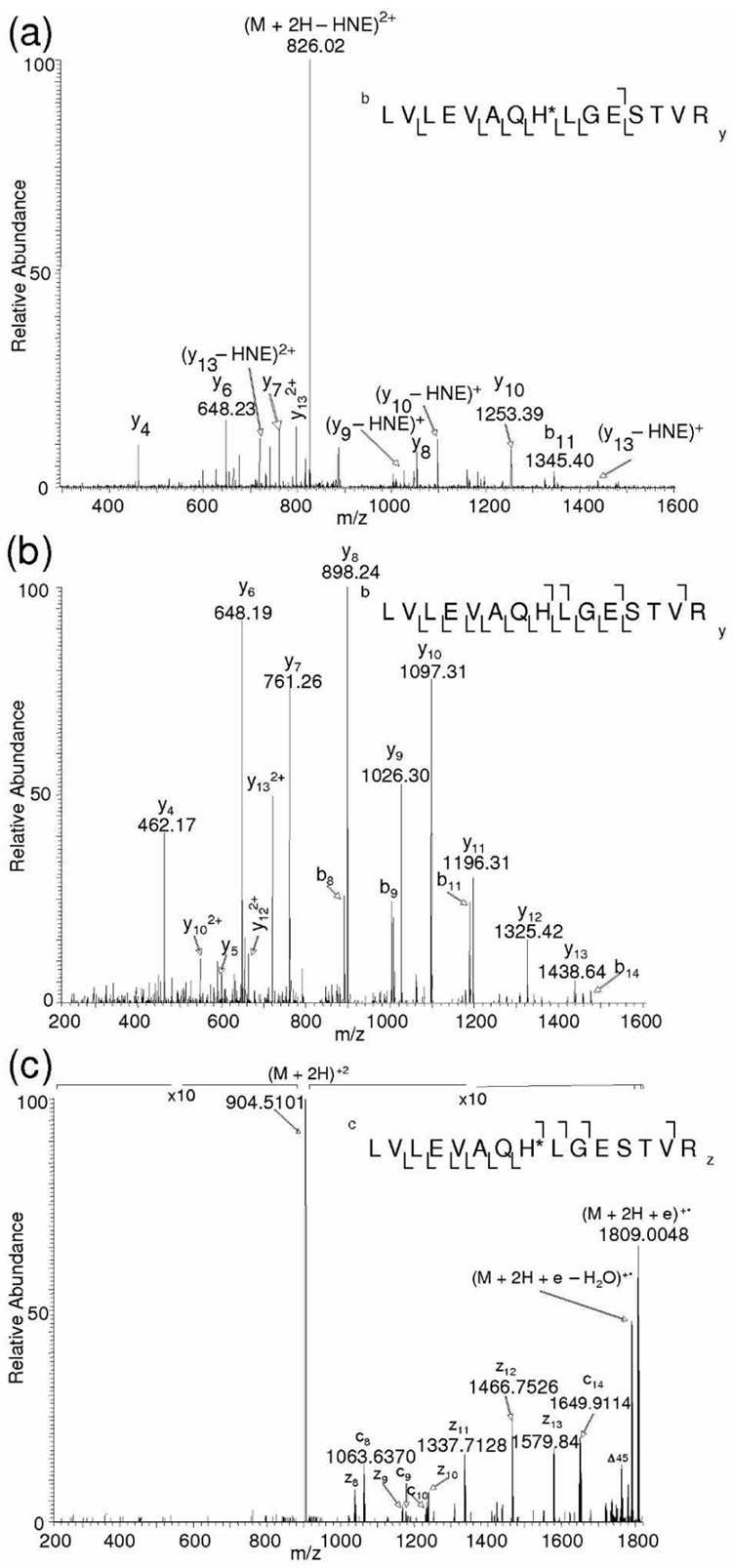
NL-MS3 and NL-ECD-MS/MS-based analyses were further employed to HNE-modified LVLEVAQHLGESTVR and IVYGHLDDPANQEIER, which correspond to tryptic peptides of ATP synthase subunit beta and aconitate hydratase, respectively. We have previously shown that His in these tryptic fragments of the proteins are susceptible to ex vivo HNE modification (Michael adducts) in the rat brain mitochondria.6 The peptide mixture was separated on-line by nanoHPLC and analyzed by NL-MS3 and NL-ECD-MS/MS as described in the experimental section. Figures 2a and b are MS/MS and MS/MS/MS spectra of the HNE-modified peptide LVLEVAQHLGESTVR obtained from NL-MS3 data-dependent acquisition. The predominant fragmentation pathway in CID MS/MS scan of the peptide is the loss of HNE from the precursor ion. The prominent neutral loss peak at m/z 826.02 obtained from CID-based fragmentation of the precursor ion m/z 903.98 (doubly charged LVLEVAQH*LGESTVR) was selected for an additional round of CID as shown in Figure 2b. In this peptide, the site of HNE modification can be assigned to His by the NL-MS3 method, as no other amino acids are known to be susceptible to HNE modification. ECD was triggered for the precursor ion m/z 904.0085 (m/zcalc 904.0198; 12.5 ppm error in mass accuracy). In the ECD spectrum, cleavage of ten out of 14 backbone amine linkage was observed (Figure 2c) and it was possible to assign the site of HNE modification based on the fragmentation pattern. Upon ECD-based MS/MS, all fragment ions that contain the His residue exhibited a +156 Da mass shift relative to the unmodified peptide. The fragment ions c8 (m/z 1063.6370) and z8 (m/z 1038.5671) confirm the presence of HNE residue at His-8 in this peptide. Thus, utilization of NL-ECD-MS/MS can undoubtedly show the HNE modification site, as ECD is capable of retaining the HNE moiety on the peptide during fragmentation. Iavarone et al. have reported that fragmentation efficiency in ECD increases with increasing precursor charge state.32 We also noted that ECD of the doubly charged precursor ion m/z 904.0085 produced a limited range of higher c (c8, c9, c10 and c14) and z (z8–z13) fragment ions.
Figure 2.
