Summary
Polyubiquitin is a diverse signal both in terms of chain length and linkage type. Lysine48-linked ubiquitin is essential for marking targets for proteasomal degradation but the significance and relative abundance of different linkages remain ambiguous. Here we dissect the relationship of two proteasome-associated polyubiquitin-binding proteins, Rpn10 and Dsk2, and demonstrate how Rpn10 filters Dsk2 interactions, maintaining proper function of the ubiquitin-proteasome system. Using quantitative mass spectrometry of ubiquitin, we found that in S. cerevisiae under normal growth conditions the majority of conjugated ubiquitin was linked via lysine48 and lysine63. In contrast, upon DSK2 induction, conjugates accumulated primarily in the form of lysine48-linkages correlating with impaired proteolysis and cytotoxicity. By restricting Dsk2 access to the proteasome, extraproteasomal Rpn10 was essential for alleviating the cellular stress associated with Dsk2. This work highlights the importance of polyubiquitin shuttles such as Rpn10 and Dsk2 in controlling the ubiquitin landscape.
INTRODUCTION
The ubiquitin-proteasome system (UPS) is by far the major regulatory mechanism for controlling cellular protein turnover (Glickman and Ciechanover, 2002). Within this pathway, a series of enzymes catalyze formation of an amide bond between the terminal carboxyl of ubiquitin (Ub) and an amino group on the substrate. This modification can be extended into a polyubiquitin (polyUb) chain by subsequent attachment of the carboxy-terminus of one Ub to a lysine residue on an earlier one. Recent mass spectrometry experiments have indicated that Ub chains in the cell can be formed through all seven lysine residues within Ub (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) as well as through its N-terminus in a head-to-tail fashion (Kirisako et al., 2006; Peng et al., 2003). Lys48-linked polyUb chains are the most extensively studied, and are documented to regulate the bulk of cellular protein turnover by targeting substrates to the 26S proteasome for degradation (Ikeda and Dikic, 2008). Lys63-linked chains are known to regulate DNA repair, signal transduction, and endocytosis, are recognized by the proteasome and can sustain substrate degradation (Ikeda and Dikic, 2008; Thrower et al., 2000), although the relative contribution of these chains to bulk degradation in cells is not known. Ub chains linked through Lys11 and Lys29 have been shown to support degradation in certain contexts (Ikeda and Dikic, 2008). There is evidence also for linkages via Lys6, Lys29, Lys33 or Lys63 of Ub, but the relative abundance and purpose of these "alternative" chains remains unclear (Ikeda and Dikic, 2008).
One essential feature of the UPS is that the proteasome must have the ability to recruit or recognize polyubiquitinated substrates. The paradigm of proteasome-associated Ub receptors is Rpn10/S5a, an intrinsic proteasome subunit that binds to polyUb chains through its hallmark ubiquitin-interacting motif (UIM) (Deveraux et al., 1994; Fu et al., 1998; van Nocker et al., 1996). Even though a significant portion of polyUb-binding capacity within purified proteasomes has been attributed to Rpn10 (Elsasser et al., 2004; Elsasser et al., 2002; Verma et al., 2004), this protein is not essential for proteasomal degradation of all ubiquitinated substrates or for viability in yeast (Mayor et al., 2007; Mayor et al., 2005), in line with other proteasome subunits participating in anchoring of Ub chains (Husnjak et al., 2008).
Another interesting property of Rpn10 is that in addition to being an intrinsic proteasome subunit, a large portion is found in a proteasome-unassociated pool (Fu et al., 1998) raising the possibility that it may also function as a polyUb shuttle. Additional proteins have been proposed to serve as shuttles as well. The most prominent ones contain a ubiquitin-like (Ubl) domain at one end and ubiquitin-associated (UBA) domain at the other, through which they bind the proteasome and ubiquitinated substrates respectively. Members of this Ubl-UBA family include Rad23/hHR23, Dsk2/hPLIC/ubiquilin and Ddi1, though most conclusions as to their mode of action arise from the research on Rad23 (Chen and Madura, 2002; Chen et al., 2001; Elsasser et al., 2002; Funakoshi et al., 2002; Kleijnen et al., 2000; Lambertson et al., 1999; Ortolan et al., 2000; Saeki et al., 2002a; Saeki et al., 2002b; Seok Ko et al., 2004; Walters et al., 2002; Wilkinson et al., 2001). Other proteins fulfilling diverse cellular functions are also known to bear a Ubl domain by which they are targeted to the proteasome, as is the case with the deubiquitinating enzyme Ubp6 or the Ub ligase Parkin (Upadhya and Hegde, 2003).
Despite suggestions that Ub shuttles support degradation, evidence from both in vitro and in vivo studies indicate that they can also, paradoxically, oppose UPS activity (Chen and Madura, 2002; Chen et al., 2001; Funakoshi et al., 2002; Hartmann-Petersen et al., 2003; Kleijnen et al., 2000; Raasi and Pickart, 2003). In vitro, Rad23 displays a concentration dependent inhibitory effect on polyUb-chain formation (Ortolan et al., 2000). Likewise, both Rpn10 and Rad23 effectively protect model substrates from proteolysis (Deveraux et al., 1995; Raasi and Pickart, 2003; Verma et al., 2004). In vivo, overexpression of RAD23 can inhibit the degradation of model substrates in yeast (Ortolan et al., 2000), while hPLIC can prevent degradation of physiological substrates including p53 and IκB in mammalian cells (Kleijnen et al., 2000). The combined data suggests that the true function of Ubl-UBA proteins may be beyond shuttling their cargo to the proteasome.
A growing body of evidence indicates that a convoluted network of physical and functional overlap exists amongst the Ub-binding proteins. Rad23 was shown to function in parallel to Rpn10, with the rad23rpn10 double deletion mutant strain exhibiting severe growth defects, sensitivity to cellular stress, cell cycle delays, accumulation of Ub conjugates and failure to degrade a Ub-fusion (UFD) pathway substrate (Elsasser et al., 2004; Lambertson et al., 1999; Verma et al., 2004). Similarly, Rad23 and Dsk2 are both involved in UFD, ER-associated degradation (ERAD), spindle pole body duplication and in delivery of polyubiquitinated substrates to the proteasome (Biggins et al., 1996; Lambertson et al., 1999; Medicherla et al., 2004; Saeki et al., 2002a). Besides the genetic and functional interactions, physical interactions have been reported between S5a and either hPLIC-2 or hHR23A (Kleijnen et al., 2003; Kleijnen et al., 2000; Walters et al., 2002; Walters et al., 2003). This regulatory complexity makes the UPS a common target for dysregulation in human disease. Thus, the individual properties and interplay between these multi-tasking proteins is a primary issue in understanding the proper function and pathological dysfunctions of the Ub-proteasome system.
Although the Ub shuttles display features indicative of functional overlap such as competition for binding to Ub-modified targets, they are not completely redundant, as no single shuttle can fully complement the loss of another (Lambertson et al., 1999; Saeki et al., 2002a; Verma et al., 2004; Wilkinson et al., 2001). Such data indicate that the receptors must display some level of substrate-specificity in the degradation process. To start unraveling this complex network, we sought to identify and exploit features which made individual receptors unique. Of particular interest, overexpression of DSK2 causes overt cytotoxicity through a yet unknown mechanism. We found that Rpn10 plays a key role in attenuating the toxicity associated with DSK2 overexpression by influencing the ability of Dsk2 to access the proteasome. Our data suggest that the biological significance of Rpn10 extends beyond the proteasome itself, defining the hierarchy of proteasome-associated Ub receptors through extraproteasomal protein-protein interactions.
RESULTS
Overdose of DSK2 dysregulates the ubiquitin-proteasome pathway
It has been previously noted that high abundance of DSK2 is toxic to cells and may interfere with cell cycle progression at metaphase (Biggins et al., 1996; Funakoshi et al., 2002; He et al., 1998). To assess whether this is a general property of proteasome-associated Ub receptors, we compared viability of cells overexpressing DSK2, RAD23 or RPN10. We found cytotoxicity of DSK2 to be a unique property not shared with overexpression of other genes encoding polyUb shuttles (Fig 1A). Only induction of full length DSK2 but not of either of its two domains individually caused this toxicity (Fig 1B), even though all versions were successfully induced (Fig 1C, and Supp Fig 1). This Dsk2-mediated toxicity coincided both with a substrate-specific effect as well as with a more general impairment of the UPS. We examined the effect of DSK2 overexpression on the turnover of CPY*, a protein shown to be a substrate of Dsk2-mediated degradation (Medicherla et al., 2004), and found the biological turnover of CPY* to be significantly delayed (Fig 1D). Global UPS impairment was observed as an increase in the high molecular weight (HMW) Ub-conjugates (Fig. 1E). These observations point to the pervasive interference of unregulated Dsk2 with the normal function of the Ub-proteasome system.
