Abstract
DNA double-strand breaks (DSBs) are critical lesions that can lead to cell death or chromosomal rearrangements. Rad51 is necessary for most mitotic and meiotic DSB repair events, although a number of RAD51-independent pathways exist. Previously, we described DSB repair in rad51Δ yeast diploids that was stimulated by a DNA region termed “facilitator of break-induced replication” (FBI) located approximately 30 kb from the site of an HO-induced DSB. Here, we demonstrate that FBI is a large inverted DNA repeat that channels repair of DSBs into the singlestrand annealing-gross chromosomal rearrangements (SSA-GCR) pathway. Further, analysis of DSB repair in rad54Δ cells allowed us to propose that the SSA-GCR repair pathway is suppressed in the presence of Rad51p. Therefore, an additional role of Rad51 might be to protect eukaryotic genomes from instabilities by preventing chromosomal rearrangements.
Keywords: recombination, yeast, double-strand DNA breaks, inverted DNA repeats, rad51Δ, rad54Δ
1.Introduction
The repair of double-strand breaks (DSBs) is an essential process in humans and other organisms. DSBs are caused by exposure of living organisms to mutagenic agents, including radiation from various sources. In addition, DSBs also frequently result from DNA breakage during various processes of DNA metabolism, such as DNA replication, transcription, and chromosomal segregation. Two major pathways, homologous recombination (HR) and non-homologous end-joining (NHEJ), have evolved to repair DSBs and are conserved from yeast to mammals [1, 2]. In the yeast Saccharomyces cerevisiae, DSBs are usually repaired by HR, which can proceed through a variety of mechanisms [3].
One HR mechanism of DSB repair is gene conversion (GC), which proceeds via invasion of two broken DNA ends into a homologous template, followed by copying of a short region of the donor chromosome in between. GC can occur with or without associated crossing-over and is considered to be a conservative HR pathway because it brings minimal change to the repaired molecule. GC depends on proteins that are members of the Rad52p epistasis group, including Rad51p, Rad52p, Rad54p, Rad55p, Rad57p, and others [4]. In yeast, Rad52p is especially important because GC is nearly abolished in its absence. Rad51p, a homolog of the bacterial RecA protein, is a second critical protein for GC, as it is needed to produce a nucleoprotein filament capable of searching for and invading homologous sequences [5]. Although the majority of known GC events are RAD51-dependent, GC events have also been observed in rad51Δ mutants. The highest level of RAD51 independency was reported for spontaneous events, wherein deletion of RAD51 led to only a 4-fold reduction of GC [6, 7]. The nature of DNA damage initiating those spontaneous events was unknown. In studies of GC repair of site-specific DSBs or of DSBs induced by DNA-damaging agents, GC was significantly more dependent on RAD51 [8]. For example, in rad51Δ mutants, ectopic GC initiated by site-specific DSBs on plasmids was reduced by two orders of magnitude [9]. Also, in studies of HO-induced DSBs, allelic GC was four orders of magnitude less successful in rad51Δ compared to RAD51 cells [10].
Other known pathways of HR DSB repair are less conservative than GC, as they cause significant change to the repaired molecule. One such pathway, single-strand annealing (SSA), proceeds by annealing between single-stranded regions of DNA [11–13]. In its classical form, SSA involves direct DNA repeats that flank a DSB. However, it has been recently suggested that SSA may also be an important player in other processes, including gene targeting, recombination between inverted DNA repeats, and inter-sister and inter-homolog recombination during repair of clustered DSBs [14, 15]. Unlike GC, SSA is RAD51-independent, as it does not require strand invasion to be mediated by a nucleoprotein filament [13].
BIR is another HR mechanism, which, according to existing models, proceeds through invasion of one broken DNA end into the intact donor molecule, followed by initiation of DNA synthesis that can proceed as far as the end of the donor chromosome [16–19]. The question of whether all forms of BIR require Rad51p remains open. BIR involving plasmid DNA was observed to be both highly efficient and largely RAD51-independent [20, 21]. Two pathways have been described for BIR between chromosomal DNA: highly efficient BIR repair that is RAD51–dependent [19, 22] and less efficient RAD51-independent BIR repair [15, 23, 24].The reported efficiencies of RAD51-independent BIR have varied greatly across studies and in various experimental systems [19, 24, 25].
In our previous studies, we described HO-induced DSB repair in rad51Δ diploids that was interpreted as RAD51-independent BIR [23, 24, 26]. These repair events were dependent upon the presence of a region called “Facilitator of BIR” (FBI), located approximately 30 kb away from the position of the HO cut site, the presence of which was required on the broken chromosome in order for BIR to occur [26]. The mechanism of FBI to facilitate BIR in rad51Δ diploids, however, remained unclear. Our later studies determined that RAD51-independent BIR frequently resulted in non-reciprocal translocations between Ty and delta sequences on the broken chromosome and homologous sequences at various ectopic sites [15]. Also, we discovered that, in our strain background, the region previously described as FBI contained two copies of Ty1 elements in inverted orientation (similar to the region referred to as FS2 in other studies; see [27] for details). Further, in our studies in haploids, as well as in rad51Δ diploids, induction of HO DSBs at MATa promoted intermolecular SSA between the inverted repeats (IRs) of Ty1 in FS2 located on different sister chromatids. These inter-sister SSA events led to formation of inverted dicentric dimers (IDs), the processing of which initiated breakage-fusion-bridge (BFB) cycles resulting in various gross chromosomal rearrangements (GCRs). The majority of these GCRs were translocations, and their formation was consistent with BIR that occurred between Ty1 or delta elements, similar to those stimulated in the presence of FBI [15]. These results led us to hypothesize that the IRs of Ty1 themselves facilitated the RAD51-independent DSB repair previously observed; i.e., the IRs of Ty1 increased the efficiency of DSB repair in rad51Δdiploids by channeling DSBs into the SSA-GCR pathway. The goal of the research presented in this paper was to test this hypothesis. We determined that the IRs of Ty1 located in the FS2 region stimulate the DSB repair previously observed in rad51Δ diploids, and that there is a direct correlation between this stimulating effect of IRs and the formation of inverted dicentric dimers (IDs). Further, based on our analysis of rad54Δ and rad54Δrad51Δ we propose that DSB repair mediated by IRs (the SSA-GCR pathway) is less efficient in the presence of Rad51p.
2. Materials and methods
2.1. Yeast strains and plasmids
The genotypes of all strains used in this study are shown in Table 1. Haploid strains AM919 [15] and YLS23 [24], and diploid strain YLS100 [24] were described previously. YLS73 and YLS36 are isogenic to AM919 and YLS23, respectively, but are rad51::LEU2, and were constructed as described in [24]. YLS105, a derivative of YLS73 containing an ars310-Δ1 deletion, was previously described [26]. Strain AM816 (a derivative of YLS73 containing a deletion of FS2) was constructed using the plasmid H9G1-1nat1. This plasmid was constructed and kindly provided by James Theis and Dr. Newlon (UMDNJ) and contained the NAT (noursothricin-resistance) gene inserted into the SwaI site of the BamH1- EcoR1 fragment of chromosome III that corresponds to positions 165317 – 172071. This plasmid is a derivative of the plasmid H9G [28] containing this region of chromosome III from a yeast strain that does not contain FS2. AM816 was constructed by transforming YLS73 with a HindIII–EcoRV restriction fragment of H9G1-1nat1, which replaced the two IRs of Ty1 at FS2 [27] with the NAT (noursothricin-resistance) gene.
Table 1.
