Abstract
Histone lysine and arginine residues are subject to a wide array of post-translational modifications including methylation, citrullination, acetylation, ubiquitination, and sumoylation. The combinatorial action of these modifications regulates critical DNA processes including replication, repair, and transcription. In addition, enzymes that modify histone lysine and arginine residues have been correlated with a variety of human diseases including arthritis, cancer, heart disease, diabetes, and neurodegenerative disorders. Thus, it is important to fully understand the detailed kinetic and chemical mechanisms of these enzymes. Here, we review recent progress towards determining the mechanisms of histone lysine and arginine modifying enzymes. In particular, the mechanisms of S-adenosyl-methionine (AdoMet) dependent methyltransferases, FAD dependent demethylases, iron dependent demethylases, acetyl-CoA dependent acetyltransferases, zinc dependent deacetylases, NAD+ dependent deacetylases, and protein arginine deiminases are covered. Particular attention is paid to the conserved active-site residues necessary for catalysis and the individual chemical steps along the catalytic pathway. When appropriate, areas requiring further work are discussed.
Keywords: methyltransferase, acetyltransferase, demethylase, deacetylase, deiminase, sirtuin
1. Introduction
Within eukaryotic organisms, the basic unit of chromosomes is the nucleosome which is composed of double-stranded DNA wrapped around a protein octamer containing two copies each of the histone proteins H2A, H2B, H3, and H4 [1, 2]. Histone proteins are subject to a wide array of post-translational modifications including methylation, citrullination (deimination), acetylation, phosphorylation, ubiquitination, and sumoylation occurring within the histone core region as well as on the N-terminal tails that protrude from the core region [3]. The combinatorial influence of these modifications in both time and space affects important DNA regulatory processes including replication, repair, and transcription [3].
Within histone proteins, lysine and arginine residues are abundant and highly post-translationally modified. Enzymes that modify these lysine and arginine residues have been correlated with a variety of human disease states such as rheumatoid arthritis [4], cancer [5], heart disease [6], diabetes [7, 8], as well as neurodegenerative disorders such as Parkinson's disease and Alzheimer's disease [8, 9]. In light of the importance of these enzymes in a large variety of human disease states, it is critical to elucidate their catalytic mechanisms.
In this review, we will focus on the kinetic and chemical mechanisms of the enzymes that perform post-translational modification of lysine and arginine residues within histone proteins. However, it is important to point out that similar reactions on non-histone proteins will proceed through the same mechanism. We will pay particular attention to the conserved active-site residues necessary for catalysis and the individual chemical steps along the catalytic pathway. Where possible we will try to incorporate both the former and new nomenclature of these enzymes designated by former name/new name (for explanation of the new nomenclature see [10]).
2. Lysine modifying enzymes
Lysine residues within histones are subject to a variety of modifications including methylation, acetylation, ubiquitination, and sumoylation on their ε-amino groups [3]. Of the enzymes that catalyze histone post-translational modification, those that modify lysine are the best understood. In general, histone lysine acetylation is correlated with gene activation whereas deacetylation is correlated with gene repression/silencing [11]. However, histone lysine methylation is correlated with either activation or repression depending on the site and degree of methylation. Given their important involvement in gene regulation, it is not surprising that the enzymes that regulate these post-translational modifications are implicated in a variety of human disease states. For example, histone lysine methyltransferases are implicated in a wide variety of cancers [12], histone acetyltransferases are associated with leukemia [13], aberrant recruitment of class I/II histone deacetylases (HDACs) are linked to leukemias and lymphomas [14], and class III HDACs are involved in age-associated diseases such as type II diabetes, obesity, Parkinson's disease, and Alzheimer's disease [7-9]. Therefore, elucidating the chemical mechanisms catalyzed by these enzymes will be important therapeutically. In fact, a small-molecule inhibitor of class I/II HDACs was recently FDA-approved for the treatment of cutaneous T-cell lymphoma [14].
2.1. Histone lysine methyltransferases
Lysine methyltransferases catalyze mono-, di-, and trimethylation of the lysine ε-amino group in an S-adenosyl-L-methionine (AdoMet) dependent manner. Histone lysine methyltransferases consist of two main classes, the SET domain containing family and the DOT1 family.
2.1.1. SET domain histone lysine methyltransferases
SET domains are found in a few bacterial proteins and a large number of eukaryotic proteins with over 60 human members [15] although not all have known histone lysine methyltransferase activity. SET domain methyltransferases exist which terminally form mono-, di-, or trimethyl-lysine. The size and bonding patterns of the active-site residues determine whether the enzyme carries out mono-, di-, or trimethylation of its target lysine residue.
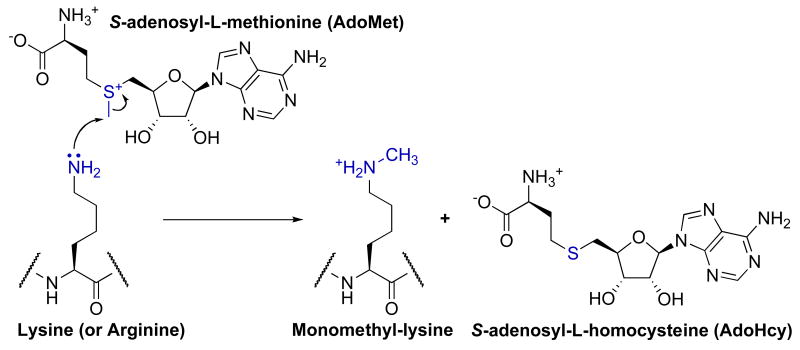
SET domain methyltransferases catalyze a sequential bi-bi kinetic mechanism in which both substrate association and product release occur in a random fashion [16, 17]. Within the active site, a conserved aspartate or glutamate hydrogen bonds to the two-ribose hydroxyls of AdoMet. In addition, a conserved arginine or lysine (Lys-294 in Set7/9/hKMT7 [10]) forms a salt bridge with the carboxylate of AdoMet. These interactions position AdoMet in a U-shaped conformation that places the methylsulfonium group at the base of a hydrophobic channel in which the lysine substrate binds. In Set7/9, the side chain hydroxyls of Tyr-245 and Tyr-305 hydrogen bond to the ε-amine of lysine, directing the lone pair towards the methyl group of AdoMet [18]. The AdoMet methyl group is positioned and activated by C–H…O hydrogen bonds between the methyl group and the side chain hydroxyl of Tyr-335 and the main chain carboxyls of Gly-264 and His-293. The energy of C–H…O hydrogen bonds is estimated to be 0.5–3 kcal/mol, which is 2–10 fold weaker than more classical N/O–H…O hydrogen bonds (see [19-21] and references therein). However, in folded proteins the contribution of C–H…O hydrogen bonds may be more significant due to a greater desolvation penalty for placing N/O–H…O compared to C–H…O hydrogen bonds within a hydrophobic protein environment [22]. Within SET domain methyltransferases these C–H…O hydrogen bonds position the two substrates such that the sulfur of AdoMet, the carbon of the transferred methyl, and the ε-nitrogen of lysine are collinear. This geometry allows SN2 nucleophilic attack of the ε-amine to transfer the methyl group from AdoMet and form the products monomethyl-lysine and S-adenosyl-L-homocysteine (AdoHcy) (Fig. 1) [23]. After one round of methyl transfer, it is hypothesized that C–H…O hydrogen-bonding between the methylammonium group and active-site residues orient monomethyl- or dimethyl-lysine either for subsequent methylation events or to disfavor subsequent methyl transfer [24].
Fig. 1.
Proposed general chemical mechanism of AdoMet-dependent histone lysine methyltransferases. SET domain, Dot1/KMT4, and protein arginine methyltransferases use a similar mechanism of methyl transfer.
For methyl transfer to occur, the ε-amine of the lysine substrate must be deprotonated. Several mechanisms of deprotonation have been proposed including deprotonation by an active-site residue (e.g. Tyr-245 [25, 26] or Tyr-335 [18, 25, 27, 28] in Set7/9), an ordered water molecule [29], bulk solvent [30], or deprotonation by decreasing the ε-amine pKa through close proximity to the cationic methylsulfonium group of AdoMet [31]. Alternatively, the observation that SET domain methyltransferases possess an unusually high pH optimum of ∼10 suggests that the lysine substrate might be deprotonated upon binding [32]. However, structural and mutagenesis studies indicate that neither Tyr-335 nor any other active-site residue are positioned to act as a general base [33, 34] and theoretical studies calculate a pKa for Tyr-335 of >13.0 [35]. Alternatively, calculations reveal that placing the positive charge of the protonated ε-amine in closely proximity to the cationic methylsulfonium lowers the pKa of the ε-amine from ≥10.9 to ≤8.2 in Set7/9 [35, 36]. As processive mechanisms have been proposed [17, 26, 29, 37] for the formation of dimethyl- and trimethyl-lysine in some SET domain methyltransferases, multiple rounds of lysine deprotonation must occur without release of methylated lysine. It has been hypothesized that an ordered water channel created upon formation of the ternary complex between Set7/9, the unmodified or methylated lysine substrate, and AdoMet allows release of protons to bulk solvent [35, 36].
For multiple methylations to occur processively, the lysine ε-carbon-nitrogen bond must rotate so that the ε-amine can be further deprotonated and the resulting lone pair aligned with the methyl-sulfur bond of AdoMet. In the monomethyl specific methyltransferase, Set7/9, Tyr-245 and Tyr-305 hydrogen bond with the lysine substrate to prevent rotation of the ε-carbon-nitrogen bond and therefore alignment of the lone pair with the methyl-sulfur bond of AdoMet [18, 23, 26, 35, 38]. Mutation of Tyr-245 or Tyr-305 of Set7/9 to phenylalanine converts the enzyme to a tri- or dimethyltransferase, respectively [26, 38]. Interestingly, in the SET domain methyltransferases DIM-5 and G9a/KMT1C [10], which are capable of trimethylation and dimethylation, respectively, the equivalent residue to Tyr-305 is a phenylalanine. Therefore, this tyrosine to phenylalanine substitution is thought to act as a switch to catalyze methylation beyond monomethylation.