Fig. 1. Overexpression of DSK2 dysregulates the UPS.
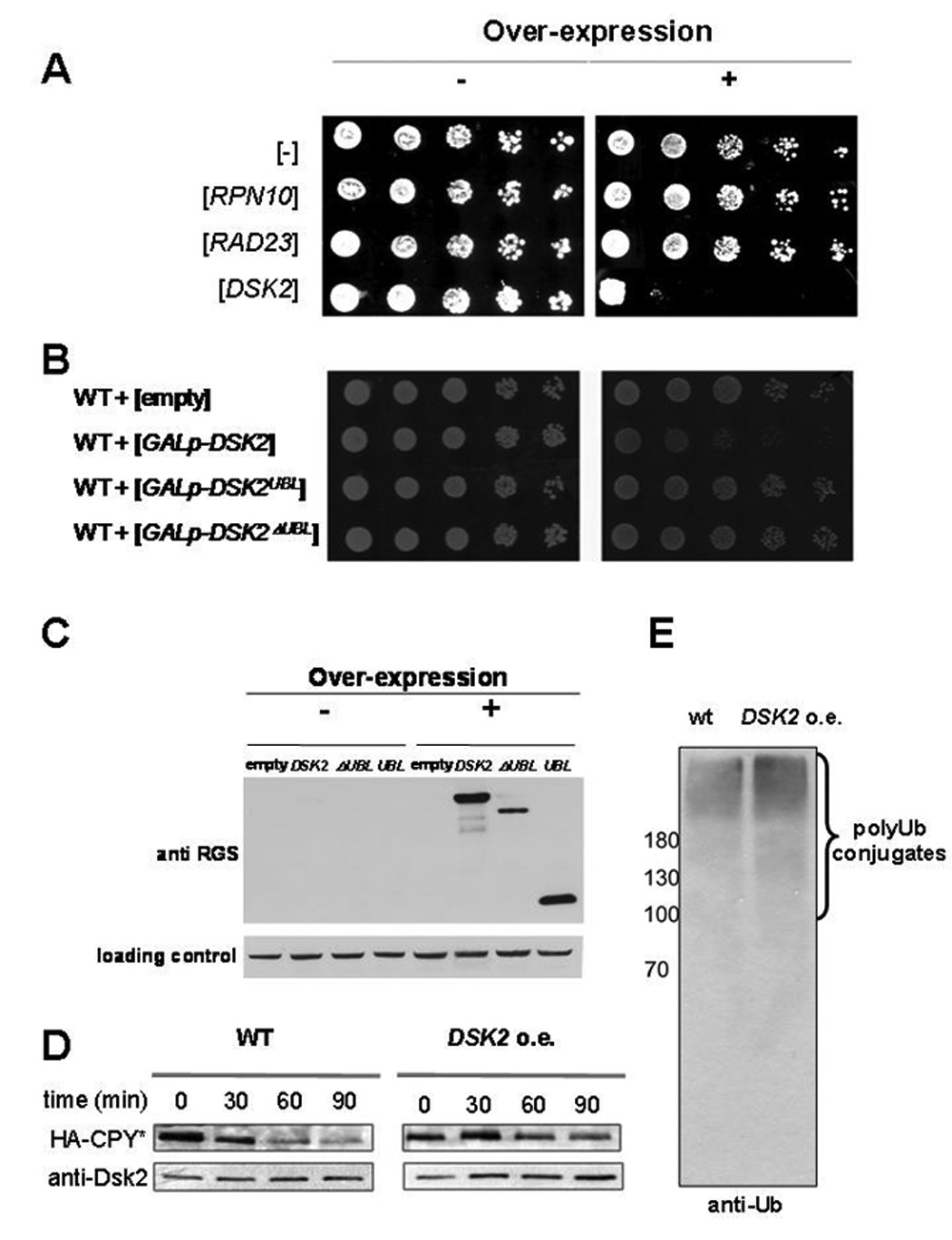
A. Unique cytotoxicity of DSK2 overexpression. Ten-fold serial dilutions of cells transformed with multicopy plasmids expressing polyUb-binding proteins from an inducible promoter: RPN10, RAD23 or DSK2. Left panel; nonexpressing conditions (glucose). Right panel; expressing conditions (galactose).
B. Full length Dsk2 is required for cytotoxicity. Cells transformed with multicopy plasmids expressing inducible wt (full length) DSK2, DSK2Ubl or DSK2ΔUbl under control of an inducible promoter (GAL1p) were plated as in panel A.
C. Cellular levels of Dsk2 variants upon induction. Extracts from cells induced for expression of RGS-His6-tagged full-length Dsk2 or either of its two truncated versions as in panel B, were immunoblotted with anti-RGS-His6 antibody. Levels of PGK serve as a loading control.
D. DSK2 overexpression leads to deceleration of proteasome substrate turnover. Degradation of HA-tagged CPY* was compared in wt and DSK2-overexpressing cells under the GAL1 promotor as in panel A –C. Three hours after induction of DSK expression, protein synthesis was blocked with 100µg/ml cycloheximide. Samples were withdrawn at indicated time points, lysed, and residual HA-CPY* analyzed by immunoblotting.
E. DSK2 overexpression correlates with an increase in HMW Ub conjugates. PolyUb levels in whole cell extract prepared from wt or DSK2 overexpressing cells were estimated by immunoblotting for Ub.
DSK2 overexpression uniquely alters cellular ubiquitin-linkage profiles
Accumulation of Ub in HMW conjugates allowed us to examine the effects of Dsk2 on the linkage profile. Recently, a method has been introduced which allows for the quantitative analysis of each of the polyUb linkage forms using mass spectrometry and stable isotope labeled internal standard peptides. This method, termed Ub-AQUA (Absolute QUAntification), has previously been used to characterize Ub conjugated to individual protein substrates (Huang et al., 2006; Kirkpatrick et al., 2006). Adapting this approach, we chose to quantify Ub linkages directly from whole cell extracts and thus characterize the Ub profile in a global rather than substrate-specific manner.
Whole cell extracts from wild type, dsk2 null or DSK2 overexpressing cells were rapidly lysed and resolved by gradient SDS-PAGE (Fig 2A). The majority of conjugated Ub was observed to migrate above 180 kDa indicating that it is in the form of HMW conjugates (as in Fig 1E). The corresponding gel region was subjected to Ub AQUA. The absolute amount of HMW conjugated Ub in wild type and Δdsk2 extracts was similar. Interestingly, a near 4-fold increase in HMW conjugated Ub was measured after 3 hours of DSK2 overexpression (Fig 2B), in accord with Ub-immunoblot results (Fig 1E).
Fig. 2. DSK2 overexpression alters cellular Ub-linkage profiles.
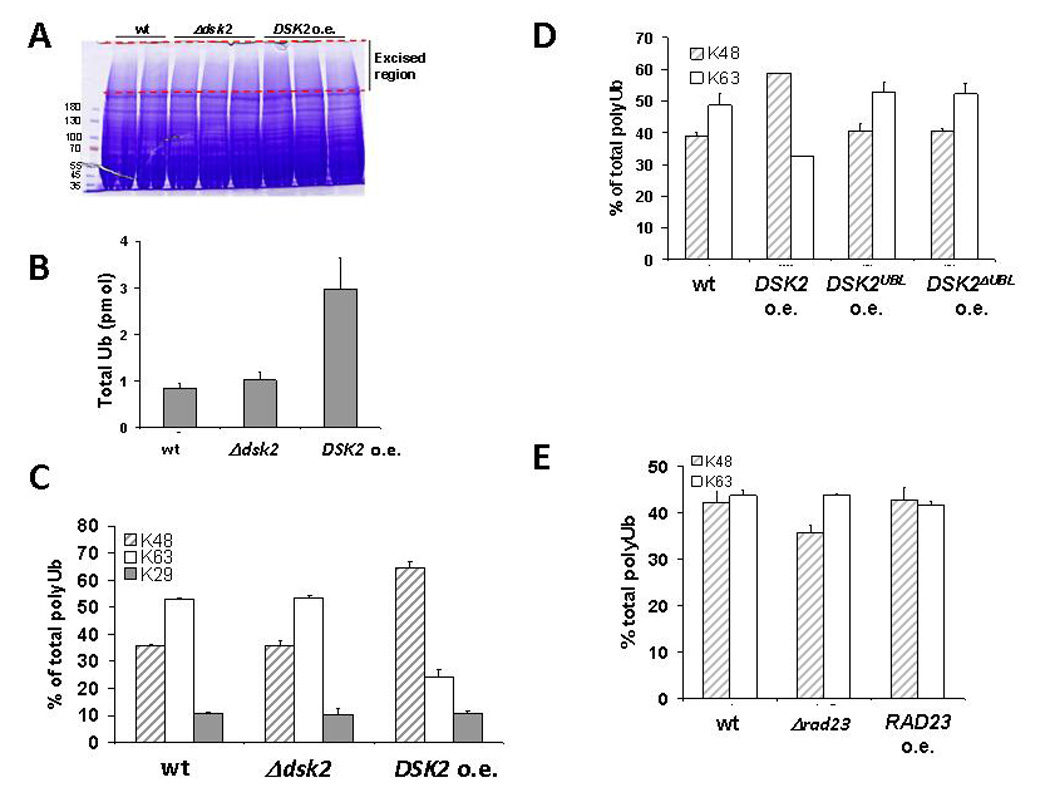
A. Whole cell extract prepared for Ub-AQUA analysis. Equal amounts of wt, Δdsk2, or DSK2 overexpressing cells grown under identical conditions were rapidly lysed, and the resulting whole cell extracts were resolved by gradient SDS-PAGE. Protein content was compared by staining with Coomassie reagent. The HMW region within the dotted lines (corresponding to the majority of the Ub signal) was prepared for Ub-AQUA analysis by tandem mass spectrometry.
