Abstract
Transforming growth factor-beta (TGF-β) elicits a variety of cellular activities primarily through a signaling cascade mediated by two key transcription factors, Smad2 and Smad3. Numerous regulatory mechanisms exist to control the activity of Smad3, thereby modulating the strength and specificity of TGF-β responses. In search for potential regulators of Smad3 through a yeast two-hybrid screen, we identified casein kinase 1 gamma 2 (CKIγ2) as a novel Smad3-interacting protein. In mammalian cells, CKIγ2 selectively and constitutively binds Smad3 but not Smad1, -2 or -4. Functionally, CKIγ2 inhibits Smad3-mediated TGF-β responses including induction of target genes and cell growth arrest, and this inhibition is dependent on CKIγ2 kinase activity. Mechanistically, CKIγ2 does not affect the basal levels of Smad proteins or activity of the receptors. Rather, CKIγ2 preferentially promotes the ubiquitination and degradation of activated Smad3 through direct phosphorylation of its MH2 domain at Ser418. Importantly, mutation of Ser418 to alanine or aspartic acid causes an increase or decrease of Smad3 activity, respectively, in the presence of TGF-β. CKIγ2 is the first kinase known to mark activated Smad3 for destruction. Given its negative function in TGF-β signaling and its reported overexpression in human cancers, CKIγ2 may act as an oncoprotein during tumorigenesis.
Keywords: Smad3, TGF-β, CKIγ2, ubiquitination, phosphorylation
Introduction
The transforming growth factor-beta (TGF-β) family of cytokines, including TGF-β s and bone morphogenetic proteins (BMPs), exerts diverse functions in many types of cells and tissues. Cell proliferation, differentiation, apoptosis, migration and extracellular matrix remodeling can all be regulated by TGF-β, although the biological outcomes often vary in different cellular contexts (Massagué, 2000; Massagué et al., 2000). During tumor progression, TGF-β has a dual function such that in the early phase of tumorigenesis, TGF-β functions as a tumor suppressor by inhibiting abnormal cell proliferation, whereas in advanced tumors, TGF-β often stimulates tumor cell invasion and metastasis, thereby acting as a tumor promoter (Bierie and Moses, 2006).
Cells perceive TGF-β signals through cell-surface proteins termed type I and type II TGF-β receptors (TβRI and TβRII), both being serine/threonine kinases. Upon TGF-β ligand binding, TβRI and TβRII form a heterotetrameric complex, within which TβRI is phosphorylated and activated by TβRII. In the canonical TGF-β pathway, TβRI in turn directly phosphorylates and activates downstream transcription factors called receptor-regulated Smad proteins (R-Smads). Two essential R-Smads, Smad2 and Smad3, specifically transduce TGF-β signals, and both Smads contain an SXS motif at their extreme C termini that serves as the phosphorylation site for TβRI. Activated Smad2 and Smad3 then interact with the common partner Smad, Smad4, and this heteromeric Smad complex becomes enriched in the nucleus, where the Smad proteins join with other transcription co-factors to regulate the expression of TGF-β target genes (reviewed by Massagué (2000); Shi and Massagué (2003); Massagué et al. (2005)).
Highly homologous in protein sequence and structure, Smad2 and Smad3 are both organized into an N-terminal MH1 domain and a C-terminal MH2 domain connected by a linker region. Numerous proteins have been shown to directly bind specific regions of the MH1 and MH2 domains and confer diverse regulations of Smad functions. In addition, the MH2 domain is critical for the nucleocytoplasmic shuttling of Smad2 and Smad3, and the MH1 domain of Smad3 (but not Smad2) contains a hairpin structure that is responsible for DNA binding (reviewed by Heldin et al. (1997); Massagué (2000); Shi and Massagué (2003)). Notably, Smad2 has a unique 30-amino-acid sequence in its MH1 domain (Dennler et al., 1999; Yagi et al., 1999), resulting in the inability of Smad2 to directly bind DNA and many other proteins (Shi et al., 1998; Jayaraman and Massagué, 2000; Guo et al., 2008). Owing to this and other differences, Smad2 and Smad3 have overlapping but sometimes distinct functions in gene regulations (Piek et al., 2001; Kretschmer et al., 2003; Dunn et al., 2005; reviewed by Weinstein et al. (2000) and Roberts et al.( 2006)).
TβRI-mediated C-terminal phosphorylation is a key step in the activation and propagation of Smad signaling, and its level reflects the overall TGF-β activity in cultured cells and in tissue samples. In vitro experiments have shown that activated Smad2 and Smad3 (P-Smad2/Smad3) can be detected with phosphospecific antibodies soon after TGF-β stimulation (5–10min), and the level of P-Smad2/Smad3 peaks at approximately 45–60min after treatment and then gradually declines (Lo and Massagué, 1999; Fukuchi et al., 2001; Inman et al., 2002; Lin et al., 2006). Research in the last decade has indicated two separate mechanisms accounting for the reduction of P-Smad2/Smad3 in the presence of persistent TGF-β stimulation (reviewed by Heldin and ten Dijke (1999) and Schilling et al. (2006)). The majority of activated Smad2 and Smad3 is dephosphorylated in the nucleus by a particular phosphatase, PPM1A (Lin et al., 2006). As a result, the deactivated Smads are recycled back to the cytoplasm for the next round of activation if the extracellular stimulus remains present (Inman et al., 2002; Xu et al., 2002; Reguly and Wrana, 2003; Schmierer and Hill, 2005; Lin et al., 2006). Besides the reversible tail phosphorylation of Smad2 and Smad3, TGF-β signaling can also be irreversibly terminated by destruction of the activated Smad proteins through the ubiquitin-dependent proteasomal pathway (Lo and Massagué, 1999; Lin et al., 2000; Fukuchi et al., 2001 and reviewed by Izzi and Attisano (2006)). Polyubiquitination of activated Smad2 (mediated by the E2 enzymes UbcH5b/c) and Smad3 (mediated by the F-Box protein β-TrCP/Fbw1a) is thought to occur in the nucleus, and the ubiquitination site is probably located in the MH2 domain of these proteins (Lo and Massagué, 1999; Fukuchi et al., 2001).
Casein kinase 1 (CKI) is a family of evolutionarily conserved serine/threonine kinases including seven known members in vertebrates (CKIα, -β, -γ1, -γ2, -γ3, -δ and -ε). The CKIs contain a typical kinase domain followed by a C-terminal tail region, which has been implicated in the regulation of CKI localization, substrate selectivity and kinase activity. A myriad of proteins have been found to be phosphorylated by CKIs, which are involved in a wide range of cellular functions including vesicular trafficking, DNA damage repair, cell cycle progression, cytokinesis and circadian rhythms (reviewed by Gross and Anderson (1998); Vielhaber and Virshup (2001); Knippschild et al. (2005)). Moreover, it is well established that CKI family members (-α, -δ/ε and -γ) modulate the activities of major signaling pathways (for example, Wnt and Shh) through several different mechanisms (Peters et al., 1999; Liu et al., 2002; Price and Kalderon, 2002; Davidson et al., 2005; Zeng et al., 2005 and reviewed by Price (2006)).