Several other means for the selective formation of distinct methylation states have been hypothesized. In Rubisco large subunit methyltransferase (LSMT), which forms trimethyl-lysine, it is thought that a more spacious lysine-binding pocket accommodates the increasing bulk of monomethyl- and dimethyl-lysine within the active site [39]. Alternatively, it has been proposed from several theoretical studies that formation of an ordered water channel (see above) is responsible for catalysis of a specific methylation state. In particular, it was proposed that methyl-lysine substrate binding to Set7/9 blocks this water channel so that the bound monomethyl-lysine cannot be deprotonated, consistent with rate-limiting deprotonation and a calculated pKa of 13.8 for protonated monomethyl-lysine [36]. In contrast, a separate theoretical study on Set7/9 indicated that methyl transfer, not lysine deprotonation, is rate limiting and that formation of monomethyl-lysine creates an ordered water channel [35, 36]. This channel results in a hydrogen bond between the lone pair of monomethyl-lysine nitrogen and the nearest water molecule in the channel, which does not allow the lone pair to react with AdoMet in a subsequent methylation step [40]. Both of these mechanisms account for the selective formation of monomethyl-lysine products by Set7/9, but future work is necessary to distinguish between these mechanisms.
2.1.2. Dot1/KMT4 histone lysine methyltransferases
Dot1/KMT4 [10] histone lysine methyltransferases do not contain a SET domain and methylate Lys-79 within the core domain of histone H3 [41-44]. In addition, Dot1/KMT4 methyltransferases only methylate nucleosomal substrates, not free histones. Structures of Dot1/KMT4 methyltransferases with AdoMet or AdoHcy bound reveal an extended AdoMet conformation that is in stark contrast to the folded U-shaped conformation found in SET domain methyltransferases [45, 46]. Sequence analysis suggests that Dot1/KMT4 methyltransferases possess AdoMet binding motifs similar to histone arginine methyltransferase family [47].
Despite lacking a SET domain, Dot1/KMT4 methyltransferases catalyze an overall similar mechanism of methyl transfer (Fig. 1). Critical contacts between AdoMet and Thr-139, Gln-168, Glu-186, Asp-161, and Asp-222 of human DOT1L/hKMT4 position the methylsulfonium group at the end of a channel in which the lysine substrate binds [45, 46]. Similar to SET domain methyltransferases, this channel aligns the methyl group of AdoMet with the ε-nitrogen of lysine for a SN2 methyl transfer reaction [45]. Also similar to SET methyltransferases, the AdoMet and lysine substrate binding sites are on separate faces of the enzyme indicating that the lysine substrate can undergo multiple rounds of methylation without being released from the enzyme, consistent with a processive mechanism. However, the fact that a mixture of unmodified, mono-, di-, and trimethylated Lys-79 exists in yeast suggests that methyl transfer proceeds distributively [44].
As no residues capable of deprotonating lysine are present in the active site of Dot1/KMT4 methyltransferases, several hypotheses have been proposed for lysine deprotonation. One hypothesis is that the carboxylate of AdoMet acts as a general base to deprotonate lysine [45]. Alternatively, it has been proposed that close proximity of the ε-amine to the positively charged methylsulfonium and/or the overall hydrophobic environment of the active site lower the pKa of the lysine residue [32]. Regardless of the mechanism of lysine deprotonation, the significantly different pH profiles between Dot1/KMT4 and SET domain methyltransferases suggests that Dot1 methyltransferases use a distinct mechanism of lysine deprotonation for AdoMet attack [28, 39, 46]. Future studies may utilize this mechanistic difference in the design specific inhibitors of human DOT1L/hKMT4. In addition, future work is needed to understand the mechanism of specific methylation of a histone core residue, Lys-79, in favor of the more exposed lysine residues located on the N-terminal histone tails within nucleosomal substrates.
2.2. Lysine specific histone demethylases
2.2.1. LSD1/KDM1 family
Until recently, it was unclear if methylation of lysine residues was a permanent modification. In 1973, Paik and Kim were the first to observe demethylation activity of calf thymus histones, but were unable to isolate the active enzyme [48]. It took another three decades for the first histone demethylase to be isolated and characterized. In 2004, a lysine-specific demethylase, LSD1/KDM1 [10], was shown to demethylate mono- and dimethylated Lys-4 of histone H3 with concomitant formation of formaldehyde. LSD1/KDM1 belongs to the amine oxidase superfamily and oxidatively demethylates lysine residues in a FAD-dependent manner.
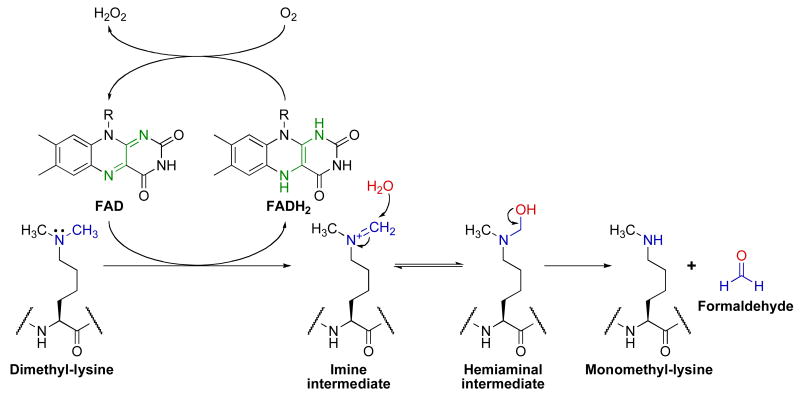
LSD1/KDM1 catalyzes the concurrent reduction of FAD to FADH2 and oxidation of the methylated lysine substrate generating an imine intermediate (Fig. 2). The formation of this imine requires a free lone pair on lysine, and accordingly, LSD1/KDM1 is only capable of demethylating mono- and dimethyl-lysine residues. While no structures of methylated lysine peptides bound to LSD1/KDM1 exist, modeling of monomethyl-lysine from a methionine residue bound within the LSD1/KDM1 active-site predicts a distance of ∼3 Å from the methyl group (the site of oxidative demethylation) to the reactive N-5 atom of FAD [49]. Once the imine is formed, water is thought to non-enzymatically attack this intermediate to form a hemiaminal that subsequently collapses to formaldehyde and amine. Although other electron acceptors have been postulated to exist [50], the FADH2 in LSD1/KDM1 is generally thought to be reoxidized to FAD by one equivalent of molecular oxygen to stoichiometrically produce hydrogen peroxide. FADH2 oxidation is likely rapid as the optical spectrum of LSD1/KDM1 reveals absorbance maxima at ∼370 and 458 nm in the resting state [51], indicative of fully oxidized FAD [52].
Fig. 2.
Proposed chemical mechanism of LSD1/KDM1. This figure was adapted from Culhane et al. [53].
The precise mechanism of imine formation in LSD1/KDM1 is controversial and may occur via hydride transfer or single electron mechanisms [53-55]. A recent study of an FAD-dependent oxidase, tryptophan 2-monooxygenase, is informative. In this study, the authors measured a large deuterium isotope effect (6.0 ± 0.5) for carbon-hydrogen bond cleavage in the conversion of amine to imine [54]. This large deuterium isotope effect is consistent with irreversible carbon-hydrogen bond cleavage, which favors a hydride transfer over a radical mechanism. Whether this is true for LSD1/KDM1 will require further investigation.
2.2.2. JHDM family
The discovery of LSD1/KDM1 opened the door towards finding other demethylases, particularly those capable of demethylating trimethyl-lysine residues, an activity that LSD1/KDM1 does not possess. In 2006, Tsukada and co-workers identified the first [56] of a large family of histone demethylases that each contain a jumonji C domain critical for demethylase activity. These enzymes have thus been designated jumonji histone demethylases (JHDMs). These enzymes are conserved from yeast to humans with approximately 30 members identified in humans [57].
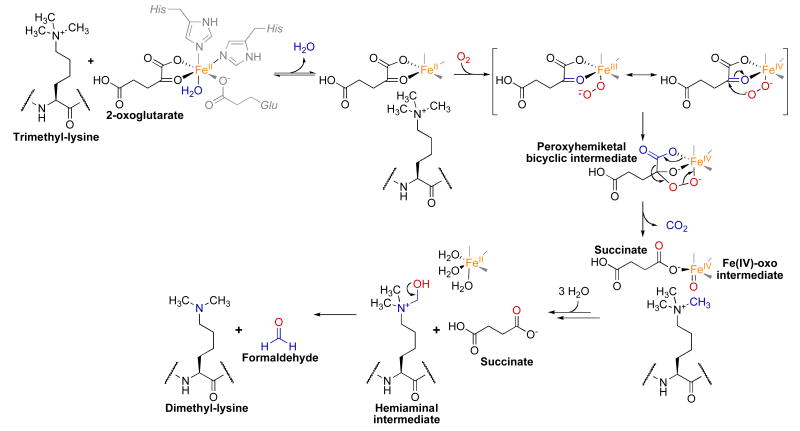
JHDMs belongs to the Fe(II)/2-oxoglutarate-dependent dioxygenase (or hydroxylase) superfamily [58], which catalyze a diverse set of reactions via Fe(IV)-oxo species [59]. In the proposed JHDM mechanism, a quaternary complex containing 2-oxoglutarate, Fe(II), and methylated lysine substrate reacts with molecular oxygen (Fig. 3). In the active-site, the conserved residues His-188, Glu-190, and His-276 (JMJD2A/KDM4A numbering [10]) chelate the Fe(II) [60-62]. In the first chemical step, an electron is transferred from Fe(II) to molecular oxygen to generate a superoxide radical and Fe(III) [57, 63]. It is hypothesized that the hydrophobic active site assists to generate high iron oxidation states [57]. Nucleophilic attack of the activated oxygen at the ketone carbon of 2-oxoglutarate results in an Fe(IV) peroxyhemiketal bicyclic intermediate [58]. Subsequent decarboxylation produces succinate, CO2, and an Fe(IV)-oxo intermediate. This intermediate oxidizes the methyl carbon of the methylated lysine producing a hemiaminal intermediate and regenerating Fe(II). As with LSD1/KDM1, this hemiaminal is thought to spontaneously decompose to demethylated lysine and formaldehyde. In contrast with LSD1/KDM1, the mechanism employed by JHDM enzymes does not require a lone electron pair on the ε-nitrogen of the methylated lysine substrate allowing demethylation of trimethyl-lysine substrates.