List of strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| AM919 | MATa ade1 ura3-52 leu2-3,112 lys5 chrIII::URA3 hmlΔ::ADE1 hmrΔ::ADE1 ade3::GAL::HO | [15] |
| YLS23 | MATα-inc ade1 met13 ura3-52 leu2-3,112 thr4 trp1 | [24] |
| YLS73 | AM919, but rad51::LEU2 | [24] |
| YLS36 | YLS23, but rad51::LEU2 | [24] |
| AM816 | YLS73, but FS2::NAT (Ty1αΔ Ty1βΔ) | This study |
| AM909 | YLS73, but Ty1α::HPH(Ty1αΔ) | This study |
| YLS105 | YLS73, but ars310-Δ1 | [26] |
| AM964 | MATa ade1 ura3-52 leu2-3,112 lys5 FS2::FEN2 hmlΔ::ADE1 hmrΔ::ADE1 ade3::GAL::HO | [15] |
| AM749 | MAT α-inc ade1 lys5 ura3 leu2 thr4 trp1 hmrΔ::NAT | This study |
| AM1070 | AM749, but YER186C::pGSKU | This study |
| AM1084 | AM1070, but YER186C::FEN2 | This study |
| YLS41 | YLS23, but rad54::LEU2 | [24] |
| AM1031 | AM919, but rad54::LEU2 | This study |
| AM1071 | AM964, but rad51::KAN | This study |
| AM1101 | AM1084, but rad51::KAN | This study |
| AM1073 | AM964, but rad54::LEU2 | This study |
| AM1100 | AM1084, but rad54::KAN | This study |
| AM1281 | AM1071, but rad54::HPH | This study |
| AM1282 | AM1101, but rad54::HPH | This study |
| AM960 | YLS73 × YLS36 | [15] |
| AM990 | AM816 × YLS36 | This study |
| AM991 | AM909 × YLS36 | This study |
| AM992 | YLS105 × YLS36 | This study |
| YLS100 | EI515 × YLS23 | [24] |
| AM993 | AM1031 × YLS41 | This study |
| AM1131 | AM964 × AM1084 | This study |
| AM1111 | AM1071 × AM1101 | This study |
| AM1112 | AM1073 × AM1100 | This study |
| AM1283 | AM1281 × AM1282 | This study |
In AM909, one Ty1 (Ty1α) of FS2 was replaced by the hygromycin B-resistance gene (HPH) by crossing YLS73 with FJL012 [27]. A meiotic segregant of this diploid was backcrossed three times to YLS73 to produce a congenic derivative.
AM964, a derivative of EI515 (described in [23], isogenic to AM919) that contains IRs of FEN2 instead of FS2, was constructed as described in [15]. AM749 was obtained through tetrad dissection of the diploid strain MY006 [22] to retrieve a strain that was MAT α-inc HML ade1 lys5 ura3 leu2 thr4 trp1 hmrΔ::NAT . Next, AM1070 was constructed by transformation of AM749 with a DNA fragment generated by PCR amplification of the plasmid pGSKU [29] using primers: 5’-TCTATGCTATCACCCACCTCTGGTAAACAGCCAATTGCGGCCTTTATGATttcgtacgctgcaggtcgac-3’ and 5’-ACGAAAACTGGAATGGTCAAGCTTCGTGTTTTCAAAGACGATCTAATATTtagggataacagggtaatccgcgcgttggccgattcat-3’. Capital letters correspond to sequences upstream and downstream of YER186C, and lower case letters correspond to the sequences specific to the pGSKU cassette. Subsequently, AM1084 was constructed by transformation of AM1070 with a DNA fragment generated by PCR amplification of the FEN2 region using genomic DNA of AM919 as a template. The primers used for the amplification were as follows: 5’-ACGAAAACTGGAATGGTCAAGCTTCGTGTTTTCAAAGACGATCTAATATTctcgctccttcttggtctcttcta-3’ and 5’-TCTATGCTATCACCCACCTCTGGTAAACAGCCAATTGCGGCCTTTATGATaagctcgtttcttccatgctacta-3’. Capital letters correspond to the sequences upstream and downstream of YER186C, and lower case letters correspond to the sequences specific to the FEN2 region.
AM1071 and AM1101 are rad51::KAN derivatives of AM964 and AM1084, respectively, and were constructed using a PCR-derived KAN-MX module flanked by short terminal sequences homologous to the sequences flanking the RAD51 gene [30]. AM1073 is a rad54::LEU2 derivative of AM964 constructed by transformation with a linearized DNA fragment derived from the pJH573 (pXRAD) plasmid [7].
AM1100 is a rad54::KAN derivative of AM1084 constructed using a PCR-derived KAN-MX module flanked by short terminal sequences homologous to the sequences flanking the RAD54 gene [30].
AM1281 and AM1282 are rad54::HPH (HYG) derivatives of AM1071 and AM1101, respectively, and were constructed using a PCR-derived HPH-MX module flanked by short terminal sequences homologous to the sequences flanking the RAD54 gene [31].
2.2. Media and growth conditions
Rich medium (yeast extract-peptone-dextrose (YEPD)), synthetic complete medium with bases and amino acids omitted as specified, and sporulation medium were made as described [32]. YEP-lactate (YEP-Lac) and YEP-galactose (YEP-Gal) contained 1% yeast extract and 2% Bacto peptone media supplemented with 3.7% lactic acid or 2% galactose, respectively. Cultures were grown at 30°.
2.3. Analysis of DNA repair
To monitor repair of HO-induced DSBs, logarithmically growing cells in YEP-Lac were harvested and plated on YEP-Gal. The resulting colonies were then replica plated onto omission media to examine the heterozygous ADE1, THR4, and URA3 markers of these strains.
Cell viability following HO induction was derived by dividing the number of colony-forming units (CFUs) on YEP-Gal by the number of CFUs on YEPD. A minimum of 3 plating experiments was used to calculate averages and standard deviations for viability.
The kinetics of DSB repair were examined as described previously [22]. To arrest cells at the G2 stage, experiments were performed in the presence of nocodazole (USB) at a concentration of 0.015 mg/mL. For pulsed-field gel electrophoresis (PFGE), chromosomal plugs were prepared using the CHEF genomic DNA plug kit (Bio-Rad). PFGE was performed using genomic DNA embedded in plugs of 1% agarose. The DNA was subsequently examined by Southern analysis. The blots were probed with ADE1 DNA fragments (see below) labeled with P32. Blots were analyzed using a Molecular Dynamics PhosphorImager.
2.4. Analysis of repair outcomes
The structures of repair outcomes were analyzed by PFGE, followed by blotting and hybridization with appropriate probes. The ADE1-specific probe was a SalI fragment from pJH879 [33]. The YER187W- and THR4 -specific probes were generated by PCR amplification using genomic DNA of AM919 [15] as a template. The location of the YER187W probe on chromosome V was 566230–566646. The location of the THR4 probe on chromosome III was 216965 – 217264. The coordinates given for both of these probes were derived from SGD.
The analysis of repair outcomes containing repair products similar in size to chromosome III was performed by allele-specific PCR using the primers described below.
To amplify HML, primer 1 (specific to the sequence located centromere-distal to HML): 5’-TCTTCCTATCGGTGGTCTTGC-3’ and primer 2 (specific to HML): 5’-CACACTGCCCTATAATGACAATATC-3’ were used. Amplification of HML led to formation of a 3-kb product.
To amplify hml::ADE1, primer 1 (see above) and primer 3 (specific to ADE1): 5’-GGTTTGAAACAACCTCAAGGACTT-3’ were used. Amplification of hml::ADE1 led to formation of a 1.2-kb product.
To amplify HMR::NAT, primer 4 (specific to the region centromere proximal to HMR): 5’-ATTAA ACTGG TCCTC ACAGT TCGC-3’ and primer 5 (specific to TEF terminator sequence (a part of the hmr::NAT construct)): 5’-CCTCGACATCATCTGCCC-3’ were used. Amplification of hmr::NAT led to formation of a 2.5-kb product.
To amplify hmr::ADE1, primer 4 (see above) and primer 3 (see above) were used. Amplification of hmr::ADE1 led to formation of a 3-kb product.
2.5. Statistical analysis
All strains were analyzed for their effect on DSB repair in at least three independent plating experiments. Results from these independent experiments were pooled if it was determined that the distributions of all events were statistically similar to each other using a chisquare test (http://www.psych.ku.edu/preacher/chisq/chisq.htm). The effects of individual conditions (alterations to the FS2 region) or mutations (for example, rad54Δ versus rad51Δ) on DSB repair were determined by comparing the resulting pooled distributions of repair outcomes obtained for different strains by chi-square tests.
