Abstract
Mastermind (Mam) is a co-activator protein of binary complexes consisting of Suppressor of Hairless (Su(H)) and Notch Intracellular Domain (NICD) proteins assembled on cis-regulatory regions of target genes activated by Notch signaling. Current evidence indicates that Mastermind is necessary and sufficient for the formation of a functional Su(H)/NICD/Mam ternary complex on at least one specific architecture of Su(H) binding sites, called the SPS element (Su(H) Paired Sites). However, using transcription assays with a combination of native and synthetic Notch target gene promoters in Drosophila cultured cells, we show here that co-activation of Su(H)/NICD complexes on SPS elements by Mam is promoter-specific. Our novel results suggest this promoter specificity is mediated by additional unknown cis-regulatory elements present in the native promoters that are required for the recruitment of Mam and formation of functional Su(H)/NICD/Mam complexes on SPS elements. Together, the findings in this study suggest Mam is not always necessary and sufficient for co-activation of binary Su(H)/NICD complexes on SPS elements.
Keywords: Mastermind, Notch, transcription, proneural, neurogenesis, promoter-specificity
Introduction
The Notch pathway is an evolutionarily conserved signaling pathway that is essential for multiple developmental processes [1; 2; 3; 4]. Notch signaling is initiated by ligand binding to the Notch receptor protein, which results in the proteolytic release of the Notch Intracellular Domain (NICD). NICD is a non-DNA binding co-activator protein that translocates to the nucleus and binds the DNA-bound protein Suppressor of Hairless (Su(H); also called CSL). Typically, NICD de-represses target genes by displacing co-repressors initially bound to Su(H) and forming an NICD/Su(H) binary complex. The binary complex further activates target gene expression by recruiting the co-activator, Mastermind, to form a ternary Notch transcription complex (NTC).
Given the multi-functional role of Notch signaling in development, it is important to understand the molecular mechanisms by which specific subsets of Notch target genes are selectively activated in the proper cell types. During Drosophila neurogenesis, Notch signaling is necessary for the cell-specific expression of several Enhancer of split Complex (E(spl)-C) genes in “proneural clusters.” These clusters of adjacent cells are defined by the expression of proneural bHLH A (basic Helix-Loop-Helix Activator) proteins, which activate target genes by heterodimerizing with the ubiquitously expressed Daughterless bHLH A protein (Da). Within proneural clusters, typically only one or a few cells become a neural precursor cell (NPC), and the remaining cells, which are non-precursor cells (non-NPCs), adopt a non-neural cell fate. In the non-NPCs, several E(spl)-C bHLH repressor (bHLH R) and Bearded-like (Brd-like) genes are specifically up-regulated during Notch-mediated lateral inhibition. The function of these E(spl)-C proteins is to repress proneural gene expression in the non-NPCs. However, the non-NPC-specific expression of several E(spl)-C genes is mediated by a strong “Notch-proneural” synergistic interaction between the bHLH A and Notch signaling pathway proteins on the cis-regulatory regions [5; 6].
Recent studies have provided insight into the precise “DNA transcription codes” that are used by Notch signaling to generate these cell-specific gene expression patterns during neurogenesis in Drosophila [5; 6; 7]. DNA transcription codes are cis-regulatory modules comprised of particular combinations and orientations of transcription factor binding sites that provide a heritable mechanism for encoding gene expression patterns [8; 9]. We have previously shown that an “SPS+A” transcription code is critical for the Notch-proneural transcriptional synergy that drives the up-regulation of the E(spl)-C m8 bHLH R gene in non-NPCs [5]. This transcription code is comprised of bHLH A binding sites (“A” sites) and an SPS DNA element, which is a precisely spaced inverted repeat of Su(H) binding sites (“S” site). A key functional feature of the SPS+A code is that the inverted orientation architecture of the SPS element is critical for transcriptional synergy between NTCs and proneural bHLH A proteins bound to SPS and A sites, respectively [5]. The SPS+A code is also predicted to mediate the non-NPC expression of the E(spl)-C m7, mγ and mδ genes [5], although a different “logic” or code regulates mα[7].
Previously we have shown that the SPS architecture is critical for co-activation of NTCs by Mam [5]. Recent structural studies have also indicated that Mam is necessary for cooperative assembly of ternary NTCs to each of the S sites in SPS elements [10]. Thus, current models of Notch signaling indicate that Mam is an essential co-activator of gene transcription regulated by all promoters containing the SPS+A transcription code. However, in this study, we show that co-activation of SPA+A modules by Mam is promoter specific and Mam does not co-activate all promoters containing functional SPS+A modules. These findings suggest, in contrast to current models, that Mam is not always necessary and sufficient for co-activation of binary NTCs bound to SPS elements.