In our early efforts to identify potential Smad3 regulators, a number of cDNAs were isolated from a yeast two-hybrid screen using Smad3 as bait. Several of the encoded proteins, such as HDAC1, JunB, WWP1 and PIAS-1, have been confirmed by us and others to directly bind Smad3 and regulate TGF-β activity (Liberati et al., 1999, 2001; Komuro et al., 2004; Long et al., 2004 and our unpublished data). Interestingly, three separate coding sequences recovered from the screen correspond to human CKIγ2 (CSNK1G2, NM_001319). One of these clones (no. 137) encodes the final 188 amino acids of CKIγ2 (aa 228–415), which was termed CKIγ2ΔN and encompasses the last third of the kinase domain and the entire tail region. CKIγ2, together with two other members, CKIγ1 and CKIγ3, belongs to the CKIγ subfamily of CKIs (Zhai et al., 1995; Kitabayashi et al., 1997). Although the function of CKIγ in TGF-β signaling had been elusive, our parallel work showed that CKIα, -δ and -ε are bona fide Smad3-binding proteins in vivo that regulate TGF-β signal transduction (Waddell et al., 2004 and our unpublished data). These findings prompted us to investigate the function of CKIγ2 in the regulation of TGF-β/Smad3 activity.
Here we demonstrate that CKIγ2 selectively interacts with Smad3 in mammalian cells and inhibits Smad3-mediated TGF-β activity. CKIγ2 directly phosphorylates Smad3 at Ser418, leading to the increased ubiquitination and proteasomal degradation of activated Smad3 following TGF-β treatment. Therefore, ubiquitination of ligand-activated Smad3 is a regulated event, and CKIγ2 is the first kinase known to control the signaling duration of Smad3 in this manner.
Results
CKIγ2 selectively binds Smad3
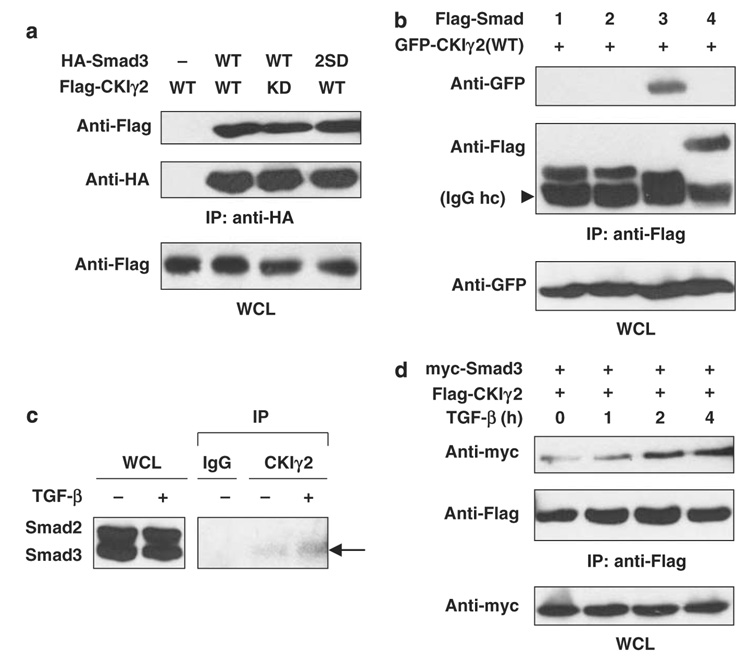
The physical interaction between CKIγ2 and Smad3 was first confirmed by in vitro binding assays. GST-CKIγ2ΔN was incubated with in vitro translated 35S-labeled Smad3, and the direct interaction between the two proteins was readily detectable following glutathione S-transferase (GST) pull-down assay (data not shown). In addition, bacterially purified full-length CKIγ2 also directly bound GST-Smad3 but not GST alone (Supplementary Figure S1A). We then expressed epitope-tagged CKIγ2 and Smad3 in 293T cells to examine their in vivo interaction and found that both CKIγ2(WT) and the kinase-deficient mutant, CKIγ2(KD), co-precipitated with Smad3 (Figure 1a). Further analysis showed that the Smad3-MH2 domain is sufficient to mediate CKIγ2 binding, whereas a partial deletion of this domain abolished Smad3–CKIγ2 interaction (Supplementary Figure S1A and B). In contrast to Smad3, overexpressed Smad1 (mediating BMP signals), Smad2 and Smad4 failed to co-immunoprecipitate with CKIγ2 (Figure 1b). This selective interaction was also seen with endogenous proteins from HaCaT cell lysates, as only Smad3 was detected from the anti-CKIγ2 precipitates (Figure 1c). Next, we examined whether the Smad3–CKIγ2 interaction is affected by TGF-β treatment, which reduces/disrupts the binding between Smad3 and some other CKI members (for example, CKIε and -α; Waddell et al. (2004) and our unpublished data). Interestingly, unlike the other CKIs, CKIγ2 remained bound to Smad3 in the presence of TGF-β (Figures 1c and d), which is consistent with its ability to bind the Smad3(2SD) mutant that mimics receptor-phosphorylated Smad3 (Figure 1a). In fact, TGF-β slightly increased the Smad3–CKIγ2 interaction. These data suggest that CKIγ2 may have a unique function in regulating activated Smad3.
Figure 1. CKIγ2 constitutively and selectively interacts with Smad3.
(a) 293T cells were transfected with the indicated constructs. Cells were lysed in universal lysis buffer supplemented with protease and phosphatase inhibitors (ULB+) at 24 h post-transfection and anti-HA immunoprecipitation was performed. 2SD, S423/425D; KD, kinase-dead; WCL, whole-cell lysate. (b) The Flag-tagged, full-length Smad constructs were co-expressed with GFP-CKIγ2(WT). Anti-Flag immunoprecipitation was performed as in (a). The arrowhead indicates the heavy chain (hc) of the Flag antibody (M2). (c) Parental HaCaT cells were pretreated with 10 µM MG-132 for 1 h before treatment with or without 100 pm TGF-β for another 2 h. Endogenous CKIγ2 was precipitated with a goat polyclonal anti-CKIγ2 antibody (C-20), and endogenous Smad proteins were blotted with a monoclonal anti-Smad1/2/3 antibody. Co-precipitated Smad3 is indicated by an arrow. An irrelevant goat polyclonal antibody (IgG) was used as a negative control. (d) Wild-type Smad3 and CKIγ2 were co-expressed in MEFs for 20h. Cells were then treated with 100pm TGF-β for the indicated time course and then lysed for anti- Flag immunoprecipitation. CKIγ2, casein kinase 1 gamma 2; HA, hemagglutinin; MEFs, mouse embryonic fibroblasts; TGF-β, transforming growth factor-beta.
CKIγ2 inhibits Smad3 transcriptional activity
To explore the functional role of CKIγ2 in the TGF-β/Smad3 pathway, we first determined by reverse transcription PCR that CKIγ2 is expressed in several human cell lines commonly used for TGF-β studies, such as HaCaT (human keratinocyte), HepG2 (hepatocellular carcinoma) and U2OS (osteosarcoma) cells (data not shown). CKIγ2 also appeared to be the major, if not the only, member of the CKIγ subfamily being expressed in those cells.