3. Results
3.1. IRs of Ty1 increase the efficiency of DSB repair in the absence of Rad51p
Previously, we examined repair in our diploid experimental system (YLS100), in which a single DSB was initiated by a galactose-inducible HO gene at the MATa locus of one copy of chromosome III, while the second copy of chromosome III contained an uncleavable MATα-inc allele and served as a template for DSB repair [24]. In addition, HML and HMR of the MATa containing chromosome were replaced by ADE1, and the two copies of chromosome III were heterozygous for THR4/thr4 centromere-distal to the DSB site (similar to the construct shown in Fig. 1A) [23, 24, 26] This analysis demonstrated that approximately 99% of RAD51 cells completed DSB repair via gene conversion, which led to formation of Ade+Thr+ or Ade+Thr+/− colonies, while rare BIR repair or GCR-forming repair (less than 2% of all colonies) resulted in Ade+Thr− or Ade+/−Thr− colonies [22–24] (see also data in Table 2). Interestingly, we also observed that repair occurred even in the absence of the primary strand invasion protein, Rad51p, and we determined that the efficiency of DSB repair was dependent upon FBI, a distant site located 30 kb proximal to MAT on chromosome III [15]. Later, we determined that the FBI region overlaps with two Ty1 elements described in [15], Ty1α and Ty1β (Fig. 1A; see also [27] for details of this structure), where they were called FS2. Here, we hypothesize that it was these Ty1 elements in FS2 that facilitated the RAD51-independent DSB repair we observed. This hypothesis predicted that deletion of one or both Ty1 elements of FS2 would reduce the efficiency of DSB repair in rad51Δ diploids.
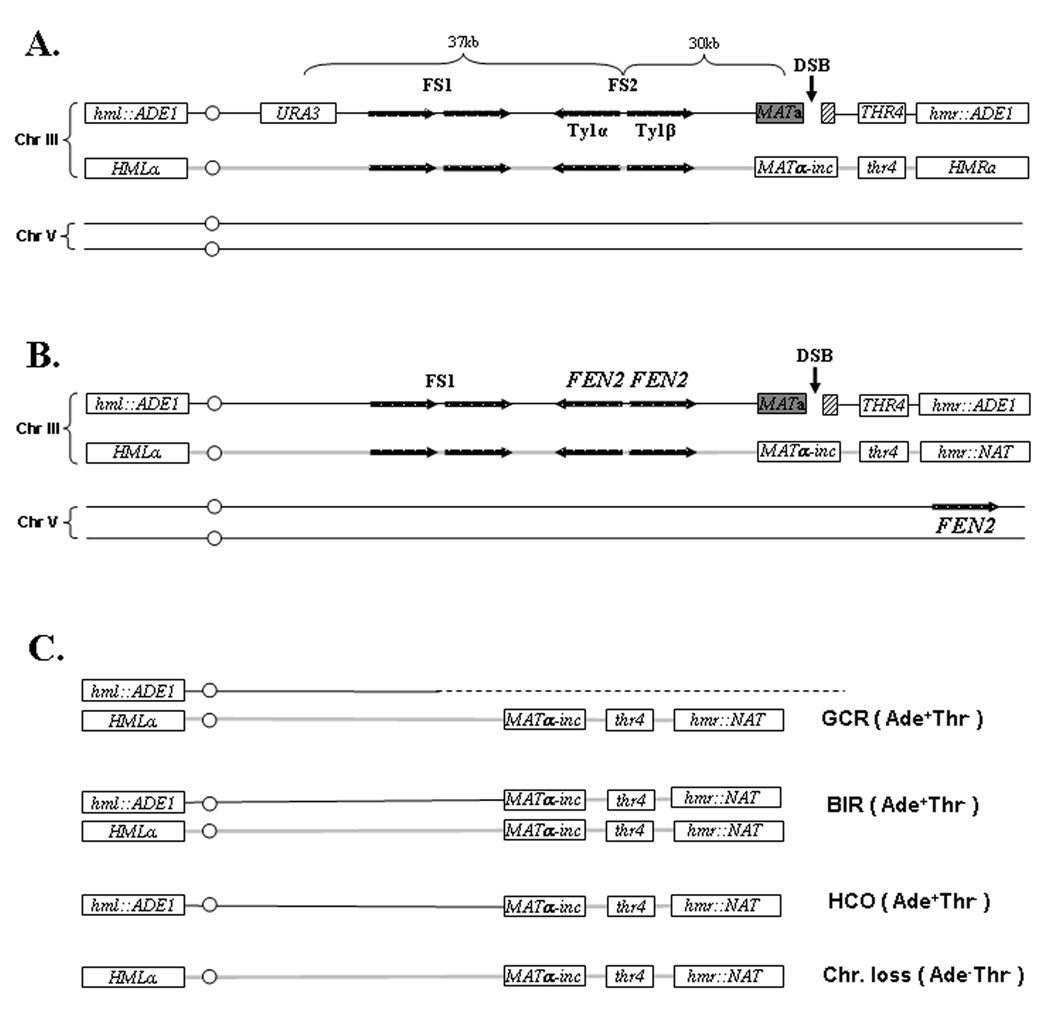
Figure 1. Arrangement of chromosome III and chromosome V markers in diploid yeast strains used in the study.
A. Diploid strains AM960 and AM993. A DSB (black vertical arrow) is induced at MATa by a galactose-inducible HO gene. HML and HMR are replaced by ADE1 in the MATa-containing chromosome (Chr) III. URA3 is inserted 133 kb from the left telomere, approximately 67 kb proximal to MATa. The other Chr III homologue contains MAT α-inc that cannot be cut by HO, has a mutant copy of thr4. FS2 consists of two Ty1 elements (labeled Ty1α and Ty1β) in inverted orientation [27] and located 30 kb proximal to MAT. FS1 consists of two Ty1 elements in direct orientation located 57 kb centromere-proximal to MATa. B. Diploid strains AM1111 and AM1112. The chromosomal arrangements in these strains are similar to those shown in A, but with an inverted repeat of FEN2 replacing FS2 in the MATa-containing chromosome. The distance between the two inverted copies of FEN2 is 1 kb. Also, HMR in this strain is replaced by NAT. In addition, one copy of Chr V contains a copy of FEN2 inserted at YER186C located approximately 13 kb from the telomere. C. The expected structure of chromosome III in various outcomes resulting from repair of HO-induced DSBs in strains shown in B. Repair outcomes for strains shown in A are identical with the exception that HMRa remains intact.
Table 2.
The effect of IRs of Ty1 on DSB repair in rad51Δ and rad54Δ diploids.
| Strain | IRa | No. Colonies tested | % Event for phenotype of coloniesb |
Viability YEP-Gal (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ade+Thr+ | Ade+Thr− | Ade+/−Thr− | Ade−Thr | |||||||
| or Ade+Thr+/− | Ura+ | Ura+/− | Ura− | Ura+/− | Ura− | |||||
|
RAD51c (YLS100) |
FS2 | 638 | 98.7 | 1.0 | 0.3 | 0.0 | 93 ± 6 | |||
|
rad51Δ (AM960) |
FS2 | 1304 | 2.2 | 7.5 | 6.3 | 4.5 | 56.9 | 9.5 | 13.1 | 92 ± 12 |
|
rad51Δ (AM990) |
Ty1αΔ Ty1βΔ |
2038 | 0.5 | 2.4 | 7.4 | 10.0 | 14.4 | 24.1 | 41.2* | 93 ± 13 |
|
rad51Δ (AM991) |
Ty1αΔ Ty1βΔ |
402 | 0.0 | 8.2 | 2.2 | 10.2 | 17.0 | 17.0 | 45.4* | 88 ± 4 |
|
rad51Δ (AM992) |
Ty1αΔ Ty1βΔ (ars310-Δ1) |
668 | 0.6 | 3.4 | 2.4 | 10.2 | 17.8 | 23.7 | 41.9* | 82 ± 6 |
|
rad54Δ (AM993) |
FS2 | 550 | 1.5 | 2.9 | 0 | 4.9 | 16.4 | 22.7 | 51.6* | 84 ± 9 |
Indicates inverted DNA repeats present in the MATa –containing copy of chromosome III undergoing DSB repair.
Some colonies were sectored for one or more nutritional markers. A colony that was half Ade+ and half Ade- is indicated as Ade+/−
Data for RAD51 diploid YLS100 (except for viability) are taken from Signon et. al. [24]. The viability for this diploid was determined in this study.
Indicates a statistically significant difference from the isogenic rad51Δ diploid strain with intact FS2 region (P<0.001).
To test this hypothesis, we employed our existing diploid system (see above and Fig. 1A) to investigate DSB repair in rad51Δ diploids that contained either intact FS2 regions (AM960; isogenic to YLS101 [24]), complete deletion of both Ty1 elements in FS2 (AM990), or deletion of Ty1α from FS2 (AM991). Genetic assays were conducted by plating AM960, AM990, or AM991 cells on a galactose-containing medium (YEP-Gal) to induce HO endonuclease, which created a DSB in MATa.