B. DSK2 overexpression results in elevated levels of Ub conjugates. Total Ub levels were quantified in the HMW region of extracts normalized for equal protein content. Error-bars represent standard deviation of independent measurements.
C. The most prevalent Ub linkage types in wt and mutant strains, via Lys48, Lys63 and Lys29, are displayed as a percentage of total polyUb. Other linkages were below the limits of detection and represent a negligible percentage of total Ub. Error-bars represent standard deviation of independent measurements.
D, E. Full-length Dsk2 alters Ub linkage profiles. Relative abundance of Lys48 and Lys63 linkages in extracts from DSK2, DSK2Ubl and DSK2ΔUbl overexpressing cells (in D) or from wt, Δrad23 or RAD23 overexpressing cells (in E) were assessed as in C.
In addition to determining the total levels of conjugated Ub, quantitative information was also obtained on each of the possible linkages. We found that under our experimental conditions the two most prevalent linkage-types in minimally treated whole cell extract were Lys48-linked and Lys63-linked chains, approximately equally abundant in extracts from wild type or Δdsk2 cells (Fig 2C). A small percentage of linkages via Lys29 of Ub was consistently found in all our samples, but did not change in response to any of the conditions we tested; therefore in the remainder of this study we focused on the ratio of Lys48 to Lys63 linkages. The prevalence of Lys48 chains was documented in preparations of captured polyUb (Bennett et al., 2007; Peng et al., 2003; Xu et al., 2006) but no such data have been reported using a direct measurement in whole cell lysates as we do herein. In response to DSK2 induction, the ratio of Lys48 to Lys63 linkages increased to greater than three, corresponding with an increase in the linkages through Lys48 (Fig 2C). These results indicate that Dsk2 acts as a potent cellular factor capable of influencing the nature of the Ub pool.
The overt effect of Dsk2 levels on Ub conjugates may be correlated with its cytotoxicity (Fig 1, 2). Consistently, cells expressing DSK2Ubl or DSK2ΔUbl displayed linkage profiles that were indistinguishable from wild type (Fig 2D). These properties appear to be somewhat specific to Dsk2, as no cytotoxicity or perturbation of cellular linkage profile was measured upon deletion or induction of RAD23 (Fig 2E).
RPN10 mitigates DSK2-mediated toxicity
The pleiotropic effects of elevated Dsk2 levels are typical of defects in the UPS. And yet, incongruously, a number of proteasome mutants are known to mitigate the cytotoxicity of high abundance Dsk2 ((Biggins et al., 1996; Funakoshi et al., 2004; Funakoshi et al., 2002) and Fig 3A). We found one proteasome mutant particularly adept in suppressing DSK2 overexpression, rpn11-1 (previously known as mpr1-1; Fig 3A). This mutant exhibits elevated levels of proteasome subunits (Fig 3B), presumably as a compensation for inherent proteolytic defects documented in this mutant strain (Rinaldi et al., 2004). Since Rpn10 is known to function as a proteasomal ubiquitin receptor, we tested whether deletion of RPN10 is also able to rescue elevated DSK2. In contrast to rpn11-1, deletion of RPN10 did not rescue these cells (Fig 3C). Rpn10 is an atypical proteasome subunit, as a portion of cellular Rpn10 resides in an extraproteasomal pool (Fu et al., 2001). Thus, we tested whether a shortage of utilizable Ub, a consequence of tying up Ub in conjugates (Fig 1, 2), may partially explain Dsk2-mediated cytotoxicity. We found that although Ub induction partially suppressed DSK2 overexpression, rescue was strictly dependent on the presence of RPN10 (Fig 3C). This result highlights a special relationship between RPN10 and DSK2.
Fig. 3. RPN10 mitigates elevated DSK2 effects.
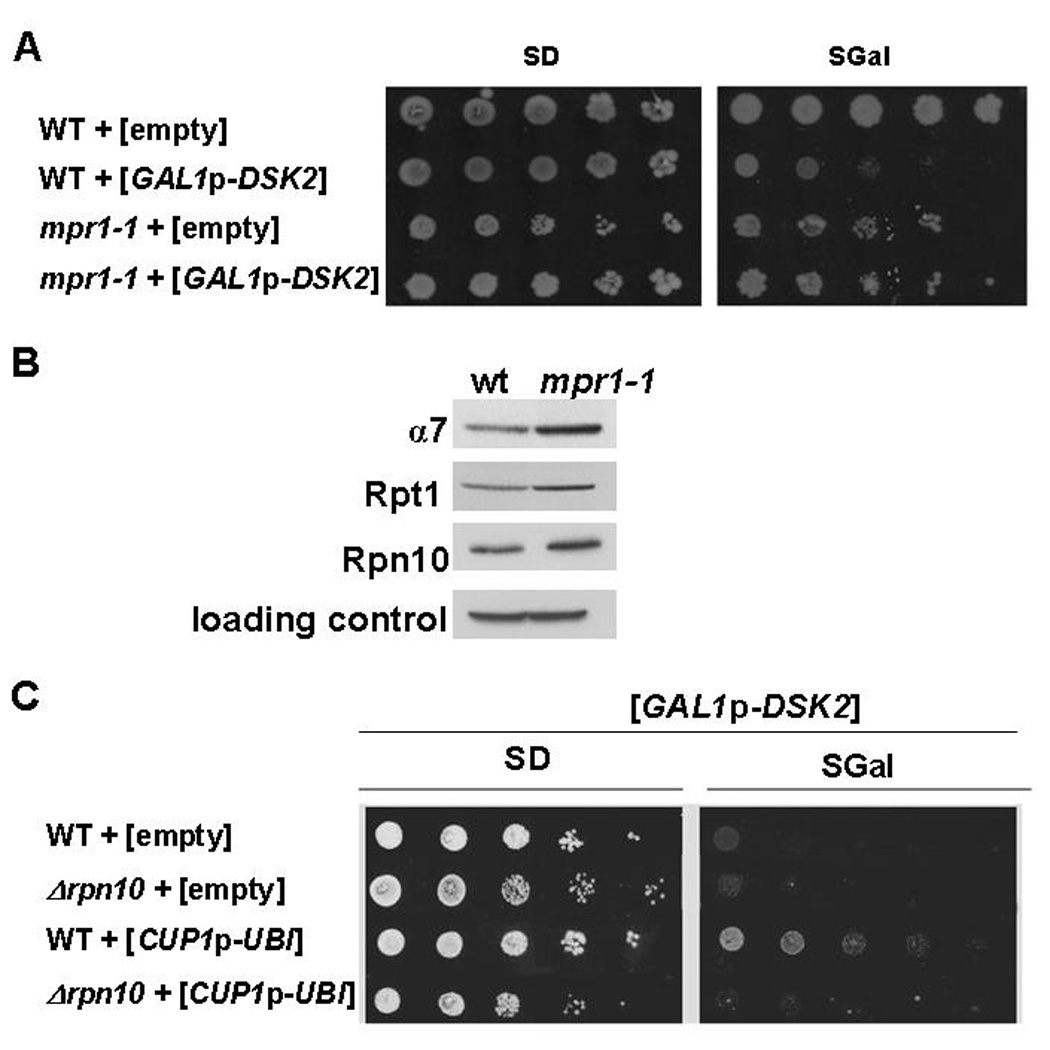
A. DSK2 overexpression growth phenotype is suppressed by proteasome mutants. Ten-fold serial dilutions of rpn11-1 and its wt parent strain bearing either a high-copy plasmid for inducible DSK2 expression or an empty vector for control were pated as in Fig 1A.