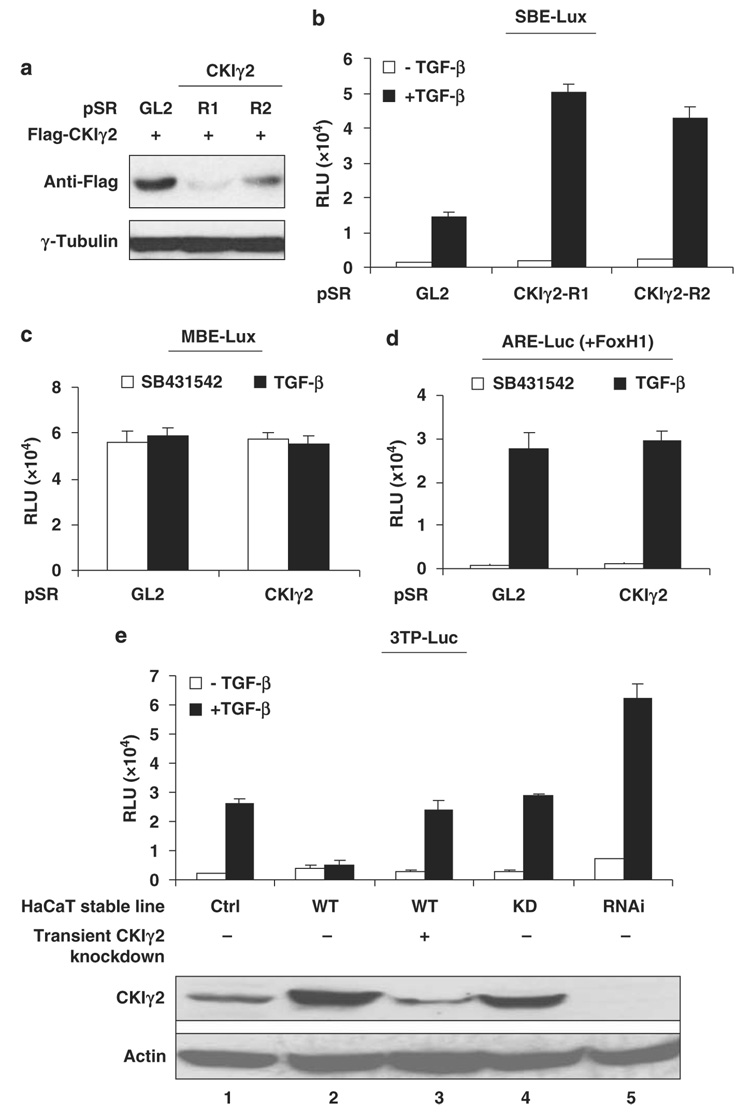
Next, we generated two short hairpin-type RNA constructs against independent coding regions of CKIγ2 (designated as R1 and R2) that effectively downregulated CKIγ2 protein (Figures 2a and e). When co-expressed with a Smad3-specific SBE-Lux luciferase reporter in HepG2 cells, both short hairpin-type RNAs caused a significant increase of Smad3 transcriptional activity but only in the presence of TGF-β (Figure 2b). CKIγ2 knockdown did not alter the expression of the MBE-Lux reporter (a mutant control for SBE, Figure 2c), nor did it affect the Smad2-specific ARE-Luc activity (Figure 2d), indicating that CKIγ2 specifically inhibits Smad3-mediated transcription.
Figure 2. CKIγ2 inhibits Smad3-mediated transcription.
(a) Validation of CKIγ2 shRNAs. Flag-CKIγ2 (1µg) was co-transfected into 293T cells with the indicated pSuperRetro plasmids (pSR, 3 µg). pSR-GL2 contains the shRNA against luciferase and was used as a non-targeting control. Cells were lysed at 24 h post-transfection and total cell lysates were blotted for Flag-CKIγ2. γ-Tubulin was used as the loading control in this and most later experiments. (b) SBE-Lux luciferase assay in HepG2 cells. The luciferase reporter (0.5 µg) was co-transfected with the indicated pSuperRetro plasmids (3 µg) before TGF-β (100 pm) or vehicle was added. Data were presented as average ± s.d. (c and d). Similar luciferase assays in HepG2 cells using the MBE-Lux reporter (c) or the ARE-Luc reporter (d, together with an equal amount of FoxH1). Cells were co-transfected with the reporter construct (1 µg) and the indicated pSuperRetro plasmids (4 µg of pSR-GL2 or a combination of pSR-CKIγ2-R1 and -R2, 2 mg each). Twenty-four hours later, cells were treated with TGF-β (100 pm) or the TβRI inhibitor, SB431542 (10 µm), for another 16 h before analysis. (e) 3TP-Luc luciferase assays in HaCaT stable lines (see Materials and methods). Cells were transiently transfected with the reporter construct (1 µg) and treated with or without TGF-β (100 pm). Note that overexpressed CKIγ2 was transiently knocked down in the CKIγ2-WT cells (lane 3) by three consecutive transfections with pSuper-CKIγ2. CKIγ2 protein levels in each HaCaT line are shown (bottom). Actin was blotted to show equal loading of lysates. CKIγ2, casein kinase 1 gamma 2; shRNA, short hairpin-type RNA; TGF-β, transforming growth factor-beta; TβRI, TGF-β receptor I.
To further gauge this inhibitory effect of CKIγ2 on Smad3 signaling, we engineered HaCaT cells that stably overexpress wild-type (WT) or kinase-dead (KD) CKIγ2, as well as a stable knockdown line (RNAi) (Figure 2e and Figure 4a). The expression level of CKIγ2 in these cell lines was confirmed by immunoblotting using a rabbit polyclonal antibody (CKIγ2–2026) generated in our laboratory. In a luciferase assay using the 3TP-Luc reporter, the overexpression of WT CKIγ2 almost completely abolished TGF-β-induced activity, whereas this inhibition was fully reversed by transient knockdown of the over-expressed CKIγ2 to the endogenous level (Figure 2e, lanes 1–3). In contrast, the KD mutant of CKIγ2 had no effect on the reporter at all, and stable downregulation of CKIγ2 led to an increased reporter activity (Figure 2e, lanes 4–5). Similar results were seen with other reporter constructs such as PAI-1-Luc and p15HD-Lux in the same HaCaT lines (data not shown). These results further demonstrate that CKIγ2 negatively regulates TGF-β/Smad3 activity in a kinase-dependent manner.
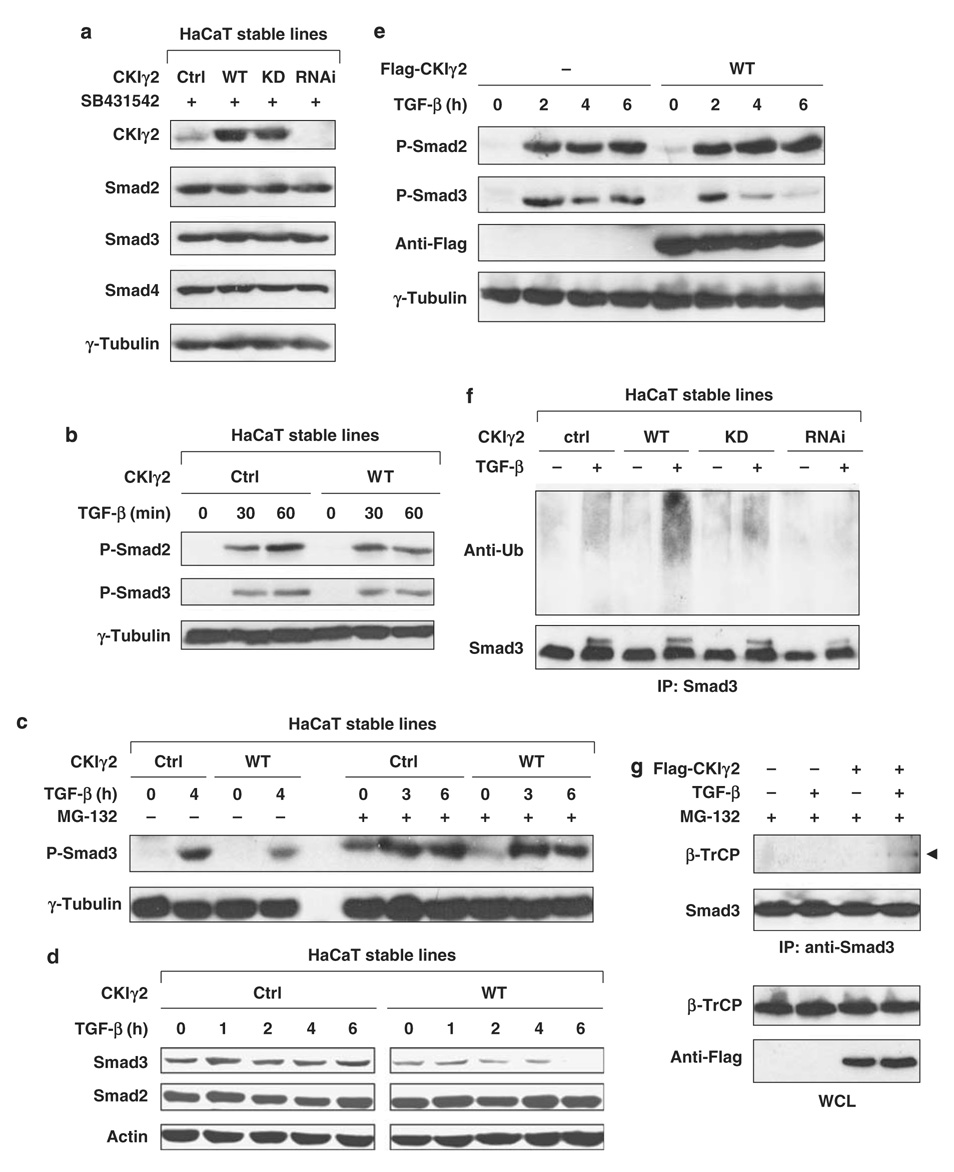
Figure 4. CKIγ2 promotes the proteasomal degradation of activated Smad3.