Fig. 3.
Proposed chemical mechanism of JHDM enzymes.
Recent structures of JMJD2A/KDM4A [60-62], which catalyzes demethylation of di- and trimethylated Lys-9 and trimethylated Lys-36 of histone H3, provide important insights into the selectivity of these enzymes for certain methylation states. In JMJD2A/KDM4A, the methylammonium binding pocket is composed of the carbonyl oxygen of Gly-170 and the side chains of Tyr-177, Glu-190, Ser-288, and Asn-290. These residues are hypothesized to participate in C–H…O hydrogen-bonding [19] with the polarized methyl group(s) of methylated lysine residues [60]. When the substrate is trimethylated, these C–H…O hydrogen bonds position one of the methyl groups towards the iron center where this methyl group can undergo oxidation. In contrast, consistent with the slow demethylation rates of JMJD2A/KDM4A with monomethyl- and dimethyl- compared to trimethyl-lysine substrates, these C–H…O hydrogen bonds direct the methyl group(s) within monomethyl- and dimethyl-lysine substrates away from the Fe(II) center. It has also been suggested that Ser-288 in JMJD2A, which is often substituted by alanine in other JHDM family members (e.g. Ala-291 in JMJD2D/KDM4D [10]), modulates the specificity of certain demethylases for trimethyl- versus dimethyl- or monomethyl-lysine substrates [60].
With the discovery of several histone lysine demethylase classes, future work is needed to address several issues. Where rates have been measured, the JHDM family catalyzes demethylation with kcat values of ∼0.013 min-1 [60], whereas rates of LSD1/KDM1 catalyzed demethylation are >200-fold faster at ∼3.1 min-1 [64]. In addition, other members of the Fe(II)/2-oxoglutarate-dependent dioxygenase superfamily possess kcat values two to four orders of magnitude greater than the JHDM family [65-68]. These slow demethylation rates suggest that JHDM demethylases may work in unidentified protein complexes or that other substrates exist in vivo. Alternatively, only a very small fraction of the in vitro characterized JHDM enzymes may be active under these conditions assayed. Another unresolved issue is that both LSD1/KDM1 and the JHDM family produce hydrogen peroxide and formaldehyde. As these compounds are known to damage DNA within the cell nucleus, pathways must exist to protect DNA. Towards this end, it is hypothesized that formaldehyde can be recycled to become the reactive methyl in AdoMet [69]. Furthermore, it is possible that the human genome encodes yet unidentified classes of histone demethylases, which work in concert with the existing LSD1/KDM1 and JHDM demethylase families.
2.3. Histone acetyltransferases
Histone acetyltransferases (HATs or KATs [10]) catalyze the transfer of the acetyl moiety from acetyl-CoA to the ε-amino of histone lysine residues resulting in acetylated lysine and CoA. HATs are grouped into three main families: GNAT (for its founding member yeast Gcn5/ScKAT2 [10]), MYST (for the founding members MOZ, Ybf2/Sas3, Sas2, and Tip60), and p300/CBP [70].
2.3.1. GNAT family
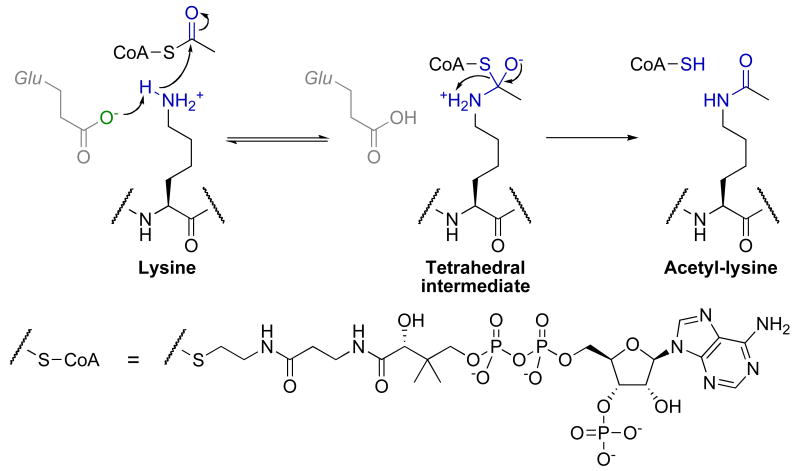
For the GNAT family, initial structural and kinetic data revealed an ordered sequential bi-bi kinetic mechanism for acetyl transfer [71]. In this mechanism, acetyl-CoA and then the lysine substrate bind to form a ternary complex. An active site glutamate (Glu-173 in yGcn5/ScKAT2) general base activates the ε-amine of lysine for direct nucleophilic attack of the carbonyl carbon within acetyl-CoA, forming a tetrahedral intermediate [72]. This intermediate then collapses to acetyl-lysine product and CoA (Fig. 4).
Fig. 4.
Proposed chemical mechanism of histone acetyltransferases.
2.3.2. MYST family
Initial structural and kinetic data on the MYST family member, yEsa1/ScKAT5 [10], by Yan et al. in 2000 revealed an active-site glutamate (Glu-338) that structurally aligned with the corresponding glutamate residue in yGcn5/ScKAT2, suggesting the MYST family utilizes a similar sequential mechanism of acetyl transfer to that of the GNAT family [73]. However, in 2002 Yan et al. published a structure of yEsa1/ScKAT5 containing a proposed acetyl-cysteine (Cys-304) intermediate, which along with some kinetic evidence suggested a ping-pong mechanism of catalysis [74]. Surprisingly, a more recent report revealed that mutation of Cys-304 to alanine or serine decreased acetyltransferase activity (kcat) by only two and ten fold, respectively [75]. This report also found no evidence of a competent acetyl-enzyme intermediate during the acetyl-transfer reaction. Steady-state kinetic analyses of yEsa1/ScKAT5 indicated a sequential mechanism in which acetyl-CoA binds prior to the lysine substrate. In addition, pH rate analysis and mutagenesis indicated Glu-338 is the general base responsible for activating the ε-amine of lysine, as was originally suggested. Therefore, the MYST family utilizes an ordered sequential bi-bi mechanism similar to the GNAT family.
2.3.3. p300/CBP family
Investigations of the p300/CBP family have led to contradictory findings with regard to their kinetic mechanism and structural relation to the GNAT and MYST families. In initial studies, a bi-substrate analog of acetyl-CoA and lysine was shown to be a potent inhibitor of p300/hKAT3B [10] (IC50 ∼ 500 nM) suggesting a sequential mechanism similar to the other HAT families [76]. However, steady-state bi-substrate kinetic experiments with p300 revealed a parallel line pattern consistent with ping-pong mechanism, which was further supported by dead-end inhibitor studies [77]. Recently, Liu and co-workers reported a crystal structure of the p300/hKAT3B HAT domain in complex with the bi-substrate analog (see above) [78], revealing that the p300/CBP structure is distantly related to that of the GNAT and MYST families. From this structure and accompanying biochemical data, the authors propose that the p300/CBP family utilizes a Theorell-Chance mechanism (a specialized case of an ordered sequential mechanism in which the ternary complex possesses a very short lifetime), which would explain the previously observed parallel line pattern. Therefore, all characterized HAT families follow an ordered sequential bi-bi kinetic mechanism where differences between families may affect substrate specificity but not the overall mechanism of catalysis. However, the catalytic mechanisms of the acetyltransferases Hat1/KAT1 and Rtt109/KAT11 [10] remain unclear.
2.4. Histone deacetylases
Histone deacetylases (HDACs) catalyze the removal of acetyl groups from the ε-amine of acetyl-lysine residues within histones. Phylogenetic analyses subdivide HDACs into four classes. Class I, II, and IV HDACs utilize an active-site metal dependent catalytic mechanism, and class I HDACs include human HDAC1-3 and HDAC8, class II include human HDAC4-7 and HDAC9-10, and class IV HDACs include human HDAC11 and are homologous to both class I and II [79]. Class III HDACs (or sirtuins) utilize a distinct nicotinamide adenine dinucleotide (NAD+) dependent catalytic mechanism and are conserved from bacteria to humans with five yeast homologs (ySir2 and Hst1-4) and seven human homologs (Sirt1-7) [80, 81].
2.4.1. Class I/II/IV histone deacetylases
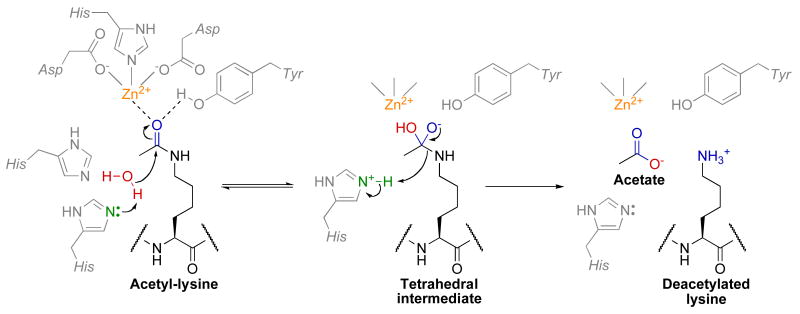
Class I/II/IV HDACs have homologous active-sites and are thought to proceed through similar catalytic mechanisms (Fig. 5) [82]. The active-site contains two active-site His-Asp dyads that are thought to work as a general acid/base catalytic pair [83-85]. Kinetic, structural, and computational data suggest a mechanism in which first an active-site metal ion and Tyr-306 (HDAC8 numbering) polarize the carbonyl by coordinating to the acetyl oxygen. For discussion purposes we will assume that this active-site metal is Zn2+, but there is significant evidence that this metal may instead be Fe2+ [86]. The Zn2+ is coordinated by Asp-178, His-180, and Asp-267 [83-85]. Structural studies have suggested that the Zn2+ also coordinates and activates the water molecule for attack of the acetyl carbonyl carbon, but theoretical calculations indicate that this water molecule is bound and activated by Asp-178, His-142, and His-143 [87]. In the first chemical step, both His-142 [83-85, 88] and His-143 [87] have been implicated as the general base that activates the water molecule for nucleophilic attack of the acetyl carbonyl carbon. Water attack results in a tetrahedral intermediate that is stabilized by Zn2+ and Tyr-306 [83-85]. Interestingly, in class IIa HDACs this tyrosine residue is replaced by histidine, which causes a significant decrease in catalytic activity compared to other HDAC classes [89]. Subsequently, the tetrahedral intermediate collapses to form acetate and lysine products, with His-143 acting as a general acid to protonate the ε-amine leaving group [83, 87, 88]. Theoretical studies indicate that simultaneous protonation of His-142 and His-143 within HDAC8 is unlikely; as a result, if His-142 is the general base in the first chemical step then His-143 must be deprotonated initially and then transfer the proton from His-142 to His-143 [88]. However, if His-143 is the general base, then the requirement of this proton transfer step is eliminated [87]. Ionizations observed from pH profiles of class I and IIb HDACs are consistent with this proposed chemical mechanism [90], but future studies are needed to unequivocally assign the individual pKa values to specific active site residues. In addition to the Zn2+ binding site, structural studies of HDAC8 have identified a second monovalent metal binding site (K+ or Na+) that is important for structural stability and may play a role in catalysis, as the metal ion is in close proximity (7.0 Å) to the active-site zinc [84]. However, future work is needed to determine the precise function of this monovalent metal binding site.
