Summary
In lower eukaryotes, Sir2 serves as a histone deacetylase and is implicated in chromatin silencing, longevity and genome stability. Here we mutated the SIRT1 gene, a homolog of yeast Sir2, in mice to study its function. We showed that a majority of SIRT1-null embryos died between E9.5–E14.5, displaying altered histone modification, impaired DNA damage response, and reduced ability to repair DNA damage. We demonstrated that SIRT1+/−;p53+/− mice developed tumors in multiple tissues, whereas activation of SIRT1 by resveratrol treatment reduced tumorigenesis. Finally, we showed that many human cancers exhibited reduced level of SIRT1 than their normal controls. Thus, SIRT1 acts as a tumor suppressor through its role in DNA damage response, genome integrity, and tumor suppression.
Significance
SIRT1 has diverse roles in various biological processes, including caloric restriction that causes changes in glucose metabolism and lifespan. The role of SIRT1 in cancer is currently under debate due to some recent different findings. It is known that calorie restriction, which activates SIRT1, extends lifespan and inhibits tumorigenesis. On the other hand, SIRT1 deacetylates p53 to decrease its activity. It was therefore hypothesized increased SIRT1 activity, although it extends lifespan, may elevate cancer risk. Here we demonstrate SIRT1 plays an important role in DNA damage response and genome integrity by maintaining proper chromatin structure and DNA damage repair foci formation. We further show that SIRT1 serves as a tumor suppressor in mice and in some types of human cancers.
Keywords: γH2AX, Brca1, DNA damage repair, tumorigenesis
Introduction
In Saccharomyces cerevisiae, Sir2 maintains genomic integrity in multiple ways. As a NAD+ dependent histone deacetylase, Sir2 was reported to regulate chromatin silencing (Blander and Guarente, 2004; Guarente, 2000). Sir2 is required for establishment and maintenance of telomeric heterochromatin (Denu, 2003; Gasser and Cockell, 2001). When over-expressed, Sir2 has been shown to extend lifespan in both budding yeast and Drosophila (Guarante, 2005). Previous reports also demonstrated that Sir2 is involved in DNA damage repair (McAinsh et al., 1999; Mills et al., 1999; Tsukamoto et al., 1997). A protein complex containing Sir2 was reported to translocate to DNA double strand breaks (McAinsh et al., 1999; Mills et al., 1999; Tsukamoto et al., 1997). In addition, Sir2-deficient yeast strains displayed defects in the Non-Homologous End-Joining (NHEJ) pathway of DNA double strand break repair (Guarente, 2000).
The mammalian Sir2 family consists of 7 members (SIRT1-7) of NAD+ dependent type III histone and protein deacetylases. These proteins share a catalytic domain of about 275 amino acids, and are primarily localized in the nucleus (SIRT1, 6 and 7), the mitochondria (SIRT3, 4 and 5), and cytoplasm (SIRT2), respectively (reviewed in (Blander and Guarente, 2004; Saunders and Verdin, 2007)). It has been shown that SIRT2 plays a role in mitotic checkpoint to arrest cells if DNA damage is detected. SIRT3 enhances acetyl-coA production by deacetylating acetyl-CoA synthetase 2. SIRT4 represses glutamate dehydrogenase to suppress insulin signaling through its ADP-ribosylase activity. SIRT6 has both ADP-ribosylase and deacetylase activity and plays a role in base excision repair. SIRT7 is involved in transcription of rRNA genes through its interaction with RNA polymerase-I (reviewed in (Baur et al., 2006; Haigis and Guarente, 2006; Saunders and Verdin, 2007; Vaquero et al., 2007)). The most extensive study, however, is conducted toward functions of SIRT1, which is the funding member of this Sirtuin family and the mammalian orthologue of yeast Sir2. SIRT1 modifies histones through deacetylation of K9 in histone H3 (H3K9) and K16 in histone H4 (H4K16) and also deacetylates many non-histone proteins that are involved in cell growth, apoptosis, neuronal protection, adaptation to calorie restriction, organ metabolism and function, cell senescence, and tumorigensis (Baur et al., 2006; Haigis and Guarente, 2006; Saunders and Verdin, 2007; Vaquero et al., 2007). A most notable target of SIRT1 is p53 that plays a critical role in cell cycle checkpoint regulation, apoptosis and tumor suppression. It has been shown that over-expression of SIRT1 deacetylates p53, leading to the suppression of p53 activity (Chen et al., 2005; Kim et al., 2008; Zhao et al., 2008).
Functions of SIRT1 were also studied at the whole organism level using targeted gene disruption. However, SIRT1 mutant mice generated by different targeting strategies exhibited distinct phenotypes (Cheng et al., 2003). Approximately 50% mice, which carry a truncation mutation through a targeted replacement of exons 5 and 6 with a hygromycin gene, died at early postnatal stages, while the remaining mice were smaller but survived to adulthood (McBurney et al., 2003b). No global defects in gene silencing in the mutant mice were detected (McBurney et al., 2003a). On the other hand, the majority (90%) of SIRT1 mutant animals carrying a deletion of exon 4 died perinatally exhibiting developmental defects of retina and heart, and the remaining 10% of the mutants were still surviving at weaning (Cheng et al., 2003). Because SIRT1 mutant cells displayed p53 hyperacetylation upon DNA damage and increased ionizing radiation-induced apoptosis in thymocytes, it was suspected that the SIRT1 deficiency might activate p53, leading to the lethality of mutant mice (Cheng et al., 2003). While these studies revealed involvement of SIRT1 in mammalian development, they have not been able to recapitulate yeast Sir2 functions in gene silencing, DNA damage repair and longevity. Of note, the role of SIRT1 in tumorigenesis is currently under debate due to some recent discrepancies. For example, the observation that SIRT1 deacetylates p53 to decrease its activity has led to the hypothesis that increased SIRT1 activity may elevate cancer risk in mammals (Chen et al., 2005). On the other hand, it was recently demonstrated that increased expression of SIRT1 reduces colon cancer formation in APCmin/+ mouse model (Firestein et al., 2008). Furthermore, resveratrol, which activates SIRT1 activity (Howitz et al., 2003), exhibits chemopreventive activity against various cancers including leukemia (Li et al., 2007), DMBA induced mammary tumors in rat (Whitsett et al., 2006), skin cancer (Aziz et al., 2005), and prostate cancer (Harper et al., 2007).
In this study, we created a SIRT1 mutant mouse model and studied the role of SIRT1 in DNA damage response and tumorigenesis. Our data provide strong evidence that mammalian SIRT1 plays an important role in DNA damage repair, genomic integrity, and inhibition of tumorigenesis.
Results
Generation of SIRT1 knockout mice
SIRT1 gene was mutated by deleting exons 5 and 6, which encodes a part of the catalytic domain (Supplementary Fig. 1A–D). Western blot analysis using an antibody to the N-terminus of SIRT1 revealed that there was no truncated protein in embryos homozygous for the mutation (Supplementary Fig. 1D), suggesting that the SIRT1 mutation we created is a candidate null mutation.
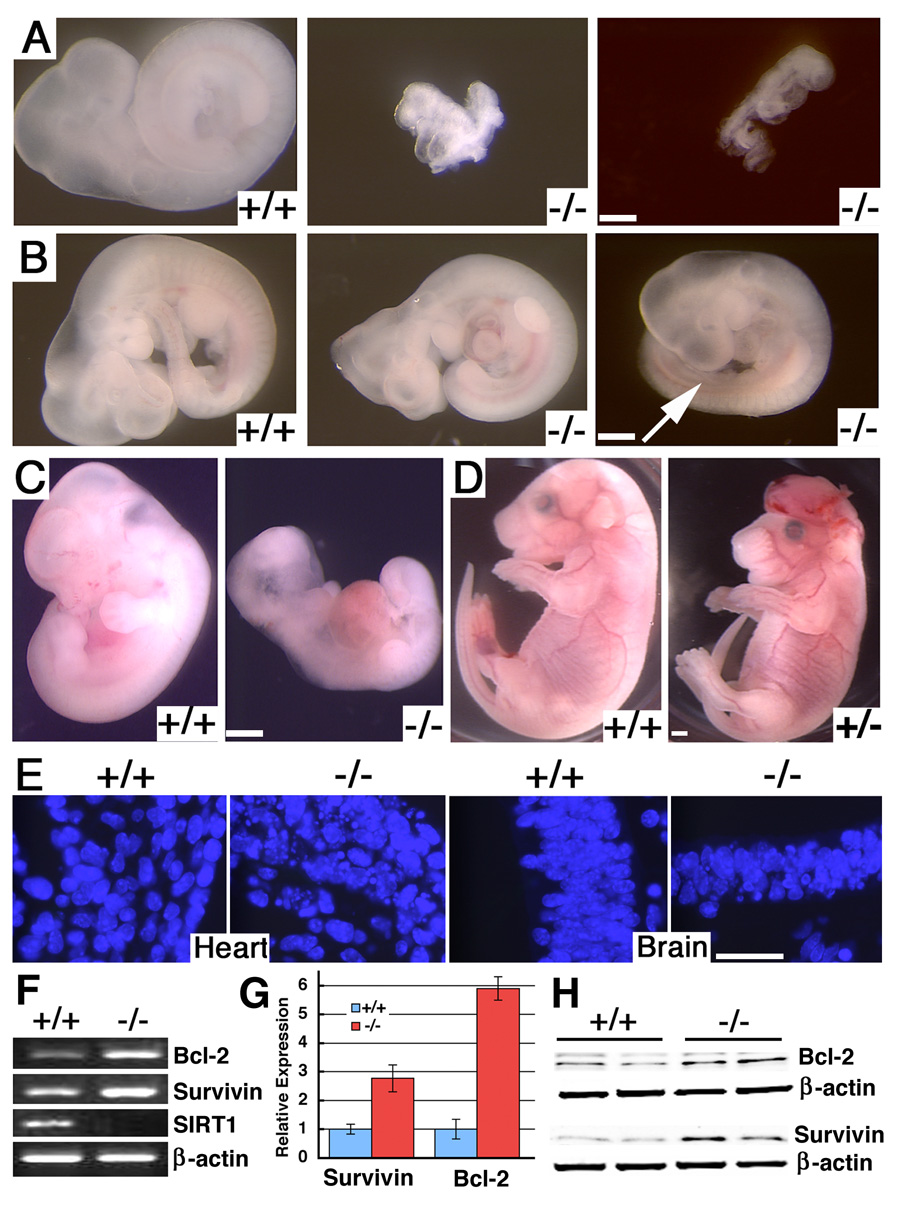
Previous investigations showed that mice carrying mutations of SIRT1 died at perinatal stages up to several months in adulthood (Cheng et al., 2003; McBurney et al., 2003a). However, we found that embryos homozygous for the mutation (SirT1−/−) started to die at embryonic (E) day 9.5 (Fig. 1A, Supplementary Table 1). Abnormal SirT1−/− embryos were also found at later stages of development (Fig. 1B, C). There was a significant decrease in SirT1−/− embryos at E14.5-16.5 and no homozygous embryos were found at E17.5–E18.5 among 69 embryos dissected. Some SirT1+/− embryos also exhibited exencenphaly (Fig. 1D). After analyzing 442 offspring, we found that the survival rate of SIRT1 homozygous mutant mice was about 1% in a 129SVEV/FVB and 8.5% in a 129SVEV/FVB/Black Swiss background (Supplementary Table 1). These observations indicated that the SirT1−/− mice exhibited more severe phenotypes than those reported previously (Cheng et al., 2003; McBurney et al., 2003b).