(a) CID-MS/MS spectrum of the ATP synthase subunit beta tryptic peptide, LVLEVAQH*LGESTVR, (M + 2H)2+ at m/z 904.03 obtained from data-dependent acquisition. (b) MS/MS/MS spectrum of the neutral loss ion m/z 826.02 obtained by NL-MS3 method. (c) ECD MS/MS spectrum of the precursor ion (M + 2H)2+, m/z 904.0085, obtained by NL-ECD-MS/ MS. An asterisk after the one-letter code of an amino acid residue denotes its HNE modification.
Supplementary Figures 1a and 1b show the CID-based MS/MS and MS/MS/MS spectra of the HNE-modified aconitate hydratase peptide (IVYGH*LDDPANQEIER) obtained by NL-MS3 data-dependent acquisition. The peak corresponding to neutral loss of HNE in the CID MS/MS spectra of the doubly charged precursor ion m/z 1013.02 during NL-MS3 was not intense (Supplementary Figure 1a) as compared to the former two peptides. Neutral loss of HNE corresponding to the ion at m/z 935.48 was nevertheless observed from the precursor ion, which therefore triggered MS3 and ECD of the neutral loss fragment ion and precursor ion, respectively, upon using the NL-MS3 and NL-ECD-MS/MS approaches (Supplementary Figure 1b and 1c). For this peptide, the CID-MS/MS spectrum of the precursor ion itself contained abundant b and y fragment ions (nine out of fifteen backbone amide bond cleavage) that distinctly localized the site of HNE addition to His-5; thus, NL-MS3 and NL-ECD-MS/MS did not provide additional information that could not be obtained from CID-MS/MS alone. The cleavage of eight of the 14 N-Cα bonds (without considering the N-terminal side of proline) in the peptide backbone of (M + 2H)2+ ions showed the fragmentation efficiency of 53%. Only the fragment ions c14–c15 and z10–z15 were produced. The mass difference of 293 Da between z12 (m/z 1576.76) and z11 (m/z 1283.59) corresponds to the mass of HNE-modified His, which permits the localization of HNE site at His-5 in the peptide IVYGHLDDPANQEIER. Although complete sequence coverage was not obtained, the observed fragments still allowed determination of the sites of modification. Nevertheless, NL-MS3 and NL-ECD-MS/MS did yield complementary information (b and y ions in the CID-based method, while several c and z ions upon employing ECD) and, in general, all the methods (CID-MS/MS, NL-MS3 and NL-ECD-MS/MS) together are necessary for the unambiguous localization of HNE-modification sites.
