Summary
Recent discoveries of histone demethylases demonstrate that histone methylation is reversible. However, mechanisms governing the targeting and regulation of histone demethylation remain elusive. Here we report that a Drosophila melanogaster JmjC domain-containing protein, dKDM4A, is a histone H3K36 demethylase. dKDM4A specifically demethylates H3K36me2 and me3 both in vitro and in vivo. Affinity purification and mass spectrometry analysis revealed that Heterochromatin Protein 1a (HP1a) associates with dKDMA4A. We found that the chromoshadow domain of HP1a and a HP1-interacting motif of dKDM4A are responsible for this interaction. HP1a stimulates the histone H3K36 demethylation activity of dKDM4A and this stimulation depends on the H3K9me binding motif of HP1a. Finally, we provide in vivo evidence suggesting that HP1a and dKDM4A interact with each other and loss of HP1a leads to increased level of histone H3K36me3. Collectively, these results suggest a function of HP1a in transcription facilitating H3K36 demethylation at transcribed and/or heterochromatin regions.
Introduction
In eukaryotic cells, DNA exists in the form of chromatin. The basic unit of chromatin is the nucleosome which consists of 146 base pairs of DNA wrapped around a histone octamer (Kornberg and Lorch, 1999). Covalent modification of histones, including acetylation, methylation, phosphorylation and ubiquitination, play an important role in regulating chromatin dynamics and gene expression (Jenuwein and Allis, 2001; Strahl and Allis, 2000). Histone lysine methylation is implicated in both gene activation and repression depending on the methylation site and the state of methylation (mono-, di- or tri-methylation) (Li et al., 2007a). For example, methylation at histone H3K4, K36 and K79 is associated with active transcription, while methylation on histone H3K9, K27 and H4K20 is linked to gene silencing (Martin and Zhang, 2005; Sims et al., 2003).
Histone methylation was long considered to be an irreversible reaction. Histone replacement or proteolysis were believed to be the possible mechanisms to remove methylation marks (Bannister et al., 2002). However, recent discoveries of several histone demethylases have brought a new perspective on how the dynamics of histone methylation can be regulated (Cloos et al., 2006; Fodor et al., 2006; Klose et al., 2006b; Shi et al., 2004; Tsukada et al., 2006; Whetstine et al., 2006; Yamane et al., 2006). LSD1 (KDM1) (Allis et al., 2007), the first histone lysine demethylase identified, demethylates di- and monomethylated histone H3K4 or K9 through a FAD-dependent oxidative reaction (Metzger et al., 2005; Shi et al., 2004). But due to the requirement of a protonated nitrogen in the reaction, LSD1 could not demethylate trimethylated lysines. More recently, a large family of JmjC domain-containing proteins were found to possess histone demethylation activity (Agger et al., 2008). Unlike LSD1, this family of proteins uses Fe (II) and α-ketoglutarate as cofactors and can demethylate all three states of methylated lysines. JHDM1 (KDM2) is the founding member of the JmjC histone demethylase family (Tsukada et al., 2006), which is evolutionarily conserved from yeast to human. Since this initial discovery, a cluster of JmjC domain-containing proteins have been identified as histone demethylases that can specifically remove methyl group from histone H3K4, K9, K27, K36, R2 and H4R3 (Agger et al., 2008)
Histone H3K36 methylation is enriched in coding regions of actively transcribed genes (Bannister et al., 2005; Pokholok et al., 2005). In S. cerevisiae, H3K36 methylation is catalyzed by a sole enzyme, Set2, which associates with the elongating form of RNA polymerase II (Krogan et al., 2003; Li et al., 2003; Schaft et al., 2003; Xiao et al., 2003). H3K36me nucleosomes are recognized by the combinatorial action of two subunits, Eaf3 and Rco1, of the Rpd3S histone deacetylase complex (Li et al., 2007b). The recruitment of Rpd3S to coding regions results in a hypoacetylated environment within ORFs to prevent cryptic transcription initiation (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). H3K36me3 is catalyzed by dSet2 (dHypb) in Drosophila (Bell et al., 2007; Larschan et al., 2007) and HYPB/Setd2 in mammals (Edmunds et al., 2008), whereas K36me2 is mediated by dMes-4 in Drosophila (Bell et al., 2007). A role for histone H3K36 in Drosophila dosage compensation has recently been reported. In this case, histone H3K36me3 helps to recruit the MSL complex to dosage compensated genes on X chromosomes through the MSL-3 subunit (Larschan et al., 2007). Moreover, knockdown of dSet2 in Drosophila cell lines results in increased level of histone H4K16 acetylation, suggesting that in flies, histone H3K36me3 has similar functions in transcriptional regulation to its yeast counterpart (Bell et al., 2007). H3K36 methylation is also subject to dynamic regulation (Klose et al., 2007; Tu et al., 2007). In yeast, Jhd1 (ScKDM2) demethylates histone H3K36me1 and me2, while Rph1 (ScKDM4) demethylates histone H3K36me2 and me3. Both demethylases promote transcription elongation by antagonizing repressive roles of histone H3K36 methylation mediated by Set2 (Kim and Buratowski, 2007).
HP1 was first identified in Drosophila melanogaster as a nonhistone chromosome binding protein, which is encoded by the Su(var)2-5 gene. HP1 is involved in the establishment and maintenance of higher order structure of heterochromatin (James et al., 1989; James and Elgin, 1986). HP1 has two critical domains, an N-terminal chromo domain (CD) and a C-terminal chromoshadow domain (CSD) (Aasland and Stewart, 1995; Paro and Hogness, 1991). The CD of HP1 recognizes methylated histone H3K9 which is important for heterochromatic silencing (Bannister et al., 2001; Lachner et al., 2001). The CSD of HP1 is responsible for interactions with HP1 binding proteins (Li et al., 2002). Recently, HP1 has been shown to be involved in transcriptional activation of some heterochromatic euchromatic genes (Cryderman et al., 2005; de Wit et al., 2007; Johansson et al., 2007; Lu et al., 2000; Piacentini et al., 2003). Moreover, histone H3K9 methylation and HP1γ are enriched at transcribed regions in mammalian cells (Vakoc et al., 2005). In addition to the role of HP1 in gene activation, HP1 has been reported to be involved in sex-specific gene regulation (Liu et al., 2005; Spierer et al., 2005). The loss of HP1a in Su(var)2-5 mutants results in bloated X chromosomes in males, suggesting a role in regulating X-linked gene expression in flies (Spierer et al., 2005). Despite these findings, the molecular mechanism by which HP1 regulates active transcription remains largely unknown.
Based on sequence homology, there are Jhd1 and Rph1 histone demethylase orthologs in Drosophila (Klose et al., 2006a). Here we report that the Drosophila Rph1 homolog, dKDM4A, is a histone demethylase specific to tri- and di-methylated histone H3K36. We demonstrate that dKDM4A interacts with HP1a through a consensus HP1 binding motif PxVxL and HP1a stimulates the demethylation activity of dKDM4A. Thus, an H3K9methyl-binding protein directs the demethylation of H3K36; a previously unidentified histone crosstalk with implications for the regulation of chromatin structure within transcribed sequences.