Figure 1. Deletion of SIRT1 resulted in embryonic lethality.
(A) E9.5 wild type (+/+) and SIRT1 null embryos (−/−). Both mutant embryos are arrested at early E8, and one (middle) has not finished turning yet. (B) E11.25 +/+ and −/− embryos. Both −/− embryos are smaller with abnormal shape of the head, or lack of hindlimb bud (arrow). (C) E12.5 +/+ and −/− embryo. (D) E18.5 +/+ and SIRT1+/− embryos. (E) Dapi staining on brain histological sections of E11.5 +/+ and −/− embryo. (F–H) Expression of Bcl2 and Survivin in E11.5 +/+ and −/− embryos revealed by regular (F), realtime RT-PCR analysis (average ± SD) (G), and Western blot analysis (H). Bars: 500 µm for A–D, and 50 µm for E.
SIRT1 deficiency results in accumulation of cells in the early phase of the mitosis
In order to understand the phenotype, we further analyzed SirT1−/− mice during embryonic development. Most E10.5–12.5 SirT1−/− embryos were abnormally small and DAPI staining showed nuclear fragmentation and cell death (Fig. 1E). However, these dead cells were negative for TUNEL assay, suggesting that the cell death might not be caused by typical apoptosis. Further analysis of the embryos demonstrated that some anti-apoptotic genes, such as Bcl-2 and Survivin were elevated in SIRT1-null embryos (Fig. 1F–H), which might have protected SirT1−/− cells from apoptosis. We next analyzed cell proliferation using BrdU labeling (Supplementary Fig. 2A–C) and phosphorylated histone H3 staining, a marker for cells at the early phase of mitosis (Supplementary Fig. 2D–F). SirT1−/− embryos had 15–20% more BrdU labeling at E10.5 and E11.5 than control embryos, but the labeling returned to normal level at E12.5 (Supplementary Fig. 2C). During the same time period, SirT1−/− embryos had 1.5–2 fold higher number of cells that are positive for phosphorylated histone H3 compared with wild type controls (Supplementary Fig. 2F). Histone H3 phosphorylation occurs in the prophase, which is normally for a short duration. The marked increase of phosphorylated histone H3 suggests that the SIRT1 deficiency caused abnormal accumulation of cells in the early phases of the mitosis.
SIRT1 deficiency causes incomplete chromosome condensation and chromosome instability
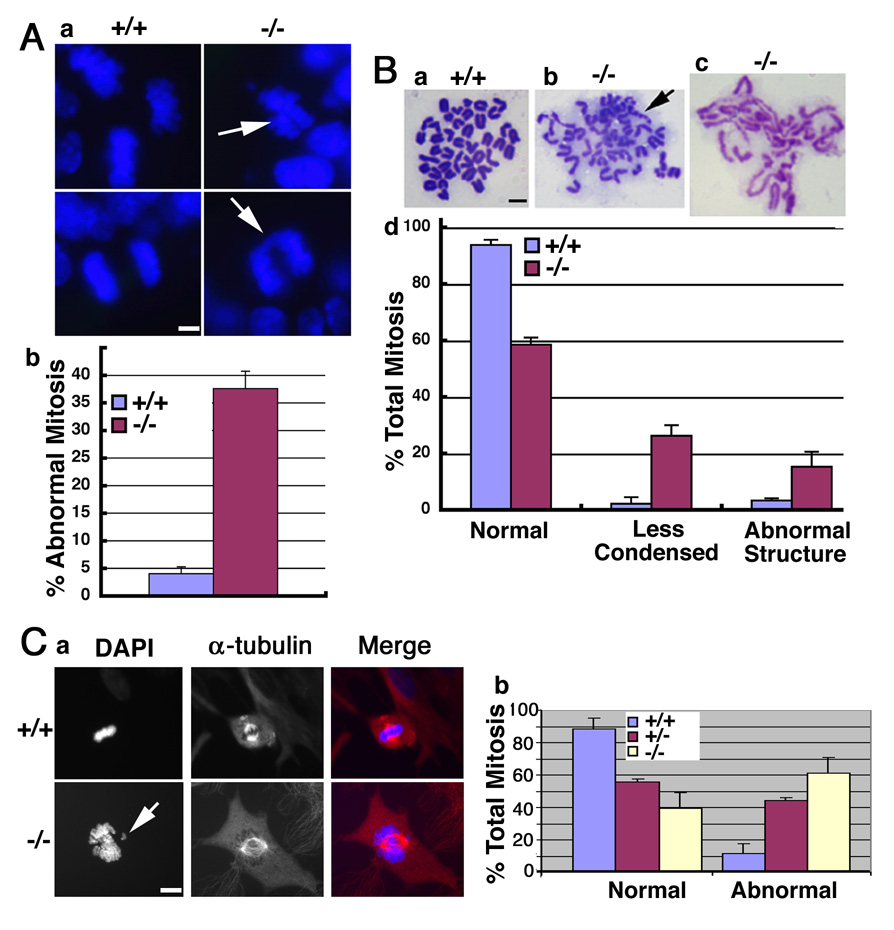
To investigate this further, we checked mitotic chromosome morphology in the embryos. Staining tissue sections of E10.5 embryos with DAPI indicated that SirT1−/− embryos exhibited chromosome abnormalities characterized by chromosome lagging and unequal segregation (Fig. 2A). These abnormal chromosome structures were found in 37% of SirT1−/− cells compared with less than 5% observed in wild-type cells. Chromosome spreads prepared from E9.5 embryos showed that about 37% of SirT1−/− cells were aneuploid and displayed a variety of structural aberrations, such as broken and decondensed chromosomes (Fig. 2B). Analysis of chromosome spreads prepared from SirT1−/− mouse embryonic fibroblast (MEF) cells detected similar abnormalities (data not shown). Next, we analyzed metaphase chromosomes in cells that were not treated with colcemid using an antibody against α-tubulin together with DAPI staining. In wild-type cells, chromosomes at the metaphase are highly condensed and aligned along with the metaphase plate. However, many SirT1−/− MEF cells at metaphase contained an partially condensed and disorganized chromosomal mass that was associated with a relatively normal spindle (Fig. 2C). Chromosome aneuploidy and breaks could, conceivably, originate from the continuous division of these mutant cells.
Figure 2. Deletion of SIRT1 causes chromosome abnormality.
(A) Dapi staining of tissue sections showing abnormal mitotic features (arrows, A-a) in a E10.5 SIRT1−/− embryo. Data (A-b) were collected from 3 pairs of embryos, 200 mitotic phases from each embryo were counted. (B) Chromosome spreads from E9.5 embryos showing normal spread (B-a), aneuploid and abnormal structure, or broken chromosome (arrow, B-b), and less condensed chromosomes (B-c). Chromosome spreads from 9 pairs of embryos were made, and all the spreads from each individual embryo were counted. (C) SIRT1 mutant MEF cells displayed incompletely condensed, and lagging chromosomes (arrow, C-a), and uneven chromosome segregation under a relative normal spindle (α-tubulin staining). Data was summarized in (C-b). Data is presented as average ±SD. Bars: 10 µm for A–B, 20 µm for C.