(a) CKIγ2 does not regulate the basal levels of Smad proteins. HaCaT stable lines were treated with SB431542 (10 µm) overnight, and endogenous Smad2, -3 and -4 were examined by western blotting. (b) The HaCaT lines were transiently stimulated with TGF-β (50pm) as indicated. C-terminal phosphorylation of endogenous Smad2 and Smad3 was measured. (c) HaCaT cells were treated with TGF-β (100 pm) for the indicated time courses in the absence (left) or presence (right) of 20 µm MG-132. The level of activated Smad3 was shown. (d) HaCaT cells were treated with TGF-β (100 pm) for the indicated time courses and the total levels of endogenous Smad2 and Smad3 were determined. (e) MEFs were transfected with or without wild-type CKIγ2 before incubation with TGF-β (50pm) for the indicated time course. C-terminal phosphorylation of endogenous Smad2 and Smad3 was measured. (f) HaCaT cells were pretreated with MG-132 (7.5 µM) for 1 h before the treatment of SB431542 (10 µm, ‘−’) or TGF-β (100 pm, ‘+’) for another 3 h. Cells were harvested in SDS lysis buffer (see Materials and methods), endogenous Smad3 was immunoprecipitated and ubiquitinated species of Smad3 was analysed by anti-ubiquitin blotting. (g) MEFs expressing either vector control or wild-type CKIγ2 were treated with SB431542 (10 mm, ‘−’) or TGF-β (100 pm, ‘+’) for 2 h in the presence of 20 µm MG-132. The interaction between endogenous Smad3 and β-TrCP was determined by co-immunoprecipitation assays. CKIγ2, casein kinase 1 gamma 2; MEFs, mouse embryonic fibroblasts; TGF-β, transforming growth factor-beta.
CKIγ2 attenuates TGF-β-induced biological responses
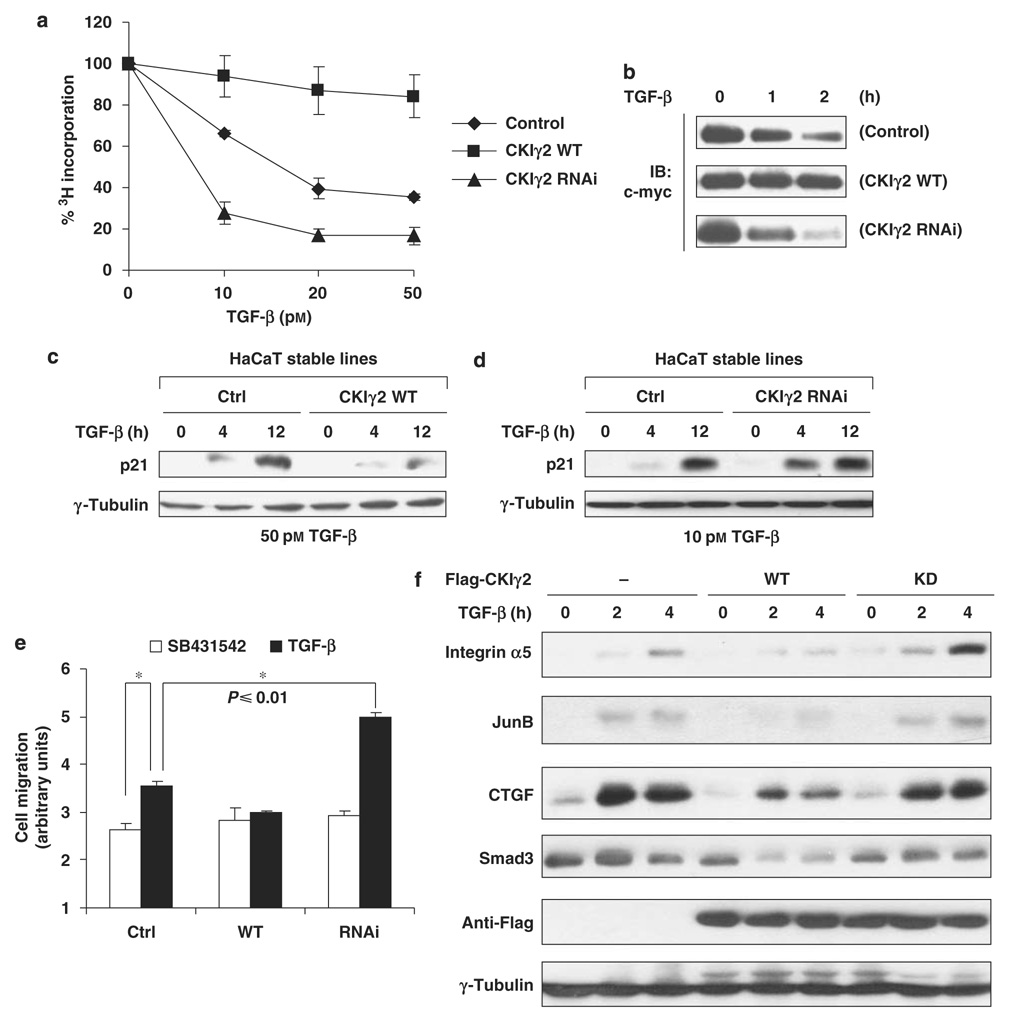
We then investigated whether CKIγ2 affects TGF-β-regulated biological activity at the cellular level. TGF-β can cause growth arrest in normal epithelial cells and keratinocytes, which is achieved through Smad3-mediated downregulation of the proto-oncogene c-myc and upregulation of cyclin-dependent kinase inhibitors such as p21 and p15 (Massagué et al., 2000). In HaCaT cells, this antiproliferative effect of TGF-β was severely impaired by CKIγ2 overexpression but markedly augmented by CKIγ2 RNAi (Figure 3a). Consistently, the downregulation of the c-myc protein by TGF-β was more prominent in the CKIγ2-RNAi cells (decreased by >50% within 1 h of TGF-β treatment) than in control cells, whereas there was barely any reduction in c-myc level in the CKIγ2-WT cells after 2 h of treatment (Figure 3b). TGF-β-induced expression of p21 was also significantly delayed and weakened in the CKIγ2-WT cells, whereas the opposite was seen in the CKIγ2-RNAi cells (Figures 3c and d). In addition to cytostasis, TGF-β-stimulated cell migration was also blocked in HaCaT cells overexpressing CKIγ2 but enhanced in the knock-down cells (Figure 3e). Note that in untreated cells, the varied levels of CKIγ2 did not produce a difference in cell proliferation or motility at the basal level. Therefore, the inhibitory function of CKIγ2 in the TGF-β/Smad3 pathway appeared to be ligand dependent.