Results
dKDM4A is a Histone H3 K36 Demethylase
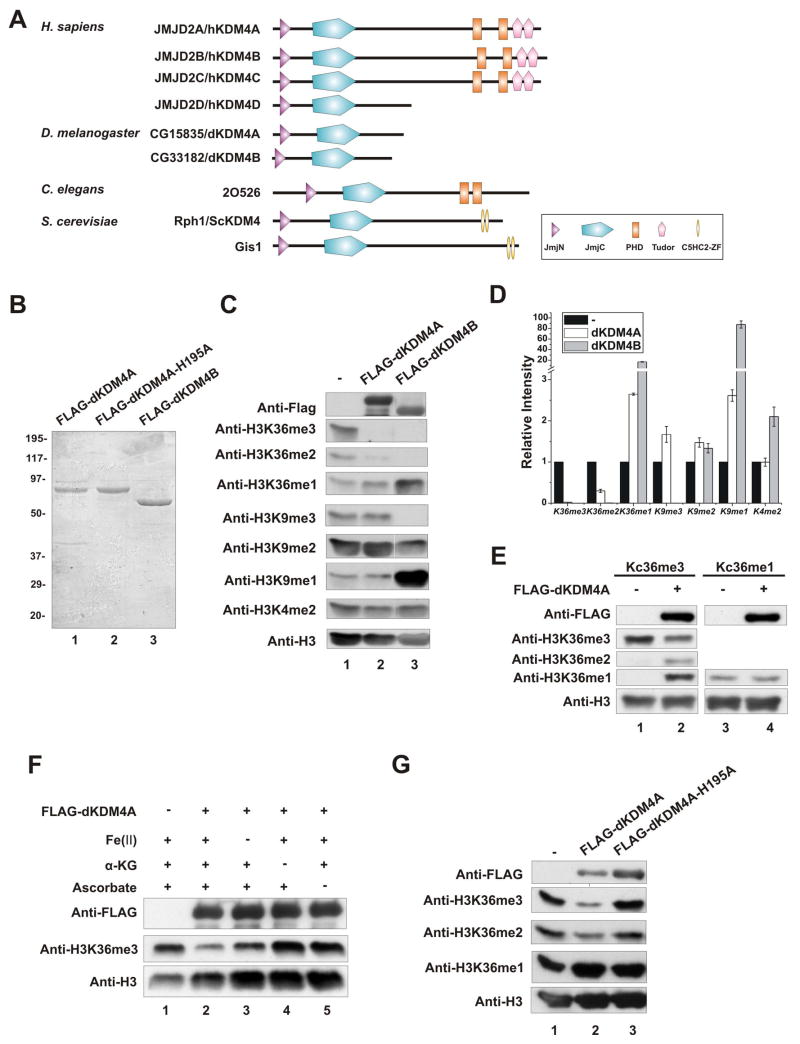
Based on sequence homology, there are two KDM4 orthologs in Drosophila, dKDM4A (CG15835) and dKDM4B (CG33182), that both contain JmjN and JmjC domains (Figure 1A). To examine whether dKDM4A has histone demethylation activity, we purified recombinant dKDM4A from baculovirus-infected Sf21 cells (Figure 1B) and tested its activity in an in vitro histone demethylation assay using HeLa core histones as substrates. As shown in Figure 1C and D, dKDM4A specifically demethylated tri- and di-methyl histone H3K36 in HeLa core histones. We also observed increasing levels of mono-methylated histone H3K36 (H3K36me1), presumably due to accumulation of the end products of the demethylation reaction of di- and tri-methylated histone H3K36. However, the level of histone H3K9 and K4 methylation remained unchanged. In contrast, we found recombinant dKDM4B, another Drosophila KDM4 ortholog, had robust demethylation activity toward both histone H3K9 and K36 (Figure 1C and 1D). To directly test the modification state preference of dKDM4A toward substrates, we utilized methyl-lysine analogs (MLAs) (Simon et al., 2007) to generate recombinant histone H3 containing tri- or mono-methylated K36. Tri- or mono-methylated histone H3 was used as substrates in demethylation assay (Figure 1E). dKDM4A displays robust activity towards K36me3 but fails to demethylate K36me1.
Figure 1. dKDM4A demethylates histone H3K36me3 and me2 in vitro.
(A) Schematic representation of KDM4 family members.
(B) Purified recombinant dKDM4A, its iron-binding mutant dKDM4A-H195A and dKDM4B from baculovirus-infected Sf21 cells were visualized by Coomassie blue staining.
(C) In vitro demethylation assay using HeLa core histones as substrates. The reaction mixtures were analyzed by wester blot using indicated histone antibodies.
(D) Quantification of western blot shown in (C). The Error bars shown are standard deviation.
(E) In vitro demethylation assay using chemically modified recombinant H3 as substrates. Tri-methyl-lysine36 analogs are used in lane 1 and 2; mono-methyl-lysine36 analogs are used in lane3 and 4.
(F) Cofactor dependence of dKDM4A. Each cofactor, Fe (II), α-ketoglutarate and ascorbate was individually excluded from histone demethylation reaction as indicated (lane 3, 4 and 5).
(G) Comparison of histone H3K36 demethylation activity of recombinant dKDM4A (lane 2) and the iron-binding mutant dKDM4A-H195 (lane 3) using HeLa core histones as substrates.
Like hKDM4A (Klose et al., 2006b), the demethylation reaction mediated by dKDM4A requires Fe (II), α-ketoglutarate and ascorbate as cofactors (Figure 1F). The slight activity of dKDM4A in the absence of Fe (II) is likely caused by trace amounts of Fe (II) associated with the purified recombinant dKDM4A. To examine the requirement of Fe (II) for the demethylation activity of dKDM4A, we purified recombinant dKDM4A in which a conserved amino acid in the iron binding site is mutated to alanine (Figure 1B). This mutant dKDM4A has no demethylation activity on histone H3K36me3 and me2, suggesting that Fe (II) is necessary for the catalytic activity of dKDM4A (Figure 1G). We conclude that dKDM4A uses an oxidative mechanism to specifically demethylate histone H3K36me3 and me2 in vitro.