SIRT1 deficiency impaired heterochromatin formation
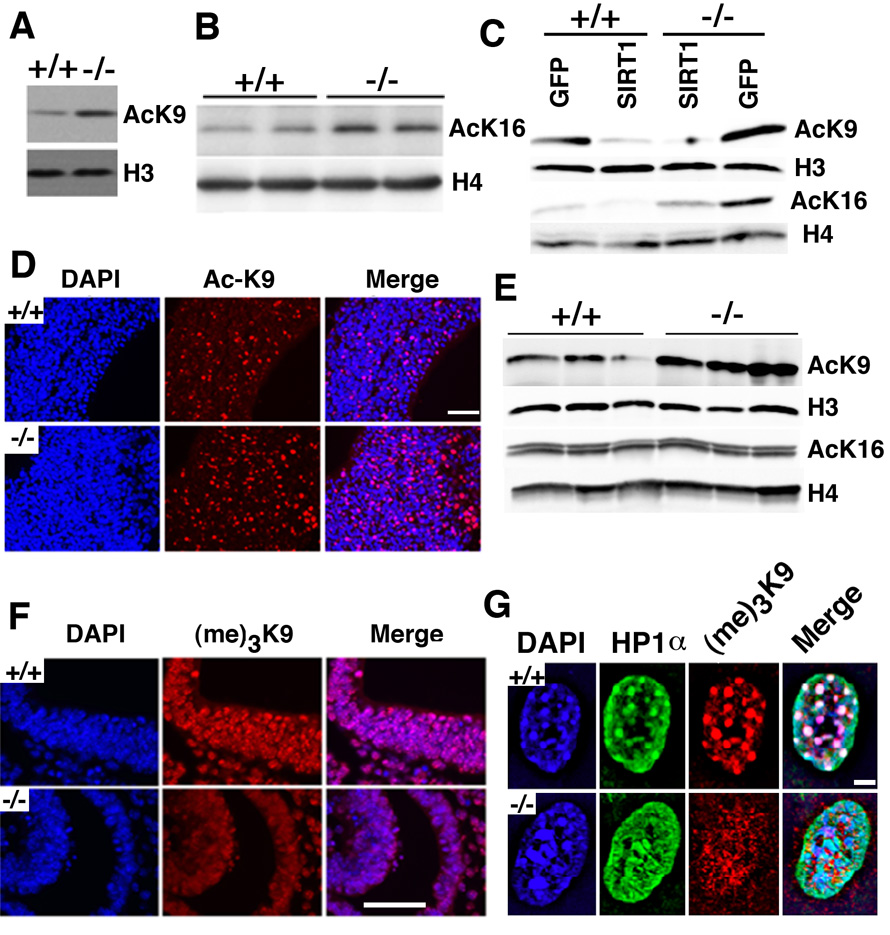
It is known that SIRT1 deacetylates K16 of histone H4 and K9 of histone H3 in yeast and in vitro cultured mammalian cells (Vaquero et al., 2007). Western blotting with antibodies against Ac-K9, and Ac-K16 revealed increased levels of both H3 Ac-K9 and H4 Ac-K16 in SirT1−/− MEFs (Fig. 3A, B), and reconstitution of SIRT1 in these cells reduced their acetylation (Fig. 3C). Because histone acetylation plays a major role in chromosome condensation, we hypothesized that the alteration in histone modification in the SirT1−/− embryos might be a cause for the chromosomal abnormalities. To investigate this, we performed immunofluorescent staining on brain sections of E11 embryos. Our data showed that SIRT1 mutants contained a much higher level of H3 Ac-K9 (Fig. 3D), while no significant alteration of H4 Ac-K16 was detected (data not shown). This finding was confirmed by Western blot analysis (Fig. 3E), demonstrating that mammalian SIRT1 is capable of modifying histones in vivo, although in the early embryos, SIRT1 seems to have stronger effects on acetylation of histone H3 K9 than histone H4 K16.
Figure 3. SIRT1 deficiency changed epigenetic modification of chromatin.
(A, B) Western blot analysis showing increased histone H3K9 (A) and H4K16 (B) levels in SIRT1−/− MEFs. (C) Reconstitution of SIRT1 in SIRT1−/− MEFs reduced histone H4K16 levels. (D, E) AC-K9 immunoflourescent staining of brain in E11 embryos. SIRT1−/− embryos displayed much more Ac-K9 staining than the +/+ embryos, which is confirmed by Western blot analysis. (F) (me)3-K9 immunoflourescent staining of embryo brain at E11. SIRT1−/− embryo displayed much less (me)3-K9 staining than the +/+ embryo. (G) In MEF cells, loss of SIRT1 impaired distribution of HP1α. Deletion of SIRT1 caused diffused localization compared with punctuated foci in +/+ embryos. Bars: 100 µm for D and F, and 10 µm for G.
Increased acetylation of K9 on histones will impair its tri-methylation level, thus affecting heterochromatin formation. To verify this point, an antibody against tri-methylated K9 was applied to brain sections from E11 embryos (Fig. 3F). SirT1−/− mutant brain contained much less tri-methylated K9 than the control brain. Similarly, a distinct reduction in tri-methylated K9 foci were also detected in SirT1−/− MEFs (Fig. 3G). A function of tri-methylated K9 is to recruit heterochromatin protein 1 (HP1α). HP1α contains a chromatin modification organizer motif that binds to histone methyltransferases in order to establish a closed chromatin configuration that represses transcription. We did not detect distinct HP1α foci in SirT1−/− cells although HP1α was diffusely present, and consequently, there was no co-localization between tri-methylated K9 and HP1α. The impaired heterochromatin formation accounts for reduced chromosome condensation, which could be a cause for genomic instability.
SIRT1 deficiency results in cell cycle abnormalities and impaired DNA damage repair
Genetic instability could also result from cell cycle checkpoint defects and impaired DNA damage repair (Deng, 2006). To comprehensively understand the effect of SIRT1 deficiency, we studied cell cycle checkpoints and DNA damage repair. Our analysis failed to detect obvious abnormalities in the G2/M cell cycle checkpoint in SirT1−/− cells (data not shown). When cells were treated with 10 Gy of γ-irradiation (IR), both WT and SirT1−/− MEF cells showed similar reduction (~60%) of cells in the S-phase, suggesting that the SirT1−/− cells have a normal G1/S cell cycle checkpoint (Fig. 4A). However, we found that SirT1−/− MEFs did not respond to lower doses of γ-irradiation (Fig. 4B), revealing abnormal G1/S checkpoint defects under these conditions. Thus, the large-scale DNA damage induced by a high-dose of IR is sufficient to activate redundant checkpoint signaling cascades in SirT1−/− cells. Of note, the H2AX mutant and Nijmegen breakage syndrome (NBS) mutant cells showed similar response, i.e. exhibiting G1/S and, or G2/M defects at low, but not at high doses of IR (Antoccia et al., 1997; Ferguson and Alt, 2001).
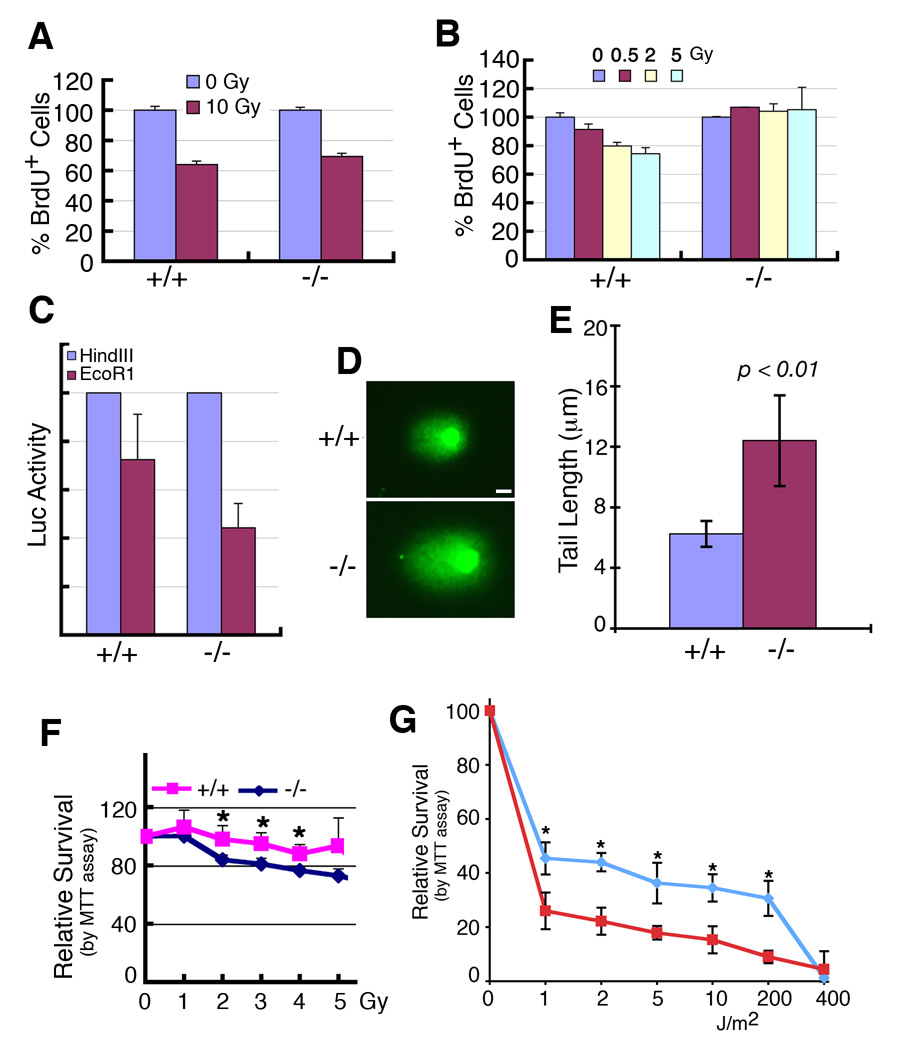
Figure 4. Deletion of SIRT1 leads to impaired DNA damage repair and radiation sensitivity.