Figure 3. CKIγ2 impairs Smad3-dependent TGF-β activity.
(a) Cell proliferation assays in HaCaT cells. The indicated HaCaT stable lines were treated with different concentrations of TGF-β for 24 h. The amounts of incorporated 3H-thymidine in untreated cells were normalized to 100%. (b) Downregulation of endogenous c-myc protein was measured in the HaCaT stable lines following TGF-β treatment as indicated (100 pm). Equal loading of samples was confirmed by anti-actin blotting (data not shown). (c and d) Induction of endogenous p21 protein by TGF-β in the HaCaT stable lines. Note that a lower concentration of TGF-β (10 pm) was used to manifest the enhanced responsiveness of the CKIγ2 knockdown cells (d). (e) Wound-healing assays in the HaCaT stable lines treated with SB431542 (10 µm) or TGF-β (100 pm). Asterisk (*), P≤0.01. (f) Wild-type or kinase-dead CKIγ2 was transiently expressed in MEFs. After treatment with TGF-β (50 pm), whole-cell lysates (in ULB+) were blotted for the indicated proteins. CKIγ2, casein kinase 1 gamma 2; MEFs, mouse embryonic fibroblasts; TGF-β, transforming growth factor-beta.
We also employed transient transfection to confirm the negative function of CKIγ2 in TGF-β responses. Mouse embryonic fibroblasts expressing identical amount of WT or KD CKIγ2 were stimulated with TGF-β, and the expression of endogenous Smad3 target genes such as integrin α5, JunB and CTGF (connective tissue growth factor) was measured. Overexpression of CKIγ2(WT), but not CKIγ2(KD), considerably attenuated the induction of those genes and also downregulated Smad3 in the presence of TGF-β (Figure 3f). These results again indicate that CKIγ2 inhibits TGF-β signaling, likely through the regulation of activated Smad3.
CKIγ2 promotes proteasomal degradation of activated Smad3
We then probed for the mechanism by which CKIγ2 regulates the TGF-β/Smad3 pathway. First, CKIγ2 did not affect the protein levels of Smad2, -3 or -4 in unstimulated HaCaT cells (Figure 4a). Second, both Smad2 and Smad3 were properly activated by shortterm TGF-β treatment (30–60 min) regardless of CKIγ2 overexpression, suggesting that CKIγ2 does not interfere with TGF-β receptor activity (Figure 4b). However, when the time course was extended to 4 or 6 h, the levels of P-Smad3 (activated) and total Smad3 became much lower in CKIγ2-overexpressing cells than in control cells (Figures 4c, d, e and Supplementary Figure S2). This effect of CKIγ2 was unlikely because of an increased phosphatase activity against P-Smad3, as P-Smad2, which is dephosphorylated by the same phosphatase PPM1A (Lin et al., 2006), was unaffected by CKIγ2 (Figures 4d, e and Supplementary Figure S2). Instead, the reduction of P-Smad3 and total Smad3 caused by CKIγ2 overexpression could be rescued by pretreating the cells with the proteasome inhibitor, MG-132 (Figure 4c and Supplementary Figure S2). This latter result suggests that CKIγ2 downregulates P-Smad3 by promoting its degradation, a notion consistent with the gradual loss of Smad3 protein after prolonged TGF-β stimulation as seen in Figure 3f. Furthermore, TGF-β-activated endogenous Smad3 was more strongly polyubiquitinated in HaCaT(CKIγ2-WT) cells than in control or CKIγ2(KD) cells, whereas this ligand-induced Smad3 ubiquitination was almost completely suppressed by CKIγ2 RNAi (Figure 4f). Lastly, the binding between endogenous Smad3 and the F-box protein β-TrCP was enhanced by CKIγ2 following TGF-β treatment (Figure 4g). All together, these data demonstrate that CKIγ2 accelerates the proteasomal degradation of activated Smad3, thereby shortening its signaling duration and inhibiting TGF-β activity.
CKγg2 directly phosphorylates Smad3
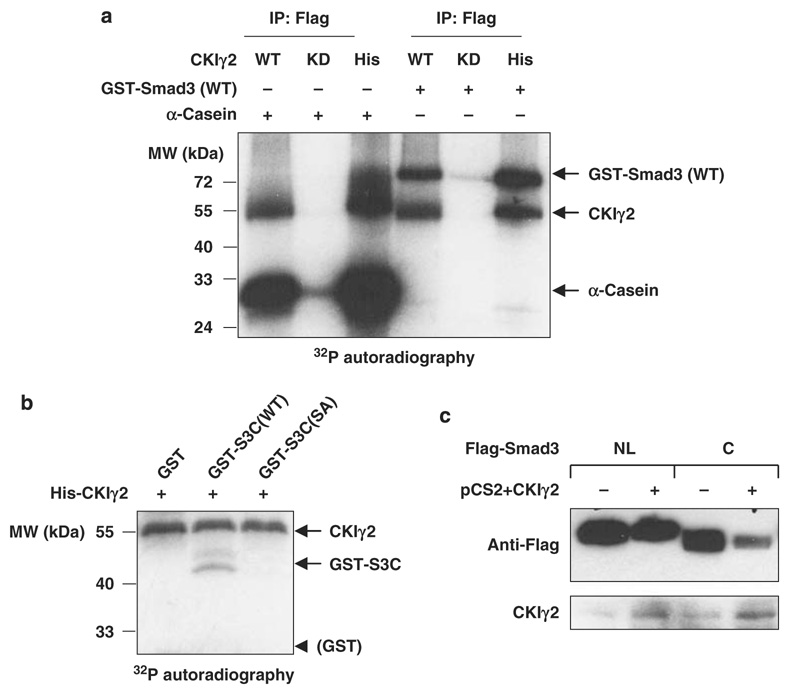
As CKIγ2 inhibits Smad3 activity in a kinase-dependent manner, we examined whether Smad3 is a direct substrate of CKIγ2. In vitro, CKIγ2 strongly phosphorylated purified GST-Smad3 (Figure 5a). Mass spectrometry analysis showed that CKIγ2 phosphorylates the MH2 domain of Smad3 at a single residue, Ser418 (recovered in peptide VLTQMGSPpSIR, see Supplementary Data for details). Indeed, purified Smad3-MH2 domain alone (termed GST-S3C) could be directly phosphorylated by CKIγ2, and the phosphorylation was blocked by mutating Ser418 to alanine (S418A) (Figure 5b). In line with this, CKIγ2 over-expression significantly decreased the protein level/stability of Flag-S3C, whereas the MH1+Linker region of Smad3 (Flag-S3NL) was unaffected (Figure 5c).
Figure 5. CKIγ2 phosphorylates Smad3 at Ser418.