B. Proteasome levels in rpn11-1. Whole cell extract prepared from wt or rpn11-1 strains were immunoblotted for different proteasome subunits (PGK levels served as a loading control).
C DSK2 overexpression is suppressed by Ub, but requires RPN10. Exogenous Ub was expressed over endogenous levels from a high-copy plasmid under a copper-induced promoter (CUP1p) in wt and Δrpn10 strains. Empty vector served as a control for Ub levels. All strains were cotransformed with a high-copy plasmid for inducible DSK2 expression under GAL1p and plated on growth media as in A.
Rpn10 interacts with Dsk2
In order to better understand the relationship between the two proteasome-associated polyUb-binding proteins, Dsk2 and Rpn10, we evaluated the genetic interactions of RPN10 and DSK2 and compared it to the analogous relationship of RPN10 with RAD23. Cells lacking both RAD23 and RPN10 were significantly more sensitive than either single deletant to a variety of stress conditions (Fig 4). This observation corroborated the reported synthetic phenotype of RPN10 and RAD23 (Lambertson et al., 1999; Wilkinson et al., 2001). Yet, we detected no such additive phenotypes for the double deletion of RPN10 and DSK2 (Fig 4). This relationship is compatible with Rpn10 and Dsk2 functioning sequentially within a defined pathway.
Fig. 4. Genetic interactions of RPN10 and genes encoding for the Ubl-UBA proteins Dsk2 or Rad23.
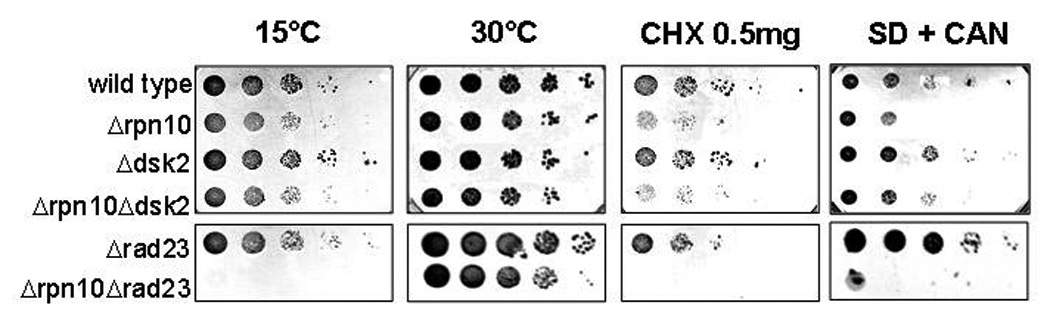
Wild-type, single- and double-deletion strains were serially diluted onto YPD as indicated and incubated at 30°C or 15°C until single colonies appeared, or plated onto media containing 0.5µg/ml of cycloheximide (CHX), or onto arginine drop-out media supplemented with 3µg/ml canavanine
To evaluate the Rpn10-Dsk2 interaction, we compared association of Rpn10 with members of the Ubl-UBA family Rad23, Dsk2, and Ddi1 (Fig 5A, B). Domain mapping showed that the main interaction between the two proteins was found to be through the UIM of Rpn10 and the Ubl domain of Dsk2 (Fig 5C). Moreover, Dsk2Ubl was potent to precipitate Rpn10 directly from whole cell extract (Fig 5D), providing this physical interaction with biological relevance. Despite similarities among Ubl domains, we conclude that the relationship of Rpn10 with Dsk2 is of a distinctive nature.
Fig. 5. Specificity of interaction between Rpn10 and the Ubl domain containing protein, Dsk2.
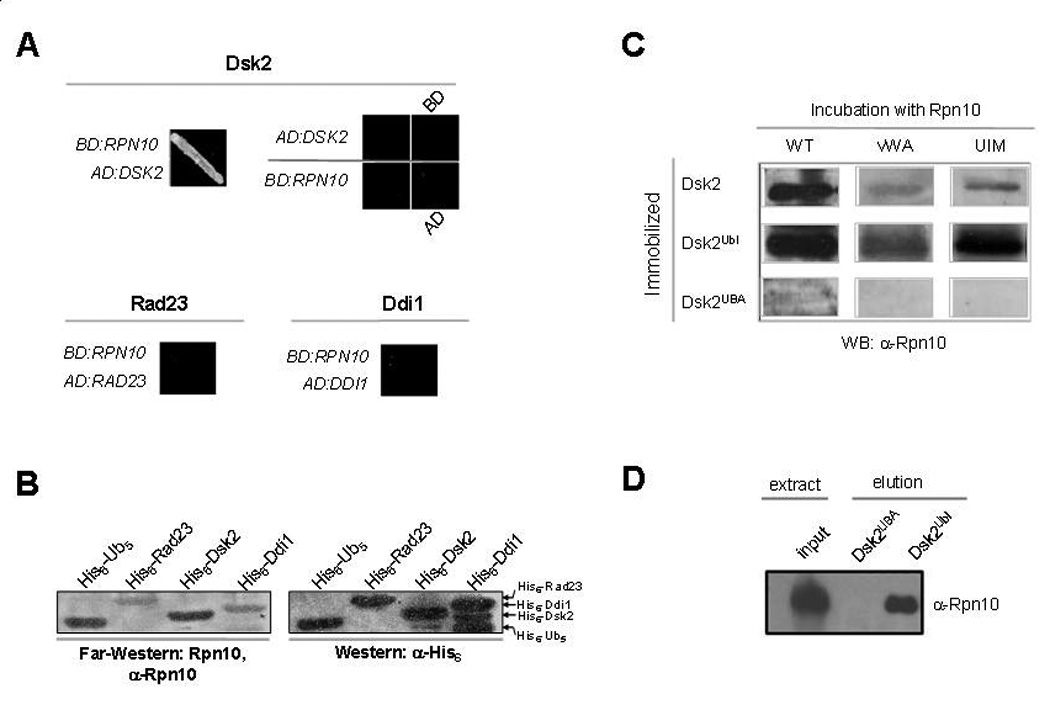
A. Dsk2 and Rpn10 interact by yeast two-hybrid. Association of Rpn10 with the Ubl-containing proteins, Dsk2, Rad23, or Ddi1 was tested in vivo by transforming all possible combinations of bait versus prey constructs and assaying for growth on –His–Trp dropout selective media supplemented with 7.5mM 3-aminotriazol. Representative results are shown. A lone combination, BD:RPN10 with AD:DSK2, supported growth.
B. Direct protein-protein interaction of Rpn10 with Dsk2. 30 pmol of Ub/Ubl-containing proteins were immobilized on a nitrocellulose membrane. Migration pattern and amounts of immobilized proteins was evaluated by immunoblotting with anti-His6 antiserum (right panel). In parallel, a similar membrane was incubated with 2µg/ml of recombinant Rpn10, washed, and immunoblotted against residually bound Rpn10 (left panel). Association of Rpn10 with polyUb is shown as a positive control.
C. The Ubl domain of Dsk2 interacts with the UIM of Rpn10. Dsk2 or its structural domains were subjected to Far-Western analysis using either full length Rpn10 or its truncated versions as in panel B.
D. The Ubl domain of Dsk2 is sufficient to precipitate Rpn10 from whole cell extract. Whole cell extract was applied to an affinity column generated from crosslinked Dsk2Ubl or Dsk2UBA coupled to activated Sepharose resin. After stringent washes, bound proteins were eluted with 8M urea and assayed from presence of Rpn10 by immunoblotting.
Rpn10 restricts Dsk2 access to the proteasome
Ubl domain-containing proteins differ also in their association with the proteasome. Whereas Rad23 and Ubp6 have been identified in purified proteasomes, no such information is available regarding Dsk2 (Gavin et al., 2002; Guerrero et al., 2006; Krogan et al., 2006; Verma et al., 2000). This result is particularly surprising given the presence of a Ubl domain in the Dsk2 protein and its ability to bind the proteasome subunit, Rpn10. Nevertheless, in vitro, exogenous Dsk2 is able to associate with purified proteasomes and even compete with Rad23 suggesting they may share a common binding site (Elsasser et al., 2002). To understand how the relationship of Dsk2 and Rpn10 (Fig 4, 5) may affect recognition of Dsk2 by the proteasome, we affinity isolated proteasome from whole cell wt or Δrpn10 extracts. Whereas the Ubl-containing proteins Rad23 and Ubp6 were both found in association with affinity purified wild-type proteasomes, Dsk2 was undetectable (Fig 6A). Proteasomes purified from the Δrpn10 strain, in contrast to wt proteasomes, contained significant amounts of the Dsk2 protein, alongside Rad23 and Ubp6.