(A, B) SIRT1 deletion caused impaired response to a low dosage of γ-irradiation (B), but not to high dosage (A) when assessed by BrdU incorporation 24 hours after the irradiation. (C) SIRT1 mutant cells exhibited impaired micro-homologous recombination, as revealed in cells transfected with a pGL2 Luc vector that was linearized with either HindIII or EcoRI. (D,E) Comet assay reveals that SIRT1−/− cells are incapable of repairing γ-irradiation induced double strand DNA damage efficiently. Comet assay was performed 2 hours after MEFs received 5Gy γ-irradiation. (F, G) −/− MEF cells are more sensitive than +/+ controls revealed by γ-irradiation (F), and UV (G). *: p< 0.05. All the data were obtained by analyzing at least 6 pairs of individual MEF cells at passage 1. Data is presented as average ±SD. Bars: 100 µm for D.
Next, we assessed DNA damage repair ability in SirT1−/− cells. To investigate this, MEF cells were transfected with a micro-homologous DNA damage repair reporter, pGL2 Luc vector, linearized with either HindIII or EcoRI. Forty-eight hours post transfection, the cells were collected and luciferase activity was quantified. With HindIII digestion, there was no significant difference in Luc activity between wild type and SirT1−/− cells. In contrast, upon EcoRI digestion, the wild type cells were able to recover about 70% of Luc activity while SirT1−/− cells recovered only 42% of Luc activity (Fig. 4C). Because EcoRI cuts within the coding sequence of the luciferase gene, the restoration of luciferase activity, therefore, requires the precise re-joining of the short protruding ends, which involves micro-homologous DNA damage repair. The reduced Luc activity in SIRT1 mutant cells indicates that the absence of SIRT1 reduced the micro-homologous DNA damage repair ability. On the other hand, HindIII cuts within the linker region between the SV40 promoter and Luc coding sequence, the restoration of luciferase activity does not require precise end joining. This data indicate the absence of SIRT1 does not interfere with end ligation if such a ligation does not require a precise ends re-joining.
To further illustrate the effect of impaired DNA damage repair, we used the Comet assay, which is an extremely sensitive assay to detect DNA damage at the single cell level. We found that SirT1−/− cells contained, on average, ~2-fold longer comet tail (12.44 µM) than that of wild type (6.74 µM) cells when quantitatively measured 2 hours post 5 Gy γ-irradiation (Fig. 4D, E). We also performed a radio-sensitivity assay using a serial increased dosage of γ-irradiation (Fig. 4F). We found that SirT1−/− cells were significantly more sensitive to radioactivity compared to wild-type cells at doses up to 5 Gy. These data are consistent with the finding that the SirT1−/− cells had impaired DNA damage repair. Because γ-irradiation primarily causes DNA double strand break, we have also treated cells with UV, which induces single strand breaks. We found that the SirT1−/− cells were also more sensitive to UV radiation than wild type cells (Fig. 4G), suggesting SIRT1 may be also involved in other types of DNA damage repair, such as base excision repair.
Defective DNA damage repair correlates with decreased γH2AX foci formation
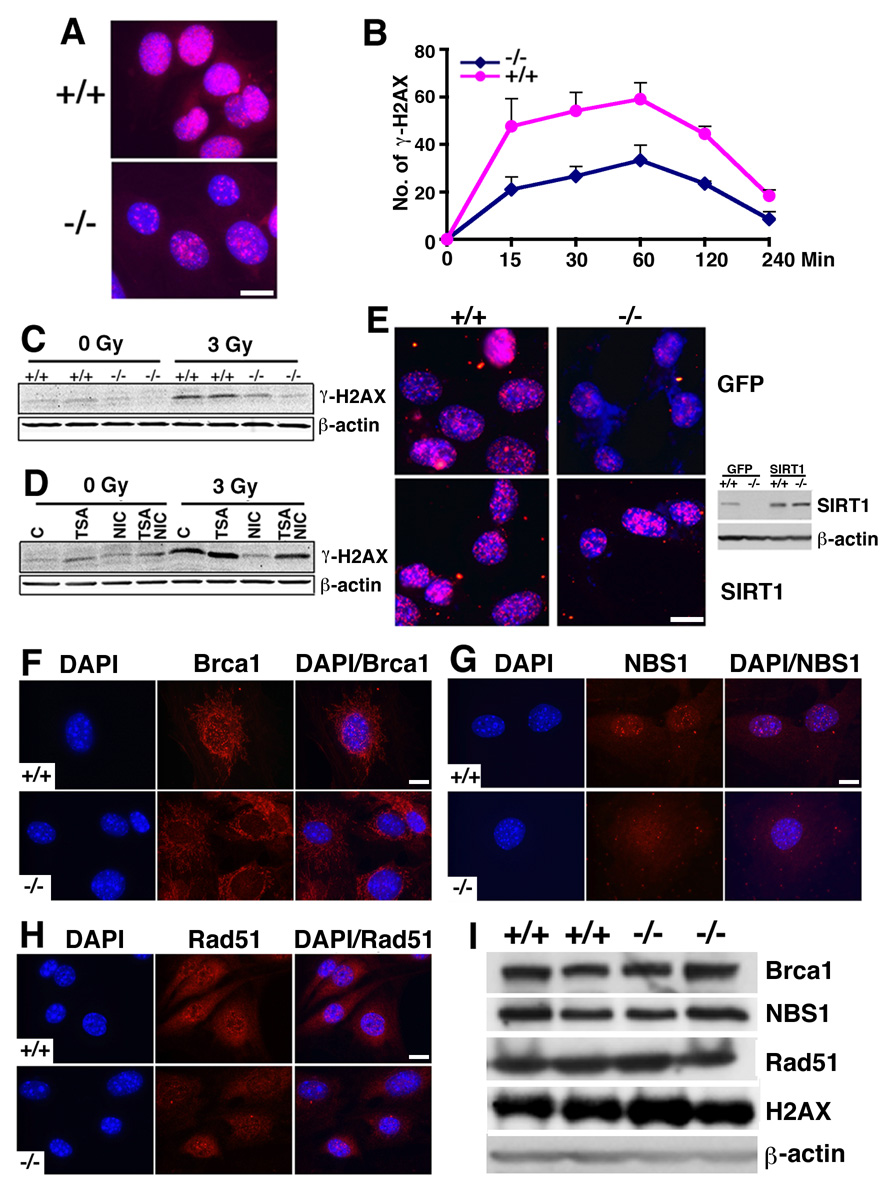
To investigate the mechanistic base of impaired DNA damage repair, we stained the cells with an antibody to γH2AX, which is a DNA damage sensor and helps maintain genome integrity (Celeste et al., 2003). Upon γ-irradiation, SirT1−/− cells showed markedly decreased γH2AX foci compared with WT cells (Fig. 5A). There are at least two possible factors resulting in a decrease in γH2AX foci formation: lack of initial phosphorylation or lack of retention of phosphorylation. To distinguish this, we irradiated the MEF cells with 3 Gy and then followed γH2AX foci formation during a time course. We found the initial γH2AX foci was significantly fewer in SirT1−/− MEF cells (20 foci/cell) than WT cells (48 foci/cell). After 15 min of IR, and both SirT1−/− and WT cells maintained a similar increase at 30 and 60 min after the treatment (Fig. 5B). This observation indicates that SIRT1 deficiency reduces initial H2AX phosphorylation, while the ability of retaining H2AX phosphorylation was not affected. The differential levels of γH2AX were also detected by Western blot analysis (Fig. 5C). To confirm this, we treated WT cells with trichostatin A (TSA) (an inhibitor of class I and II histone deacetylase), and/or nicotinamide, (an inhibitor for class III histone deacetylase) followed by western blot with an antibody to γH2AX. This data confirmed that the inhibition of class III, but not class I and II histone deacetylases, inhibited H2AX phosphorylation (Fig. 5D). Indeed, inhibition of class I and II histone deacetylases increased level of γH2AX, and it happened even in the presence of nicotinamide. This data suggests that class I/II and class III histone deacetylases have opposite roles in H2AX phosphorylation, and the negative effect of class I/II histone deacetylases could supersede the positive effect of class III histone deacetylases if both deacetylases are inhibited.
Figure 5. SIRT1 deficiency impairs γH2AX foci formation.
(A) γH2AX foci formation was reduced 2 hrs post 3 Gy irradiation. (B) Time course showing reduced initiation of γH2AX foci in SIRT1 mutant cells. Six pairs of MEF cells at Passage 1 were irradiated with 3 Gy, and γH2AX foci number was counted in each individual cell. One hundred cells from each MEF cell were counted in both untreated and 3 Gy irradiated cells. *: p< 0.05. Data is presented as average ±SD. (C) Western blot showing significantly reduced γH2AX levels in SIRT1−/− than +/+ cells. (D) Treatment of nicotinomide diminished γH2AX levels in SIRT1+/+ cells. (E) Transfection of a vector carrying a wild type SIRT1 (pUse-SIRT1, Upsate), but not a GFP control, restored γH2AX foci formation in SIRT1−/− cells. SIRT1 expression levels were shown by Western blot analysis. (F–H) Immunofluorescent staining of Brca1, Rad51 and NBS1 in SIRT1+/+ or −/− MEF cells. Nuclear foci formation is reduced in the mutant cells. (I) Western blots showing no alteration in total protein levels of Brca1, Rad51, NBS1 and H2AX. Bars: 100 µm for A and E, and 10 µm for F–H.