(a) CKIγ2 in vitro kinase assay. Flag-tagged CKIγ2 (WT or KD) immunoprecipitated from 293T cell lysates as well as bacterially purified His-CKIγ2 were individually incubated with α-casein (positive control) or GST-Smad3(WT) in the presence of [32P]- γ-ATP. Phosphorylated proteins were visualized by autoradiography. Note that CKIγ2 underwent significant autophosphorylation. (b) His-CKIγ2 was incubated with GST alone, GST-S3C(WT) or GST-S3C( S418A) in a similar kinase assay as in (a). Equal loading of protein substrates was confirmed by Coomassie Blue staining (data not shown). CKIγ2 does not phosphorylate the GST moiety. (c) The indicated Flag-tagged Smad3 fragments were co-expressed with either a vector control or wild-type CKIγ2 in MEFs without inhibition of proteasomal function. Total cell lysates were analysed for the levels of Flag-S3NL (MH1 + Linker) and Flag-S3C (MH2). CKIγ2, casein kinase 1 gamma 2; GST, glutathione S-transferase; KD, kinase-dead; MEFs, mouse embryonic fibroblasts; WT, wild-type.
Ser418 phosphorylation controls Smad3 stability/activity
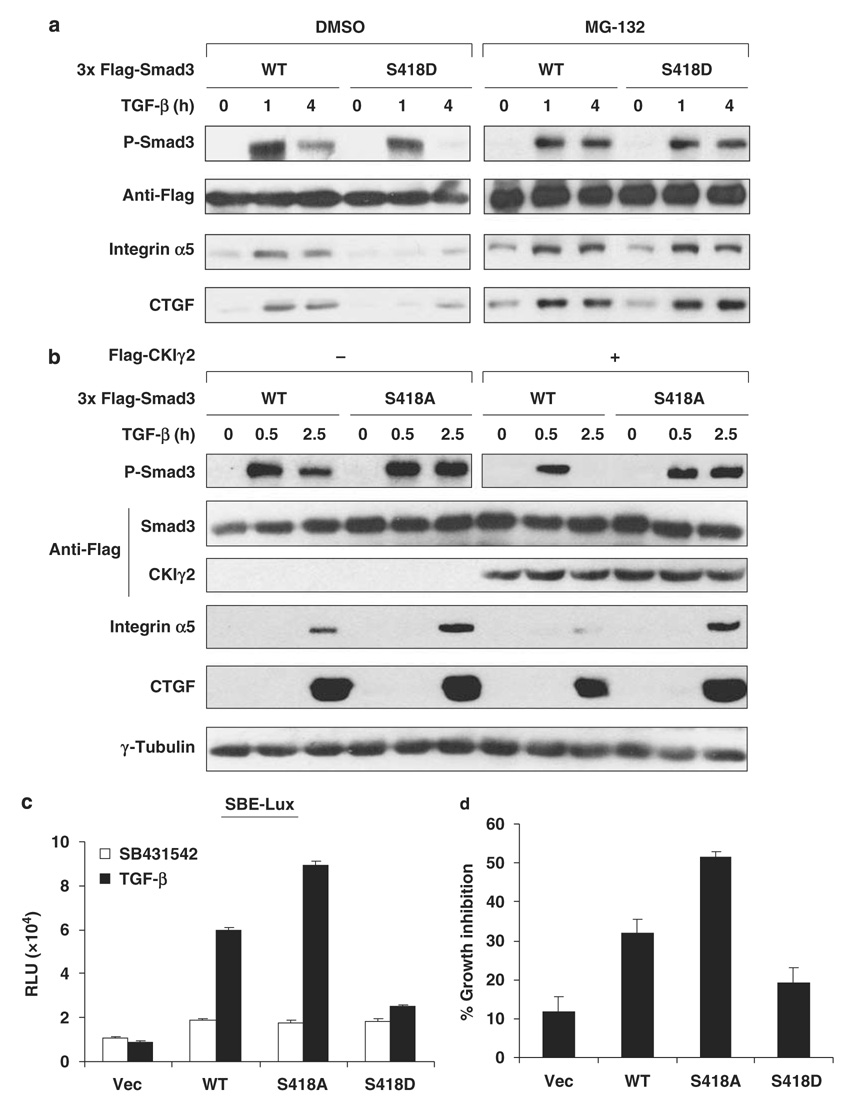
To understand the functional role of Ser418 phosphorylation by CKIγ2 in cells, we compared the stability and activity of Smad3(WT), Smad3(S418A, phosphodeficient) and Smad3(S418D, phosphomimetic). Ser418 mutations did not affect Smad3 expression in untreated cells or the early-phase activation of Smad3 by shortterm (0.5–1 h) TGF-β treatment (Figures 6a and b). However, after prolonged TGF-β stimulation, the S418D mutant showed a much lower level of C-terminal phosphorylation than Smad3(WT) (Figure 6a, left), consistent with our postulation that CKIγ2-mediated phosphorylation downregulates the activated form of Smad3. Owing to the rapid removal of activated Smad3(S418D), TGF-β-induced upregulation of Smad3 target genes such as integrin α5 and CTGF was greatly compromised in cells expressing this mutant (Figure 6a, left). It is noteworthy that pretreating cells with MG-132 effectively stabilized C-terminally phosphorylated Smad3(S418D) and fully restored its activity (Figure 6a, right). This result indicates that S418 phosphorylation does not perturb the intrinsic transactivating ability of Smad3 but attenuates Smad3 activity by lowering its protein stability in the presence of TGF-β. Indeed, the S418D mutation or CKIγ2 overexpression did not affect the DNA-binding ability of Smad3 or its affinity to Smad4 in the presence of MG-132 (Supplementary Figure S4). On the other hand, the phosphodeficient S418A mutant was more resistant to CKIγ2 overexpression than WT Smad3 (Figure 6b), further supporting that Ser418 is a critical site for CKIγ2 to inhibit Smad3 function. We also expressed these Smad3 variants in SMAD3−/− mouse embryonic fibroblasts and HepG2 cells to measure the SBE-Lux activity and growth inhibition, respectively (Figures 6c and d). In both the assays, the Smad3 mutants exhibited almost identical basal activity as WT Smad3 in the absence of TGF-β. However, upon activation, Smad3(S418A) was more potent, whereas Smad3(S418D) was much weaker, than Smad3(WT) in activating downstream responses (Figures 6c and d). Taken together, we conclude that CKIγ2 inhibits TGF-β signaling by destabilizing the activated form of Smad3 through direct phosphorylation of Ser418.
Figure 6. Ser418 phosphorylation regulates Smad3 stability and activity following TGF-β treatment.
(a) MEFs expressing Smad3(WT or S418D) were treated with TGF-β (50 pm) for the indicated time course in the absence or presence of 20 µm MG-132. Induction of Smad3 target genes and Smad3 tail phosphorylation are shown. (b) MEFs were transfected with Smad3(WT or S418A) and CKIγ2 (or a vector control). After TGF-β (50 pm) treatment for the indicated time course, induction of Smad3 target genes and Smad3 tail phosphorylation were analysed. (c) Smad3-null MEFs were co-transfected with the SBE-Lux reporter and a vector control or the indicated Smad3 variants. Luciferase activity was measured after treatment with SB431542 or 100 pm TGF-β. (d) HepG2 cells expressing a vector control or the indicated Smad3 variants were treated with or without TGF-β (200 pm) for 16 h. The extent of growth inhibition (with reference to untreated cells) under each condition is shown. MEFs, mouse embryonic fibroblasts; TGF-β, transforming growth factor-beta; WT, wild-type.
Discussion
One striking feature of TGF-β/Smad signaling is the combination of versatility and accuracy, which is ensured by a plethora of regulatory proteins that control every step of the TGF-β pathway. Many Smad3-interacting proteins are of yet unknown identities and functions. In this study, we demonstrated that CKIγ2 is a novel Smad3-binding protein that targets activated Smad3 to ubiquitination and degradation, thus revealing another mechanism for the regulation of TGF-β signaling.