dKDM4A has Histone H3K36 Demethylation Activity in vivo
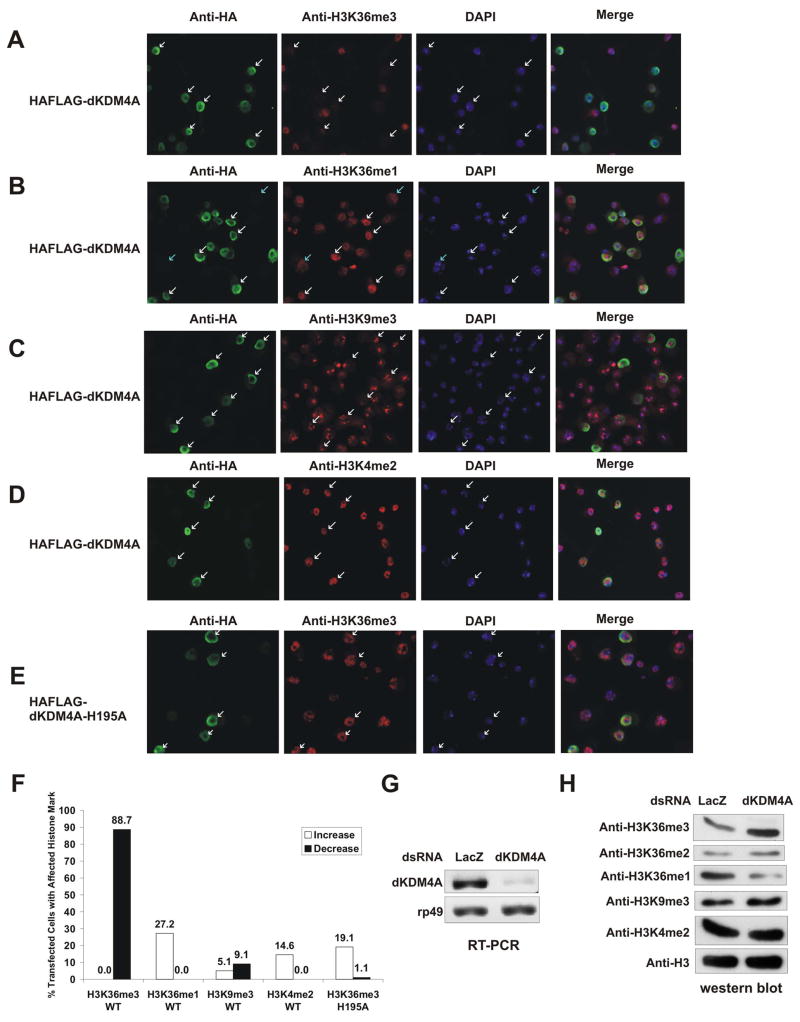
To determine whether dKDM4A functions as a histone H3K36 demethylase in vivo, we established a stable cell line in which epitope-tagged dKDM4A is under the control of a copper inducible promoter. The level of histone methylation was then examined by immunofluorescence analysis (Figure 2A–F). We found that cells containing high level of dKDM4A display significantly reduced level of histone H3K36me3 (Figure 2A). Similarly to our in vitro observations, the level of histone H3K36me1 increases in those cells, possibly due to the accumulation of demethylation products (Figure 2B). Overexpression of dKDM4A seems to only lead to demethylation of histone H3K36, since the level of histone H3K9me3 (Figure 2C) and K4me2 (Figure 2D) remains unchanged. A recent paper reported that dKDM4A can demethylate both histone H3K36 and K9 when overexpressed in S2 cells (Lloret-Llinares et al., 2008). However, we did not observe significant decrease of histone H3K9me3 level in our assays (Figure 2C), and our result is consistent with our observations in vitro (Figure 1C). Lastly, overexpression of a mutant form of dKDM4A that has no histone demethylation activity in vitro, does not significantly change the level of histone H3K36me3 (Figure 2E). Collectively, these data suggest that dKDM4A is a histone H3K36 demethylase in vivo.
Figure 2. dKDM4A has histone H3K36 demethylation activity in vivo.
Drosophila S2 cells were transfected with dKDM4A or dKDM4A-H195A constructs. The stable cell lines were induced by addition of 100 mM CuSO4 and stained with anti-HA and anti-H3K36me3 (A, E), anti-H3K36me1 (B), anti-H3K9me3 (C) and anti-H3K4me2 (D) antibodies. The green corresponds to anti-HA staining, the red corresponds to anti-histone methylation specific antibodies, and the blue corresponds to DAPI staining. White arrows point to the dKDM4A or dKDM4A-H195A positive-staining cells, while blue arrows indicate non-expressing cells.
(F) Quantification of the percentage of anti-HA stained cells that have increased or decreased histone modification in (A) to (E). More than 90 cells were counted for each condition as representative results are shown above.
(G) The mRNA level of dKDM4A was examined by RT-PCR with primers specific for dKDM4A and rp49 (internal control).
(H) Acid-extracted bulk histones from RNAi treated samples were analyzed by western blot using indicated antibodies.
To further examine the demethylation activity of dKDM4A in vivo, we knocked down endogenous dKDM4A in S2 cells using double-stranded RNA against dKDM4A. RT-PCR analysis showed that the mRNA level of dKDM4A decreased in S2 cells after 4 days of RNAi treatment (Figure 2G). Under these conditions, the level of histone H3K36me3 and me2 increased while the level of histone H3K36me1 decreased (Figure 2H). Therefore, dKDM4A is responsible for maintaining proper level of H3K36 methylation in vivo.
HP1a Associates with dKDM4A
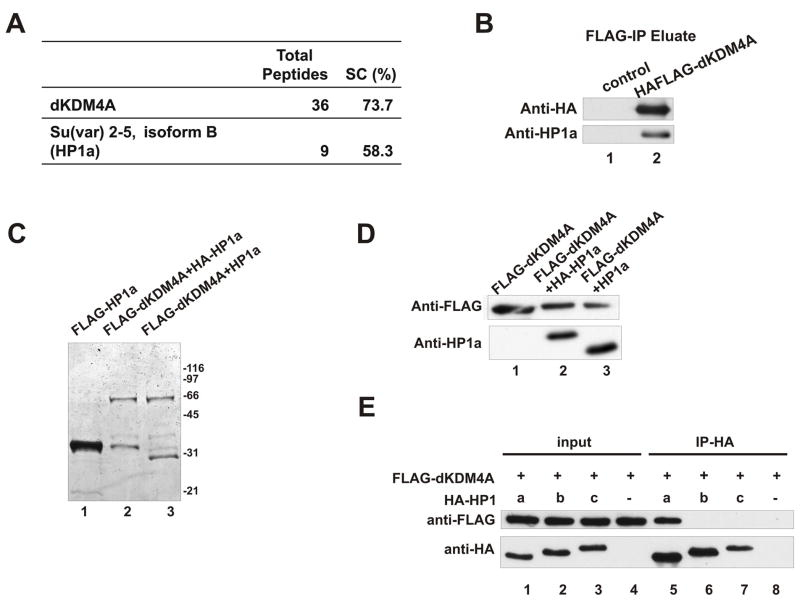
We next sought to identify protein factors that associate with dKDM4A. Using an established stable cell line expressing epitope tagged dKDM4A, we performed affinity purification. Proteins in the eluate were then identified through MudPIT analysis (Washburn et al., 2001). Surprisingly, we found that the product of Su(var)2-5, Drosophila HP1a, co-purifies with dKDM4A (Figure 3A). HP1a is the 2nd most abundant protein behind the tagged protein dKDM4A, except for some common contaminants. We then performed western blotting analysis using an antibody against HP1a to confirm this interaction. Indeed, HP1a is associated with dKDM4A (Figure 3B). To further examine this interaction in another cellular system, we co-infected Sf21 cells with baculovirus encoding FLAG-tagged dKDM4A and HA-tagged or non-tagged HP1a. Anti-FLAG antibody-conjugated agarose beads were used to immunoprecipitate FLAG-dKDM4A. Both Coomassie blue staining (Figure 3C) and western blot (Figure 3D) shows that HP1a co-purifies with dKDM4A in this system. To test if HP1a directly interacts with dKDM4A, we carried out an in vitro binding assay by mixing recombinant dKDM4A and HP1a, followed by anti-HA immunoprecipitation. The results shown in Figure 3E (lane 5) indicate that these two proteins directly bind to each other. To further demonstrate the specificity of HP1a-dKDM4A interaction, we purified recombinant versions of the other two isoforms of HP1, HP1b and HP1c. As shown in Fig 3E (lane 6 and 7), HP1b and HP1c fail to interact with dKDM4A in the in vitro binding assay, suggesting that dKDM4A only associates with HP1a, but not HP1b or HP1c.