Materials and Methods
Details of the S2 cell culture transfection protocol have been described elsewhere [5]. All protein expression plasmids for S2 cell culture were constructed using pAc 5.1/V5-HisA plasmids (Invitrogen). The Mam expression plasmid used in this study was generated by isolating the Mam cDNA from pNB40 (B. Yedvobnick, Emory Univ.) using NheI and BamHI, and then ligating this cDNA into pAc 5.1/V5-HisA which had been digested with XbaI and BamHI. The MamΔC expression plasmid was generated by digesting the Mam pAc 5.1/V5-HisA plasmid with SacI and then religating the plasmid. The MamΔC-VP16 fusion protein was generated by isolating the VP16 activation domain coding sequence from pVP16 (Clontech) and inserting this coding sequence, in frame, with the MamΔC pAc 5.1/V5-HisA expression plasmid. Construction of all other expression plasmids used for S2 cell culture have been previously described [5].
All transcription reporters for S2 cell culture were constructed with pGL2-basic luciferase plasmids (Promega). Details about the construction of the native m8 and SPS-4A promoters have been given elsewhere [5]. Insertion of the second S site to create the ac-SPS promoter was generated by PCR. The mγ promoter contained nucleotides –319 to +84 of the native gene. This mγ promoter was isolated from a pGL2-basic plasmid containing –1210 and +84 of the native mγ gene (C. Delidakis, Inst. Mol. Biol. and Biotech.). Proper construction of all plasmids in this study were confirmed by DNA sequencing.
Results and Discussion
We have previously shown that Mam can strongly enhance Notch-proneural transcriptional synergy on the native m8 promoter in transcription assays with cultured S2 cells (Figure 1B cf. expts. 7 vs. 8) [5]. We have now confirmed our prediction [5] that Mam also strongly enhances Notch-proneural transcriptional synergy on the native mγ promoter (Figure 1B cf. expts. 15 vs. 16), which contains an SPS+A module and is specifically expressed in the non-NPCs of proneural clusters by Notch signaling.
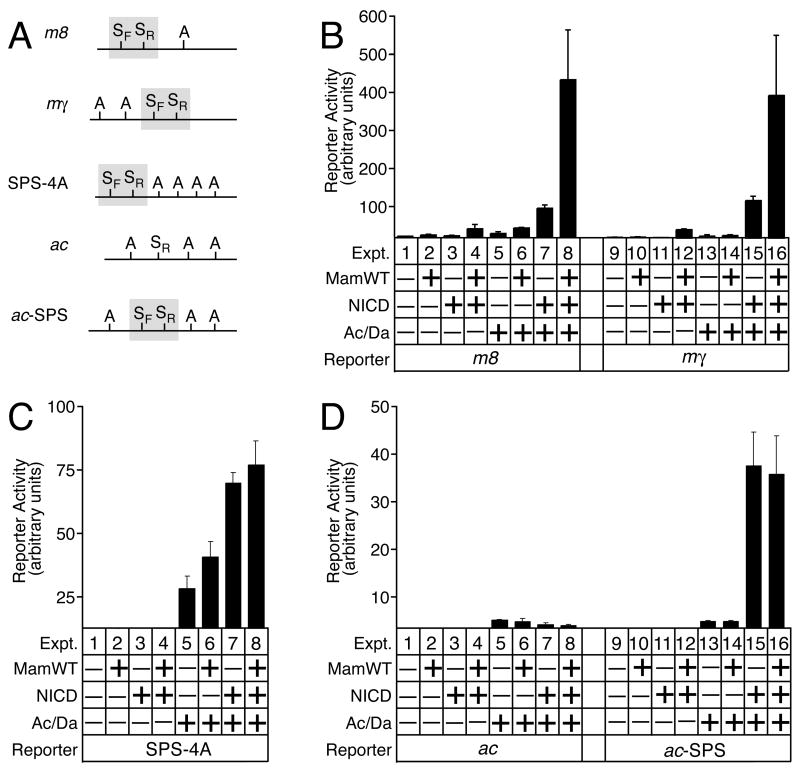
Figure 1.