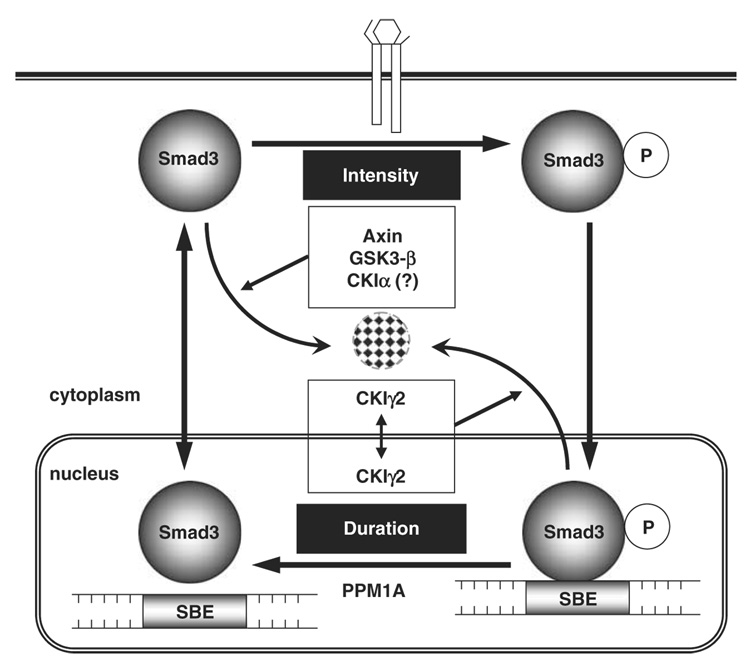
Most Smad proteins can be polyubiquitinated in either a ligand-dependent or a ligand-independent manner (reviewed by Izzi and Attisano (2006)). As for R-Smads, ubiquitination and degradation at the steady state (without ligand stimulation) lead to lowered cellular sensitivity to TGF-β ligands and weakened signal intensity, whereas ubiquitination of activated R-Smads shortens the duration of signaling and irreversibly shuts down TGF-β activity (Figure 7). Multiple E2 (conjugating enzyme) and E3 (ubiquitin ligase) enzymes have been found to conduct Smad ubiquitination. However, our knowledge about the molecular events that precede and trigger those ubiquitination reactions remains incomplete. Very recently, we reported that ubiquitination and degradation of non-activated Smad3 is synergistically regulated by the scaffolding protein Axin and its associated kinase, GSK3-β (Guo et al., 2008; Figure 7). Our unpublished results also indicate a requirement for CKIα, a CKI family member, in the basal degradation of Smad3. In this study, we have demonstrated that ubiquitination and degradation of TGF-β-activated Smad3 is controlled by a different CKI, CKIγ2. Therefore, two CKI family members can induce Smad3 ubiquitination and negatively regulate TGF-β activity but through different mechanisms (Figure 7).
Figure 7. The Smad3 ‘life cycle’ and regulators of Smad3 ubiquitination.
Smad3 ubiquitination has important functions in determining both the intensity and the duration of TGF-β signals. Steady-state Smad3 is constantly degraded by the proteasome under the regulation of Axin, GSK3-β and probably CKIα. On the other hand, proteasomal degradation of activated Smad3 requires CKIγ2 phosphorylation, which may happen in the cytoplasm or in the nucleus or both. CKIγ2, casein kinase 1 gamma 2; TGF-β, transforming growth factor-beta.
TGF-β stimulates the translocation of Smad2 and Smad3 into the nucleus, where they can be assembled into protein complexes for transcriptional regulation, or be dephosphorylated and exported to the cytoplasm, or undergo polyubiquitination (Shi and Massagué, 2003). Therefore, when these Smads need to be eliminated as a way to swiftly and irreversibly turn off TGF-β responses, additional signals must be given to specifically direct them down the degradation pathway. In regard to Smad3, one of such signals, as we demonstrated in this study, is its phosphorylation by CKIγ2 at Ser418. This extra step of regulation determines whether an activated Smad3 molecule is to be labeled for destruction, thereby representing an important mechanism to ensure the proper signaling duration of TGF-β.
One interesting question, however, is where exactly in the cell CKIγ2 phosphorylates Smad3. Our fluorescence microscopy and biochemical fractionation analyses showed that CKIγ2 is localized in the cytoplasm, in the nucleus and at the plasma membrane (see Supplementary Data for details). Although the plasma membrane association appears to be critical for CKIγ2 to regulate Wnt/β-catenin signaling (Davidson et al., 2005), it is not required for Smad3 regulation. On the other hand, different CKIγ2 mutants that are either unable to enter the nucleus or highly enriched in the nucleus all inhibited Smad3 activity and reduced P-Smad3 stability in a similar manner as WT CKIγ2 (Supplementary Figures S5 and S6). Therefore, Smad3 may be phosphorylated by CKIγ2 in the cytoplasm or in the nucleus or both, and this phosphorylation may occur independently of TGF-β stimulation in view of the constitutive binding between CKIγ2 and Smad3. However, CKIγ2 does not affect the steady-state level of Smad3, and no difference was seen in the protein stability or activity of the Smad3 Ser418 mutants in untreated cells (Figure 4 and Figure 6). The inhibitory effect of CKIγ2 is manifested only when Smad3 is activated (Figure 2–Figure 4). These observations strongly support the notion that Smad3 regulation by CKIγ2 is dependent on TGF-β, which is necessary to place Smad3 in the right environment (that is, the nucleus) and in an optimal conformation such that the CKIγ2-mediated phosphorylation can be recognized by the appropriate ubiquitination machinery (Figure 4G and Supplementary Figure S4C).
CKIγ2 phosphorylates Smad3 at serine 418, and Ser418 mutants exhibit altered protein stability, activity and sensitivity to CKIγ2 following TGF-β stimulation (Figure 5 and Figure 6). Ser418 is conserved in Smad3 of human, mouse, rat and frog origins. Interestingly, close to this site is a lysine residue (Lys409) that is also evolutionarily conserved. Among the four lysine residues present in the Smad3-MH2 domain, only the K409R mutation leads to an increase in TGF-β-induced Smad3 activity (Imoto et al., 2005). This fact raises the tempting possibility that Lys409 might be the ubiquitination site of activated Smad3, as β-TrCP-mediated polyubiquitination is thought to occur in the MH2 domain (Fukuchi et al., 2001). It would be even more interesting to determine whether Ser418 phosphorylation by CKIγ2 is necessary and/or sufficient to trigger Smad3 ubiquitination at Lys409, a hypothesis that we are currently investigating in the laboratory.
It is noteworthy that the BMP Smads (Smad1, -5 and -8) and Smad4 do not possess a site equivalent to Smad3-Ser418, nor do they interact with CKIγ2 (Figure 1). As a result, CKIγ2 does not seem to inhibit BMP signaling (data not shown). Interestingly, although Smad2 contains a residue (Ser460) that corresponds to Smad3-Ser418, it is not associated with or regulated by CKIγ2 (Figure 1, Figure 2 and Figure 4). The inability of Smad2 to bind CKIγ2 may result from the unique structure of the Smad2-MH1 domain, as substitution of the MH1 domain of Smad3 with that of Smad2 resulted in a loss of CKIγ2 binding to the Smad2/3 chimeric protein (Supplementary Figure S1B). This is similar to our recent findings that Smad3 and Smad2 are differentially regulated by the Axin/GSK3-β complex because of structural differences of their MH1 domains (Guo et al., 2008). In addition, Fukuchi et al. (2001) showed that overexpression of β-TrCP cannot induce Smad2 ubiquitination in the nucleus. Therefore, with respect to ubiquitination, Smad2 and Smad3 are differentially regulated. It remains to be seen whether TGF-β-dependent Smad2 ubiquitination is also regulated through a similar mechanism as what we hereby demonstrated for Smad3 but by a different kinase.