Fig. 6. Extraproteasomal Rpn10 restricts Dsk2 access to the proteasome.
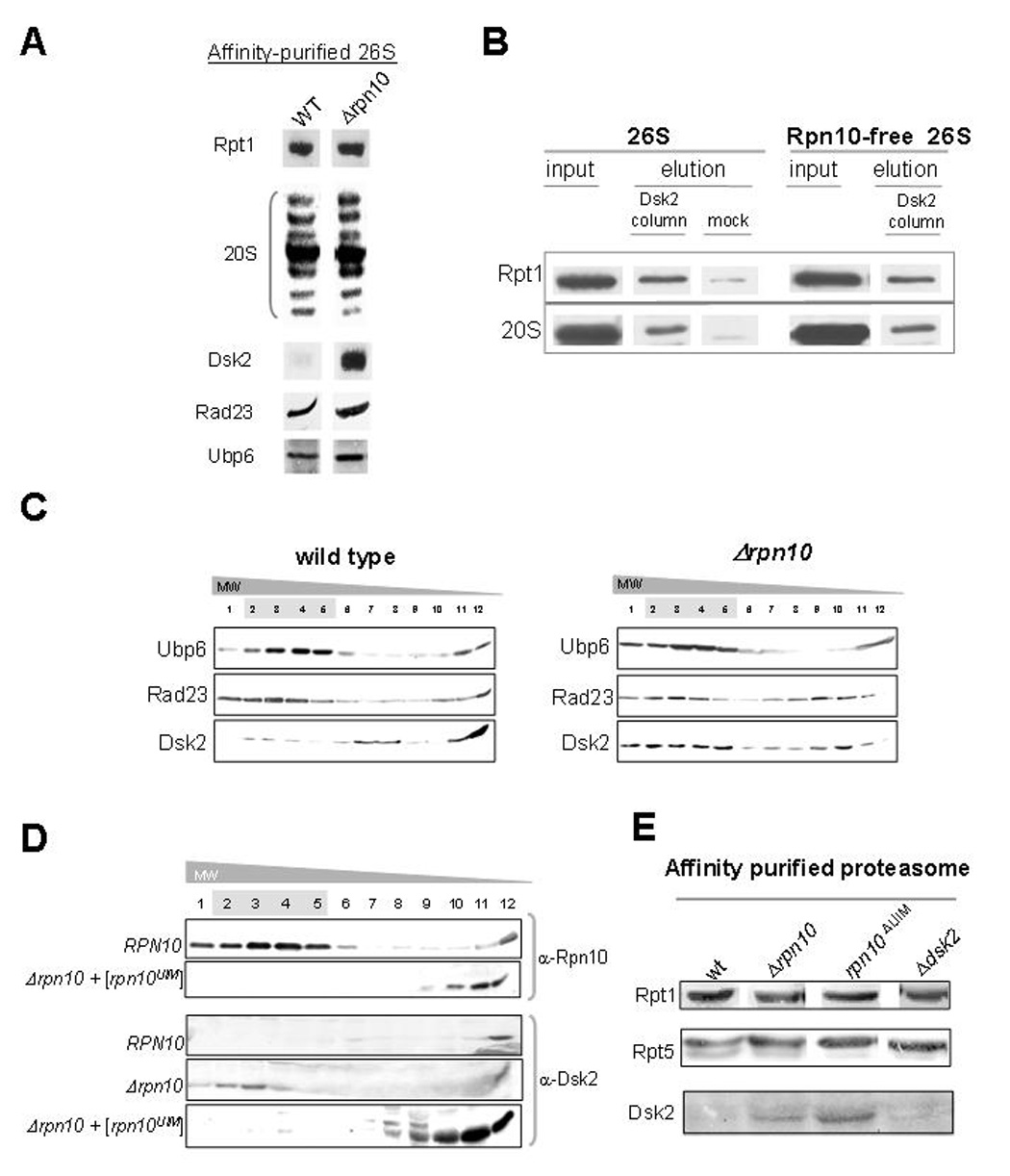
A. Significant enrichment of Dsk2-associated with affinity-purified Δrpn10 proteasomes. Protein A-tagged proteasomes were affinity purified from wt or Δrpn10 backgrounds and analyzed for presence of Ubl-domain containing proteins. Proteasomes were normalized according to integral subunit content as indicated, and immunoblotted for presence of copurifying Dsk2, Rad23, or Ubp6.
B. Affinity of Dsk2 for isolated proteasome is Rpn10-independent. Purified wt or Rpn10-free proteasomes were passed over Dsk2 immobilized on Ni-NTA resin. After stringent washing, Dsk2 was eluted with imidazole, and associated proteasomes were evaluated. Binding of proteasomes to mock Ni-NTA resin is shown as a negative control.
C. Partitioning of Dsk2 between proteasome-bound and unbound forms depends on Rpn10. Whole cell extract prepared from wt (left) or Δrpn10 (right) was fractionated by glycerol gradient centrifugation. Resulting fractions were assayed for proteasomes by peptidase activity (highlighted in grey), and for presence of Ubl-domain containing proteins by immunoblotting as indicated.
D. Rpn10UIM is sufficient to alter Dsk2 partitioning. Whole cell extracts bearing full length Rpn10 or a truncated version encompassing the C-terminal UIM motif (Rpn10UIM) were fractionated as in C. Distribution of full length Rpn10 or Rpn10UIM is indicated, as well as partitioning of Dsk2 as a function of Rpn10 in the relevant strains.
E. UIM of Rpn10 is required to sequester Dsk2 from associating with 26S proteasomes. Affinity purified protein A-tagged proteasomes from wt, Δrpn10, rpn10ΔUIM and Δdsk2 strains were analyzed for presence of Dsk2 in comparison to intrinsic proteasome subunits.
In order to gain insight into the negative effect that Rpn10 has on proteasome-incorporation of Dsk2, we measured Dsk2 binding to isolated proteasomes. Proteasomes were stringently purified from wild type or Δrpn10 strains (removing most auxiliary factors as documented (Glickman and Coux, 2001)) and subjected to immobilized recombinant Dsk2. Both proteasome preparations were equally competent to retain Dsk2. Apparently, proteasome-incorporated Rpn10 did not change the affinity of proteasome for Dsk2 (Fig 6B).
Next, we measured the effect of Rpn10 in whole cell extract on the MW partitioning of Dsk2. In wild type extract, the majority of Dsk2 was found in low MW fractions, with only traces migrating in proteasome-containing HMW fractions (Fig 6, C left panel). In contrast, other Ubls, such as Ubp6 or Rad23, were observed migrating with proteasomes. (Fig 6, C left panel). In extract from Δrpn10, partitioning of Dsk2 shifted markedly toward HMW proteasome-containing fractions (Fig 6, C right panel). Behavior of Ubp6 or Rad23 remained unchanged. In summary, individual members of the Ubl family display differential proteasome association: among the Ubls we checked, only distribution of Dsk2 was Rpn10-dependent.
To further investigate the role of extraproteasomal Rpn10, we took advantage of a mutant form of Rpn10 that is able to bind Dsk2, yet cannot incorporate into proteasomes, e.g. Rpn10UIM (Fu et al., 2001). We confirmed that Rpn10UIM is not self-sufficient to incorporate into proteasomes (Fig 6D). Then, MW distribution of Dsk2 was assayed as a function of Rpn10 or of its truncated UIM variant (Fig 6D, bottom). Expression of just the UIM fragment was sufficient to block Dsk2 from associating with proteasomes, rendering most Dsk2 to low MW fractions. This result indicates that Rpn10 outside of the proteasome competes against the proteasome for Dsk2 binding. The converse was also true: elimination of the UIM from Rpn10 was sufficient to elevate levels of Dsk2 in proteasome samples (Fig 6E). Taken together, extraproteasomal Rpn10 limits association of Dsk2 with the proteasome.