Figure 3. dKDM4A specifically interacts with HP1a.
(A) The MudPIT analysis of native dKDM4A complex purified from HAFLAG-tagged
dKDM4A-expressing stable cells under 100 mM CuSO4 induction. The table lists the number of non-redundant spectra (total peptides) and the amino acid sequence coverage (SC).
(B) The eluate of affinity purification from wild-type S2 cells (control) and dKDM4A-expressing cells was analyzed by western blot using anti-HA and anti-HP1a antibody to detect the tagged dKDM4A and HP1a respectively.
(C) FLAG-tagged recombinant proteins were purified from Sf21cells infected with baculovirus encoding FLAG-HP1a (lane 1), or co-infected with baculovirus encoding FLAG-dKDM4A and HA-HP1a (lane 2) or non-tagged HP1a (lane 3). The eluate from anti-FLAG beads was visualized by Coomassie blue staining.
(D) Recombinant dKDM4A (lane 1) and dKDM4A-HP1a heterodimers (lane 2 and 3)purified from Sf21 insect cells were analyzed by western blot.
(E) HP1a specifically interacts with HP1a but not HP1b and HP1c. Recombinant dKDM4A and HP1a, HP1b or HP1c were mixed with 500μg of Sf21 cell lysate to reduce background binding. The resulting complexes were immunoprecipitated using anti-HA agarose beads. The entire immunoprecipitated material and 2% of input were loaded on a gel and analyzed by western blot using anti-FLAG and anti-HA antibodies.
HP1a Stimulates Demethylation Activity of dKDM4A
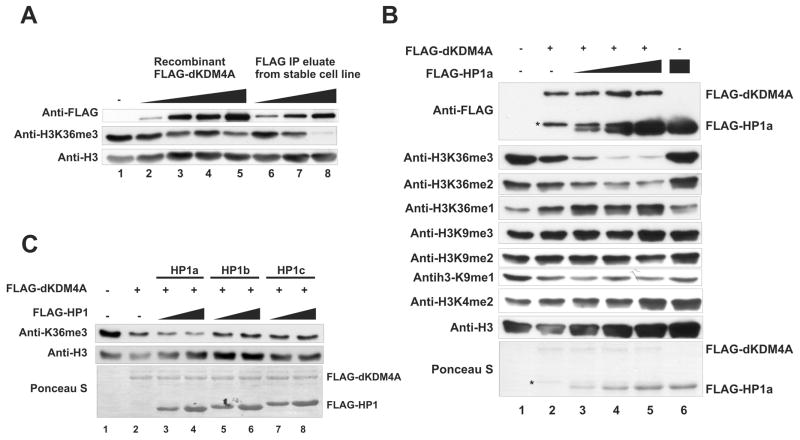
While examining the demethylation activity of purified native dKDM4A complex, we noticed that the native complex displays stronger specific activity on histone H3K36me3 compared to recombinant dKDM4A (Figure 4A, compare lane 8 with lane 3–5), albeit containing less dKDM4A (anti-FLAG in Figure 4A). This result suggests that protein factors associated with dKDM4A may enhance dKDM4A enzymatic activity. Since HP1a binds dKDM4A, we asked whether HP1a stimulates dKDM4A demethylation activity in vitro. Increasing amounts of HP1a were titrated into a recombinant dKDM4A-mediated demethylation assay. The demethylation activity of dKDM4A on H3K36me3 and me2 is enhanced in the presence of HP1a (Figure 4B). The level of histone H3K9 and K4 methylation remains unchanged in the same reaction, and HP1a alone does not affect histone methylation levels (Figure 4B, lane6). Furthermore, no enhancement of dKDM4A activity is observed when HP1b and HP1c are added to the demethylation reactions (Figure 4C). This suggests that the stimulation of dKDM4A demethylation activity is specific to HP1a.
Figure 4. HP1a stimulates the histone demethylation activity of dKDM4A.
(A) In vitro demethylation assay using recombinant dKDM4A or the dKDM4A complex that were purified from HAFLAG-dKDM4A-expressing stable cells. HeLa core histones were used as substrates, and the reactions were analyzed by western blot.
(B) In vitro demethylation assay using recombinant dKDM4A with addition of HP1a. HeLa core histones were used as substrates, and the reactions were analyzed by western blot. The molar ratio of dKDM4A and HP1a is 1:1, 1:2 and 1:4 in lane 3, 4 and 5. HP1a was added to the reaction without dKDM4a as a control in lane 6. Asterisks indicate the degradation products of recombinant dKDM4A.
(C) In vitro demethylation assay using recombinant dKDM4A with addition of HP1a, HP1b or HP1c. The molar ratio of dKDM4A and HP1 is 1:1 in lane 3, 5 and 7 and 1:2 in lane 4, 6, and 8.
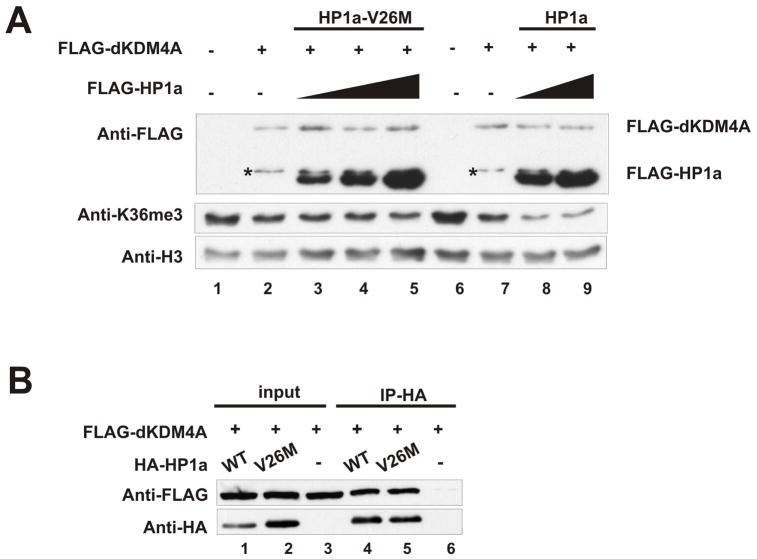
Since HP1 is known to recognize methylated H3K9 through its CD (Bannister et al., 2001; Lachner et al., 2001), we wondered whether the CD is important for HP1a to stimulate dKDM4A demethylation activity. To this end, we generated recombinant protein with a mutation in the HP1a CD (V26M) that has been shown previously to abolish HP1a binding to histone H3K9me (Jacobs et al., 2001). As shown in Figure 5A (lane1–5), this mutant fails to enhance the demethylation activity of dKDM4A on histone H3K36me3. It is likely that this defect is due to a reduced interaction of HP1a with histone H3 because its interaction with dKDM4A was unaffected by the mutation (Figure 5B).