To assess whether the reduced γH2AX foci formation is a direct consequence of SIRT1 loss, we transfected SIRT1−/− cells with a SIRT1 expression vector. Our data indicated that the SIRT1 reconstituted SirT1−/− cells contained relatively equal numbers of γH2AX foci compared with wild type controls upon 3Gy treatment (Fig. 5E). Since γH2AX foci formation serves as a sensor for DNA damage (Kobayashi, 2004; Paull et al., 2000), impairment at this step may affect the downstream response to DNA damage. To investigate this, we examined foci formation of Rad51, Brca1 and Nbs1 following γ-irradiation. Our data revealed marked reduction in nuclear foci formation of these proteins in SIRT1 mutant mice compared with wild type MEF cells (Fig. 5F, G, H). Of note, our Western blot analysis did not reveal changes in protein levels (Fig. 5I), suggesting that these proteins could not be efficiently recruited to DNA damage sites in SIRT1 mutant cells.
H2AX can be phosphorylated by Ataxia-Telangiectasia Mutated (ATM) upon γ-irradiation. Therefore, we studied ATM phosphorylation and its downstream substrates. Immunofluorescence staining with an antibody to pi-ATM did not reveal an significant changes of pi-ATM foci in SirT1−/− MEF cells post γ-irradiation (Supplementary Fig. 3A). Western blots showed that after γ-irradiation, levels of phosphorylated CHK2, and p53ser20 showed no obvious difference between wild type and SirT1−/− MEF cells (Supplementary Fig. 3B, C). Altogether, these observations suggest that SIRT1 deficiency impaired DNA damage response and this effect is independent of the ATM/CHK2/p53 pathway.
Haploinsufficiency of SIRT1 facilitates tumorigenesis
Genetic instability is a major cause for tumor formation (Deng, 2001). However a role of SIRT1 in tumorigenesis could not be determined due to embryonic lethality. Since the absence of SIRT1 increased p53 activity, it was suspected that embryonic lethality of SIRT1 mutant embryos is due, at least in part, to p53 activation (Cheng et al., 2003). To test this, we introduced a p53-null mutation into SIRT1 mutant mice. We failed to obtain SirT1−/−;p53−/− mice among 429 pups generated from interbreeding between SirT1+/−;p53+/− mice, although three (3/429=0.7%) SirT1−/−;p53+/− mice were found (Supplementary Table 2). Thus, absence of p53 did not rescue embryonic lethality associated with SIRT1 deficiency, suggesting that the embryonic lethality associated with SIRT1 deficiency was not caused by p53 activation.
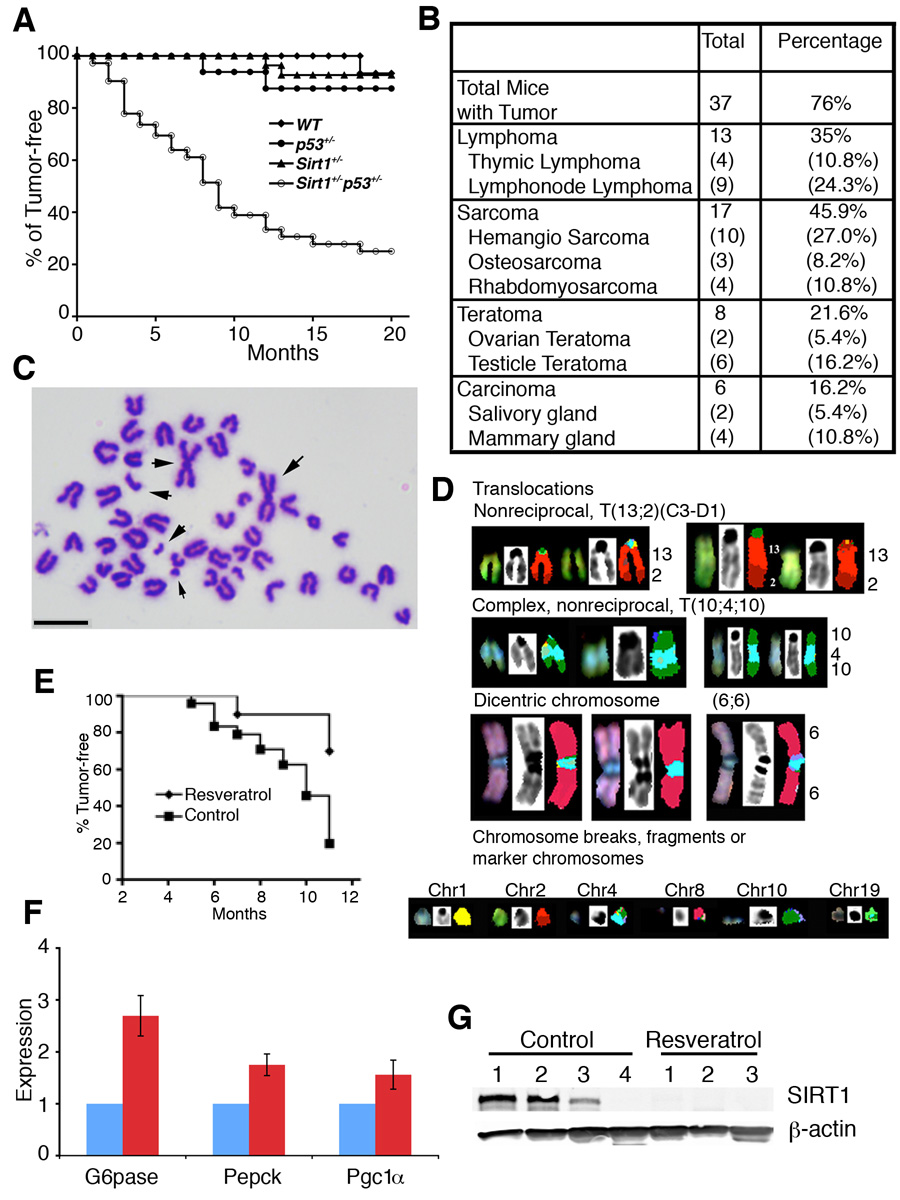
SirT1+/−;p53+/− mice were healthy, however, they started to develop spontaneous tumors from about 5 months of age, and reached about 76% by 20 months of age, while only 2 out of 21 SirT1+/− mice and 3 out of 22 p53+/− mice developed tumors, respectively, during the same period of time (Fig. 6A). The tumors occurred in SirT1+/−;p53+/− mice were primarily sarcomas (45.9%), lymphomas (35%), teratomas (21.6%), and carcinomas (16.2%) (Fig. 6B and Supplementary Fig. 4). Chromosome spreads from primary tumors (11 tumors) showed extensive aneuploidy (83.8%) and chromosomal aberrations, notably translocations, chromosome breaks, deletions, dicentric chromosomes (Fig. 6C).
Figure 6. SIRT1 deficiency causes genomic instability and tumor formation.
(A) Tumor free curve of different genotypes mice, including SIRT1+/−p53+/− (n=49), p53+/− (n=12), SIRT1+/− (n=37), and WT (n=18). (B) Types and percentage of tumors developed in SIRT1+/−p53+/− mice. (C) Chromosome spread from a mammary tumor. Regardless of the tumor type, general events are aneuploidy, numerous structural chromosomal aberrations, and premature chromosome segregation. Arrow points to abnormally long chromosome created by end fusion. (D) SKY analysis on metaphase spreads from a primary tumor, showing non-reciprocal translocation (T(13;2)(C3-D1)), a complex non-reciprocal translocation (T(10;410)), dicentric chromosomes, and a variety of chromosomal fragments from chromosomes 1,2, 4, 8, 10 and 19 respectively. (E) Treatment of resveratrol reduced tumor incidence in SIRT1+/−p53+/− mice. Resveratrol group treated group consisted of 10 female mice. The control ground contained 10 DMSO treated and 16 untreated female mice. Logrank test: p<0.01. (F) Treatment of resveratrol caused altered expression of several known downstream genes of SIRT1. Data is presented as average ±SD. (G). Western blot analysis showing that all three tumors developed in the resveratrol treated SIRT1+/−p53+/− mice lost SIRT1 expression, while only one out of four tumors in the mock treated mice lost SIRT1 expression. Bar: 10 µm for C.
Spectral karyotyping analysis (SKY) was performed on metaphase spreads derived from early passages of two primary tumors, 841A (mammary gland carcinoma) and 785S (hemangio sarcoma) (Fig. 6D). SKY analyses on metaphase spreads derived from primary cells at early passages of tumor 785S showed a variety of clonal aberrations. The non-reciprocal translocation involved chromosome 13 and chromosome 2 giving rise to a T(13;2) translocation (30%) and a complex translocation involving an insertion of chromosome 4 into chromosome 10 resulting in a T (10;4;10) translocation or T(4;10) (30%) (Fig. 6D, Supplementary Fig. 5A). Dicentric chromosomes were observed involving two copies of chromosome 6 in a Dic (6; 6) (40%) (Fig. 6D, Supplementary Fig. 5B). Numerous chromosome breaks, both acentric and centric chromosome fragments from chromosomes 1, 2, 4, 8, 10 and 19 were identified (Fig. 6D). Furthermore, 50% of the spreads lost chromosome 10, 70% of the spreads lost chromosome 7, and 60% of the spreads lost chromosome 12. Extensive chromosome abnormality has been observed in cells from 841A carcinomas (Supplementary Fig. 5A). All of these clonal aberrations recorded from 841A and 785S metaphase spreads were able to be confirmed by fluorescence in situ hybridizations (FISH) (Supplemental Fig. 5B–E). Both these tumor cells showed random gains and losses of whole chromosomes. However, a consistent and recurrent gain of chromosome 3 (>90%) and loss of chromosome 7 (55%) was observed in spreads from both tumors. Taken together these results demonstrate that SIRT1 deficiency severely impairs genome integrity or stability, and could be one of the causes for spontaneous tumorigenesis in SIRT1 and p53 double heterozygote animals.