Effect of Charge State on Neutral Loss and ECD Fragmentation of HNE-modified Peptides
DeGnore et al. have reported that the tendency of phosphopeptides to lose the phosphate moiety in CID-based fragmentation in an ion trap depends strongly on the charge state.33 Unlike loss of H3PO4 from phosphopeptides, the tendency of HNE neutral loss is independent of the charge states of the peptides, at least for doubly or triply charged precursor ions. Supplementary Figures 2a–d show the CID MS/MS spectra of triply charged HNE-modified AGT I, AGT II, LVLEVAQHLGESTVR and IVYGHLDDPANQEIER, respectively. The fragment ion from neutral loss was observed in all but the MS/MS of the triply charged peptide IVYGH*LDDPANQEIER.
ECD fragmentation was found to be more efficient in triply charged peptides; hence, multiply charged precursors are generally preferred for this technique.34 Supplementary Figure 3a–c shows ECD mass spectra of the HNE-modified triply charged peptides DRVYIHPFH*L, DRVYIH*PF and LVLEVAQH*LGESTVR obtained by NL-ECD-MS/MS. Intensities of the HNE-neutral loss fragment ions during CID MS/MS of these peptides were high enough that the original precursor ions passed the defined selection criteria for another stage of mass spectrometric analysis, i.e., ECD MS/MS. The ECD MS/MS spectra were of improved quality as evident by the longer peptide sequence tags (consecutive cleavages) than CID MS/MS spectra. The peptide IVYGH*LDDPANQEIER didn’t show any neutral loss for the 3+ charge state; hence, the 3+ precursor ion of this peptide was not selected for ECD. It is observed from the ECD spectrum that the (M + 2H)2+ ion preferentially produces large c and z fragment ions, but ECD of 3+ charge states of same peptides produces both large and small fragment ions. This tendency to shift toward generation of lower mass fragment ions with ECD of peptides of increasing charge state has been observed.32 It was also observed that the ECD spectra of 3+ precursor ions in the peptides DRVYIHPFH*L and DRVYIH*PF showed a neutral loss of HNE from charge-reduced species (m/z 648.84, 2+ and 1296.68, 1+ for DRVYIHPFH*L and m/z 523.76, 2+ and 1046.50, 1+ for DRVYIH*PF) and also from some fragment ions (e.g., c7 m/z 898.46 and z5 m/z 660.32, z6 m/z 759.39 in DRVYIH*PF). Overall, it appears that the sequence of the peptide will determine its tendency towards neutral loss of HNE from the precursor ion and until the correlation of primary structure with fragmentation behavior is well-characterized, both CID and ECD methods of dissociation for successful sequence identification and modification site elucidation will be necessary.