Fig. 5.
Proposed chemical mechanism of class I/II/IV HDACs.
Within class I/II/IV HDACs, future work will be necessary to determine the similarities and differences between individual HDAC enzymes in terms of their catalytic mechanism and substrate specificity. Current knowledge is limited as the majority of in vitro mechanistic studies have been performed with HDAC8 due to the difficultly in purifying other HDACs in an enzymatically active form [91]. This difficultly likely stems from the requirement of other accessory proteins for full HDAC activity.
2.4.2. Sirtuin (class III) histone deacetylases
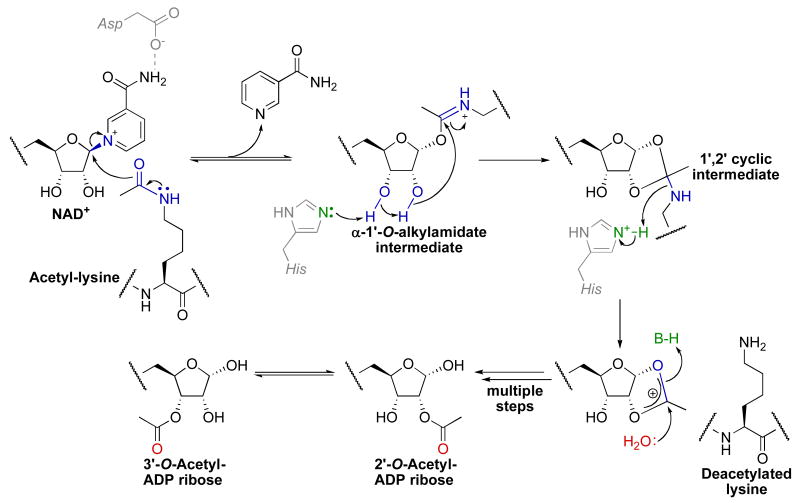
In stark contrast to class I/II/IV HDACs, sirtuins require NAD+ in a 1:1 stoichiometry with acetyl-lysine for deacetylation, producing nicotinamide and 2′-O-acetyl-ADP-ribose (OAADPr) [92, 93] in addition to deacetylated lysine as products (Fig. 6). Kinetic studies indicate that sirtuins catalyze an ordered sequential bi-ter catalytic mechanism in which acetyl-lysine substrate binds first followed by NAD+ to form a ternary complex [94]. Structural evidence suggests that acetyl-lysine binding positions the nicotinamide ring of NAD+ within a highly conserved enzyme pocket where the carboxamide of NAD+ hydrogen bonds to a conserved aspartate residue (Asp-118 in Hst2) [95, 96]. Once the ternary complex is formed, the initial chemical step involves cleavage of the nicotinamide-ribosyl bond of NAD+ and the attack of the acetyl oxygen to form the α-1′-O-alkylamidate intermediate and the first product, nicotinamide. Several lines of evidence support the formation of this O-alkylamidate intermediate. First, in the presence of both acetyl-lysine peptide and NAD+, exogenously added 14C-nicotinamide is rapidly incorporated to form 14C-NAD+ in a process termed the nicotinamide exchange reaction [97-99]. The acetyl-lysine substrate is absolutely required for efficient cleavage of the nicotinamide-ribosyl bond in both the nicotinamide exchange and deacetylation reactions. Replacement of NAD+ with 2′-deoxy-2′-fluoro-NAD+ halted the deacetylation reaction but not the nicotinamide exchange reaction [97] indicating that the first chemical step occurs at the 1′-carbon of the nicotinamide ribose. Second, oxygen labeling studies revealed transfer of an 18O label from the acetyl oxygen to the 1′-hydroxyl in OAADPr consistent with the formation of the α-1′-O-alkylamidate intermediate [93, 100]. Third, replacement of the acetyl oxygen in an acetyl-lysine histone peptide with sulfur yielded a slow sirtuin substrate that permitted mass spectral detection of a species consistent with an α-1′-S-alkylamidate intermediate [101].
Fig. 6.
Proposed chemical mechanism of class III HDACs (or sirtuins).
Although there is significant evidence for the O-alkylamidate intermediate, the mechanism of its formation is a matter of debate. In particular, some studies suggest an SN1 mechanism in which nicotinamide-ribosyl bond cleavage occurs in a distinct step from acetyl-lysine attack producing a distinct oxacarbenium intermediate [96], while other studies suggest an SN2 mechanism where nicotinamide-ribosyl bond cleavage and acetyl-lysine attack occur in the same step [102-104]. Consistent with (but not proof of) an SN2 mechanism, electron-withdrawing fluorinated acetyl-lysine analog substrates displayed significantly slower single-turnover rates of nicotinamide formation and steady-state deacetylation rates (kcat) suggesting that nicotinamide-ribosyl bond cleavage step is directly tied to the nucleophilicity of the acetyl oxygen [104]. The actual NAD+ cleavage mechanism may lie somewhere on the continuum between these two extremes. In the subsequent chemical step, a conserved histidine (His-135 in Hst2) general base activates the 2′-hydroxyl for attack of the O-alkylamidate imidate carbon to generate the 1′,2′-cyclic intermediate. Mutagenesis of this histidine to alanine disables deacetylation but not nicotinamide exchange [97], indicating acetyl-transfer and cleavage of the nicotinamide-ribosyl bond are two distinct chemical steps. This two-step mechanism is further supported by rapid quenching studies with Hst2 that demonstrated the nicotinamide–ribosyl bond is cleaved at a rate of 8 s-1, while the rate of subsequent acetyl-transfer to the 2′-hydroxyl occurred at a rate of 2 s-1 [94]. Once the 1′,2′-cyclic intermediate is formed, multiple steps involving water attack on the 1′,2′-cyclic intermediate and general acid catalyzed elimination of the ε-amine of lysine yields the final two products 2′-OAADPr and deacetylated lysine. Although 2′-OAADPr is the immediate enzymatic product, non-enzymatic interconversion between 2′-OAADPr and 3′-OAADPr is observed in solution [92, 93].
While much progress has been made to elucidate the sirtuin catalytic mechanism, further studies are needed to determine the mode of initial acetyl-lysine attack. Utilizing kinetic isotope effects, bacterial toxin ADPr-transfer reaction mechanisms have been determined to proceed through oxacarbenium/SN1 mechanisms of NAD+ cleavage [105-110]. Similar kinetic isotope effects performed with sirtuins would be valuable in determining the transitions state(s) leading up to O-alkylamidate formation. It is also intriguing that NAD+ is utilized in a deacetylation reaction that is already thermodynamically favored. One possible reason is that utilization of NAD+ allows the coupling of a metabolic parameter (NAD+ levels) to gene silencing. There is also significant evidence that the product OAADPr possesses biologically important functions [111-114].
Several reports have suggested that in addition to deacetylase activity, some sirtuins possess NAD+-dependent ADP-ribosyltransferase activity [81, 115-119]. However, these studies have yet to demonstrate that sirtuins are capable of multiple rounds of ADP-ribosylation, putting into question the biological relevance of this activity. Additionally, only limited evidence suggests that this is indeed an ADP-ribosylation reaction as the majority of evidence relies on the detection of a 32P label transferred from 32P-NAD+ to the protein. To date, the specific amino acid site modified and the nature of the linkage to the ADP-ribose portion of NAD+ remain unclear. A recent study suggests two main mechanisms for the observed ADP-ribosylation, both of which require NAD+ and an acetyl-lysine substrate [120]. In the first mechanism, acetyl-lysine and NAD+ react within the sirtuin active site to form the O-alkylamidate intermediate. It has been shown previously that this intermediate can react with exogenous alcohols suggesting that amino acid side chains may react with this intermediate to yield protein ADP-ribosylation [100]. In the second mechanism, sirtuins react with acetyl-lysine and NAD+ to form OAADPr, which then reacts non-enzymatically with a protein resulting in ADP-ribosylation. In cases where rates of both activities have been determined, deacetylase activity was three to five orders of magnitude greater than ADP-ribosylation activity [120, 121]. The large difference in catalytic efficiency as well as the observed non-enzymatic reaction of OAADPr put into question the physiological significance of ADP-ribosylation by sirtuins that also possess deacetylase activity. Interestingly, human Sirt4 and Sirt6 have been suggested to possess ADP-ribosyltransferase activity [117, 118]. No deacetylase activity has been observed for Sirt4, while there is only one report of Sirt6 displaying deacetylase activity [122]. Future studies are needed to determine if these sirtuins can catalyze bona fide ADP-ribosylation.
2.5. Other histone lysine post-translational modifications
While methylation and acetylation remain the predominant modifications present on histone lysine residues, other modifications have been reported, including ADP-ribosylation, ubiquitination, sumoylation, biotinylation, propionylation, and butyrylation [123-127]. Enzymes that are responsible for the cleavage of some of these modifications are also known, including ubiquitin hydrolases and poly(ADP-ribose)glycohydrolases. In addition, in vitro evidence indicates that class III HDACs are capable of depropionylation and debutyrylation in addition to deacetylation [104, 128, 129]. Because modifications on histone lysine residues are mutually exclusive (except perhaps monomethylation and acylation), it will be critical to determine under what conditions each of these modifications occur and what physiological response each initiates.