Promoter-specific co-activation by Mastermind. A, a diagram of the organization of bHLH A and Su(H) sites (“A” and “S”, respectively) in the promoters used in this study. S sites and their respective “forward” and “reverse” orientations are indicated by “SF” and “SR” respectively. The SF and SR orientations are defined as 5’-GTGNGAA’3’ and 5’-TTCNCAC-3’ on the DNA strand containing the “ATG” start codon. Promoter size is not shown to scale. B, Mam selectively activates the native E(spl)-C m8 and mγ promoter, which are activated in vivo by Notch signaling. C, by contrast, the synthetic SPS-4A promoter is not co-activated by Mam, even though this promoter can mediate strong Notch-proneural transcriptional synergy. D, the native ac promoter does not mediate either Notch-proneural transcriptional synergy or co-activation by Mam, but this promoter is also not an in vivo target gene for Notch signaling even though it has an SR site in its proximal promoter. Creation of an SPS element in the native ac promoter (generating the ac-SPS promoter) allows for Notch-bHLH A transcription synergy, but not co-activation by Mam. The observed Notch-proneural transcriptional synergy on all the promoters containing SPS elements indicates that functional Su(H)/NICD complexes have formed on the S sites of the SPS elements. However, the promoter-specificity of co-activation by Mam suggests that additional cis-regulatory elements, present only in the m8 and mγ promoters, are necessary to mediate co-activation by Mam.
We next used rationally-designed synthetic reporters, which are useful for addressing whether specific transcription factor binding sites hypothesized to function are actually sufficient for promoter activation. The SPS-4A synthetic promoter contains an SPS element placed adjacent to 4 A sites and a minimal Hsp70 basal promoter (Figure 1A). As a result of this design, there should be no other functional binding sites for other transcription factors that could influence or contribute to Notch-proneural transcriptional synergy and co-activation by Mam.
The SPS-4A synthetic promoter mediates strong Notch-proneural synergy when NICD and Ac/Da are co-expressed (Figure 1C cf. expts. 3, 5 and 7), as previously shown [5]. By contrast, co-expression of Mam did not co-activate a synthetic SPS-A promoter (Figure 1C cf. expts. 7 vs. 8). The ability of co-expressed NICD and bHLH A proteins to synergistically activate the SPS-4A promoter confirmed that the Su(H)/NICD binary complexes bound to the SPS element were functional. However, the inability of Mam to co-activate this Notch-proneural synergy on the SPS-4A promoter was unexpected since current models predict that Mam should co-activate all functional Su(H)/NICD binary NTCs. These new findings indicate that the SPS+A module is sufficient to mediate Notch-proneural transcriptional synergy, but not to mediate co-activation by Mam.
To further examine the promoter specificity of co-activation by Mam on SPS+A modules, we tested the ability of Mam to co-activate the achaete (ac) promoter. The ac gene encodes a proneural bHLH A protein that is not activated by Notch signaling in proneural clusters even though the ac proximal promoter contains an single S site (Figure 1A). In fact, ac is indirectly repressed in vivo by NICD via recruitment of E(spl)-C bHLH R proteins [11; 12]. In S2 cells, expression of NICD or Mam either separately or together did not synergistically activate the ac promoter when co-expressed with the Ac/Da bHLH A proteins (Fig. 1D cf. expts. 5–8). These results are consistent with our previous findings that single S sites do not mediate NTC-bHLH transcriptional synergy [5]. By contrast, if an additional S site is added to the ac promoter in order to create an SPS element (Figure 1A), then the resulting ac-SPS promoter is strongly and synergistically activated by co-expression of NICD and Ac/Da (Fig. 1D cf. expts. 11, 13 and 15). However, similar to the SPS-4A promoter, co-expression of Mam did not generate additional ac-SPS promoter activity despite the presence of functional NTCs that can mediate transcriptional synergy with bHLH A proteins (Figure 1D cf. expts 15 vs. 16).
Together, these studies show that Mam co-activated only the native m8 and mγ promoters, which are activated by Notch signaling in vivo. By contrast, Mam did not co-activate either the synthetic SPS-4A promoter, the native ac promoter or the modified ac-SPS promoter, which are not activated by Notch signaling in vivo. These results indicate that co-activation by Mam is promoter-specific. This promoter-specificity of co-activation was unexpected since all of the promoters containing an SPS+A module were sufficient to mediate strong Notch-proneural transcriptional synergy, which indicated that functional NTC complexes had assembled on the SPS elements. Thus, these novel findings indicate that there are promoter contexts in which Mam does not co-activate of NTCs bound to SPS elements.