RT-PCR analysis of tumor tissues demonstrated that 73% (11/15) of the tumors examined lost p53 expression, and 27% (4/15) of the tumors lost SIRT1 expression (Supplementary Fig. 6A). Southern blot showed 77% (10/13) lost heterozygosite (LOH) of p53 (Supplementary Fig. 6B). Next, we examined SIRT1 protein levels in tumor tissues by Western blot analysis and found 44% (7/16) tumors had no or low levels of SIRT1 (Supplementary Fig. 6C). Meanwhile, Southern blot analysis revealed 18% (3/17) tumors showed LOH of SIRT1 (Supplementary Fig. 6D).
The observation that the majority of tumors maintained one wild-type allele of SIRT1 suggests that SIRT1 serves as a haploid tumor suppressor gene. To test this, we investigated whether this SIRT1 allele is functional by using resveratrol, an activator of SIRT1 (Howitz et al., 2003). SirT1+/−;p53+/− female mice were randomized into two groups. One group (n=10) was provided with resveratrol supplemented drinking water (7.5 µg/ml), the other group was either provided with (n=10) /without (n=16) carrier (DMSO) supplemented drinking water. The treatment started at 2 months of age, and went on for 9 months. During this period, 21 out of 26 (80%) mice in the control group developed tumors, in contrast, only 3 out of 10 (30%) mice in the resveratrol treated group developed tumors (1 hemangio sarcoma and 2 thymic lymphoma). We noticed that the partial inhibition of tumor formation was also correlated with a delay in tumor onset, i.e. the first tumor incidence developed in the control group at 5 months of age, while the first tumor in the treated group occurred at 7 months of age (Fig. 6E).
Next, we investigated whether the reduced tumor formation was due to SIRT1 activation. We first checked expression of several SIRT1 downstream genes, including G6pase, Pepck and PGC1α (Lagouge et al., 2006; Rodgers et al., 2005). We detected significantly increased levels of these genes in the resveratrol treated, compared with control, SirT1+/−;p53+/− mice (Fig. 6F), suggesting that the remaining copy of SIRT1 in the SirT1+/−;p53+/− mice is activated. We showed earlier that 18% tumors showed LOH of SIRT1 and up to 44% tumors had no or low levels of SIRT1. Resveratrol should not inhibit growth of tumors if they are negative for SIRT1. To see if this is the case, we performed Western blot analysis on SIRT1 expression in these tumors. We showed that one of four tumors developed in the DMSO treated group lost SIRT1 protein, while all three tumors developed in resveratrol treated mice did not contain SIRT1 protein (Fig. 6G). The loss of SIRT1 in these tumors explains why resveratrol did not inhibit the formation of these tumors. However, the cause for the loss of SIRT1 protein in these tumors remains elusive as no LOH of SIRT1 was detected by Southern blot analysis (Supplementary Fig. 6E).
Expression levels of SIRT1 in clinical cancer samples
Our view that SIRT1 may serve as a tumor suppressor seems, at least on the surface, contradictory to the current reports that SIRT1 is expressed in certain primary tumors and cell lines at high levels. This includes prostate cancer (Huffman et al., 2007), acute myeloid leukemia (Bradbury et al., 2005), nonmelanoma skin cancers (Hida et al., 2007), and colon cancer (Stunkel et al., 2007). To investigate this, we compared SIRT1 levels in the available datasets and, surprisingly, we found that SIRT1 levels are actually lower in many cancers than normal tissues, including glioblastoma, bladder carcinoma, prostate carcinoma and various forms of ovarian cancers (Supplementary Fig. 7). To provide our own assessment for this issue, we performed the following three experiments.
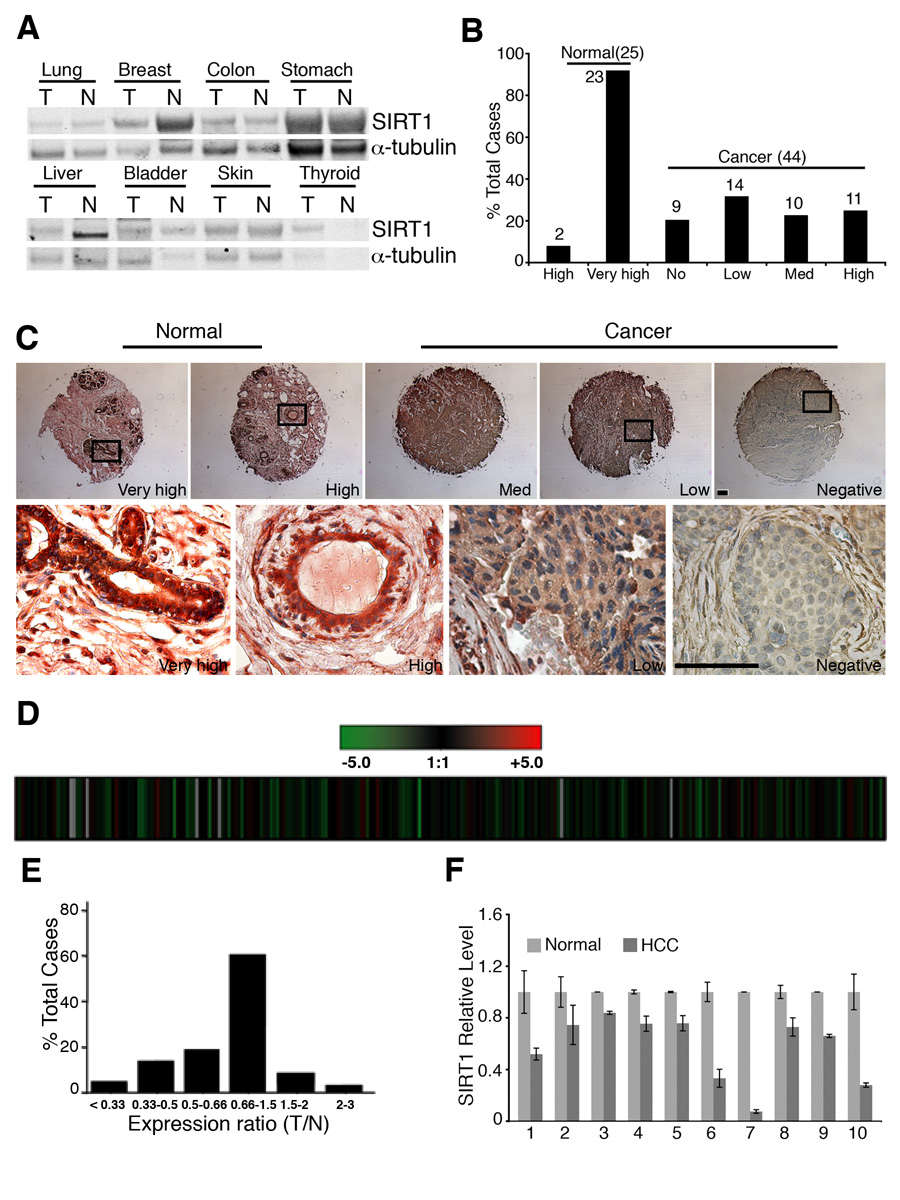
First, we performed Western blot analysis of 8 types of tumors, including lung, breast, colon, stomach, liver, bladder, skin, and thyroid. Our data revealed reduced SIRT1 levels in breast cancer and hepatic cell carcinoma (HCC) than their normal controls, while slightly increase (thyroid) or no change (lung, colon, stomach, bladder, and skin) of SIRT1 levels were detected in other tumors (Fig. 7A).
Figure 7. SIRT1 gene expression in human clinical cancers.
(A) Levels of SIRT1 in 8 different cancers and their normal controls revealed by Western blot analysis. (B,C) SIRT1 protein levels between 44 breast cancers and 25 normal breast tissues revealed by tissue array. Levels of SIRT1 staining were classified as very high, high, medium, low and negative (B), and the actual immunochemical images were shown (C) The boxed region showed high levels of SIRT1 in normal epithelium and lowered levels in cancers. (D–F) SIRT1 expression levels in microarray data of 263 HCC samples, presented as raw log2 ratio (T/N) using previously described dataset (GEO accession number, GSE5975) (D), and bars (E). Realtime RT-PCR of 10 pairs of samples was also presented (F). Data shown is average ±SD. Bars: 100 µm for C.
Next, we performed tissue array to compare SIRT1 protein levels between 44 breast cancers and 25 normal breast tissues. Our data detected significantly higher levels of SIRT1 in all normal breast tissues than cancers (Fig. 7B, C). We have also analyzed SIRT1 expression levels in microarray data of 263 HCC samples. Our data indicated that SirT1 reduced by 2 fold in 42/263 tumors and increased by 2 fold in 4 tumors, and have no change in the remaining 217 tumors (Fig. 7D,E). To provide a validation of the microarray data, we randomly picked 10 HCC samples that showed reduced level of SIRT1 and performed real-time RT-PCR. Our data revealed the reduction of SIRT1 in all the samples compared with their normal controls (Fig. 7F). Altogether, the observation that SIRT1 levels are reduced in many human cancers and that SIRT1+/−;p53+/− mice develop tumors in multiple tissues provides strong evidence that SIRT1 may function as a tumor suppressor in mice and in some human tissues.