Figure 5. Stimulation of the demethylation activity of dKDM4A depends on the CD of HP1a.
(A) In vitro demethylation assay using recombinant dKDM4A with addition of HP1a or HP1a-V26M mutant. HeLa core histones were used as substrates. The molar ratio of dKDM4A and HP1a is 1:1, 1:2 and 1:4 in lane 3, 4 and 5; 1:2 and 1:4 in lane 8 and 9. Asterisks indicate the degradation products of recombinant dKDM4A.
(B) Recombinant dKDM4A and HP1a or HP1a-V26M were mixed with 500μg of Sf21 cell lysate and immunoprecipitated using anti-HA agarose beads. The entire immunoprecipitated material and 2% of input were analyzed by western blot using anti-FLAG and anti-HA antibodies.
The CSD of HP1a and a Consensus HP1-interacting PxVxL Motif in dKDM4A are Responsible for the HP1a-dKDM4A Interaction
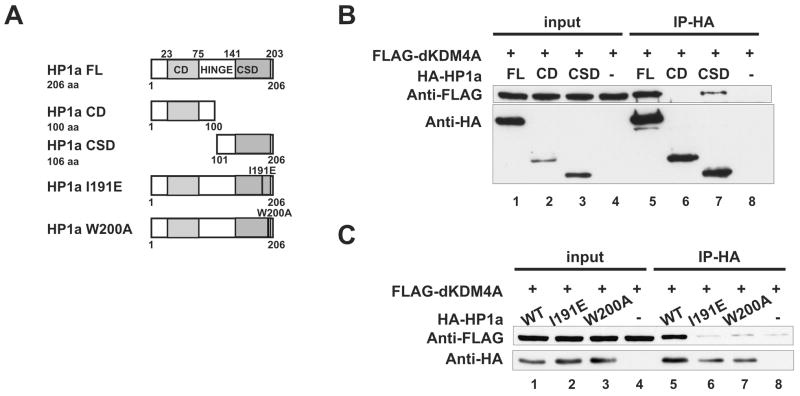
To map the domain of HP1a that mediates the direct interaction with dKDM4A, we purified truncated forms of HP1a that contain either the CD or the CSD alone (Figure 6A) and tested them in the in vitro binding assay. As shown in Figure 6B, the CSD is sufficient for the binding of HP1a to dKDM4A, while the CD does not interact with dKDM4A under the same conditions.
Figure 6. An intact CSD dimerization interface of HP1a is required for its interaction with dKDM4A.
(A) Schematic representation of HP1a truncation mutants (CD and CSD) and two critical residues that are predicted to disrupt either its dimerization (I191E) or its target-binding interface (W200A). Full-length HP1a (FL) contains a chromo domain (CD), a hinge domain and a chromoshadow domain (CSD).
(B) HP1a CSD but not CD binds to dKDM4A. Recombinant dKDM4A and full-length HP1a (FL), truncation mutants HP1a CD, or HP1a CSD were mixed with 500μg of Sf21 cell lysate and immunoprecipitated using anti-HA agarose beads. The entire immunoprecipitated material and 2% of input were analyzed by western blot using anti-FLAG and anti-HA antibodies.
(C) HP1a mutants I191E and W200A fail to interact with dKDM4A.
To dissect the interaction between dKDM4A and HP1a CSD further, we introduced two point mutations at conserved residues within CSD, I191E and W200A. These mutations have been shown to disrupt the dimerization of CSD and its interaction with HP1 binding proteins (Brasher et al., 2000; Thiru et al., 2004). As expected, recombinant HP1a-I191E and W200A both fail to interact with dKDM4A (Figure 6C), suggesting that an intact CSD dimerization interface is required for the HP1-dKDM4A interaction.
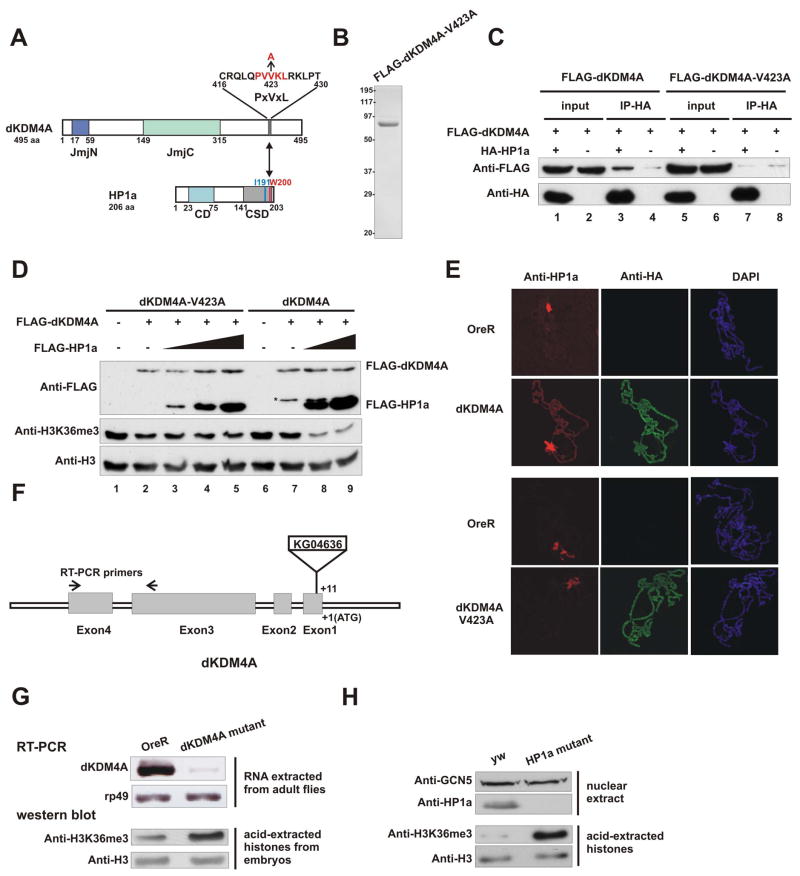
The CSD of HP1 recognizes a consensus peptide pentamer, PxVx [M/L/V], in most HP1-interacting proteins (Smothers and Henikoff, 2000; Thiru et al., 2004). We found that the C-terminal region of dKDM4A contains a PxVxL motif, PVVKL (amino acid 421 to 425) (Figure 7A). To examine whether HP1a associates with dKDM4A through this motif, we generated a mutant in which the critical valine 423 was mutated to alanine. Recombinant dKDM4A-V423A was purified from baculovirus-infected Sf21 cells (Figure 7B). This mutant protein could no longer stably associate with HP1a (Figure 7C, lane 5–8). Thus, HP1a associates with dKDM4A through the conserved PxVxL motif.