Enrichment of HNE-Modified Peptides by Solid-Phase Hydrazide (SPH) Strategy for NL-Driven Tandem Mass Spectrometry
The reduction of sample complexity prior to mass spectrometric analysis of PTMs is advantageous, since it increases the chance of detecting those modifications that are sub-stoichiometric in nature and/or affecting low-abundance proteins. Taking this into consideration, we tested the efficiency of the NL-MS3 and NL-ECD-MS/MS approach for the analysis of HNE-modified peptides that were spiked into and enriched from a complex protein digest by the SPH method described by Roe et al.7 Three HNE-modified synthetic peptide (DRVYIH*PF, LVLEVAQH*LGESTVR and IVYGH*LDDPANQEIER) were spiked into mouse brain tryptic digest and the resulting mixture was subjected to SPH enrichment. Figure 3a shows the base peak chromatogram of the brain protein tryptic digest spiked with standard peptides obtained by NL-MS3 data-dependent acquisition, together with the full-scan FTMS corresponding to the elution time of peptide DRVYIH*PF. The complex fullscan FTMS shows that too many ions were concurrently presented to the mass spectrometer along with the doubly protonated peptide DRVYIH*PF (m/z 601.84, 2+) at the elution time of the latter. Figure 3b corresponds to the base peak chromatogram of the SPH-enriched HNE modified peptides obtained by NL-MS3 data-dependent acquisition and full-scan FTMS upon the elution of DRVYIH*PF. The spectrum clearly shows the efficiency of SPH enrichment technique in selectively enriching HNE-modified peptide, DRVYIH*PF. The other two spiked HNE-modified peptides (LVLEVAQH*LGESTVR and IVYGH*LDDPANQEIER) were also successfully enriched by SPH beads from the complex matrix of brain protein digest (data not shown). MS/MS of m/z 601.84 (DRVYIH*PF, 2+) and MS3 of the neutral loss ion (m/z 524.08) corresponding to the loss of HNE from doubly charged peptide of m/z 601.84 was triggered in NL-MS3 analysis of the peptide in the complex mixture before and after SPH enrichment. However, compared with the MS3 spectra from the enriched fraction, the low quality of the MS3 spectra of the peptide in complex mixture can be observed (Supplementary Figure 4a–b online). This may be because enrichment techniques increase the number of ion counts for fragmentation as the MS/MS and MS3 spectra of m/z 524.08 in non-enriched fraction were triggered with an ion count of normalization level 14900 and 210, respectively, whereas the ion count normalization level was 22100 and 420 after SPH enrichment. The quality of MS/MS spectrum of peptide LVLEVAQH*LGESTVR was somewhat compromised and no neutral loss ion of significant intensity was visible as the peptide co-eluted with DRVYIH*PF. The retention time (RT) of DRVYIH*PF was around 72 min and the RT of LVLEVAQH*LGESTVR was 73.5 min. It is noteworthy that performing NL-MS3 data-dependent acquisition from brain protein digest spiked with HNE-modified peptides recorded approximately 310 MS3 spectra, whereas MS3 was triggered on only two ions, m/z 349.76 and 524.08 corresponding to neutral loss peaks from triply and doubly charged DRVYIH*PF, after SPH enrichment. Therefore, the enrichment technique also reduces the acquisition of false MS3 spectra triggered due to isobaric fragment ions produced during CID. Modified peptides may also ionize less efficiently than unmodified peptides by electrospray and MALDI depending on the chemical properties of the modification.21 By reducing potential suppression effects through the removal of unmodified peptides, SPH-based enrichment of HNE-modified species could therefore increase the relative intensity of precursor ion signal for efficient fragmentation by CID or ECD and for subsequent application of NL-driven tandem mass spectrometry.
Figure 3.
SPH enrichment of HNE-modified peptide standards, DRVYIH*PF, LVLEVAQH*LGESTVR and IVYGH*LDDPANQEIER, from a mouse brain tryptic digest. (a) Base peak chromatogram from NL-MS3 data-dependent acquisition showing the complex mixture of mouse brain tryptic peptide spiked with the three standard peptides together with the full-scan FTICR spectrum displaying the doubly protonated ion of DRVYIH*PF peptide at m/z 601.84, but other co-eluting peptides present in the complex mixture of mouse brain tryptic digest as well; (b) Base peak chromatogram showing the peptides enriched by solid-phase hydrazide beads (HNE-modified LVLEVAQH*LGESTVR co-eluted with HNE-modified DRVYIH*PF), along with the full-scan FTICR spectrum revealing only peptide DRVYIH*PF after its release from the SPH beads by 10% formic acid.