Extraproteasomal Rpn10 acts as a filter mitigating effects of Dsk2 overabundance
To put the Rpn10-filter hypothesis to test, we assayed whether extraproteasomal Rpn10 could attenuate DSK2-induced stress. Cell viability was assessed upon DSK2-induction in the presence or absence of Rpn10 or its UIM fragment. Expressing RPN10UIM was able to suppress DSK2-mediated toxicity to a similar extent as the full length protein (Fig 7A). Although Rpn10UIM fragment does not associate with the 26S proteasome (Fig 6D), it may mask the Ubl of Dsk2, competing for association with the proteasome. The outcome may mimic the effect of inducing Dsk2 deleted for its Ubl domain (Fig 1, 2). We conclude that the mechanism of Dsk2-mediated toxicity involves association of Dsk2 and its cargo with the proteasome.
Fig. 7. UIM of Rpn10 is required to suppress Dsk2-mediated effects.
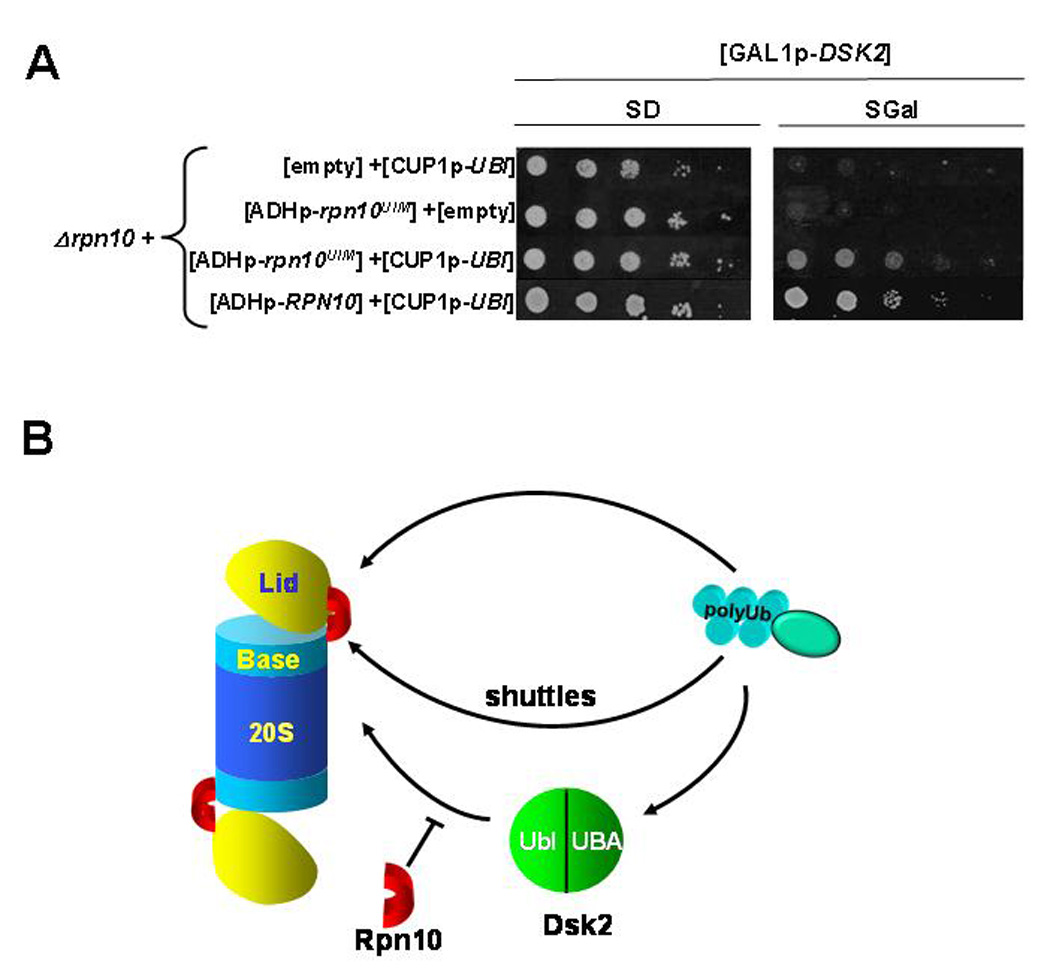
A. Sequestering Dsk2 from proteasome by Rpn10UIM fragment suppresses cytotoxicity. A Δrpn10 strain was co-transformed with combinations of high copy plasmids for expressing GAL1p-DSK2, CUP1p-UBI, and either ADH1p-RPN10UIM, ADH1p-RPN10 or empty vectors for control where indicated. Three-fold serial dilutions of the resulting strains were spotted onto selective media (glucose; left) or media for DSK2 induction (galactose; right).
B. A model describing regulation of Dsk2-escorted cargo destined for the proteasome. Ub-modified substrates may reach the proteasome directly, binding to internal Ub-receptors (top trajectory). Alternatively, recruitment of Ub-conjugates may be mediated by Ub-delivery proteins such as Dsk2, Rad23, Ddi1, and possibly Rpn10 or Cdc48 (middle arrow). The Dsk2-mediated trajectory (bottom) is subject to a filter in the form of extraproteasomal Rpn10 that limits the availability of Dsk2. Rpn10 plays a critical role in filtering Dsk2 and its substrates from the proteasome and therefore in prioritization of proteolysis and in maintaining proper equilibrium of cellular Ub linkages.
DISCUSSION
The first polyUb-binding protein to be identified was Rpn10 (Deveraux et al., 1994). It was naturally supposed to play an essential role in the proteasome-recruitment of ubiquitinated substrates. However, approximately 90% of all Ub-tagged targets were later estimated to reach the proteasome and be degraded independently of Rpn10UIM (Mayor et al., 2007), suggesting that proteasomes have other intrinsic Ub receptors (as has been proposed for Rpn13 (Husnjak et al., 2008)). Here we describe an essential role of RPN10UIM in suppressing the lethality associated with another polyUb-binding protein, DSK2, and in doing so we provide the first evidence for a biological role of extraproteasomal Rpn10.
In principle, any factor with affinity for polyUb species has the capacity to alter their fate and influence the global Ub landscape. Among such proteins, proteasome-associated Ub shuttles are likely to be key players. Published studies highlight the critical need to fine-tune cellular levels of Ub shuttles as either diminished or elevated levels interfere with efficiency of the UPS (Chen and Madura, 2002; Elsasser et al., 2004; Hartmann-Petersen et al., 2003; Lambertson et al., 1999; Raasi and Pickart, 2003; Saeki et al., 2002a; Verma et al., 2004). At least as far as Dsk2 is concerned, elevated levels stabilize Ubconjugates and inhibit proteasome-dependent degradation (Fig 1). Since Dsk2 is apparently one of the more potent Ub binders (Funakoshi et al., 2002; Hartmann-Petersen et al., 2003; Raasi et al., 2005; Saeki et al., 2002a; Wilkinson et al., 2001) the rationale for a mechanism restricting its association with the degradation machinery becomes apparent. We found that Dsk2-induced stress can be partially overcome by expanding the pool of utilizable Ub, nevertheless, cell viability under elevated Dsk2 levels is strictly dependent on the presence of its proteasomal filter (Fig 3, 7). The ability of Rpn10UIM to interfere with the Dsk2-proteasome interaction and attenuate the severity of DSK2-associated phenotypes (Fig 6, 7) stresses the importance of extraproteasomal Rpn10 as a filter for Dsk2. As a direct consequence, some of the substrates whose degradation have been found to be altered in absence of functional RPN10 (Mayor et al., 2007) may reflect an involvement of Dsk2.
In this study we focused on an interaction of Dsk2 with Rpn10, however, we expect that other Ubl domain-containing proteins may be under similar regulation. For instance, human S5a contains two UIMs: UIM1 and UIM2 with slightly different binding specificities (Fujiwara et al., 2004; Hofmann and Falquet, 2001; Miller et al., 2004; Wang et al., 2005). UIM1, which resembles the single UIM of Rpn10, has been shown to associate with polyUb and with the Ubl of hPLIC (the mammalian ortholog of Dsk2) in line with the Rpn10-Dsk2 interaction we reported herein (Fig 5). UIM2 was found to preferentially bind the Ubl of hHR23 (Fujiwara et al., 2004; Kleijnen et al., 2000; Ryu et al., 2003; Seok Ko et al., 2004; Walters et al., 2002). The combinatorics of multiple spliced versions of S5a (Kikukawa et al., 2002) may provide modular filters for a wide array of substrates by possessing alternate binding specificities. Other proteins with affinities for Ubls could provide competing binding partners. For example Pth2 has also been shown to bind Dsk2, affecting is cellular availability (Ishii et al., 2006). Beyond the Ubl-UBA shuttles, it would be interesting to study conditions that govern the cellular distribution of other Ubls such as the E3 ligase Parkin (Sakata et al., 2003), the co-chaperone Bag1, or the DUB Ubp6/USP14. Not only are there many pathways a substrate can take to reach the proteasome, but in many cases, these trajectories may be subject to differential regulation (Fig 7B).