In AM960, which contained an intact FS2 region, viability associated with plating on YEP-Gal was 92%, which was not different from the viability observed in the RAD51 strain (YLS100; Table 2). In AM960, approximately 13% of colonies displayed the Ade-Thr- phenotype indicative of failure of the broken chromosome to repair (Table 2, Fig. 1C). Repair of the broken chromosome III, identified phenotypically as Ade+Thr−, occurred in approximately 18% of the colonies (Table 2). Approximately 66% of the colonies were sectored Ade+/− and fully Thr− (Table 2), which were likely to represent cases where one of two sister chromatids completed repair while the second chromatid was left unrepaired and lost in the next cell division. Alternatively, these sectored Ade+/−Thr− colonies may have resulted from cases in which the broken chromosome was replicated and inherited without repair for one or more divisions, after which some of the broken chromosomes were lost and others were repaired. Thus, the overall distribution of repair outcomes was similar to that previously observed in YLS101 [24]. Also, among cells that repaired the broken chromosome (from either Ade+Thr− or Ade+/−Thr− colonies), the majority retained the URA3 marker inserted 133 kb from HML (67 kb centromere proximal to MATa; Table 2; Fig. 1A). These repair events occurred centromere-distal to URA3. Such Ade+Ura+Thr− outcomes were obtained previously from similar experiments [15, 26] and were subjected to PFGE and microarray analyses [15]. These analyses showed that at least 90% of Ade+Ura+Thr− repair events were GCRs facilitated by recombination involving Ty1 elements located on different chromosomes [15].
Next, we analyzed rad51Δ diploids with alterations in the FS2 region on the MATa chromosome. Importantly, neither AM990, with FS2 wholly deleted (including Ty1α and Ty1β), nor AM991, with deletion of only Ty1α, showed any significant difference in viability after DSB induction compared with AM960 (Table 2). However, loss of the broken chromosome (Ade−Thr−) was increased in both of the strains containing FS2 alterations. While only 13% of rad51Δ colonies lost the broken chromosome when FS2 was intact (AM960), chromosome loss increased to 41% and 45% with full (AM990) or partial (AM991) deletion of FS2, respectively (Table 2). Similarly, in rad51Δ diploids, the frequency of partially repaired colonies (Ade+/−Thr−) was significantly reduced (P<0.001) to less than 40% in strains with either a full (AM990) or partial (AM991) deletion of FS2, while 66% of colonies showed partial repair with intact FS2 (AM960) (Table 2). The number of colonies that fully repaired the broken chromosome (Ade+Thr−) was similar among strains with altered or unaltered FS2; however, the preservation of URA3 varied. Among cells that repaired the broken chromosome (either among Ade+Thr− or Ade+/−Thr− colonies), significantly more repaired chromosomes retained URA3 in cells with intact FS2 (AM960) compared to strains with full (AM990) or partial (AM991) deletion of FS2 (P < 0.005; Table 2). These findings suggest that IRs improve the efficiency of DSB repair in rad51Δ diploids, as well as facilitate repair closer to the site of the DSB.
The finding of reduced DSB repair efficiency in rad51Δ diploids lacking FS2 were similar to previous results in a strain with full deletion of FBI (AM992, isogenic to ars310-Δ1 with a large chromosomal region surrounding ARS310 deleted [26]; Table 2). Because cells with a deletion of either FS2 or FBI are phenotypically indistinguishable (based on a similar distribution of all repair classes), we propose that the IRs of Ty1 that comprise FS2 promote DSB repair in yeast rad51Δ diploids, and that FS2 is thus responsible for the FBI activity reported previously [26].
Recently we demonstrated that induction of HO-created DSBs at MATa led to SSA between IRs of Ty1, resulting in formation of inverted dicentric dimers (IDs). Mitotic breakage of IDs initiated BFB cycles, leading to GCRs. Because this pathway, which we termed the “SSA-GCR” pathway, could be responsible for the IR-stimulated DSB repair we observed in rad51Δ diploids (AM960), we tested whether the efficiency of DSB repair in rad51Δ diploids correlated with the efficiency of ID formation. The formation of IDs was followed in time-course experiments (see Materials and Methods) in the original rad51Δ diploid strain (AM960), as well as in its derivatives containing various alterations of the FS2 region (AM990 and AM991). In rad51Δ strains containing intact FS2 (AM960), formation of the ID was found to be efficient (Fig. 2A) and to occur with kinetics similar to those previously observed in RAD51 cells [15]. However, in rad51Δ diploids with either full or partial deletion of FS2 (AM990 and AM991, respectively), as well as in diploids containing ars310-Δ1 (AM992), formation of the ID was eliminated (Fig. 2B, 2C, 2D). Therefore, we conclude that efficient formation of IDs correlates with efficient DSB repair, which supports our hypothesis that IRs promote DSB repair in rad51Δ diploids by channeling it into the SSA-GCR pathway.
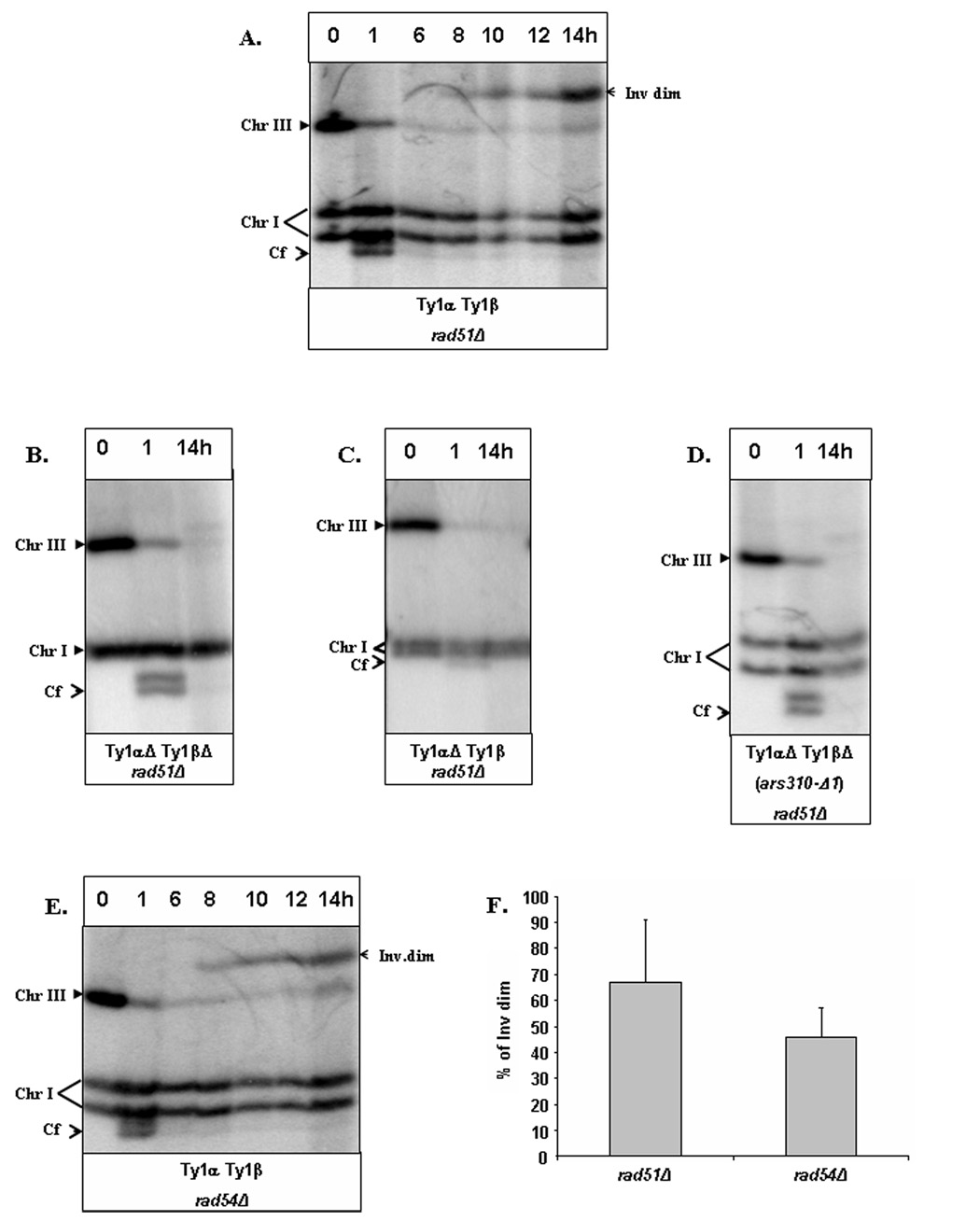
Figure 2. Formation of IDs resulting from SSA between IRs of Ty1.