3. Arginine modifying enzymes
Arginine residues within histones are subject to methylation and citrullination (deimination) of their guanidinium side chains, catalyzed by protein arginine methyltransferases and protein arginine deiminases, respectively. Regulation of histone arginine methylation has been linked to a variety of important cellular processes including transcriptional regulation, translation, and DNA repair [130]. Although less characterized than histone lysine methylation, histone arginine methylation can be correlated with repression or activation depending on the site and degree of methylation [131]. The biological effects of histone citrullination are less clear, but citrullination has been suggested to be involved in terminal differentiation and apoptosis [132]. In addition to the important roles in normal cell function, protein arginine methyltransferases are overexpressed in human prostate carcinoma [133, 134] and implicated in implicated in coronary heart disease [6]. Protein arginine deiminases have been linked to multiple sclerosis [135] and rheumatoid arthritis [4]. Given these essential roles in both normal cell function as well as pathogenesis, it is important to understand the precise chemical mechanisms catalyzed by these enzymes.
3.1. Protein arginine methyltransferases
Protein arginine methyltransferases (PRMTs) catalyze methyl transfer from S-adenosyl-L-methionine (AdoMet) to the guanidinium side chain of arginine residues resulting in methylated arginine and S-adenosyl-L-homocysteine (AdoHcy). PRMTs are conserved from yeast to humans, but are not found in bacteria. Mammals have at least nine members of this family (PRMT1-9), and are separated into two main types. Whereas both types catalyze the formation of monomethylarginine (MMA), type I PRMTs (e.g. PRMT1, PRMT3, CARM1/PRMT4, PRMT6, and PRMT8) continue to form asymmetric N,N′-dimethylarginine (ADMA) and type II PRMTs (e.g. PRMT5, PRMT7, and FBXO11/PRMT9) continue of form symmetric N,N-dimethylarginine (SDMA). PRMT2 was identified by sequence homology, but no methyltransferase activity is known.
Structural evidence suggest that PRMT enzymes catalyze an ordered sequential bi-bi kinetic mechanism in which AdoMet binds prior to the arginine substrate [136]. A conserved glutamate and arginine residue (Glu-100 and Arg-54 in PRMT1) interacts with the two ribose hydroxyls and carboxylate of AdoMet, respectively. Similar to histone lysine methyltransferases, the methylsulfonium group of AdoMet is positioned at the base of a channel in which the arginine substrate binds. Two invariant glutamate residues (Glu-144 and Glu-153 in PRMT1) hydrogen bond to and position the guanidinium side chain of the arginine substrate. These hydrogen bonds are hypothesized to localize the positive charge to one terminal nitrogen of the guanidinium side chain leaving a lone pair on the other nitrogen to attack the methylsulfonium of AdoMet [137]. Both of these glutamate residues are critical for activity as mutation of either to glutamine reduces methyltransferase activity by ≥3000-fold and mutation to aspartate reduces activity by ≥10-fold [138]. Similar to histone lysine methyltransferases, methyl transfer proceeds through an SN2 mechanism (Fig. 1). The proton elimination step after methyl transfer is hypothesized to occur through a His-Asp proton relay system [137].
As mentioned above, type I PRMTs catalyze the formation of ADMA whereas type II PRMTs catalyze the formation of SDMA. In the formation of ADMA or SDMA, available in vitro data is most consistent with a partially processive mechanism in which AdoHcy release and AdoMet binding occurs on the same time scale as release of MMA containing products. In other words, the dimethyl-arginines are produced processively without release of MMA between methylation steps but not in an obligate fashion as MMA products are also released in vitro [139, 140]. However, PRMT substrates isolated in vivo are almost completely dimethylated [141-144]. The available PRMT crystal structures (rat PRMT1, rat PRMT3, yeast RMT1/hmt1, and CARM1/PRMT4) provide some insight into the processive formation of dimethyl-arginines. All structurally characterized PRMTs form a ringlike dimer, and dimer formation of PRMTs is thought necessary for methyltransferase activity [137, 138, 145, 146]. In addition, PRMT7 and PRMT8 contain two conserved catalytic core regions each containing an AdoMet binding motif essentially constituting a dimer [32, 147]. While dimerization influences AdoMet binding [32], another potential function is to allow processive production of ADMA or SDMA. Therefore, it is possible that this dimer structure exhibits processivity by allowing the product of the first methylation reaction to enter the active site of the second molecule of the dimer without releasing MMA. In support of this hypothesis, when the site of PRMT dimerization is deleted [138], replaced with alanine residues [146], or otherwise mutated [148], these PRMTs lack methyltransferase activity. Additionally, PRMT1 can also form oligomers in vitro and in vivo [138, 149], which could contribute to processive mechanisms on a larger scale. Since the N-terminal tails of histones contain multiple arginines, a PRMT1 oligomer could methylate multiple arginines simultaneously, a different form of processivity.
3.2. Histone arginine demethylases
Recently, Chang et al. reported that JMJD6, a homolog of the JHDM family of histone lysine demethylases, possesses histone arginine demethylase activity in lieu of lysine demethylase activity [150]. In particular, the authors observed a reduction in dimethylated Arg-2 of histone H3 (H3R2) and dimethylated Arg-3 of histone H4 (H4R3) with concurrent formation of 3H-formaldehyde in the presence of JMJD6. However, no change in dimethylated Arg-17 (H3R17) and Arg-26 of H3 (H3R26) was observed suggesting the effects of JMJD6 were specific. Consistent with the in vitro dimethylation results, HeLa cells overexpressing JMJD6 also revealed a reduction in the levels of dimethylated H3R2 and H4R2 but not dimethylated H3R17 or H3R26. Demethylase activity was dependent on the presence of Fe(II) and 2-oxoglutarate, and the three residues predicted to bind Fe(II). In addition, mass spectrometry of a dimethyl-arginine containing peptide incubated with JMJD6 and then immunoprecipitated with a monomethylarginine specific antibody suggested the loss of one methyl group from the dimethylated substrate. Given the predicted conservation of structural elements and key catalytic residues, JMJD6 is hypothesized to possess a similar demethylation mechanism to JHDM lysine demethylases (Fig. 3).
While this first report of arginine demethylase activity is exciting, several issues should be resolved before bone fide arginine demethylase activity is assigned. Of primary importance is establishing a catalytic rate of JMJD6 arginine demethylation that exhibits saturation with respect to the arginine substrate and comparable to JHDM family lysine demethylase activity. The requirement of an immunoprecipitated peptide prior to mass spectral analysis suggests a very slow rate of catalysis and that only a small percentage of the substrate was demethylated in the reported assays. In addition, these immunoprecipitated peptides revealed significant oxidation of two lysine residues, suggesting the oxidation activity of JMJD6 is not specific for methylated arginine residues. The physiological significance of this lysine oxidation will also need to be addressed.
3.3. Protein arginine deiminases
Protein arginine deiminases (PADs) catalyze the hydrolysis of guanidinium side chains of histone arginine residues to form citrulline and ammonia. PADs belong to a larger group of guanidinium-modifying enzymes termed the amidinotransferase superfamily and thus are related to non-peptidyl arginine deiminase (ADI) and dimethyl-arginine dimethylaminohydrolase (DDAH). To date, five human PAD homologs have been identified designated as PAD1-4 and PAD6.
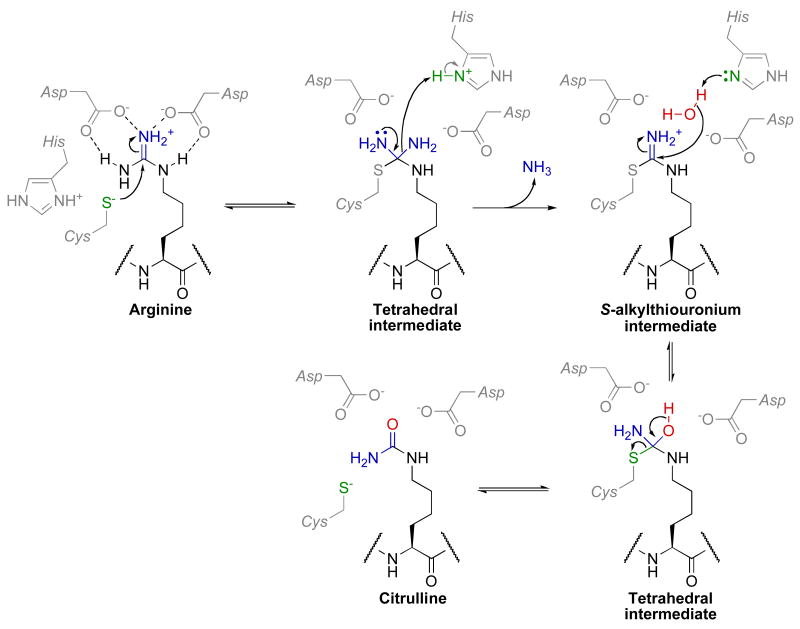
Within PAD enzymes, the guanidinium side chain is held by hydrogen bonding interactions with two conserved aspartate residues (Asp-350 and Asp-473 in PAD4) (Fig. 7). In the first chemical step, a conserved thiolate (Cys-645) whose charge is stabilized through an ion pair with His-471 undergoes nucleophilic attack at the guanidinium carbon to form a tetrahedral intermediate. This tetrahedral intermediate has been observed in a crystal structure of ADI co-crystallized with arginine [151]. Haloacetamine mechanism-based inhibitors that covalently modify Cys-645 corroborate nucleophilic attack by Cys-645 [152, 153]. The pH profile of PAD4 reveals two key ionizable groups of pKa 7.3 and 8.2 that must be deprotonated and protonated for activity, respectively. These ionizations are consistent with a reverse protonation mechanism in which Cys-645 is deprotonated and His-471 is protonated prior to substrate binding [154]. After formation of this tetrahedral intermediate, general acid catalyzed elimination of ammonia by His-471 results in the S-alkylthiouronium intermediate. A structure of the S-alkylthiouronium intermediate bound to ADI demonstrates that the hydrogen-bonding network between the two aspartate residues and reaction intermediates is maintained throughout the reaction [151, 155]. Once the S-alkylthiouronium intermediate is formed, an ordered water molecule (observed in the ADI/S-alkylthiouronium structure) attacks this intermediate passing through a tetrahedral intermediate to then generate citrulline as the second product. While the mechanism of water activation is a matter of debate, available data is most consistent with His-471 acting as the general base [154, 156, 157]. However, mechanisms in which the arginine substrate [155] or the ammonia product [157] acts as the general base have also been proposed. While most mechanistic studies on PADs have been performed with PAD4, it will be of interest to determine if other PADs catalyze similar mechanisms or possess distinct in vivo substrates.