CONCLUSIONS
The current report demonstrates the synergistic role of CID and ECD methods of fragmentation to detect and localize HNE modification in various peptide models. It was observed that a loss of 156 Da from the peptide precursor ion following CID indicates the presence of HNE but precludes localization of the site of modification depending on the quality of the MS/MS spectrum. The benefits of implementation of ECD for the characterization of various posttranslational modifications are being realized gradually and we have shown that the ECD method of fragmentation will facilitate unambiguous assignment of HNE adduct sites in peptides.19 To this end, we have explored the benefit of NL-ECD-MS/MS in characterization of HNE modification as this technique was efficient in characterizing phosphorylated peptides.30
This study has demonstrated that NL-ECD-MS/MS provides a useful tool to analyze HNE-modified peptides as ECD fragmentation will allow for the retention of the HNE moiety on the product ions and, in addition, generally produces more meaningful peptide fragmentation than CID-based MS/MS analysis. Since CID and ECD yield complementary fragment ions, the combination of the two increases the specificity of sequence information36 and, hence, their combined use is beneficial.11 Zubarev et al. have discussed the limitations associated with using CID or ECD alone and the authors argue that the best would be to implement both techniques together for obtaining sequence information of peptides.36 The inclusion of ECD can reduce the uncertainty associated with CID alone by providing additional sequence information. On the other hand, most search engines work best for CID mass spectra.36 We have shown here that an NL-ECD-MS/MS approach utilizing both CID and ECD and synergistically combining them to enable enhanced MS/MS analyses also has benefits for the interrogation of posttranslational protein modifications. Since this method does not involve a long duty cycle normally associated with full ECD-based data-dependent acquisition, it is less likely that HNE-modified peptides will be overlooked with this method.
SPH enrichment of HNE-modified peptides7 prior to data-dependent LC-MS/MS is expected to increase the chances of detecting low abundance proteins that are susceptible to substoichiometric HNE modification during oxidative stress. We have also demonstrated the value of this selective enrichment methodology for subsequent NL-driven tandem mass spectrometric acquisitions.
Supplementary Material
ACKNOWLEDGEMENT
This research has been supported by the grant AG025384 from the National Institutes of Health. Laszlo Prokai is the Robert A. Welch Professor at the University of North Texas Health Science Center (endowment BK-0031).
REFERENCES
- 1.Esterbauer H, Schaur RJ, Zollner H. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Aldiniq G, Dalle-Donne I, Facino RM, Milzani A, Carini M. Med. Res. Rev. 2007;27:817–868. doi: 10.1002/med.20073. [DOI] [PubMed] [Google Scholar]
- 3.Uchida K. Prog. Lipid Res. 2003;42:318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 4.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Drug Metab. Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 5.Carbone DL, Doorn JA, Kiebler Z, Ickes BR, Petersen DR. J. Pharmacol. Exp. Ther. 2005;315:8–15. doi: 10.1124/jpet.105.088088. [DOI] [PubMed] [Google Scholar]
- 6.Stevens SM, Jr, Rauniyar N, Prokai L. J. Mass Spectrom. 2007;42:1599–1605. doi: 10.1002/jms.1349. [DOI] [PubMed] [Google Scholar]
- 7.Roe MR, Xie H, Bandhakavi S, Griffin TJ. Anal. Chem. 2007;79:3747–3756. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- 8.Guan Z, Yates NA, Bakhtiar R. J. Am. Soc. Mass Spectrom. 2003;14:605–613. doi: 10.1016/S1044-0305(03)00201-0. [DOI] [PubMed] [Google Scholar]