Figure 7. The interaction between HP1a and dKDM4A is important for the demethylation activity of dKDM4A both in vitro and in vivo.
(A–E) dKDM4A interacts with HP1a through a conserved HP1a-binding PxVxL motif (A) Schematic representation of consensus HP1a binding motif within dKDM4A. The amino acid sequence from 421 to 425 of dKDM4A contains an HP1a binding PxVxL motif, which is colored red. The critical residue (V423) was mutated into alanine as indicated.
(B) Recombinant dKDM4A-V423A was purified from baculovirus-infected Sf21 cells and visualized by Coomassie blue staining.
(C) HP1a directly associates with dKDM4A through a consensus HP1 binding motif. Recombinant HP1a and dKDM4A or V423A mutant were mixed and immunoprecipitated using anti-HA agarose beads.
(D) In vitro demethylation assay using recombinant dKDM4A-V423A or wild-type dKDM4A in the presence of HP1a. HeLa core histones were used as substrates. The molar ratio of dKDM4A and HP1a is 1:1, 1:2 and 1:4 in lane 3, 4 and 5; 1:2 and 1:4 in lane 8 and 9. Asterisk indicates the degradation of recombinant dKDM4A.
(E) Overexpression of dKDM4A induces HP1a spreading into euchromatin. Salivary glands from wild type were placed on the same slide as those prepared from either the dKDM4A-HAFLAG overexpressing line or the dKDM4A-V423A-HAFLAG-overexperssing line. Each combination of glands were squashed together, and resulting polytene chromosomes were stained with antibodies against HP1a and HA. Images from each slide were taken on a confocal laser scanning microscope using the exact same setting. The red corresponds to anti-HP1a staining, the green corresponds to anti-HA staining of dKDM4A-HAFLAG, which was used to distinguish between OreR or overexpressing chromosomes and the blue corresponds to DAPI staining.
(F) Schematic representation of the insertion site of P element KG04636. The arrows indicate the location of primers used in RT-PCR.
(G) RNA was extracted from adult flies of OreR and dKDM4A mutant (KG04636). The mRNA level of dKDM4A was then examined by RT-PCR. The rp49 probe served as an internal control. Acid-extracted histones from embryos of OreR and dKDM4A mutant were analyzed by western blot using indicated antibodies.
(H) Loss of HP1a significantly increases the level of histone H3K36me3 in Drosophila larvae. Nuclear extracts (upper panel) or acid-extracted histones (lower panel) from third instar larvae of yw and the HP1a null mutant (Su(var)2-504/Su(var)2-505) were subjected to western blot using indicated antibodies. The levels of GCN5 and histone H3 were used as loading controls.
We next asked whether the stimulation of dKDM4A activity by HP1a relies on the direct physical association between these two proteins. Recombinant dKDM4A-V423A, which fails to bind HP1a, was used in the in vitro demethylation assay. We first demonstrate that this mutation has minimal effect on intrinsic enzymatic activity of dKDM4A (compare lane2 and lane7 in Figure 7D). We then titrated in increasing amount of HP1a to the reaction, and found that HP1a fails to stimulate the demethylation activity of the dKDM4A-V423A mutant (Figure 7D, lane 3–5). Taken together, these results indicate that the association of HP1a with dKDM4A regulates the histone H3K36 demethylation activity of dKDM4A.
To explore the biological function of HP1a-dKDM4A interaction, we crossed transgenic flies, UAS-Kdm4A-HA1FLAG2 or UAS-Kdm4A-V423A-HA1FLAG2, with Sgs3-GAL4 to overexpress dKDM4A in salivary glands. We first performed immunofluorescence analysis of polytene chromosomes from the larvae overexpressing wild type dKDMA4A in salivary glands. Salivary glands from wild type (OreR) and dKDM4A-overexpressing larvae were squashed on the same slide to minimize any procedural variation. Indeed, we found that overexpression of dKDM4A induces HP1a to spread into chromosome arms (Figure 7E upper panel, and Figure S1). This pattern is in contrast to that of HP1a in wild type flies, in which it is mainly located at the chromocenter (Figure 7E, upper panel). This result is in agreement with a recent paper using a similar system (Lloret-Llinares et al., 2008). We then tested if the spreading of HP1a is directly related to its interaction with dKDM4A using transgenic larvae that overexpress dKDM4A mutant (V423A) in salivary glands. We observed a very similar staining pattern of the mutant dKDM4A (Figure 7E, lower panel, anti-HA) compared to wild type dKDM4A (Figure 7E, upper panel, anti-HA). However, consistent with the fact that dKDM4A-V423A does not bind to HP1a in vitro, we found that the spreading of HP1a was significantly reduced in the larvae overexpressing dKDM4A-V423A (Figure 7E, lower panel and Figure S1). This result supports the notion that the binding of HP1a to chromosome arms is helped through its interaction with overexpressed dKDM4A.
HP1a Regulates Histone H3K36 Methylation in Drosophila Larvae
Our biochemical data suggest that HP1a collaborates with dKDM4A to regulate the level of H3K36me. Thus, we wondered whether mutations disrupting HP1a or dKDM4A expression might share a similar phenotype. To this end, we obtained a fly stock containing the P-element KG04636 inserted within the coding region of dKDM4A (Figure 7F). This insertion abrogated the endogenous transcript of dKDM4A as detected by RT-PCR (Figure 7G, upper panel). Although the mutant is homozygous viable, the P-element insertion elevates the bulk level of histone H3K36me3 in mutant embryos (Figure 7G, lower panel). We next wanted to test if loss of HP1a gives rise to similar changes of K36me3. A previous study showed that chromatin bound HP1a was not detectable in the Su(var)2-504/Su(var)2-505 (Fanti et al., 1998). We thus examined the level of histone H3K36me3 in third instar larvae of this mutant. As shown in Figure 7H (upper panel), we failed to detect any HP1a signal in nuclear extracts from Su(var)2-504/Su(var)2-505 larvae. However, the level of histone H3K36me3 increased significantly compared to that of wild type (Figure 7H, lower panel). This result supports the notion that HP1a is required for the demethylation of H3K36 mediated by dKDM4A in vivo.
Discussion
In this study, we identified one of the JmjC domain-containing KDM4 orthologs in Drosophila, dKDM4A. The in vitro demethylation assay shows that dKDM4A demethylates histone H3K36me3 and me2 using an oxidative demethylation mechanism which requires Fe (II) and α-ketoglutarate as cofactors. Overexpression of dKDM4A in Drosophila S2 cells reduces the level of histone H3K36me3, whereas knockdown of endogenous dKDM4A increases the level of histone H3K36me3 and me2. These results together demonstrate that dKDM4A is a bona fide histone H3K36 demethylase in vivo.