Fig. 7.
Proposed chemical mechanism of PAD enzymes. This figure was adapted from Thompson et al. [159].
In addition, PAD enzymes are activated by calcium, with PAD4 exhibiting positive cooperativity with calcium concentrations in the mid to high micromolar range [157]. The structures of calcium-free wild-type PAD4 and a calcium-bound PAD4 C645A mutant with and without arginine substrate reveal that calcium binding induces the formation of the active site cleft and thus is essential for catalysis [156]. This suggests a potential connection between PAD activity and apoptosis when tight control of calcium concentrations is lost [158].
It has been hypothesized that PADs might serve an analogous function to protein arginine demethylases in the removal of methyl groups (demethylimination) within histones by converting methylated arginines to citrulline. This hypothesis has been strengthened by the observation that the sites of deimination by PAD4 overlap with the sites of arginine methylation within histones (see [159] and references therein). However, in vivo and in vitro studies into PAD catalyzed demethylimination are contradictory. In vivo evidence reveals an increase in histone citrulline levels that correlates with a decrease in methylated arginine levels [160, 161]. In contrast, in vitro studies using synthetic substrates containing MMA, ADMA, or SDMA are processed by PAD2, PAD3, and PAD4 with rates two to four orders of magnitude slower than substrates containing unmodified arginine [157, 159, 162]. Therefore, PAD enzymes are relatively inefficient demethyliminases in vitro. However, it is possible that accessory proteins or post-translational modifications enhance PAD activity toward methylated arginine substrates.
Consistent with a lack of demethyliminase activity, structural and kinetic data obtained with PAD4 and the related enzyme DDAH, which is highly selective for methylated arginine substrates, lend insight into the ability of PAD4 to discriminate between substrates differing by one methyl group. A structural overlay of the PAD4 and DDAH active sites reveal one notable difference in the guanidinium-binding pocket [159]. PAD4 hydrogen bonds to the two terminal nitrogens via Asp-473, whereas DDAH contains a lysine in this position forming a part of a pocket that accommodates N-methyl groups. In PAD4, this Lys-to-Asp substitution along with changes in the positioning of Val-469, Glu-474, and Asn-588 are thought to help prevent N-methyl binding within the active-site, resulting in selectivity of PAD4 for unmethylated arginine substrates.
In vivo evidence that histone H3 and H4 citrulline levels rise and fall suggests that citrullination may be a reversible modification. Variations in citrulline levels could be due to histone tail clipping, epitope occlusion, or nucleosome displacement. Alternatively, these variations could be due to an undiscovered aminotransferase that converts citrulline to arginine within histones. Precedence for such a ‘decitrullinase’ exists for the conversion of nonpeptidyl citrulline to arginine by arginosuccinate synthase and argininosuccinate lyase in the urea cycle.
4. Conclusions and Perspectives
While the last decade has seen significant progress in the diverse and complex chemical mechanisms catalyzed by histone lysine and arginine modifying enzymes, there is still much to be discovered. For example, many of these enzymes form complexes with other proteins containing histone-binding domains specific for a particular post-translational modification or contain these domains within their primary sequence. Therefore, determining how these binding domains affect catalytic activity, substrate specificity, and enzyme targeting will be essential. The combinatorial action of these binding and enzymatic domains may enact part of the “histone code”, which suggests histone modifications act in a combinatorial and sequence-dependent fashion to yield specific downstream events [163, 164].
The involvement of these histone lysine and arginine modifying enzymes in human disease indicates that study of these enzymes is useful therapeutically. With this in mind, it will be interesting to follow small-molecules targeting these enzymes currently in clinical trials to determine if these compounds will be efficacious or if more specific compounds are desired. In the latter case, further elucidation of differences in mechanism and substrate specificity between these enzymes will be important. Furthermore, it is possible that other homologous enzymes or enzymes that catalyze new chemistries on histones (e.g. conversion of citrulline to arginine) have yet to be discovered. Even though many questions still remain, it is clear that the post-translational modification of histone lysine and arginine residues is a critical signal-transduction pathway reminiscent of other well-established cascades [165].
Acknowledgments
This work was supported by National Institutes of Health grant GM065386 (to J.M.D.) and by National Institutes of Health Biotechnology Training Grant NIH 5 T32 GM08349 (to B.C.S.). We thank Christopher Berndsen, William Hallows, and Kelly Hoadley for contributive discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–96. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–51. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki A, Yamada R, Yamamoto K. Citrullination by peptidylarginine deiminase in rheumatoid arthritis. Ann N Y Acad Sci. 2007;1108:323–39. doi: 10.1196/annals.1422.034. [DOI] [PubMed] [Google Scholar]
- 5.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–32. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Niroomand F, Liu Z, Zankl A, Katus HA, Jahn L, Tiefenbacher CP. Expression of nitric oxide related enzymes in coronary heart disease. Basic Res Cardiol. 2006;101:346–53. doi: 10.1007/s00395-006-0592-5. [DOI] [PubMed] [Google Scholar]
- 7.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–68. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Milne JC, Denu JM. The Sirtuin family: therapeutic targets to treat diseases of aging. Curr Opin Chem Biol. 2008 doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 9.Anekonda TS, Reddy PH. Neuronal protection by sirtuins in Alzheimer's disease. J Neurochem. 2006;96:305–13. doi: 10.1111/j.1471-4159.2005.03492.x. [DOI] [PubMed] [Google Scholar]
- 10.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–6. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 11.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–73. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 12.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27:396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 13.Van Beekum O, Kalkhoven E. Aberrant forms of histone acetyltransferases in human disease. Subcell Biochem. 2007;41:233–62. [PubMed] [Google Scholar]
- 14.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 15.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–4. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin HG, Patnaik D, Esteve PO, Jacobsen SE, Pradhan S. Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: participation of the chromodomain in enzymatic catalysis. Biochemistry. 2006;45:3272–84. doi: 10.1021/bi051997r. [DOI] [PubMed] [Google Scholar]
- 17.Patnaik D, Chin HG, Esteve PO, Benner J, Jacobsen SE, Pradhan S. Substrate specificity and kinetic mechanism of mammalian G9a histone H3 methyltransferase. J Biol Chem. 2004;279:53248–58. doi: 10.1074/jbc.M409604200. [DOI] [PubMed] [Google Scholar]
- 18.Guo HB, Guo H. Mechanism of histone methylation catalyzed by protein lysine methyltransferase SET7/9 and origin of product specificity. Proc Natl Acad Sci U S A. 2007;104:8797–802. doi: 10.1073/pnas.0702981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derewenda ZS, Lee L, Derewenda U. The occurrence of C-H…O hydrogen bonds in proteins. J Mol Biol. 1995;252:248–62. doi: 10.1006/jmbi.1995.0492. [DOI] [PubMed] [Google Scholar]
- 20.Scheiner S, Kar T, Gu Y. Strength of the Calpha H‥O hydrogen bond of amino acid residues. J Biol Chem. 2001;276:9832–7. doi: 10.1074/jbc.M010770200. [DOI] [PubMed] [Google Scholar]
- 21.Park H, Yoon J, Seok C. Strength of Calpha-H…O=C hydrogen bonds in transmembrane proteins. J Phys Chem B. 2008;112:1041–8. doi: 10.1021/jp077285n. [DOI] [PubMed] [Google Scholar]
- 22.Scheiner S, Kar T. Effect of solvent upon CH…O hydrogen bonds with implications for protein folding. J Phys Chem B. 2005;109:3681–9. doi: 10.1021/jp0446736. [DOI] [PubMed] [Google Scholar]
- 23.Hu P, Zhang Y. Catalytic mechanism and product specificity of the histone lysine methyltransferase SET7/9: an ab initio QM/MM-FE study with multiple initial structures. J Am Chem Soc. 2006;128:1272–8. doi: 10.1021/ja056153+. [DOI] [PubMed] [Google Scholar]
- 24.Couture JF, Hauk G, Thompson MJ, Blackburn GM, Trievel RC. Catalytic roles for carbon-oxygen hydrogen bonding in SET domain lysine methyltransferases. J Biol Chem. 2006;281:19280–7. doi: 10.1074/jbc.M602257200. [DOI] [PubMed] [Google Scholar]
- 25.Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, Kim YC, Lee J, Cho Y. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. Embo J. 2003;22:292–303. doi: 10.1093/emboj/cdg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–85. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs SA, Harp JM, Devarakonda S, Kim Y, Rastinejad F, Khorasanizadeh S. The active site of the SET domain is constructed on a knot. Nat Struct Biol. 2002;9:833–8. doi: 10.1038/nsb861. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Tamaru H, Khan SI, Horton JR, Keefe LJ, Selker EU, Cheng X. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell. 2002;111:117–27. doi: 10.1016/s0092-8674(02)00999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dirk LM, Flynn EM, Dietzel K, Couture JF, Trievel RC, Houtz RL. Kinetic manifestation of processivity during multiple methylations catalyzed by SET domain protein methyltransferases. Biochemistry. 2007;46:3905–15. doi: 10.1021/bi6023644. [DOI] [PubMed] [Google Scholar]
- 30.Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ, Wilson JR. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 2005;19:1444–54. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westheimer FH. Coincidences, decarboxylation, and electrostatic effects. Tetrahedron FIELD Full Journal Title:Tetrahedron. 1995;51:3–20. [Google Scholar]
- 32.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct. 2005;34:267–94. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci. 2006;63:2755–63. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao B, Wilson JR, Gamblin SJ. SET domains and histone methylation. Curr Opin Struct Biol. 2003;13:699–705. doi: 10.1016/j.sbi.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Bruice TC. Histone lysine methyltransferase SET7/9: formation of a water channel precedes each methyl transfer. Biochemistry. 2007;46:14838–44. doi: 10.1021/bi7014579. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Bruice TC. Mechanism of Product Specificity of AdoMet Methylation Catalyzed by Lysine Methyltransferases: Transcriptional Factor p53 Methylation by Histone Lysine Methyltransferase SET7/9. Biochemistry. 2008;47:2743–8. doi: 10.1021/bi702370p. [DOI] [PubMed] [Google Scholar]