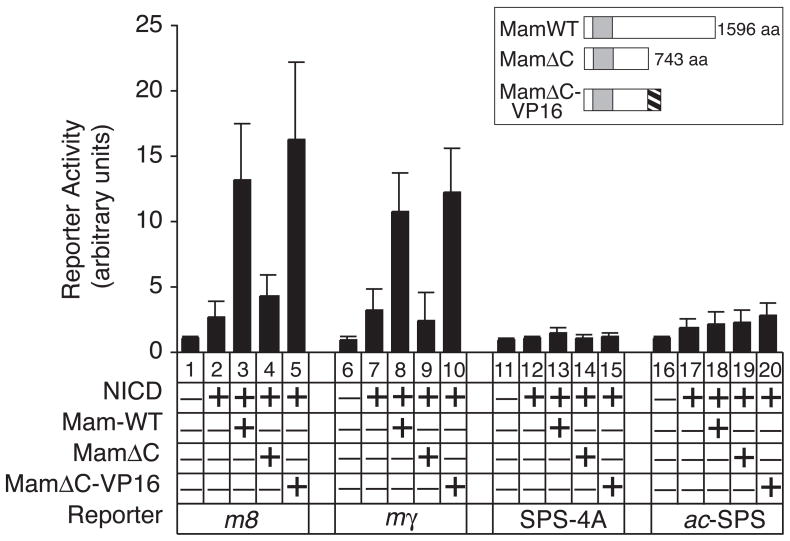
Previous studies have reported that Mam mediates cooperative assembly and binding of Su(H)/NICD/Mam ternary NTCs on SPS elements in the absence of combinatorial cofactors, such as the proneural proteins [13]. Consistent with these findings, co-expression of NICD and Mam activated the m8 and mγ promoters in the absence of Ac/Da proteins (Figure 2 expts. 3 and 8). These findings indicate that the Su(H)/NICD/Mam ternary complex formed on these native promoters and activated reporter gene transcription. However, formation of the ternary complex in absence of bHLH A proteins mediates substantially lower levels of reporter gene expression than when bHLH A are co-expressed (cf. Figure1 expts. 4 vs. 8 and 12 vs. 16). In contrast to the native m8 and mγ promoters, neither the SPS-4A or ac-SPS promoters were activated when NICD and Mam were co-expressed in the absence of bHLH A proteins (cf. Figure 2 expts 3 and 8 vs. 13 and 18). Although the functional Su(H)/NICD complexes can form on the SPS-4A and ac-SPS promoters and mediate NTC-bHLH transcriptional synergy (Figure 1C and 1D), these findings suggest that functional Su(H)/NICD/Mam ternary complexes do not form on these promoters.
Figure 2.
Evidence for promoter-specific recruitment of Mam. In the absence of bHLH A proteins, co-expression of NICD with either wild-type Mam (MamWT) or a MamΔC-VP16 fusion protein activated the m8 and mγ promoters. By contrast, co-expression of these proteins did not activate the SPS-4A or ac-SPS promoters. The ability of all these promoters to mediate Notch-proneural transcriptional synergy indicated that functional Su(H)/NICD complexes can form on the respective SPS elements of these promoters (Figure 1). However, the promoter-specific co-activation by Mam in the absence of bHLH A proteins suggests that recruitment of Mam and the formation of functional Su(H)/NICD/Mam ternary complexes occurs on the native m8 and mγ promoters. Diagrams of the Mam proteins expressed are shown in the insert. The region within Mam required for binding both Su(H) and NICD is indicated by the shaded box.
The promoter-specific formation of functional Su(H)/NICD/Mam ternary NTCs was due either to Mam being in an inactive state when bound the Su(H)/NICD binary complex on the SPS-4A and ac-SPS promoters or the lack of recruitment of Mam on to the SPS-4A and ac-SPS promoters. To test these possibilities, we removed the previously identified Mam transcription activation (TA) domain [14; 15] and replaced it with the strong constitutively active viral VP16 protein TA domain to create a MamΔC-VP16 fusion protein (Figure 2A). Co-expression of MamΔC, which lacked the entire Mam TA domain, showed no co-activation on either the m8 and mγ promoters (Figure 2B, expts. 4 and 9). However, co-expression of MamΔC-VP16 and NICD co-activated the m8 and mγ promoters at levels comparable to those by MamWT (cf. Figure 2 expts 3 vs. 5 and expts 8 vs. 10). By contrast, MamΔC-VP16 did not co-activate either the SPS-4A or ac-SPS promoters (Figure 2). Together, these results suggest that Mam is selectively recruited by Su(H)/NICD binary complex on the m8 and mγ promoters, but not on the SPS-4A or ac-SPS promoters, even though binary NTC complexes are present and functional for mediating strong Notch-proneural transcription synergy on all four promoters.