The existence of multiple delivery proteins may reflect a need to selectively target ubiquitinated cargo either at the level of the substrate or of the Ub signal. In this study we mapped the conjugated Ub landscape and demonstrated that Lys48 and Lys63-linkages make up the bulk of HMW polyUb chains in S. cerevisiae grown under normal conditions (Fig 2). Induction of DSK2 uniquely led to accumulation of Lys48-linkages. This raises the question of why increased Dsk2 promotes build-up of Lys48-linked chains, and why these targets are not degraded? In one scenario, Dsk2 binding to the proteasome sterically impedes the binding of Ubl-containing shuttles or of Lys48-linked chains directly by competing for an intrinsic Ubl-receptor on the proteasome (such as Rpn1; (Elsasser et al., 2002; Rosenzweig et al., 2008; Saeki et al., 2002b; Seeger et al., 2003)) or by imposing steric hindrance on a Lys48-recognition unit within the 26S (theoretically, Rpn13). In such a model, Dsk2-mediated toxicity would stem from the prevention of substrate flux through the proteasome, with the altered composition of Ub linkages (enrichment of Lys48 linkages) reflecting the consequence of deficient substrate processing (Fig 2). This model has appeal since either deleting or masking the Ubl domain of Dsk2 attenuated its deleterious effects (Fig 1, 7). Nevertheless, Dsk2 lacking cargo does not seem to clog the proteasome since expression of the Dsk2 Ubl domain alone had no discernable effects (Fig 1, 2). Another model would invoke Dsk2 preference for Lys48 chains, which in absence of sufficient binding sites on the proteasome sequesters conjugates from editing or degradation by the proteasome. That expression of the UBA alone does not lead to substrate accumulation or polyUb accumulation is not easy to rationalize in this case, though it may indicate that the Ubl domain has a stabilizing effect on the ability of Dsk2 to trap its cargo. One model that has not been formally ruled out is that Dsk2 preferentially accelerates the turnover of substrates modified by alternate polyUb chains, thereby delaying their turnover over "canonical" targets conjugated to Lys48-linked chains. Currently we are attempting to experimentally distinguish between these possibilities and identify substrates whose biological turnover is specifically affected by Dsk2.
Maintaining alternate routes to the proteasome is beneficial for prioritization of substrates when the system is at full capacity. Accumulation of polyUb conjugates often correlates with decreases in available free Ub, which may explain the need for extra Ub during periods of cellular stress (Chernova et al., 2003; Hanna et al., 2003; Hanna et al., 2007; Swaminathan et al., 1999) (see also Fig 3). A recent report suggested that a cellular Ub sensor is in place to detect the amount of available free Ub and to direct proteasome remodeling in order to compensate for deficits (Hanna et al., 2007). We propose that Ub-sensing is a broader phenomenon that takes into account Ub in multiple forms including conjugated Ub. Although it is not yet known whether the specific accumulation of Lys48-polyUb chains in DSK2 mutants is the initiator or consequence of Ub stress, it is likely that excessive Lys48-linkages disrupts the default proteolysis prioritization scheme marking the cell’s attempt to increase proteolytic efficiency by producing higher-priority substrates. This model is in accord with what has recently been observed in Huntington's disease pathology (Bennett et al., 2007).
Experimental Procedures
Yeast strains, E coli plasmids, growth media, and Canavanine, heat-shock or cycloheximide phenotypic analysis
See details in accompanying supplementary material online.
Cycloheximide-chase assay
The assay was performed as described previously (Hanna et al., 2003).
Yeast two-hybrid assay
Physical interactions between RPN10 cloned into pGBKT7 (Clontech) and DSK2, RAD23, DDI1 ORFs cloned into pGADT7 (Clontech) were assayed by yeast two-hybrid as described previously (Fu et al., 2001).
Construction of plasmids
The genes were subcloned either into bacterial or into yeast expression vectors as described in Supplementary material (Table I).
Expression and purification of His6-tagged protein
All His-tagged proteins were cloned into pQE30 (Qiagen), expressed in bacteria and purified on Ni-NTA according to manufacturers recommendations. Details in supplementary material online.
Far-Western Assay
30pmole of proteins were resolved by SDS-PAGE and transferred to 0.2µ nitrocellulose membranes. Membranes were blocked with 10% nonfat milk for 1hr, and then incubated in the presence of 1.5µg/ml native recombinant Rpn10 derivatives in PBST solution for 2hrs and washed. Membranes were immunoblotted with anti-Rpn10.
Binding proteins from whole cell extracts to Dsk2UBA or Dsk2Ubl column
Dsk2UBA or Dsk2Ubl column preparation
6His-tagged Dsk2UBA or Dsk2Ubl were purified by Ni-NTA as described in Expression and purification of 6His-tagged protein. The proteins were dialyzed against PBS buffer and conjugated to Activated CH-Sepharose (Sigma) according to manufacturer’s instructions.
Binding proteins from whole cell extracts to the columns
2 ml extracts of BY4742 prepared in Lysis buffer (50mM Tris pH 7.5, 150mM KCl, 0.1% (v/v) Triton X-100, 2mM DTT supplemented with Protease Cocktail Inhibitor) were incubated with 150µL agarose-immobilized Dsk2ΔUBA or Dsk2ΔUbl for 1hr at 25°C, washed with Lysis buffer followed by 50mM Tris pH 7.5, 1M KCl, 2mM DTT. Proteins were eluted with 50mM Tris pH 7.5, 8M Urea.
Fractionation of extracts by glycerol gradients
Whole cell extract was fractionated over 11-ml 10% and 40% glycerol gradients as described (Fu et al., 1998). Details in supplementary material online.
Purification of the 26S proteasome and Peptidase assay for proteasome activity
Proteasomes were purified from yeast lysates as described previously (Glickman and Coux, 2001). Details in supplementary material online.
Pull-down of the wt/Rpn10-free proteasome by His6-Dsk2
Extract of E.coli expressing His6-Dsk2 was applied to Ni-NTA resin pre-equilibrated with lysis buffer (10mM Imidazole, 150mM NaCl, 50 mM Tris pH 7.4). The mixture was incubated for 2hr at 4°C. The beads were rinsed with buffer A (50mM NaCl, 150 mM Tris pH 7.4, 10mM MgCl2, 1mM β-mercaptoethanol, 1mM ATP, 0.05% Tween-20) and incubated either with purified wt 26S or with Rpn10-free 26S in the buffer A overnight. After washing with buffer-B (buffer A + 100mM NaCl), the proteins were eluted with Laemmli buffer. A typical experiment contained 40µL His6-Dsk2 immobilized on 40µL Ni-NTA resin with 75–80µg of purified 26S proteasome in 150µL buffer-A.
Tandem affinity purification (TAP) of the 26S proteasome from yeast lysates
Cells carrying proteasome TAP-tagged at one of its 20S subunits were used for extract preparation using buffer 1 (50mM Tris pH 8.0, 1mM EDTA, 10mM MgCl2, 5mM ATP, 100mM NaCl, 10%(v/v) glycerol). The extracts were applied to 1ml IgG Superose-6 Fast Flow resin (Amersham Biosciences). The mixture was tumbled for 2hrs at 4°C. After the wash with buffer 2 (50mM Tris pH 7.4, 1mM EDTA, 10mM MgCl2, 5mM ATP, 100mM NaCl, 10%(v/v) glycerol), the proteasomes were eluted using 100units of TEV protease (Stratagen) in buffer 3 (50mM Tris pH8.0, 0.5mM EDTA, 10mM MgCl2, 1mM ATP, 10%(v/v) glycerol).
Ub-AQUA analysis
Yeast cell extract preparation for AQUA analysis
5-ml cell culture was grown to stationary phase; OD was measured at 595nm and cell density normalized. Samples were prepared by TCA lysis and precipitation (sup. file) and separated on 4–12% Tris-Bis SDS PAGE.
Gel regions were excised as indicated, and prepared for Ub-AQUA by liquid-chromatography selected reaction monitoring (LC-SRM) in a TSQuantum Ultra (ThermoElectron, San Jose, CA) as published (Kirkpatrick et al., 2005) and detailed in the accompanying supplementary material file online.