Repair of DSBs in yeast diploids was examined by PFGE. A. Formation of inverted dimers (inv dim) in rad51Δ diploids (AM960) containing intact FS2. DNA was prepared for PFGE 0, 1, 6, 8, 10,12 and 14 hours after induction of DSBs at MATa in AM960. Southern blots were probed with ADE1, which hybridizes with its normal locus on chromosome (Chr) I and to ADE1 insertions on Chr III (Fig. 1A). Cf represents the chromosome fragment resulting from cleavage by HO. An additional Cf band observed one hour after addition of galactose corresponds to the processed (partially single-stranded) form of the cut fragment (VanHulle and Malkova, unpublished observation). B, C, D. Formation of inv dims is defective in strains containing disruptions of FS2. An experiment similar to the one described in A, but with fewer time points, was conducted in strains isogenic to AM960, but with deletion of one or both Ty1 elements of FS2 on the MATa-containing Chr III. E. Formation of inv dims is normal in rad54Δ diploids. An experiment similar to the one described in A was performed in strains isogenic to AM960 but RAD51 rad54Δ All strains shown in A, B, C, D, and E are isogenic to each other and contain two copies of Chr I that are different in size from each other. The varied pattern of Chr I during PFGE analysis is the result of different running conditions. F. The amount of inv dim in AM960 and AM993 strains formed at the 14-hour time point calculated as the percentage of the amount of initial (unbroken) Chr III.
3.2. IRs of FEN2 increase the efficiency of DSB repair in the absence of Rad51p
We sought to determine whether the ability to stimulate DSB repair in rad51Δ diploids was a property specific to IRs of Ty1 elements, or if it is a common property of other IRs. To this end, we analyzed DSB repair in rad51Δ diploids where FS2 on the MATa chromosome was substituted by IRs of FEN2 to create a 2-kb IR with a spacer of 1-kb (AM1111; Fig. 1B; similar to the IRs of FEN2 described in [15]). First, we observed that IDs accumulated in the presence of IRs of FEN2 (in AM1111) with kinetics similar to the kinetics of ID formation in the presence of FS2 (in AM960); i.e., between 6 and 14 hours after galactose induction of the DSB (Fig. 2A, 3A).
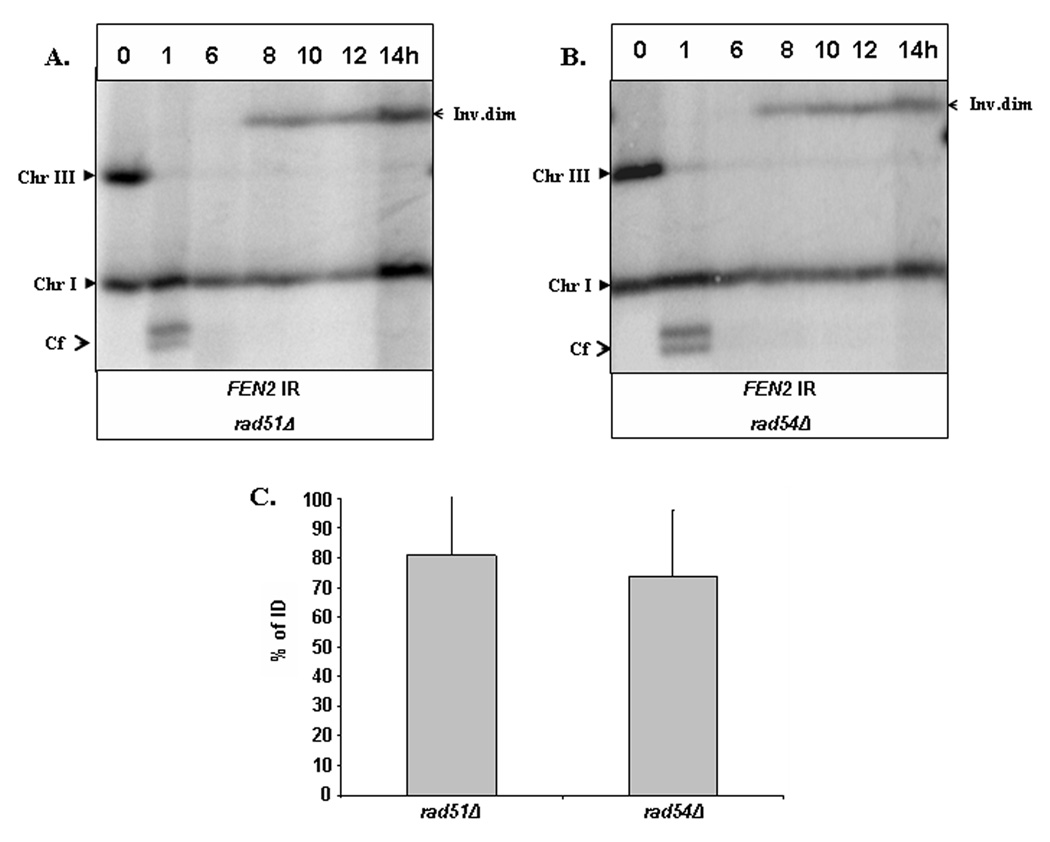
Figure 3. Formation of IDs mediated by IRs of FEN2.
A. An experiment similar to the one described in Fig. 2A was performed in a strain isogenic to AM960 that contained IRs of FEN2 instead of FS2 (AM1111). The strain shown in B is similar, but in addition is RAD51 rad54Δ (AM1112). C. The amount of inv dim in AM1111 and AM1112 formed at the 14-hour time point calculated as a percentage of the amount of initial (unbroken) Chr III.
Second, genetic analysis of DSB repair in rad51Δ diploids with IRs of FEN2 (AM1111) showed that the number of Ade−Thr− colonies indicative of chromosome loss was approximately 24% in AM1111 (Table 3), which was significantly lower (P < 0.001 for all comparisons) than the approximately 40% of chromosome loss observed in rad51Δ diploids containing no IRs (AM990, AM991, AM992; Table 2). In addition, the majority (approximately 75%) of colonies in diploids with IRs of FEN2 (AM1111) were Ade+Thr− or Ade+/−Thr− (Table 3), indicating that at least some cells repaired the broken chromosome, while less than 60% of colonies showed either of these phenotypes in the absence of IRs (AM990, AM991, AM992; P < 0.001 for all comparisons; Table 2). As with the intact or altered FS2 strains studied, the viability of rad51Δ diploids with IRs of FEN2 (AM1111) following HO induction was not significantly compromised compared to the rad51Δ strain with intact FS2 (AM960; Table 2, Table 3). Overall, the results presented here indicate that IRs of FEN2 increase the efficiency of DSB repair in rad51Δ diploids and, therefore, suggest that the FBI property seems to be common to IRs other than Ty1 elements.
Table 3.
DSB repair in rad51Δ and rad54Δ containing IRs of FEN2.
| Strain | IRa | Total Colonies analyzed | % Event for phenotype of coloniesb |
Viability YEP-Gal (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Ade+ Thr+ | Ade+/− Thr+/− | Ade+ Thr− | Ade+/−Thr− | Ade−Thr− | ||||
| RAD51 | FEN2 | 400 | 98.0 | 0.0 | 2.0 | 0.0 | 0.0 | 99 ± 6 |
| (AM1131) | ||||||||
| rad51Δ | FEN2 | 1031 | 1.3 | 1.3 | 3.6 | 71.5 | 23.6 | 90 ± 5 |
| (AM1111) | ||||||||
| rad54Δ | FEN2 | 701 | 0.4 | 0.3 | 2.7 | 29.7 | 66.9* | 94 ± 7 |
| (AM1112) | ||||||||
| rad51Δ | FEN2 | 620 | 0.7 | 0 | 2.6 | 74.8 | 21.9 | 88 ± 13 |
| rad54Δ | ||||||||
| (AM1283) | ||||||||
Indicates inverted DNA repeats present in the MATa –containing copy of chromosome IIIundergoing DSB repair
Some colonies were sectored for one or more nutritional markers. A colony that was half Ade+ and half Ade− is indicated as Ade+/−
Indicates a statistically significant difference from the isogenic rad51Δ diploid strain (P<0.001).