- 37.Esteve PO, Patnaik D, Chin HG, Benner J, Teitell MA, Pradhan S. Functional analysis of the N- and C-terminus of mammalian G9a histone H3 methyltransferase. Nucleic Acids Res. 2005;33:3211–23. doi: 10.1093/nar/gki635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–6. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 39.Trievel RC, Flynn EM, Houtz RL, Hurley JH. Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. Nat Struct Biol. 2003;10:545–52. doi: 10.1038/nsb946. [DOI] [PubMed] [Google Scholar]
- 40.Hu P, Wang S, Zhang Y. How Do SET-Domain Protein Lysine Methyltransferases Achieve the Methylation State Specificity? Revisited by Ab Initio QM/MM Molecular Dynamics Simulations. J Am Chem Soc. 2008 doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–8. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 42.Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–4. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 43.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–27. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–56. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 45.Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–23. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 46.Sawada K, Yang Z, Horton JR, Collins RE, Zhang X, Cheng X. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J Biol Chem. 2004;279:43296–306. doi: 10.1074/jbc.M405902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–35. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paik WK, Kim S. Enzymatic demethylation of calf thymus histones. Biochem Biophys Res Commun. 1973;51:781–8. doi: 10.1016/0006-291x(73)91383-1. [DOI] [PubMed] [Google Scholar]
- 49.Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J Biol Chem. 2007;282:20070–4. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- 50.Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579:2203–7. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 51.Forneris F, Binda C, Dall'Aglio A, Fraaije MW, Battaglioli E, Mattevi A. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. J Biol Chem. 2006;281:35289–95. doi: 10.1074/jbc.M607411200. [DOI] [PubMed] [Google Scholar]
- 52.Ghisla S. Fluorescence and optical characteristics of reduced flavins and flavoproteins. Methods Enzymol. 1980;66:360–73. doi: 10.1016/0076-6879(80)66481-7. [DOI] [PubMed] [Google Scholar]
- 53.Culhane JC, Cole PA. LSD1 and the chemistry of histone demethylation. Curr Opin Chem Biol. 2007;11:561–8. doi: 10.1016/j.cbpa.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ralph EC, Anderson MA, Cleland WW, Fitzpatrick PF. Mechanistic studies of the flavoenzyme tryptophan 2-monooxygenase: deuterium and 15N kinetic isotope effects on alanine oxidation by an L-amino acid oxidase. Biochemistry. 2006;45:15844–52. doi: 10.1021/bi061894o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silverman RB, Hoffman SJ, Catus WB., III A mechanism for mitochondrial monoamine oxidase catalyzed amine oxidation. J Am Chem Soc FIELD Full Journal Title:Journal of the American Chemical Society. 1980;102:7126–8. [Google Scholar]
- 56.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 57.Anand R, Marmorstein R. Structure and mechanism of lysine-specific demethylase enzymes. J Biol Chem. 2007;282:35425–9. doi: 10.1074/jbc.R700027200. [DOI] [PubMed] [Google Scholar]
- 58.Hausinger RP. Fe(II)/alpha -ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol FIELD Full Journal Title:Critical Reviews in Biochemistry and Molecular Biology. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 59.Ozer A, Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat Chem Biol. 2007;3:144–53. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- 60.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol. 2007;14:689–95. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 61.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, Rose NR, Offer J, Scheinost JC, Borowski T, Sundstrom M, Schofield CJ, Oppermann U. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 62.Chen Z, Zang J, Kappler J, Hong X, Crawford F, Wang Q, Lan F, Jiang C, Whetstine J, Dai S, Hansen K, Shi Y, Zhang G. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc Natl Acad Sci U S A. 2007;104:10818–23. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell FIELD Full Journal Title:Molecular Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 64.Szewczuk LM, Culhane JC, Yang M, Majumdar A, Yu H, Cole PA. Mechanistic analysis of a suicide inactivator of histone demethylase LSD1. Biochemistry. 2007;46:6892–902. doi: 10.1021/bi700414b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grzyska PK, Ryle MJ, Monterosso GR, Liu J, Ballou DP, Hausinger RP. Steady-state and transient kinetic analyses of taurine/alpha-ketoglutarate dioxygenase: effects of oxygen concentration, alternative sulfonates, and active-site variants on the FeIV-oxo intermediate. Biochemistry. 2005;44:3845–55. doi: 10.1021/bi048746n. [DOI] [PubMed] [Google Scholar]
- 66.Roy TW, Bhagwat AS. Kinetic studies of Escherichia coli AlkB using a new fluorescence-based assay for DNA demethylation. Nucleic Acids Res. 2007;35:e147. doi: 10.1093/nar/gkm1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolfe MD, Altier DJ, Stubna A, Popescu CV, Munck E, Lipscomb JD. Benzoate 1,2-dioxygenase from Pseudomonas putida: single turnover kinetics and regulation of a two-component Rieske dioxygenase. Biochemistry. 2002;41:9611–26. doi: 10.1021/bi025912n. [DOI] [PubMed] [Google Scholar]
- 68.Ryle MJ, Padmakumar R, Hausinger RP. Stopped-flow kinetic analysis of Escherichia coli taurine/alpha-ketoglutarate dioxygenase: interactions with alpha-ketoglutarate, taurine, and oxygen. Biochemistry. 1999;38:15278–86. doi: 10.1021/bi9912746. [DOI] [PubMed] [Google Scholar]
- 69.Tyihak E, Trezl L, Szende B. Formaldehyde cycle and the phases of stress syndrome. Ann N Y Acad Sci FIELD Full Journal Title:Annals of the New York Academy of Sciences. 1998;851:259–270. [Google Scholar]
- 70.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 71.Tanner KG, Langer MR, Kim Y, Denu JM. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J Biol Chem. 2000;275:22048–55. doi: 10.1074/jbc.M002893200. [DOI] [PubMed] [Google Scholar]
- 72.Tanner KG, Trievel RC, Kuo MH, Howard RM, Berger SL, Allis CD, Marmorstein R, Denu JM. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–60. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 73.Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell. 2000;6:1195–205. doi: 10.1016/s1097-2765(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 74.Yan Y, Harper S, Speicher DW, Marmorstein R. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat Struct Biol. 2002;9:862–9. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 75.Berndsen CE, Albaugh BN, Tan S, Denu JM. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry. 2007;46:623–9. doi: 10.1021/bi602513x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sagar V, Zheng W, Thompson PR, Cole PA. Bisubstrate analogue structure-activity relationships for p300 histone acetyltransferase inhibitors. Bioorg Med Chem. 2004;12:3383–90. doi: 10.1016/j.bmc.2004.03.070. [DOI] [PubMed] [Google Scholar]
- 77.Thompson PR, Kurooka H, Nakatani Y, Cole PA. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J Biol Chem. 2001;276:33721–9. doi: 10.1074/jbc.M104736200. [DOI] [PubMed] [Google Scholar]
- 78.Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–50. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- 79.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 80.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–8. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 81.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 82.Hodawadekar SC, Marmorstein R. Chemistry of acetyl transfer by histone modifying enzymes: structure, mechanism and implications for effector design. Oncogene. 2007;26:5528–40. doi: 10.1038/sj.onc.1210619. [DOI] [PubMed] [Google Scholar]
- 83.Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, Tang J, Sang BC, Verner E, Wynands R, Leahy EM, Dougan DR, Snell G, Navre M, Knuth MW, Swanson RV, McRee DE, Tari LW. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–34. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 84.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinkuhler C, Di Marco S. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci U S A. 2004;101:15064–9. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, De Francesco R, Steinkuhler C, Di Marco S. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep. 2007;8:879–84. doi: 10.1038/sj.embor.7401047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gantt SL, Gattis SG, Fierke CA. Catalytic activity and inhibition of human histone deacetylase 8 is dependent on the identity of the active site metal ion. Biochemistry. 2006;45:6170–8. doi: 10.1021/bi060212u. [DOI] [PubMed] [Google Scholar]
- 87.Corminboeuf C, Hu P, Tuckerman ME, Zhang Y. Unexpected deacetylation mechanism suggested by a density functional theory QM/MM study of histone-deacetylase-like protein. J Am Chem Soc. 2006;128:4530–1. doi: 10.1021/ja0600882. [DOI] [PubMed] [Google Scholar]
- 88.Vanommeslaeghe K, De Proft F, Loverix S, Tourwe D, Geerlings P. Theoretical study revealing the functioning of a novel combination of catalytic motifs in histone deacetylase. Bioorg Med Chem. 2005;13:3987–92. doi: 10.1016/j.bmc.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 89.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 90.Schultz BE, Misialek S, Wu J, Tang J, Conn MT, Tahilramani R, Wong L. Kinetics and comparative reactivity of human class I and class IIb histone deacetylases. Biochemistry. 2004;43:11083–91. doi: 10.1021/bi0494471. [DOI] [PubMed] [Google Scholar]
- 91.Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 92.Jackson MD, Denu JM. Structural identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD+-dependent histone/protein deacetylases. J Biol Chem. 2002;277:18535–44. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 93.Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–63. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 94.Borra MT, Langer MR, Slama JT, Denu JM. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry. 2004;43:9877–87. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- 95.Avalos JL, Boeke JD, Wolberger C. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol Cell. 2004;13:639–48. doi: 10.1016/s1097-2765(04)00082-6. [DOI] [PubMed] [Google Scholar]
- 96.Zhao K, Harshaw R, Chai X, Marmorstein R. Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases. Proc Natl Acad Sci U S A. 2004;101:8563–8. doi: 10.1073/pnas.0401057101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem. 2003;278:50985–98. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- 98.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–11. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–56. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 100.Smith BC, Denu JM. Sir2 protein deacetylases: evidence for chemical intermediates and functions of a conserved histidine. Biochemistry. 2006;45:272–82. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smith BC, Denu JM. Mechanism-based inhibition of sir2 deacetylases by thioacetyl-lysine Peptide. Biochemistry. 2007;46:14478–86. doi: 10.1021/bi7013294. [DOI] [PubMed] [Google Scholar]