Through MudPIT analysis of the affinity-purified native dKDM4A complex, we found that HP1a associates with dKDM4A. More importantly, we demonstrate that HP1a stimulates the demethylation activity of dKDM4A, while the HP1a CD mutant V26M, that cannot bind methyl-K9 histone H3, fails to stimulate dKDM4 activity. In addition, dKDM4A directly binds to the HP1 CSD and this binding requires an intact HP1 CSD dimer interface. A consensus HP1 binding PxVxL motif of dKDM4A is responsible for its interaction with CSD of HP1a. Interestingly, overexpression of dKDM4A causes the spread of HP1a to euchromatin regions, presumably through this specific interaction, and dKDM4A-V423A, which does not bind to HP1a, failed to localize HP1a to euchromatin. These data suggest HP1a-dKDM4A is a euchromatic H3K36 demethylase complex.
Set2 mediated histone H3K36 methylation is an important mark on chromatin during transcription elongation (Li et al., 2007a). In fungi, such as S. cerevisiae, S. pombe, and N. crassa, a sole histone lysine-methyltransferase Set2 is responsible for all three methylation states of H3K36 (Adhvaryu et al., 2005; Morris et al., 2005; Strahl et al., 2002). In Drosophila, histone H3K36 methylation is catalyzed by two enzymes, dSet2 and dMes-4 (Bell et al., 2007; Larschan et al., 2007). Although yeast Set2 is the only histone methyltransferase that catalyzes methylation of histone H3K36, two histone H3K36 demethylases, Jhd1 and Rph1, are responsible for demethylation of histone H3K36 at different modification states in budding yeast (Kim and Buratowski, 2007; Klose et al., 2007; Tu et al., 2007). In Drosophila, there are three histone demethylases that govern demethylation of histone H3K36. dKDM2 has been identified as a histone H3K36me2 demethylase (Lagarou et al., 2008). We demonstrate here that dKDM4A is a histone H3K36me3 and me2 demethylase, and dKDM4B has demethylation activity on both histone H3K9 and K36me3/me2 in vitro. Therefore, histone H3K36 methylation in Drosophila is likely regulated by highly specific enzymes in both directions. Since both modification and de-modification enzymes possess high modification state specificity, histone H3K36 may be subjected to much more sophisticated regulation in higher eukaryotes than in yeast.
Purification of the dKDM4A complex from S2 cells revealed a specific association of HP1a with dKDM4A. Three of the HP1-like chromatin proteins (HP1a, HP1b, HP1c) in Drosophila share high amino acid sequence similarity. Both HP1a and HP1b localize to euchromatin and heterochromatin, while HP1c is found only in euchromatin (Smothers and Henikoff, 2001). It is unclear whether these HP1-like chromatin proteins have specific or redundant functions in transcription regulation. However, we demonstrate here that dKDM4A specifically interacts with HP1a, but not HP1b and HP1c. Furthermore, HP1b and HP1c cannot stimulate dKDM4A demethylation activity in vitro. A previous study showed that the yeast homolog of KDM4, Rph1 (ScKDM4), did not stably associate with any other protein (Klose et al., 2007). It was speculated that the C-terminal ZF domain of Rph1, which can potentially bind to DNA, allows Rph1 to function without associated factors (Klose et al., 2007). Unlike other proteins in the KDM4 family which commonly contain PHD, tudor or ZF domains (Figure 1A), dKDM4A only has JmjN and JmjC domains. In this article, we found that HP1a stably associates with dKDM4A and stimulates its demethylation activity. Since the H3K9 binding motif is required for this stimulation, we propose that the CD of HP1a might contribute to target dKDM4A to specific loci, particularly to H3K9me enriched regions, to regulate gene expression.
In S. pombe, the HP1 homolog recruits a JmjC domain-containing protein Epe1 to heterochromatin loci where they function together to counteract repressive chromatin (Zofall and Grewal, 2006). Here we show that HP1a directly interacts with dKDM4A through a consensus binding motif PxVxL. Most importantly, the presence of HP1a stimulates histone demethylation activity of dKDM4A in vitro, and HP1a is required for maintaining normal level of H3K36me3 in vivo as well. Since Epe1 on its own seems to have no histone demethylation activity (Tsukada et al., 2006), it would be interesting to see whether a similar scenario also occurs in S. pombe, in which Swi6 may stimulate enzymatic activity of Epe1 towards other non-histone substrates.
HP1 has been reported to associate with actively transcribed euchromatin regions (Cryderman et al., 2005; de Wit et al., 2007; Piacentini et al., 2003; Vakoc et al., 2005). Mammalian HP1γ and histone H3K9 methylation are enriched at the coding region of active genes, implying that they may play a role during transcription elongation (Vakoc et al., 2005). In yeast, histone H3K36me3 appears to be a repressive mark at coding region of actively transcribed genes (Li et al., 2007a). In higher eukaryotes, histone H3K9 methylation, which is absent in the budding yeast, might replace the role of K36 methylation in the coding regions of transcribed genes (Berger, 2007). However, the mechanism by which HP1 functions in active transcription is largely unknown. Our findings here suggest a possible role of HP1a in recruitment of the histone H3K36me3/me2 demethylase dKDM4A to transcribed regions to remove histone H3K36 methylation. The formation of the HP1a-dKDM4A complex may help to release HP1a from heterochromatin regions, thus targeting it to specific gene loci. It is also possible that dKDM4A, which targets histone modification marks within the 3′ ORF of actively transcribed genes, recruits HP1a to euchromatic regions. We currently favor a model in which HP1a facilitates recruitment of dKDM4A, because the HP1a CD mutant, V26M, fails to stimulate dKDM4A activity. This result suggests that HP1a binding to histone H3 is required for the enhancement of dKDM4A demethylation activity. We speculate that HP1a-mediated histone demethylation may serve as a regulatory mechanism to control chromatin states during active transcription elongation. Alternatively, a similar mechanism might also apply to maintaining silenced states of heterochromatin.
Experimental Procedures
Fly stocks and crosses
The full length cDNAs of dKDM4A or dKDM4A-V423A were cloned into pUAST vector containing a C-terminal HA and FLAG tag. Transgenic fly lines, UAS-Kdm4A-HA1FLAG2 (w; P{w[+mC]=[UAS-Kdm4A-HA1FLAG2]}) and UAS-Kdm4A-V423A-HA1FLAG2 (w; P{w[+mC]=[UAS-Kdm4A-V423A-HA1FLAG2]}) were generated by Genetic Services. To overexpress HAFLAG-tagged dKDM4A or dKDM4A-V423A in salivary glands, transgenic flies were crossed to the Sgs3-GAL4 (w[1118]; P{w[+mC]=Sgs3-GAL4.PD}TP1) stock (Bloomington stock number 6870) (Brand and Perrimon, 1993). The KG04636 P element insertion mutant, P{SUPor-P}CG15835KG04636, was obtained from Bloomington Stock Center at Indiana University (stock number 13828). Fly stocks Su(var)2-504/Cyo-GFP and Su(var)2-505/Cyo-GFP were provided by Sarah Elgin (Washington University, St. Louis, MO).