3.3. IR-mediated DSB repair is less efficient in rad54Δ compared to rad51Δ
The efficiency of DSB repair was assayed genetically in rad54Δ diploid cells (AM993) that contained intact FS2 and were isogenic to AM960. The number of Ade−Thr− colonies indicative of chromosome loss increased from 13% in rad51Δ (AM960) to approximately 52% in rad54Δ (AM993) (Table 2). Further, the number of events indicative of at least partial BIR repair (Ade+Thr− and Ade+/−Thr− events combined) decreased from 85% in rad51Δ to only 47% in rad54Δ (Table 2) This suggests a lower efficiency (P < 0.001) of DSB repair leading to retention of at least a part of the broken chromosome III in rad54Δ compared to rad51Δ Similar results were obtained for DSB repair in rad54Δ diploids containing IRs of FEN2 (AM1112; Table 3). In this strain, the number of Ade−Thr− colonies indicative of chromosome loss was 67%, which was significantly higher than in rad51Δ cells (AM1111), where only 24% of colonies lost the broken chromosome. Further, the number of colonies that exhibited at least partial BIR repair (Ade+Thr− and Ade+/−Thr− combined) was 32% in rad54Δ (AM1112), compared to 75% in rad51Δ (AM1111).
According to our SSA-GCR model [15], repair of DSBs in chromosomes containing IRs involves two steps: SSA between IRs leading to formation of IDs, followed by BIR-mediated telomere acquisition to stabilize the chromosome. Thus, the reduced efficiency of DSB repair observed in rad54Δ could result either from a defect in SSA or from a defect in BIR. The former possibility was investigated by analyzing ID formation in rad54Δ cells containing IRs of Ty1 (AM993) or IRs of FEN2 (AM1112), and comparing the efficiency and kinetics of ID formation in these strains with those in the corresponding rad51Δ strains (AM960 and AM1111, respectively). Time course experiments were conducted in the presence of nocodazole to prevent dissociation of IDs during mitosis (see Materials and Methods), and samples were analyzed by PFGE. IDs accumulated between 6 and 14 hours in all four strains tested (Fig. 2A, 2E, Fig. 3A, 3B). The intensity of the ID in rad54Δ diploids with intact FS2 (AM993) reached 45.6% ± 11.5% of the intensity of the unbroken chromosome III 14 hours after induction of the DSB (Fig. 2E, 2F), which was not significantly different from the intensity of the ID observed in the corresponding rad51Δ strain (AM960), which reached 67.1% ± 24.2 % (Fig. 2A, 2F). Similarly, the intensity of the ID in rad54Δ diploids with IRs of FEN2 (AM1112) reached 73.5% ± 22.8 % and was similar to the intensity observed in the corresponding rad51Δ strain (AM1111), where it reached 81% ± 25.7% (Fig. 3 A, B, and C). Based on these results, rad54Δ cells are proficient at the first step of the SSA-GCR pathway (formation of IDs by SSA), suggesting the reduced efficiency of DSB repair in these mutants most likely results from a defect in the second step of the SSA-GCR pathway, BIR-mediated chromosome stabilization.
3.4. The frequency of GCR repair outcomes is higher in rad51Δ compared to rad54Δ
Diploids of rad51Δ (AM1111) and rad54Δ (AM1112) containing IRs of FEN2 were plated on YEP-Gal, and repaired chromosomes from Ade+Thr− or Ade+/−Thr− colonies were examined by PFGE using ADE1 as a hybridization probe. The results of PFGE distinguished two classes of repair outcomes that hybridized to the ADE1 probe: 1) repair events with a normal-sized chromosome III (Fig. 4C, D), and 2) repair events that contained a chromosome III of altered size (Fig. 4A, B). The former events were likely to result from recombination between the broken chromosome and the second copy of chromosome III, while the latter events were likely to represent GCRs resulting from recombination between the broken chromosome and ectopic donors. Further analysis of a subset of rad51Δ repair outcomes (AM1111) showed that the majority were GCRs (57% of Ade+Thr+ and 89% of Ade+/−Thr−, respectively; Table 4). Conversely, GCRs represented a smaller fraction of analyzed events in rad54Δ (AM1112; 29% of Ade+Thr− and 46% of Ade+/−Thr− outcomes; Table 4). Statistical analysis showed that GCR formation among Ade+/−Thr− events was significantly decreased in rad54Δ compared to rad51Δ (P < 0.005; Table 4). (The rarity of Ade+Thr− events in both mutants precluded statistical analysis to determine whether the numerical increase in GCRs in rad51Δ compared to rad54Δ was significant.) The GCR data obtained for the Ade+/−Thr− class were used to estimate the efficiency of these GCRs among all repair outcomes. Results of these calculations (Table 4) suggest that GCRs are reduced in rad54Δ (AM1112) to only 14% of survivors compared to an estimated 64% of all survivors in rad51Δ(AM111) Thus, we conclude that the frequency of DSB repair leading to GCRs is reduced in rad54Δ compared to rad51Δ.
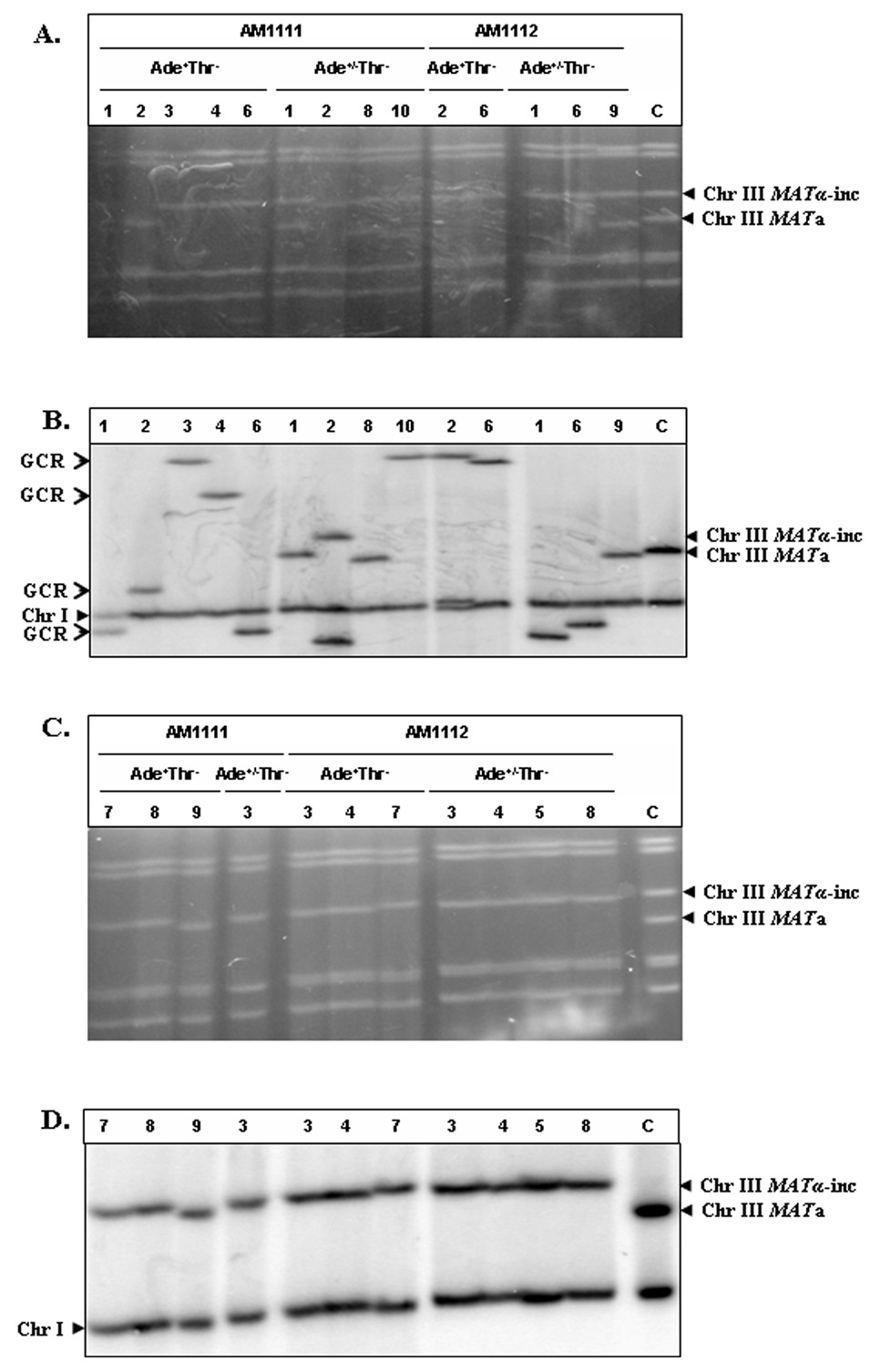
Figure 4. Structural analysis of Ade+Thr− and Ade+/−Thr− repair outcomes.