- 9.Crabb JW, O'Neil J, Miyagi M, West K, Hoff HF. Protein Sci. 2002;11:831–840. doi: 10.1110/ps.4400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 11.Zubarev RA. Curr. Opin. Biotechnol. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Tsybin YO, Ramstrom M, Witt M, Baykut G, Hakansson P. J. Mass Spectrom. 2004;39:719–729. doi: 10.1002/jms.658. [DOI] [PubMed] [Google Scholar]
- 13.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han H, Xia Y, Yang M, McLuckey SA. Anal. Chem. 2008;80:3492–3497. doi: 10.1021/ac7022734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turecek F. J. Am. Chem. Soc. 2003;125:5954–5963. doi: 10.1021/ja021323t. [DOI] [PubMed] [Google Scholar]
- 16.Turecek F, Syrstad EA, Seymour JL, Chen X, Yao C. J. Mass Spectrom. 2003;38:1093–1104. doi: 10.1002/jms.527. [DOI] [PubMed] [Google Scholar]
- 17.Jones JW, Sasaki T, Goodlett DR, Turecek F. J. Am. Soc. Mass Spectrom. 2007;18:432–444. doi: 10.1016/j.jasms.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Bakhtiar R, Guan Z. Biotechnol. Lett. 2006;28:1047–1059. doi: 10.1007/s10529-006-9065-z. [DOI] [PubMed] [Google Scholar]
- 19.Rauniyar N, Stevens SM, Jr, Prokai L. Anal. Bioanal. Chem. 2007;389:1421–1428. doi: 10.1007/s00216-007-1534-2. [DOI] [PubMed] [Google Scholar]
- 20.Zubarev RA, Haselmann KF, Bogdan B, Kjeldsen K, Jensen F. Eur. J. Mass Spectrom. 2002;8:337–349. [Google Scholar]
- 21.Stensballe A, Jensen ON, Olsen JV, Haselmann KF, Zubarev RA. Rapid Commun. Mass Spectrom. 2000;14:1793–1800. doi: 10.1002/1097-0231(20001015)14:19<1793::AID-RCM95>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 22.Shi SD, Hemling ME, Carr SA, Horn DM, Lindh I, McLafferty FW. Anal. Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 23.Haselmann KF, Nielsen PF, Zubarev RA. J. Am. Soc. Mass Spectrom. 2005;16:548–552. doi: 10.1016/j.jasms.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Mirgorodskaya E, Roepstorff P, Zubarev RA. Anal. Chem. 1999;71:4431–4436. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- 25.Medzihradszky KF, Guan S, Maltby DA, Burlingame AL. J. Am. Soc. Mass Spectrom. 2007;18:1617–1624. doi: 10.1016/j.jasms.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Ramstrom M, Hagman C, Tsybin YO, Markides KE, Hakansson P, Salehi A, Lundquist I, Hakanson R, Bergquist J. Eur. J. Biochem. 2003;270:3146–3152. doi: 10.1046/j.1432-1033.2003.03690.x. [DOI] [PubMed] [Google Scholar]
- 27.Palmblad M, Tsybin YO, Ramstrom M, Bergquist J, Hakansson P. Rapid Commun. Mass Spectrom. 2002;16:988–992. doi: 10.1002/rcm.667. [DOI] [PubMed] [Google Scholar]
- 28.Davidson W, Frego L. Rapid Commun. Mass Spectrom. 2002;16:993–998. doi: 10.1002/rcm.666. [DOI] [PubMed] [Google Scholar]
- 29.Cooper HJ, Akbarzadeh S, Heath JK, Zeller M. J. Proteome Res. 2005;4:1538–1544. doi: 10.1021/pr050090c. [DOI] [PubMed] [Google Scholar]
- 30.Sweet SM, Creese AJ, Cooper HJ. Anal. Chem. 2006;78:7563–7569. doi: 10.1021/ac061331i. [DOI] [PubMed] [Google Scholar]
- 31.Cooper HJ, Hudgins RR, Hakansson K, Marshall AG. J. Am. Soc. Mass Spectrom. 2002;13:241–249. doi: 10.1016/S1044-0305(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 32.Iavarone AT, Paech K, Williams ER. Anal. Chem. 2004;76:2231–2238. doi: 10.1021/ac035431p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeGnore JP, Qin J. J. Am. Soc. Mass Spectrom. 1998;9:1175–1188. doi: 10.1016/S1044-0305(98)00088-9. [DOI] [PubMed] [Google Scholar]
- 34.Cooper HJ, Hakansson K, Marshall AG. Mass Spectrom. Rev. 2005;24:201–222. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen ML, Savitski MM, Zubarev RA. Mol. Cell. Proteomics. 2005;4:835–845. doi: 10.1074/mcp.T400022-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Zubarev RA, Zubarev AR, Savitski MM. J. Am. Soc. Mass Spectrom. 2008;19:753–761. doi: 10.1016/j.jasms.2008.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.