Discussion
We have shown that the majority of our SirT1−/− mice died at E9.5–14.5. This phenotype is more severe than the other two SIRT1 mutant mice reported previously (Cheng et al., 2003; McBurney et al., 2003b). A number of possible reasons could account for this difference. First, all these mutant mice were studied in different genetic backgrounds. McBurney’s mice were generated by using R1 ES cells, which were derived from embryos of 129SV X 129 SV-CP (Nagy et al., 1993). Homozygous SIRT1 mutant mice in 129SV-CP background were smaller and died invariably within one month after birth, while in 129/CD1 mixed background, the mutant more often survived to adulthood although their stature was smaller than that of their littermates (McBurney et al., 2003b). Cheng’s mice (Cheng et al., 2003) and our mice were generated by using TC1 ES cells, which were derived from embryos of 129SVEV mice (Deng et al., 1996). In the 129SVEV/B6C57 background, about 90% of mutant mice died at perinatal or early postnatal stages, and most of the remaining surviviors died three months after birth (Cheng et al., 2003). In our study, the majority of the SIRT1−/− (deletion of exons 5 and 6) mice died at middle stages of embryonic development, while about 1% in 129SVEV/FVB background and 8% in the 129SEVE/FVB/Black Swiss background survived to adulthood. These observations indicate that the genetic background has a profound effect on the phenotypes of all three SirT1 mutant stains. Furthermore, these mutant mice also contained distinct targeted mutations, i.e. a replacement of exons 5 and 6 with a hygromycin gene (McBurney et al., 2003b), and a deletion of exon 4 (Cheng et al., 2003), or a deletion of exons 5 and 6 (this study) using the Cre-loxP mediated approach. The potential effect of these different mutations on phenotypes of SIRT1 mutant mice is currently unclear as no truncated SIRT1 protein was observed in these mutant mice.
Absence of SIRT1 causes genetic instability
What is the cause(s) for the early lethality of SIRT1 mutant embryos? Our data revealed several major defects that may contribute to embryonic lethality. Mutant embryos analyzed at E10.5–12.5 exhibited an altered pattern of histone modification, i.e. reduced level of H3mK9, and increased acetylation of H3K9. These data provide in vivo evidence for the function of SIRT1 in histone modification previously found in yeast and cultured cells (Vaquero et al., 2007). A consequence of altered acetylation is reduced chromosome condensation, which may account for the reason why mutant embryos contained a 1.5 to 2 fold higher amount of cells in the prophase of mitosis. In addition, we also detected loosely compacted chromosomes in the metaphase. This abnormality may interfere with normal progression of the remaining phases of mitosis, and consequently lead to the formation of chromosome bridges, chromosome breaks, unequal chromosome segregation, and aneuploidy. The extensive genetic instability may serve as a primary reason for the death of mutant embryos.
In yeast, Sir2 plays a role in DNA damage repair (McAinsh et al., 1999; Mills et al., 1999; Tsukamoto et al., 1997). This function, however, has not been demonstrated in its mammalian homolog, SIRT1. Our study detected the impaired micro-homology mediated DNA damage repair and double strand break repair in SIRT1 mutant cells. The marked reduction of γH2AX, Brca1, NBS1 and Rad51 foci formation upon DNA damage could serve as a cause for the reduced efficiency of DNA damage repair. Furthermore, a recent study indicated that SIRT1 acetylates Ku70 (Jeong et al., 2007), and NBS1 (Yuan et al., 2007). All these proteins are involved in the regulation of cellular response for DNA double strand break, and/or DNA damage repair, although the involvement of any other factors need to be investigated.
SIRT1 is a haploinsufficiency tumor suppressor gene
A role of SIRT1 in cancer is currently under debate. SIRT1 is over expressed in several types of human cancers (Bradbury et al., 2005; Hida et al., 2007; Huffman et al., 2007; Stunkel et al., 2007). However, it is unclear whether SIRT1 simply serves as a marker for tumorigenesis, indeed, affects tumor growth (Lim, 2006). The regulation of the cellular response from damaged DNA, and maintainance of genetic stability may indicate that SIRT1 inhibits tumor formation (Stunkel et al., 2007). Consistent with this, it was recently demonstrated that increased expression of SIRT1 reduces colon cancer formation in APCmin/+ mouse model (Firestein et al., 2008). On the other hand, data also uncovers a role of SIRT1 in deacetylating p53, leading to p53 inhibition (Chen et al., 2005). More recently, it was shown that inhibition of SIRT1 using a specific inhibitor causes p53 hyperacetylation and increases p53-dependent transcription activity (Lain et al., 2008).
On the other hand, previous studies revealed that p53 could be activated by targeted mutation of several genes, such as Brca1 (Xu et al., 2001), leading to embryonic lethality, which can be suppressed by introduction of a p53-null mutation. Therefore we reasoned that if the lethality of SIRT1 mutant mice is attributed to the activation of p53, the simultaneous mutation of p53 should have some impact on SIRT1 phenotypes. To test whether embryonic lethality was caused by p53 activation, we crossed the SIRT1 mutant mice with p53-null mice (Donehower et al., 1992). Our data detected no rescue of embryonic lethality, suggesting that activation of p53 in SIRT1 mutant mice is not a major reason for the embryonic lethality.
Moreover, our analysis of public datasets revealed that SIRT1 expression levels are lower than their normal controls in five types of tumors, including glioblastoma, bladder carcinoma, prostate carcinoma and ovarian cancers. Our own analysis of 44 breast cancers and 263 HCC also revealed marked reduced expression of SIRT1 in these tumors. These data suggest that SIRT1 may not serve as an oncogene, instead it could act as a tumor suppressor in these tissues. Consistent with this notion, we demonstrate that the SirT1+/−;p53+/− mice developed cancers in multiple tissues. Southern blot and Western blot analyses indicated that most tumors still maintained one wild type allele of SIRT1, suggesting that a proper dose of SIRT1 is critical for inhibiting tumorigenesis. We further showed that that activation of SIRT1 by resveratrol could paritially inhibit tumor formation. It has been demonstrated that resveratrol has chemopreventive activity against various cancers including leukemia, DMB-induced mammary tumors in rat, skin cancer, and prostate cancer (reviewed in (Aggarwal et al., 2004; Delmas et al., 2006)). We also found that resveratrol treatment activates SIRT1 and significantly inhibits growth of BRCA1 associated cancers (Wang and Deng unpublished data).
In summary, our analysis of SIRT1 mutant mice has yielded insights regarding the functions of SIRT1. SIRT1 plays an important role in maintenance of heterochromatin structure through deacetylation of histones in vivo. SIRT1 also has an important role in modulating γH2AX, Brca1, Rad51 and NBS1 foci formation that are involved in DNA damage repair and cell cycle checkpoint. Finally, our observations that impaired SIRT1 function results in tumor formation in a p53-null background and that activation of SIRT1 by resveratrol reduced tumorigenesis provide compelling evidence that SIRT1 serves as a tumor suppressor gene in mice and some human cancers.
Materials and Methods
Mating and genotyping mice
Chimeric mice, obtained by injecting the targeted SIRT1+/− ES cells into blastocysts, were mated with NIH Blackswiss or C57B6 females to screen for germline transmission. Male mice bearing germline transmission were mated with female FVB EII-Cre mice (Lakso et al., 1996) to generate complete deletion of SIRT1 exon 5–6. The animals were genotyped with either southern blot or PCR using the following primers: 1: 5’tccttgccacagtcactcac 3’, 2: 5’acagtcccattcccatacc 3’; and 3: 5’cat cta aac ttt gtt ggc tgc 3’. Primers 1 and 3 are located within intron 4 and amplify the wild-type allele (660bp). Primer 2 is located within exon 7, the combination of primer 1 and 2 amplify the deleted allele (716bp). All experiments were approved by the Animal Care and Use Committee of the National Institute of Diabetes, Digestive and Kidney Diseases (ACUC, NIDDK).
Proliferation assays on embryos
SirT1+/− mice were mated to generate wild-type (+/+) and mutant (−/−) embryos. At E10.5, E11.5 and E12.5, the females were injected with BrdU. Two hours post injection, the females were euthanized and embryos were collected, fixed with formalin and genotyped. Sections of five µm from paraffin embedded samples were processed either with BrdU staining (BrdU labeling Kit, Zymed) or stained with antibody against phosphorylated histone H3 at ser10 (pi-H3, Upstate). BrdU and pi-H3 positive signals were counted from 20 different areas, and analyzed by student t-test.
Chromosome spreads from embryos, primary tumors and MEF cells
Chromosome spreads from embryos were performed as described (Deng and Xu, 2004; Shen et al., 1998). Briefly, the embryos were incubated with 100 ng/ml colcemid for 2 hours. The hypotonic treatment was carried out for 20 min at RT in 0.56% KCl. The embryos were transferred to methanol:acetic acid (3:1) for fixation. Then the embryos were disaggregated under a dissection microscope in 60% acetic acid. The disaggregated embryos were spun down, suspended with methanol:acetic acid, and dropped onto slides. All the chromosome spreads were stained with Giemsa and chromosome number and morphology were assessed under a Leica microscope with a100X objective and camera from Olympus with software Magnafire.
Immuno-flourescence staining
Methanol fixed MEF cells were stained with antibodies against BRCA1 (Turner et al., 2004), Rad51 (Ab-1, Calbiochem), NBS1 (C-19, Santa Cruz), α-tubulin (Sigma), and γ-H2AX, (Me)3-K9 of histone H3, and HP1α (Upstate), with methods described previously (Wang et al., 2004). The images were taken with either 63X or 100X lens on an Olympus X81 microscope, and processed with Slidebox software. De-paraffinized brain sections from embryos were cooked with Retriever (Cat. 62700-10, Electronic Microcopy Science) in buffer A (Citrate buffer, pH5.0), followed by staining with antibodies against acetyl-K9 of hisone H3, and/or acetyl-K16 of histone H4 (Upstate).