A. Ethidium bromide-stained PFGE gel of GCR outcomes (Ade+Thr− and Ade+/− Thr−) obtained from AM1111 (rad51Δ) and from AM1112 (rad54Δ). B. Southern blot analysis of the PFGE gel shown in A using an ADE1-specific probe that hybridizes to chromosome (Chr) III and to Chr I. Lane C contained DNA from AM1111 in which the HO site was not cleaved. C. Ethidium bromide-stained PFGE gel of Chr III-sized Ade+Thr− and Ade+/−Thr− repair outcomes obtained from AM1111 (rad51Δ) and from AM1112 (rad54Δ). D. Southern blot analysis of the PFGE gel shown in C using an ADE1-specific probe. Lane C contained DNA from AM1111 in which the HO site was not cleaved.
Table 4.
Analysis of repair outcomes obtained from strains containing IRs of FEN2.
| Strain | Ade+Thr− |
Ade+/−Thr− |
||||
|---|---|---|---|---|---|---|
| Total analyzeda | % GCRb | % Chr III | Total analyzeda | % GCRb (% GCR est)d |
% Chr IIIc (% Chr III est)d |
|
| rad51Δ (AM1111) | 15 | 57.0 | 43.0 | 41 | 89.0 (63.6) |
11.0 (7.9) |
| rad54Δ (AM1112) | 14 | 28.6 | 71.4 | 13 | 46.2 (13.7) |
53.8 (16.0) |
The number of colonies from the corresponding class analyzed by PFGE
The percent of GCRs among analyzed repair outcomes
The percent of chromosome III-sized outcomes among analyzed repair outcomes
The estimated percent of different classes of events (GCRs and chromosome III-sized outcomes) among all survivors
Further characterization of GCRs from rad54Δ and rad51Δ diploids containing IRs of FEN2 (AM1112 and AM1111, respectively) was conducted using a THR4-specific probe specific to the region centromere-distal to MATa (Fig. 1A). We observed that the majority (44/45 and 9/10 analyzed GCRs for rad51Δ and rad54Δ, respectively) did not hybridize to the THR4-specific probe, which suggested that these events resulted from recombination between the broken chromosome and an ectopic donor. (The two GCR outcomes that hybridized to the THR4 probe most likely resulted from recombination between the two copies of chromosome III that were initiated at a non-allelic location.)
Further, according to the SSA-GCR model, formation of all GCRs in AM1111 and AM1112 was initiated through SSA between the IRs of FEN2; thus, we expected that, at least in some cases, formation of GCRs would proceed via strand invasion of the broken end (resulting from breakage of the FEN2-mediated ID) into the copy of FEN2 located in these strains on chromosome V at YER186C (Fig. 1B). This would be followed by BIR-mediated acquisition of all sequences centromere-distal to FEN2 through the telomere to produce a GCR. Such outcomes could be identified as a repaired chromosome that hybridized to an ADE1 probe, as well as to a YER187W probe specific to the region of chromosome V centromere-distal to FEN2 (Fig. 1B). Alternatively, the strand invasion could be mediated by Ty1 or delta elements such as those located in FS1, and in this case were expected not to hybridize to the YER187W probe. Out of 45 GCRs analyzed from rad51Δ diploids (AM1111), 2 (4.4%) were determined to proceed through invasion into FEN2 on chromosome V, while 1 out of 10 (10%) GCRs analyzed from rad54Δ diploids was confirmed to have been produced through this mechanism. The remaining GCRs hybridized to the ADE1 probe, but did not hybridize to the YER187W probe. Based on our previous characterization of the SSA-GCR pathway [15], we expect that these other events were likely mediated by invasion between Ty1 or delta elements on chromosome III and other Ty1 or delta elements at ectopic locations. The higher frequency of Ty1-mediated versus FEN2- mediated invasions could possibly be explained by the higher number of Ty1 and delta repetitive elements within the genome compared with the number of copies of FEN2 in our strain.
Repair events similar in size to the normal chromosome III were further analyzed by allelespecific PCR. Four reactions were designed to amplify fragments of HML, hml::ADE1, hmr::ADE1, or hmr::NAT that were present in the initial diploids (AM1111 and AM1112) in heterozygous combinations (see Materials and Methods and Table 1 for details). When the DNA purified from the original diploids was used as templates, the reactions led to formation of products consistent with the presence of all four alleles. This same result was expected in cases of DSB repair through inter-homolog GC. Repair outcomes resulting from inter-homolog or ectopic BIR were expected to preserve heterozygosity at HML (hml::ADE1/HML), but to become homo- or hemizygous for hmr::NAT, respectively, thus producing bands for only three of the four allele-specific PCR reactions. Repair outcomes resulting from half-crossovers or chromosome loss were expected to contain only one of two HML and one of two HMR alleles and, therefore, to produce only two out of four PCR products. Analysis of five chomosome III-sized repair outcomes obtained from rad51Δ (AM1111; four Ade+Thr− and one Ade+/−Thr−) demonstrated that one of them (Ade+Thr−) was consistent with BIR (hml::ADE1/HML heterozygous and homo-/hemizygous for hmr::NAT). The other four events contained only hml::ADE1 and hmr::NAT, which indicated formation through a half-crossover between the two copies of chromosome III. Analysis of 10 repair outcomes from rad54Δ (AM1112; four Ade+Thr− and six Ade+/−Thr−) showed all 10 outcomes to contain only hml::ADE1 and hmr::NAT, which was consistent with formation of a half-crossover.
3.5. DSB repair in rad54Δ is compromised due to presence of Rad51p
The results presented above suggested that rad54Δ cells are proficient at the first step of the SSA-GCR pathway (formation of IDs by SSA), but have a problem to produce repair outcomes that require BIR using Ty1 or delta sequences as sites of invasion. At least two explanations of this result are possible. The first possibility is that Rad54p is directly required for strand invasion in the absence of Rad51p. The second possibility is that the chromosomal stabilization observed in rad51Δ proceeds through a repair pathway that is normally suppressed by Rad51p. To distinguish between these two possibilities, we analyzed DSB repair in rad51Δrad54Δ homozygous diploids (AM1283, Table 3). We observed that the efficiencies of DSB repair (the sum of Ade+Thr− and Ade+/−Thr−) and chromosome loss (Ade−Thr−) in rad51Δrad54Δ double mutants were similar to the efficiencies observed in rad51Δ strains, but significantly different from the respective efficiencies observed in rad54Δ (P < 0.001). Overall, these results support the hypothesis that the presence of Rad51p suppresses DSB repair in rad54Δ
4. Discussion
4.1. IRs increase efficiency of DSB repair in the absence of Rad51p
The data presented in this paper demonstrate that IRs located in the vicinity of a DSB promote its repair. This finding explains the nature of a cis-factor (FBI) that was described in our previous work using the same experimental system [26]. We propose that IRs channel DSB repair into the SSA-GCR pathway [15], which is initiated by SSA between IRs located on different sister chromatids, leading to formation of IDs. Mitotic breakage of IDs is followed by BFB cycles that create GCRs resulting from recombination between Ty1 and delta elements. In our system, efficient DSB repair correlated with the efficient formation of IDs, which underscores the importance of the SSA step for successful SSA-GCR repair. Overall, we believe that DSB repair is usually inefficient in rad51Δ cells, and broken chromosomes are frequently degraded and lost. However, initiation of SSA between IRs resulting in formation of IDs stabilizes chromosomal fragments, at least temporarily. After chromosome stabilization, BFB cycles can maintain broken chromosomes through cell generations, thus increasing their chances for successful repair through RAD51-independent recombination (see below).