Summary and conclusions
The findings in this study indicate that co-activation of binary Su(H)/NICD complexes on SPS elements by Mam is promoter specific. The MamΔC-VP16 data suggest that selective recruitment and formation of functional Su(H)/NICD/Mam ternary NTCs underlie this promoter specificity. Since all the promoters in this study were tested in an identical cellular context, the differential co-activation by wild-type Mam is not likely to be due to a post-translational mechanism. Rather, the promoter-specificity of co-activation by Mam is likely mediated by DNA sequences specific to the native m8 and mγ promoters that contain cryptic cis-regulatory binding sites for additional combinatorial cofactor proteins. These cryptic sites are missing from the synthetic SPS-4A promoter as well as the ac-SPS promoter, which is not a native Notch target gene in vivo.
The apparent requirement for additional regulatory elements to mediate co-activation by Mam was unexpected since in vitro studies have indicated that the Su(H)/NICD/Mam ternary NTC can self-assemble on SPS elements [10]. However, our findings suggest that in a cellular context additional factors are required to either recruit or stabilize functional ternary NTC complexes containing Mam. Furthermore, the ability of the SPS-4A and ac-SPS promoters to mediate Notch-proneural transcriptional synergy in the absence of co-activation by Mam indicates that the functional Su(H)/NICD binary complexes can assemble and bind the SPS element without Mam. These findings differ from recent mammalian studies reporting that Mam is required for the assembly and binding of homologous Su(H)/NICD complexes on SPS elements in the absence of proneural proteins [10]. It is possible that physical interactions between Su(H) and the bHLH A Da protein programmed by assembly on an SPS module can stabilize the Su(H)/NICD complex bound to the SPS element in the absence of Mam [5].
Together, the findings in this study indicate that co-activation of SPS+A modules by Mam is promoter specific, and requires additional unknown cofactors that are bound to unknown sites in the native promoters. Our results also provide preliminary evidence that Mam is not always necessary to co-activate Su(H)/NICD binary complexes, and that other cofactors, such as Ac/Da, can co-activate these binary NTCs in the absence of Mam. Thus, these findings suggest that Mam is not always necessary and sufficient for co-activation of Su(H)/NICD complexes bound to SPS elements. These results suggest that the mechanisms regulating cell-specific regulation of Notch target gene expression are more complex than the current models, and involve additional unknown combinatorial cofactors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–73. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 3.Schweisguth F. Notch signaling activity. Curr Biol. 2004;14:R129–38. [PubMed] [Google Scholar]
- 4.Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–65. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 5.Cave JW, Loh F, Surpris JW, Li X, Caudy MA. A DNA transcription code for cell-specific gene activation by Notch signaling. Curr Biol. 2005;15:94–104. doi: 10.1016/j.cub.2004.12.070. [DOI] [PubMed] [Google Scholar]
- 6.Cooper MT, Tyler DM, Furriols M, Chalkiadaki A, Delidakis C, Bray S. Spatially restricted factors cooperate with notch in the regulation of Enhancer of split genes. Dev Biol. 2000;221:390–403. doi: 10.1006/dbio.2000.9691. [DOI] [PubMed] [Google Scholar]
- 7.Castro B, Barolo S, Bailey AM, Posakony JW. Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development. 2005;132:3333–44. doi: 10.1242/dev.01920. [DOI] [PubMed] [Google Scholar]
- 8.Markstein M, Levine M. Decoding cis-regulatory DNAs in the Drosophila genome. Curr Opin Genet Dev. 2002;12:601–6. doi: 10.1016/s0959-437x(02)00345-3. [DOI] [PubMed] [Google Scholar]
- 9.Michelson AM. Deciphering genetic regulatory codes: a challenge for functional genomics. Proc Natl Acad Sci U S A. 2002;99:546–8. doi: 10.1073/pnas.032685999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci U S A. 2007;104:2103–8. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 1994;8:2743–55. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- 12.Van Doren M, Bailey AM, Esnayra J, Ede K, Posakony JW. Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes Dev. 1994;8:2729–42. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- 13.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–83. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 14.Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16:1397–411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helms W, Lee H, Ammerman M, Parks AL, Muskavitch MA, Yedvobnick B. Engineered truncations in the Drosophila mastermind protein disrupt Notch pathway function. Dev Biol. 1999;215:358–74. doi: 10.1006/dbio.1999.9477. [DOI] [PubMed] [Google Scholar]