- 102.Hoff KG, Avalos JL, Sens K, Wolberger C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure. 2006;14:1231–40. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 103.Khan AN, Lewis PN. Use of substrate analogs and mutagenesis to study substrate binding and catalysis in the Sir2 family of NAD-dependent protein deacetylases. J Biol Chem. 2006;281:11702–11. doi: 10.1074/jbc.M511482200. [DOI] [PubMed] [Google Scholar]
- 104.Smith BC, Denu JM. Sir2 deacetylases exhibit nucleophilic participation of acetyl-lysine in NAD+ cleavage. J Am Chem Soc. 2007;129:5802–3. doi: 10.1021/ja070162w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Berti PJ, Blanke SR, Schramm VL. Transition State Structure for the Hydrolysis of NAD+ Catalyzed by Diphtheria Toxin. Journal of the American Chemical Society. 1997;119:12079–12088. doi: 10.1021/ja971317a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Parikh SL, Schramm VL. Transition State Structure for ADP-Ribosylation of Eukaryotic Elongation Factor 2 Catalyzed by Diphtheria Toxin. Biochemistry. 2004;43:1204–1212. doi: 10.1021/bi035907z. [DOI] [PubMed] [Google Scholar]
- 107.Rising KA, Schramm VL. Transition state analysis of NAD+ hydrolysis by the cholera toxin catalytic subunit. Journal of the American Chemical Society. 1997;119:27–37. [Google Scholar]
- 108.Scheuring J, Berti PJ, Schramm VL. Transition-State Structure for the ADP-Ribosylation of Recombinant Gia1 Subunits by Pertussis Toxin. Biochemistry. 1998;37:2748–2758. doi: 10.1021/bi972594x. [DOI] [PubMed] [Google Scholar]
- 109.Scheuring J, Schramm VL. Pertussis toxin: transition state analysis for ADP-ribosylation of G-protein peptide alphai3C20. Biochemistry. 1997;36:8215–23. doi: 10.1021/bi970379a. [DOI] [PubMed] [Google Scholar]
- 110.Scheuring J, Schramm VL. Kinetic isotope effect characterization of the transition state for oxidized nicotinamide adenine dinucleotide hydrolysis by pertussis toxin. Biochemistry. 1997;36:4526–34. doi: 10.1021/bi962841h. [DOI] [PubMed] [Google Scholar]
- 111.Borra MT, O'Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J Biol Chem. 2002;277:12632–41. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- 112.Grubisha O, Rafty LA, Takanishi CL, Xu X, Tong L, Perraud AL, Scharenberg AM, Denu JM. Metabolite of SIR2 reaction modulates TRPM2 ion channel. J Biol Chem. 2006;281:14057–65. doi: 10.1074/jbc.M513741200. [DOI] [PubMed] [Google Scholar]
- 113.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–5. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 114.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–27. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 115.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–9. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 116.Garcia-Salcedo JA, Gijon P, Nolan DP, Tebabi P, Pays E. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. Embo J. 2003;22:5851–62. doi: 10.1093/emboj/cdg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 118.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–20. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 119.Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–45. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 120.Kowieski TM, Lee S, Denu JM. Acetylation-dependent ADP-ribosylation by Trypanosoma brucei Sir2. J Biol Chem. 2008;283:5317–26. doi: 10.1074/jbc.M707613200. [DOI] [PubMed] [Google Scholar]
- 121.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–82. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008 doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–9. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–41. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 125.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–69. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 127.Hassan YI, Zempleni J. Epigenetic regulation of chromatin structure and gene function by biotin. J Nutr. 2006;136:1763–5. doi: 10.1093/jn/136.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Garrity J, Gardner JG, Hawse W, Wolberger C, Escalante-Semerena JC. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J Biol Chem. 2007;282:30239–45. doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- 129.Smith BC, Denu JM. Acetyl-lysine Analog Peptides as Mechanistic Probes of Protein Deacetylases. J Biol Chem. 2007;282:37256–65. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 130.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–72. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 131.Wysocka J, Allis CD, Coonrod S. Histone arginine methylation and its dynamic regulation. Front Biosci FIELD Full Journal Title:Frontiers in Bioscience. 2006;11:344–355. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 132.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003;25:1106–18. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 133.Hong H, Kao C, Jeng MH, Eble JN, Koch MO, Gardner TA, Zhang S, Li L, Pan CX, Hu Z, MacLennan GT, Cheng L. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004;101:83–9. doi: 10.1002/cncr.20327. [DOI] [PubMed] [Google Scholar]
- 134.Majumder S, Liu Y, Ford OH, 3rd, Mohler JL, Whang YE. Involvement of arginine methyltransferase CARM1 in androgen receptor function and prostate cancer cell viability. Prostate. 2006;66:1292–301. doi: 10.1002/pros.20438. [DOI] [PubMed] [Google Scholar]
- 135.Mastronardi FG, Wood DD, Mei J, Raijmakers R, Tseveleki V, Dosch HM, Probert L, Casaccia-Bonnefil P, Moscarello MA. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J Neurosci. 2006;26:11387–96. doi: 10.1523/JNEUROSCI.3349-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. Embo J. 2007;26:4402–12. doi: 10.1038/sj.emboj.7601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang X, Zhou L, Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. Embo J. 2000;19:3509–19. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–20. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–43. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 140.Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry. 2007;46:13370–81. doi: 10.1021/bi701558t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Klein S, Carroll JA, Chen Y, Henry MF, Henry PA, Ortonowski IE, Pintucci G, Beavis RC, Burgess WH, Rifkin DB. Biochemical analysis of the arginine methylation of high molecular weight fibroblast growth factor-2. J Biol Chem. 2000;275:3150–7. doi: 10.1074/jbc.275.5.3150. [DOI] [PubMed] [Google Scholar]
- 142.Lischwe MA, Cook RG, Ahn YS, Yeoman LC, Busch H. Clustering of glycine and NG,NG-dimethylarginine in nucleolar protein C23. Biochemistry. 1985;24:6025–8. doi: 10.1021/bi00343a001. [DOI] [PubMed] [Google Scholar]
- 143.Lischwe MA, Ochs RL, Reddy R, Cook RG, Yeoman LC, Tan EM, Reichlin M, Busch H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG,NG-dimethylarginine. J Biol Chem. 1985;260:14304–10. [PubMed] [Google Scholar]
- 144.Smith JJ, Rucknagel KP, Schierhorn A, Tang J, Nemeth A, Linder M, Herschman HR, Wahle E. Unusual sites of arginine methylation in Poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J Biol Chem. 1999;274:13229–34. doi: 10.1074/jbc.274.19.13229. [DOI] [PubMed] [Google Scholar]
- 145.Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J. Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. Embo J. 2007;26:4391–401. doi: 10.1038/sj.emboj.7601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Weiss VH, McBride AE, Soriano MA, Filman DJ, Silver PA, Hogle JM. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat Struct Biol. 2000;7:1165–71. doi: 10.1038/82028. [DOI] [PubMed] [Google Scholar]
- 147.Miranda TB, Miranda M, Frankel A, Clarke S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem. 2004;279:22902–7. doi: 10.1074/jbc.M312904200. [DOI] [PubMed] [Google Scholar]
- 148.Higashimoto K, Kuhn P, Desai D, Cheng X, Xu W. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc Natl Acad Sci U S A. 2007;104:12318–23. doi: 10.1073/pnas.0610792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tang J, Gary JD, Clarke S, Herschman HR. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J Biol Chem. 1998;273:16935–45. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]
- 150.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–7. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 151.Das K, Butler GH, Kwiatkowski V, Clark AD, Jr, Yadav P, Arnold E. Crystal structures of arginine deiminase with covalent reaction intermediates; implications for catalytic mechanism. Structure. 2004;12:657–67. doi: 10.1016/j.str.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 152.Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, Sato M, Thompson PR. Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry. 2006;45:11727–36. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Luo Y, Knuckley B, Lee YH, Stallcup MR, Thompson PR. A fluoroacetamidine-based inactivator of protein arginine deiminase 4: design, synthesis, and in vitro and in vivo evaluation. J Am Chem Soc. 2006;128:1092–3. doi: 10.1021/ja0576233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Knuckley B, Bhatia M, Thompson PR. Protein arginine deiminase 4: evidence for a reverse protonation mechanism. Biochemistry. 2007;46:6578–87. doi: 10.1021/bi700095s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Galkin A, Lu X, Dunaway-Mariano D, Herzberg O. Crystal structures representing the Michaelis complex and the thiouronium reaction intermediate of Pseudomonas aeruginosa arginine deiminase. J Biol Chem. 2005;280:34080–7. doi: 10.1074/jbc.M505471200. [DOI] [PubMed] [Google Scholar]
- 156.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol. 2004;11:777–83. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 157.Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry. 2005;44:10570–82. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 158.Dong Z, Saikumar P, Weinberg JM, Venkatachalam MA. Calcium in cell injury and death. Annu Rev Pathol. 2006;1:405–34. doi: 10.1146/annurev.pathol.1.110304.100218. [DOI] [PubMed] [Google Scholar]
- 159.Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chem Biol. 2006;1:433–41. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 160.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 161.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 162.Raijmakers R, Zendman AJ, Egberts WV, Vossenaar ER, Raats J, Soede-Huijbregts C, Rutjes FP, van Veelen PA, Drijfhout JW, Pruijn GJ. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol. 2007;367:1118–29. doi: 10.1016/j.jmb.2007.01.054. [DOI] [PubMed] [Google Scholar]
- 163.Cosgrove MS, Wolberger C. How does the histone code work? Biochem Cell Biol. 2005;83:468–76. doi: 10.1139/o05-137. [DOI] [PubMed] [Google Scholar]
- 164.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 165.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–8. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]