4.2. The SSA-GCR repair pathway is suppressed by Rad51p
The results presented in this paper clarify the first step of DSB repair observed in rad51Δ diploids; i.e., SSA between IRs. However, questions remain concerning the final steps of the process leading to chromosome stabilization in the absence of Rad51p. According to our original hypothesis, this stabilization occurs via RAD51-independent BIR, which often takes place at positions of Ty1 and delta elements located at various places in the genome. Here we observed that repair was less efficient in rad54Δ than in rad51Δ despite the normal level of ID formation in both strains. Moreover, DSB repair in rad51Δrad54Δ was similar to repair in rad51ΔBased on these results, we propose that the chromosomal stabilization observed in rad51Δ proceeds through a pathway different from the one used by cells in the presence of Rad51p in the following ways. First, the RAD51-independent pathway does not require Rad54p. Second, in the absence of Rad51p, strand invasion is allowed to proceed at shorter regions of homology at ectopic positions (at Ty1 or delta elements), which is discouraged when strand invasion is mediated by a Rad51p filament. Alternatively, repair in rad51Δ diploids might proceed by SSA involving regions of the genome that transiently become single-stranded as a result of transcription, replication, or various other processes. Repair events of this kind might be inhibited by Rad51p. In support of this hypothesis are several studies that document suppression of SSA by Rad51p [14, 34].
When Rad51p is present, the majority of chromosomal stabilization is channeled into the RAD51-dependent pathway, a pathway that is also RAD54-dependent. In this pathway, Rad54p has been proposed to play many roles, including stabilization of the Rad51p filament [35], stimulation of Rad51p-mediated strand invasion [35–41], chromatin remodeling [42–44], disassembly of various protein-DNA complexes [45], and branch migration [46]. We propose that the decreased repair efficiency in rad54Δ in our system is explained by an accumulation of unprocessed recombination intermediates that require Rad54p for their processing. This is consistent with a “late” recombination defect documented in vivo for rad54Δ by several other studies [47, 48].
4.3. Analysis of rad51Δ and rad54Δ repair outcomes
Based on our previous analysis of GCRs in rad51Δ diploids, it appeared that the same Ty1 elements that participated in SSA also mediated recombination with Ty1 repeats located on other chromosomes [15]. Here we tested whether this could be generalized to non-Ty1 IRs and came to a negative conclusion. In diploids where SSA occurred between IRs of FEN2, we observed that the resultant GCRs rarely occurred by recombination involving FEN2; rather, they occurred more often through recombination between Ty and delta elements located on chromosome III and other chromosomes. This could suggest that the efficiency of RAD51-independent recombination is directly affected by copy number, as we observed a significantly higher amount of recombination in rad51Δ cells between repetitive DNA elements compared to recombination events between unique DNA sequences.
The second class of repair outcomes resulted from recombination between the homologous copies of chromosome III. Among rad51Δ outcomes, inter-homolog repair was extremely rare and precluded a thorough analysis to determine the actual distribution of the kinds of inter-homolog recombination events. However, the analysis performed did confirm that both BIR and half-crossover events occurred. In comparison, inter-homolog repair outcomes from rad54Δ were easier to obtain because they were slightly more frequent compared to rad51Δ (Table 4), and the frequency of GCR events was also lower. All of the analyzed events from these cells were confirmed as half-crossovers. Half-crossovers were previously reported in rad52Δ cells [49], including diploids isogenic to the strains described here, wherein HO-induced half-crossovers were observed at frequencies close to 1% [23]. Recently, we observed that interruption of break-induced replication in pol32Δ mutants increased the frequency of halfcrossovers to up to 20% [50]. Because the defect of rad54Δ is also localized to the stage of HR repair following strand invasion [48], one might predict that it should also increase half-crossovers in comparison to rad51Δ. While our data are supportive of this hypothesis, a thorough comparison of rad51Δ and rad54Δ in a system that makes it easier to follow inter-homolog repair outcomes by eliminating competition with the SSA-GCR pathway (i.e., a system without DSB-proximal IRs) is required to strengthen this argument.
Acknowledgements
We thank James E. Haber for his support and suggestions (the project was originally started in his laboratory, where it was supported by NIH grant GM20056 to J.E.H.). We are thankful to Carol Newlon and James Theis for their analysis of the structure of chromosome III and for helpful discussion pertinent to this subject. This work was supported by an IUPUI RSFG grant and NIH grant 1R15GM074657-01A1 (including 3R15GM074657-01A1S1) to A.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 2.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 3.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 5.Colaiacovo MP, Paques F, Haber JE. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics. 1999;151:1409–1423. doi: 10.1093/genetics/151.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rattray AJ, Symington LS. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics. 1994;138:587–595. doi: 10.1093/genetics/138.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rattray AJ, Symington LS. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fasullo M, Giallanza P, Dong Z, Cera C, Bennett T. Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology-directed translocations. Genetics. 2001;158:959–972. doi: 10.1093/genetics/158.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartsch S, Kang LE, Symington LS. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol Cell Biol. 2000;20:1194–1205. doi: 10.1128/mcb.20.4.1194-1205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pohl TJ, Nickoloff JA. Rad51-independent interchromosomal double-strand break repair by gene conversion requires Rad52 but not Rad55, Rad57, or Dmc1. Mol Cell Biol. 2008;28:897–906. doi: 10.1128/MCB.00524-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishman-Lobell J, Rudin N, Haber JE. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugawara N, Haber JE. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storici F, Snipe JR, Chan GK, Gordenin DA, Resnick MA. Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol Cell Biol. 2006;26:7645–7657. doi: 10.1128/MCB.00672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanHulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, McCullough C, Bellinger M, Lobachev K, Petes TD, Malkova A. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol. 2007;27:2601–2614. doi: 10.1128/MCB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrow DM, Connelly C, Hieter P. "Break copy" duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosco G, Haber JE. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics. 1998;150:1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 19.Davis AP, Symington LS. RAD51-dependent break-induced replication in yeast. Mol Cell Biol. 2004;24:2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol Cell Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang LE, Symington LS. Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:9162–9172. doi: 10.1128/mcb.20.24.9162-9172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol. 2005;25:933–944. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci U S A. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Signon L, Malkova A, Naylor ML, Klein H, Haber JE. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol. 2001;21:2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galgoczy DJ, Toczyski DP. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol Cell Biol. 2001;21:1710–1718. doi: 10.1128/MCB.21.5.1710-1718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malkova A, Signon L, Schaefer CB, Naylor ML, Theis JF, Newlon CS, Haber JE. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev. 2001;15:1055–1060. doi: 10.1101/gad.875901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 28.Newlon CS, Lipchitz LR, Collins I, Deshpande A, Devenish RJ, Green RP, Klein HL, Palzkill TG, Ren RB, Synn S, et al. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and location of ARS elements. Genetics. 1991;129:343–357. doi: 10.1093/genetics/129.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 30.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 31.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 32.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- 33.Leung W, Malkova A, Haber JE. Gene targeting by linear duplex DNA frequently occurs by assimilation of a single strand that is subject to preferential mismatch correction. Proc Natl Acad Sci U S A. 1997;94:6851–6856. doi: 10.1073/pnas.94.13.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. Rad51 protein controls Rad52-mediated DNA annealing. J Biol Chem. 2008;283:14883–14892. doi: 10.1074/jbc.M801097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazin AV, Alexeev AA, Kowalczykowski SC. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J Biol Chem. 2003;278:14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- 36.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 37.Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 38.Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 39.Sigurdsson S, Van Komen S, Petukhova G, Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J Biol Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- 40.Van Komen S, Petukhova G, Sigurdsson S, Sung P. Functional cross-talk among Rad51, Rad54, and replication protein A in heteroduplex DNA joint formation. J Biol Chem. 2002;277:43578–43587. doi: 10.1074/jbc.M205864200. [DOI] [PubMed] [Google Scholar]
- 41.Mazin AV, Bornarth CJ, Solinger JA, Heyer WD, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 42.Jaskelioff M, Van Komen S, Krebs JE, Sung P, Peterson CL. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J Biol Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 43.Alexiadis V, Kadonaga JT. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 2002;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 45.Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- 46.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 47.Houston PL, Broach JR. The dynamics of homologous pairing during mating type interconversion in budding yeast. PLoS Genet. 2006;2:896–905. doi: 10.1371/journal.pgen.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 49.Haber JE, Hearn M. Rad52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics. 1985;111:7–22. doi: 10.1093/genetics/111.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deem A, Barker K, VanHulle K, Downing B, Vayl A, Malkova A. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics. 2008;179 doi: 10.1534/genetics.108.087940. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]