Supplementary Material
Acknowledgments
MG was supported by grants from the Israel Science Foundation (ISF) and the USA-Israel Binational science foundation (BSF). OK was partially funded by an ISF postdoctoral fellowship (VATAT). SG is supported by NIGMS GM67945. We thank Ray DeShaies, Dan Finley, Hongyong Fu, Rasmus Hartmann-Petersen, Colin Gordon, Kay Hofmann, and Thibault Mayor for insightful ideas. None of the authors of this work have a financial interest related to this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bennett EJ, Shaler TA, Woodman B, Ryu K-Y, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Biggins S, Ivanovska I, Rose MD. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J Cell Biol. 1996;133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Shinde U, Ortolan TG, Madura K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001;2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova TA, Allen KD, Wesoloski LM, Shanks JR, Chernoff YO, Wilkinson KD. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due todepletion of free ubiquitin pool. J Biol Chem. 2003 doi: 10.1074/jbc.M310283200. M310283200. [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26S subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Deveraux Q, van Nocker S, Mahaffey D, Vierstra R, Rechsteiner M. Inhibition of Ubiquitin-mediated proteolysis by the Arabidopsis 26S protease subunit s5a. J Biol Chem. 1995;270:29660–29663. doi: 10.1074/jbc.270.50.29660. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 Serve as Alternative Ubiquitin Receptors for the Proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- Fu H, Sadis S, Rubin DM, Glickman MH, van Nocker S, Finley D, Vierstra RD. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26S proteasome subunit Mcb1. J Biol Chem. 1998;273:1970–1989. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- Fu HY, Reis N, Lee Y, Glickman MH, Vierstra R. Subunit interaction maps for the regulatory particle of the 26s proteasome and the cop9 signalosome reveal a conserved core structure. EMBO J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Tenno T, Sugasawa K, Jee J-G, Ohki I, Kojima C, Tochio H, Hiroaki H, Hanaoka F, Shirakawa M. Structure of the Ubiquitin-interacting Motif of S5a Bound to the Ubiquitin-like Domain of HR23B. J Biol Chem. 2004;279:4760–4767. doi: 10.1074/jbc.M309448200. [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Li X, Velichutina I, Hochstrasser M, Kobayashi H. Sem1, the yeast ortholog of a human BRCA2-binding protein, is a component of the proteasome regulatory particle that enhances proteasome stability. J Cell Sci. 2004;117:6447–6454. doi: 10.1242/jcs.01575. [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc Natl Acad Sci USA. 2002;99:745–750. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The Ubiquitin-proteasome Proteolytic Pathway: Destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Coux O. Current Protocols in Protein Science. John Wiley & Sons: New York; 2001. Purification and characterization of proteasomes from Saccharomyces cerevisiae; pp. 21.25.21–21.25.17. [DOI] [PubMed] [Google Scholar]
- Guerrero C, Tagwerker C, Kaiser P, Huang L. An Integrated Mass Spectrometry-based Proteomic Approach: Quantitative Analysis of Tandem Affinity-purified in vivo Cross-linked Protein Complexes (qtax) to Decipher the 26 s Proteasome-interacting Network. Mol Cell Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- Hanna J, Leggett DS, Finley D. Ubiquitin Depletion as a Key Mediator of Toxicity by Translational Inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Meides A, Zhang DP, Finley D. A Ubiquitin Stress Response Induces Altered Proteasome Composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- Hartmann-Petersen R, Hendil KB, Gordon C. Ubiquitin binding proteins protect ubiquitin conjugates from disassembly. FEBS Letters. 2003;535:77–81. doi: 10.1016/s0014-5793(02)03874-7. [DOI] [PubMed] [Google Scholar]
- He X, Jones MH, Winey M, Sazer S. Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe. J Cell Sci. 1998;111:1635–1647. doi: 10.1242/jcs.111.12.1635. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal degradation systems. Trends Biochem Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Funakoshi M, Kobayashi H. Yeast Pth2 is a UBL domain-binding protein that participates in the ubiquitin-proteasome pathway. The EMBO Journal. 2006;25:5492–5503. doi: 10.1038/sj.emboj.7601418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikukawa Y, Shimada M, Suzuki N, Tanaka K, Yokosawa H, Kawahara H. The 26S proteasome Rpn10 gene encoding splicing isoforms: evolutional conservation of the genomic organization in vertebrates. Biol Chem. 2002;383:1257–1261. doi: 10.1515/BC.2002.139. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. Embo J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Kleijnen MF, Alarcon RM, Howley PM. The Ubiquitin-associated Domain of hPLIC-2 Interacts with the Proteasome. Mol Biol Cell. 2003;14:3868–3875. doi: 10.1091/mbc.E02-11-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijnen MF, Shiih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. The hPLIC proteins may provide a link between the Ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Lambertson D, Chen L, Madura K. Pleiotropic defects caused by loss of the proteasomal-interacting factors Rad23 and Rpn10 of S. cerevisiae. Genetics. 1999;153:69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor T, Graumann J, Bryan J, MacCoss MJ, DeshaieS RJ. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol Cell Proteomics. 2007 doi: 10.1074/mcp.M700264-MCP200. M700264-MCP700200. [DOI] [PubMed] [Google Scholar]
- Mayor T, Russell Lipford J, Graumann J, Smith GT, Deshaies RJ. Analysis of poly-ubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol Cell Proteomics. 2005 doi: 10.1074/mcp.M400220-MCP200. M400220-MCP400200. [DOI] [PubMed] [Google Scholar]
- Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:1–6. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SLH, Malotky E, O'Bryan JP. Analysis of the role of UIMs in ubiquitin-binding and ubiquitylation. J Biol Chem. 2004;279:33528–33537. doi: 10.1074/jbc.M313097200. [DOI] [PubMed] [Google Scholar]
- Ortolan TG, Tangaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2:601–607. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Raasi S, Pickart CM. Rad23 Ubiquitin-associated Domains (UBA) Inhibit 26 S Proteasome-catalyzed Proteolysis by Sequestering Lysine 48-linked Polyubiquitin Chains. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Pick E, Gambadoro A, Zilli S, Maytal-Kivity V, Frontali L, Glickman MH. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu K-S, Lee K-J, Bae S-H, Kim B-K, Kim K-A, Choi B-S. Binding Surface Mapping of Intra- and Interdomain Interactions among hHR23B, Ubiquitin, and Polyubiquitin Binding Site 2 of S5a. J Biol Chem. 2003;278:36621–36627. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Saitoh A, Toh-e A, Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem Biophys Res Commun. 2002a;293:986–992. doi: 10.1016/S0006-291X(02)00340-6. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Sone T, Toh-e A, Yokosawa H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem Biophys Res Commun. 2002b;296:813–819. doi: 10.1016/s0006-291x(02)02002-8. [DOI] [PubMed] [Google Scholar]
- Sakata E, Yamaguchi Y, Kurimoto E, Kikuchi J, Yokoyama S, Yamada S, Kawahara H, Yokosawa H, Hattori N, Mizuno Y, et al. Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep. 2003;4:301–306. doi: 10.1038/sj.embor.embor764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Hartmann-Petersen R, Wilkinson CRM, Wallace M, Samejima I, Taylor MS, Gordon C. Interaction of the Anaphase-promoting Complex/Cyclosome and Proteasome Protein Complexes with Multiubiquitin Chain-binding Proteins. J Biol Chem. 2003;278:16791–16796. doi: 10.1074/jbc.M208281200. [DOI] [PubMed] [Google Scholar]
- Seok Ko H, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Letters. 2004;566:110–114. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. M Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart C. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya SC, Hegde AN. A potential proteasome-interacting motif within the ubiquitin-like domain of parkin and other proteins. Trends in Biochemical Sciences. 2003;28:280–283. doi: 10.1016/S0968-0004(03)00092-6. [DOI] [PubMed] [Google Scholar]
- van Nocker S, Sadis S, Rubin DM, Glickman MH, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin chain binding protein Mcb1 is a component of the 26S proteasome in S. cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;11:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal Proteomics: Identification of Nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin Chain Receptors Define a Layer of Substrate Selectivity in the Ubiquitin-Proteasome System. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural Studies of the Interaction between Ubiquitin Family Proteins and Proteasome Subunit S5a. Biochemistry. 2002;41:1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- Walters KJ, Lech PJ, Goh AM, Wang Q, Howley PM. DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci USA. 2003;100:12694–12699. doi: 10.1073/pnas.1634989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Young P, Walters KJ. Structure of S5a Bound to Monoubiquitin Provides a Model for Polyubiquitin Recognition. Journal of Molecular Biology. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multiubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- Xu P, Cheng D, Duong DM, Rush J, Roelofs J, Finley D, Peng J. A Proteomi Strategy for Quantifying Polyubiquitin Chain Topologies. 2006;46:171–182. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