Western blot
Western blot was carried out by Licor (Lincoln, Nebraska) with antibodies against BRCA1 (Turner et al., 2004), Rad51 (Ab-1, Calbiochem), NBS1 (C-19, Santa Cruz), H2AX (Cat. 07-627, Upstate), γ-H2AX (Cat. 05-636, Upstate), Chk2 (Cat. 611571, BD), p53ser20 (Cat. 9287, Cell Signaling), p53 (DO-1, BD), SIRT1 (Cat. 07-131, Upstate), Bcl2 (Sc-783, Santa Cruz), Survivin (NB500-201, Novus), Ac-K9 of histone H3 (Cat. 07-352, Upstate), histone H3 (Cat. 06-755, Upstate), Ac-K16 of histone H4 (Cat. 06-762, Upstate), histone H4 (Cat. 07-108, Upstate), β-actin (A5441, Sigma), α-tubulin (T-6074, Sigma).
FACS analysis on γ-irradiated MEF cells
MEF cells at passage 1 were irradiated with different dosages, and labeled with BrdU for 24 hours. The cells were fixed with 70% ethanol, stained with anti-BrdU antibodies (Becton Dickenson) and then counterstained with 25 µg/ml propidium iodide. The stained cells were analyzed with the FACSCalibur (Becton Dickenson). The percentage of BrdU positive cells in the control group was set as 100%.
Comet Assay
Primary MEF cells at passage 1 were irradiated with 5 Gy and incubated for 2 hours. Cells were then collected and processed as described in manufacture’s protocol (Trevigen, CometAssay 4250-050-K).
RT-PCR
Total RNA was isolated from cells or tissues with STAT-60™ following manufacture’s protocol (TEL-TEST, INC). cDNA was synthesized with Cells-to-cDNA™II (Ambion Inc). Primer sequences are as following: Mouse Gapdh: forward 5'-ACA GCC GCA TCT TCT TGT GC-3', reverse 5'-CAC TTT GCC ACT GCA AAT GG-3'; mouse SIRT1: forward 5’ TTG TGA AGC TGT TCG TGG AG 3’, reverse 5’ GGC GTG GAG GTT TTT CAG TA 3’; mouse Survivin: forward 5’ GTT TGA GTC GTC TTG GCG GAG 3’, reverse 5’ GTC TCC TTC TCT AAG ATC CTG 3’; mouse BCL-2: forward 5’ ATA CCT GGG CCA CAA GTG AG 3’, reverse 5’ TCT TGT AGG CAC CTG CTC CT 3’; mouse p53: forward 5’ CAC GTA CTC TCC TCC CCT CA 3’; reverse 5’ CTT CTG TAC GGC GGT CTC TC 3’.
Resveratrol treatment
Resveratrol (AKSci, Cat. No. 60512A) treatment was carried out with 20 female SirT1+/−;p53+/− mice. The mice were randomized into 2 groups, 10 mice/group. At 2 months of age, one group of mice was treated with resveratrol drinking water (7.5 µg/ml) daily; another group was treated with carrier drinking water (DMSO 0.015%) daily. The drinking water was kept away from light and changed every 3 days. Resveratrol treatment was maintained for over 9 months.
Spectral karyotyping analysis
SKY was performed as described (Padilla-Nash et al., 2006). FISH 2D-FISH was performed on metaphase spreads from 785SS and 841A by hybridization with whole chromosome paints against selected target chromosomes that appeared clonally in two or more instances out of 10 spreads from SKY analyses.
Clinical specimens
Tumor and matched normal sample lysates were purchased from Protein Biotechnologies (http://www.proteinbiotechnologies.com/index.html). Twenty µg of protein lysates were loaded on gel for Western blot analysis. The western blot was performed with both SIRT1 and tubulin antibodies. Tissue array of breast cancer samples was purchased from US Biomax (Cat. BR1002). Immunohistochemical staining against SIRT1 (Cat. 07-131, Upstate) was carried out with Histomouse ™-SP (AEC) kit (Cat. 95-9544, Zymed Laboratories). cDNA microarray analysis for 263 hepatic cell carcinomas was previously described (Yamashita et al., 2008). The validation of microarray data was carried out by qRT-PCR with the following primers:
SIRT1: forward 5’ GCA GAT TAG TAG GCG GCT TG 3’, reverse 5’ TCA TCC TCC ATG GGT TCT TC 3’;
18S: forward 5’ CGG CTA CCA CAT CCA AGG AA 3’, reverse 5’ GCT GGA ATT ACC GCG GCT 3’.
Use of these (human) tissues was approved by the NIH Office of Human Subjects Research.
Supplementary Material
Acknowledgement
We thank members of Dr. Deng lab for their critical discussion of this work. We gratefully acknowledge Hesed Padilla Nash and Nicole McNeil for help with SKYanalyses. This research was supported by the Intramural Research Program of the National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal BB, Bhardwaj A, Aggarwal RS, Seeram NP, Shishodia S, Takada Y. Role of resveratrol in prevention and therapy of cancer: preclinical and clinical studies. Anticancer Res. 2004;24:2783–2840. [PubMed] [Google Scholar]
- Antoccia A, Ricordy R, Maraschio P, Prudente S, Tanzarella C. Chromosomal sensitivity to clastogenic agents and cell cycle perturbations in Nijmegen breakage syndrome lymphoblastoid cell lines. Int J Radiat Biol. 1997;71:41–49. doi: 10.1080/095530097144409. [DOI] [PubMed] [Google Scholar]
- Aziz MH, Afaq F, Ahmad N. Prevention of ultraviolet-B radiation damage by resveratrol in mouse skin is mediated via modulation in survivin. Photochem Photobiol. 2005;81:25–31. doi: 10.1562/2004-08-13-RA-274. [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Bradbury CA, Khanim FL, Hayden R, Bunce CM, White DA, Drayson MT, Craddock C, Turner BM. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19:1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas D, Lancon A, Colin D, Jannin B, Latruffe N. Resveratrol as a chemopreventive agent: a promising molecule for fighting cancer. Curr Drug Targets. 2006;7:423–442. doi: 10.2174/138945006776359331. [DOI] [PubMed] [Google Scholar]
- Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- Deng CX. Tumorigenesis as a consequence of genetic instability in Brca1 mutant mice. Mutat Res. 2001;477:183–189. doi: 10.1016/s0027-5107(01)00119-1. [DOI] [PubMed] [Google Scholar]
- Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response, and cancer evolution. Nucleic Acids Res. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CX, Xu X. Generation and analysis of Brca1 conditional knockout mice. Methods Mol Biol. 2004;280:185–200. doi: 10.1385/1-59259-788-2:185. [DOI] [PubMed] [Google Scholar]
- Denu JM. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem Sci. 2003;28:41–48. doi: 10.1016/s0968-0004(02)00005-1. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Ferguson DO, Alt FW. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene. 2001;20:5572–5579. doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser SM, Cockell MM. The molecular biology of the SIR proteins. Gene. 2001;279:1–16. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- Guarante LP. Regulation of Aging by SIR2. Ann N Y Acad Sci. 2005;1055:222. [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Harper CE, Patel BB, Wang J, Arabshahi A, Eltoum IA, Lamartiniere CA. Resveratrol suppresses prostate cancer progression in transgenic mice. Carcinogenesis. 2007;28:1946–1953. doi: 10.1093/carcin/bgm144. [DOI] [PubMed] [Google Scholar]
- Hida Y, Kubo Y, Murao K, Arase S. Strong expression of a longevity-related protein, SIRT1, in Bowen's disease. Arch Dermatol Res. 2007;299:103–106. doi: 10.1007/s00403-006-0725-6. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Kobayashi J. Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J Radiat Res (Tokyo) 2004;45:473–478. doi: 10.1269/jrr.45.473. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, McCarthy A, Appleyard V, Murray KE, Baker L, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13:454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Fan GX, Wang W, Yuan YK. Resveratrol induces apoptosis, influences IL-6 and exerts immunomodulatory effect on mouse lymphocytic leukemia both in vitro and in vivo. Int Immunopharmacol. 2007;7:1221–1231. doi: 10.1016/j.intimp.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Lim CS. SIRT1: Tumor promoter or tumor suppressor? Med Hypotheses. 2006 doi: 10.1016/j.mehy.2006.01.050. [DOI] [PubMed] [Google Scholar]
- McAinsh AD, Scott-Drew S, Murray JA, Jackson SP. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr Biol. 1999;9:963–966. doi: 10.1016/s0960-9822(99)80424-2. [DOI] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Bieman M, Th'ng J, Lemieux M. The absence of SIR2alpha protein has no effect on global gene silencing in mouse embryonic stem cells. Mol Cancer Res. 2003a;1:402–409. [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003b;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Nash HM, Barenboim-Stapleton L, Difilippantonio MJ, Ried T. Spectral karyotyping analysis of human and mouse chromosomes. Nat Protoc. 2006;1:3129–3142. doi: 10.1038/nprot.2006.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L, Guan XY, Ried T, Deng CX. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–3124. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- Stunkel W, Peh BK, Tan YC, Nayagam VM, Wang X, Salto-Tellez M, Ni B, Entzeroth M, Wood J. Function of the SIRT1 protein deacetylase in cancer. Biotechnol J. 2007;2:1360–1368. doi: 10.1002/biot.200700087. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV, Barrett JC, Burgoyne PS, Deng CX. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- Wang RH, Yu H, Deng CX. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc Natl Acad Sci U S A. 2004;101:17108–17113. doi: 10.1073/pnas.0407585101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett TG, Jr, Carpenter DM, Lamartiniere CA. Resveratrol, but not EGCG, in the diet suppresses DMBA-induced mammary cancer in rats. J Carcinog. 2006;5:15. doi: 10.1186/1477-3163-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA, Harris CC, Deng CX. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.