Histone demethylation assay
HeLa core histones were incubated with recombinant dKDM4A or native complex in histone demethylation assay buffer (50 mM HEPES-KOH pH7.9, 100 uM Fe(NH4)2(SO4)2, 1 mM α-ketoglutarate, 2 mM Ascorbate) in a final volume of 10 μl for 1 hour at 37 °C. The reaction mixture was analyzed by western blot using histone methylation specific antibodies. Chemically modified histone H3 used in Figure 1E was prepared as described in (Simon et al., 2007).
in vitro binding assay
Recombinant HA-HP1a, HP1b, HP1c or HP1a mutants and FLAG-dKDM4A or its mutant FLAG-dKDM4A-V423A were mixed in the buffer containing 50mM HEPES-NaOH (pH 7.9), 150 mM NaCl, 2 mM MgCl2, 0.05% Triton X-100, 10% (v/v) glycerol, 0.5mM EDTA, 1mM PMSF and 0.1 μg/μl BSA or 500μg of Sf21 cell lysate overnight at 4°C and then incubated with anti-HA agarose beads for 2 hour at 4°C. The beads were washed 4 times using the same buffer described above and eluted by boiling in SDS-PAGE sample buffer. The eluate and 2% of the input were analyzed by western blot using anti-FLAG and anti-HA antibodies.
RNAi knockdown of dKDM4A in S2 cells
Primers containing T7 sequence tagged at the 5′ end were used to amplify dKDM4A and LacZ fragments. PCR products were gel-purified and served as templates to generate dsRNA with MEGAscript T7 kit (Ambion) following manufacturer’s instruction. 1μg of dsRNA was transfected into S2 cells using Effectene (Qiagen). After 4 days of RNAi treatment, histones were acid-extracted from S2 cells and analyzed by western blot.
Immunostaining of polytene chromosomes
The third instar larvae were dissected in PBS supplemented with 0.1% TritonX-100. Salivary glands were fixed first in solution 1 (phosphate-buffered saline (PBS), 3.7% paraformaldehyde and 1% Triton X-100) and then in solution 2 (3.7% paraformaldehyde, 50% acetic acid). The chromosomes were spread on poly-L-lysine coated microscope slides. Anti-HP1 antibody was used at 1:50 and anti-HA antibody was used at 1:100. Cy3 and Cy3-conjugated secondary antibodies were used at 1:400. Images were taken on a confocal laser scanning microscope (LSM-510 META, Zeiss).
Supplementary Material
Acknowledgments
We would like to thank Lori Wallrath for contributing anti-HP1a monoclonal antibody (C1A9) to DSHB and Sarah Elgin for kindly sharing fly stocks. We also thank Matthew Simon for advice on making chemically modified histones. We are grateful to Vikki Weake and Tamaki Suganuma for technical suggestions, So Hee Kwon for HP1 cDNA and other members of Workman lab for helpful discussions. This work was supported by a grant from the National Institute of General Medical Sciences to J.L.W. and funding from Stowers Institute for Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aasland R, Stewart AF. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic acids research. 1995;23:3168–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhvaryu KK, Morris SA, Strahl BD, Selker EU. Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryotic cell. 2005;4:1455–1464. doi: 10.1128/EC.4.8.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Current opinion in genetics & development. 2008 doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. The Journal of biological chemistry. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. The EMBO journal. 2007;26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Cambridge, England) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Cryderman DE, Grade SK, Li Y, Fanti L, Pimpinelli S, Wallrath LL. Role of Drosophila HP1 in euchromatic gene expression. Dev Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet. 2007;3:e38. doi: 10.1371/journal.pgen.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. The EMBO journal. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Molecular cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- Fodor BD, Kubicek S, Yonezawa M, O’Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes & development. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. The EMBO journal. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. European journal of cell biology. 1989;50:170–180. [PubMed] [Google Scholar]
- James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Molecular and cellular biology. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science New York, NY . 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johansson AM, Stenberg P, Pettersson F, Larsson J. POF and HP1 Bind Expressed Exons, Suggesting a Balancing Mechanism for Gene Regulation. PLoS Genet. 2007;3:e209. doi: 10.1371/journal.pgen.0030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Molecular cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim T, Buratowski S. Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. The Journal of biological chemistry. 2007;282:20827–20835. doi: 10.1074/jbc.M703034200. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Gardner KE, Liang G, Erdjument-Bromage H, Tempst P, Zhang Y. Demethylation of histone H3K36 and H3K9 by Rph1: a vestige of an H3K9 methylation system in Saccharomyces cerevisiae? Molecular and cellular biology. 2007;27:3951–3961. doi: 10.1128/MCB.02180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006a;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006b;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Molecular and cellular biology. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes & development. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Molecular cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007a;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science New York, NY. 2007b;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. The Journal of biological chemistry. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- Li Y, Kirschmann DA, Wallrath LL. Does heterochromatin protein 1 always follow code? Proceedings of the National Academy of Sciences of the United States of America . 2002;99(Suppl 4):16462–16469. doi: 10.1073/pnas.162371699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LP, Ni JQ, Shi YD, Oakeley EJ, Sun FL. Sex-specific role of Drosophila melanogaster HP1 in regulating chromatin structure and gene transcription. Nature genetics. 2005;37:1361–1366. doi: 10.1038/ng1662. [DOI] [PubMed] [Google Scholar]
- Lloret-Llinares M, Carre C, Vaquero A, de Olano N, Azorin F. Characterization of Drosophila melanogaster JmjC+N histone demethylases. Nucleic acids research. 2008;36:2852–2863. doi: 10.1093/nar/gkn098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nature reviews. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Morris SA, Shibata Y, Noma K, Tsukamoto Y, Warren E, Temple B, Grewal SI, Strahl BD. Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryotic cell. 2005;4:1446–1454. doi: 10.1128/EC.4.8.1446-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. The Journal of cell biology. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Schaft D, Roguev A, Kotovic KM, Shevchenko A, Sarov M, Shevchenko A, Neugebauer KM, Stewart AF. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic acids research. 2003;31:2475–2482. doi: 10.1093/nar/gkg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Smothers JF, Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- Smothers JF, Henikoff S. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Molecular and cellular biology. 2001;21:2555–2569. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spierer A, Seum C, Delattre M, Spierer P. Loss of the modifiers of variegation Su(var)3-7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. Journal of cell science. 2005;118:5047–5057. doi: 10.1242/jcs.02623. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Molecular and cellular biology. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiru A, Nietlispach D, Mott HR, Okuwaki M, Lyon D, Nielsen PR, Hirshberg M, Verreault A, Murzina NV, Laue ED. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. The EMBO journal. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Tu S, Bulloch EM, Yang L, Ren C, Huang WC, Hsu PH, Chen CH, Liao CL, Yu HM, Lo WS, et al. Identification of histone demethylases in Saccharomyces cerevisiae. The Journal of biological chemistry. 2007;282:14262–14271. doi: 10.1074/jbc.M609900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Molecular cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature biotechnology. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes & development. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Molecular cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.