In mesangial cells, Smad3 is critical in mediating TGF-β-induced fibrosis (reviewed by Wang et al. (2005)). Long-term TGF-β treatment of these cells causes a significant decrease in the protein level of Smad3 but not Smad2 (Poncelet et al., 2007), which might well result from the action of CKIγ2. Therefore, it would be interesting to determine whether CKIγ2 is relevant to the fibrotic responses in the kidney under normal and pathological conditions.
Although the majority of activated Smad3 normally undergoes dephosphorylation and relocates back to the cytoplasm, our study suggests that a significant portion of Smad3 molecules can be shunted toward the proteasomal pathway because of increased CKIγ2 activity. Indeed, overexpression of CKIγ2 mRNA has been found in several types of cancer (http://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.651905), in which the cytostatic response to TGF-β is often lost. In addition, CKIγ2 not only inhibits TGF-β function but also positively regulates the Wnt pathway (Davidson et al., 2005). Thus, overexpression of CKIγ2 is likely to promote the proliferation of epithelial cells and may be advantageous for tumor growth. Although this notion remains to be firmly established, CKIγ2 may act as an oncoprotein and the development of specific CKIγ2 inhibitors can be potentially valuable for anticancer therapeutics.
Materials and methods
Plasmids and sequences
Epitope-tagged Smad2 and Smad3 (WT and 2SD) have been used earlier (Guo et al., 2008). pRK5-Smad1-Flag and pRK5-Smad4-Flag were provided by Rik Derynck (University of California, San Francisco). The cDNA of human CKIγ2 in the pMOSBlue vector was a gift of Jun Kusuda and has been described earlier (Kitabayashi et al., 1997). pSuperRetro-GL2 was described earlier (Guo et al., 2008). pSuper- and pSuperRetro-CKIγ2 were generated according to the manufacturer’s protocol (Oligoengine, Seattle, WA, USA). The sequences of primers used for subcloning, mutagenesis and RNAi are described in Supplementary Data.
Cell culture, transfection and retroviral infection
Growth of HaCaT, HepG2, 293T and mouse embryonic fibroblasts has been described (Guo et al., 2008). HaCaT cells stably overexpressing CKIγ2 were generated by transfection of pcDNA3-CKIγ2(WT or KD) using LipofectAmine 2000 (Invitrogen Inc., Carlsbad, CA, USA), followed by G418 selection (800 µg/ml, Invitrogen). Retroviral infection of HaCaT cells with pSuperRetro-CKIγ2(R1 + R2) was performed as reported earlier (Guo et al., 2008). Transient transfection of HaCaT cells was achieved using the FuGene 6 reagent (Roche, Indianapolis, IN, USA). 293T, HepG2 and mouse embryonic fibroblasts were transfected as reported earlier (Guo et al., 2008).
Antibodies and other reagents
The anti-CKIγ2(2026) polyclonal antibody was raised by immunizing a rabbit with GST-CKIγ2ΔN at Duke vivarium according to a standard protocol. Serum from the fourth immunized bleed was centrifuge cleared and used for western blotting of endogenous CKIγ2. Integrin α5 polyclonal anti-body was provided by Jun-Lin Guan (University of Michigan). Other antibodies used for immunoblotting: Smad3, Smad2 and β-TrCP (Zymed/Invitrogen, Carlsbad, CA, USA); Flag(M2), GFP and γ-tubulin (Sigma, St Louis, MO, USA); P-Smad2 and P-Smad3(S423/425) (Cell Signaling, Danvers, MA, USA); actin(C-2), HA(F-7), c-myc(9E10), JunB(C-11), p21(F-5), Smad1/2/3(H-2), CTGF(L-20) and Ubiquitin(P4D1) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The antibodies used for immunoprecipitation were HA(Y-11), CKIγ2(C-20) (Santa Cruz Biotechnology) and Smad3 (Zymed). TGF-β1 was obtained from R&D Systems (Minneapolis, MN, USA). SB431542 was purchased from Tocris (Ellisville, MO, USA) and MG-132 from BioMol (Plymouth Meeting, PA, USA).
Western blotting and immunoprecipitation
The protocols for western blotting and immunoprecipitation have been described earlier (Guo et al., 2008).
Transcriptional reporter assay
SBE-Lux and MBE-Lux were gifts from Xin-Hua Feng (Baylor College of Medicine, originally from Bert Vogelstein at Johns Hopkins University). ARE-Luc and FoxH1 were kindly provided by Anita Roberts (National Cancer Institute). 3TP-Luc has been described (Waddell et al., 2004). Luciferase assays were performed as reported earlier (Guo et al., 2008). Chemiluminescence was measured by the Victor3 system (PerkinElmer, Waltham, MA, USA).
Cell proliferation and wound-healing assays
Cell proliferation and wound-healing assays were performed exactly as described earlier (Guo et al., 2008).
In vitro kinase assay
Flag-CKIγ2 overexpressed in 293T cells was immunoprecipitated with an anti-Flag antibody (M2) in radioimmuno precipitation assay buffer (Guo et al., 2008). The beads were washed twice with radioimmuno precipitation assay buffer, once with high-salt buffer (100mM Tris-HCl, 500mM NaCl (pH 7.4)) and once with 1 × CKI kinase buffer (30 mM 4-(2-hydroxyethyl)- 1-piperazineethanesulfonic acid, 7mM MgC12, 1mM dithiothreitol (pH 7.5)). The immunoprecipitated kinase was then resuspended in 2× CKI kinase buffer and incubated with 2 mg of GST fusion proteins, 50 µM unlabeled ATP and 20 µCi [32P]-γ-ATP at 37°C for 30min. Reactions were terminated by boiling samples in Laemmli sample buffer for 5min. Phosphorylated proteins were separated by SDS–polyacrylamide gel electrophoresis and visualized by autoradiography. His-CKIγ2 (PV3499) was purchased from Invitrogen (Carlsbad, CA) and used under the same reaction conditions.
Supplementary Material
Acknowledgements
We thank Drs Jun Kusuda, Joan Massagué, Xin-Hua Feng, Rik Derynck, Jun-Lin Guan, James Woodgett and Anita Roberts for valuable reagents. We appreciate the Wang laboratory members for insightful scientific discussions and excellent technical support. We thank Natalie Ahn, Kathryn Resing and Will Old for MS facility and support. This work was supported by NIH grants DK064113 and GM083000 to X-F W, and an NIH Grant GM083172 to XL. DSW was supported by Department of Defense Breast Cancer Predoctoral Fellowship DAMD17-00-1-0299. NTL was supported by a National Science Foundation Predoctoral Fellowship.
References
- Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, et al. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- Dennler S, Huet S, Gauthier JM. A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene. 1999;18:1643–1648. doi: 10.1038/sj.onc.1202729. [DOI] [PubMed] [Google Scholar]
- Dunn NR, Koonce CH, Anderson DC, Islam A, Bikoff EK, Robertson EJ. Mice exclusively expressing the short isoform of Smad2 develop normally and are viable and fertile. Genes Dev. 2005;19:152–163. doi: 10.1101/gad.1243205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi M, Imamura T, Chiba T, Ebisawa T, Kawabata M, Tanaka K, et al. Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of ROC1 and associated proteins. Mol Biol Cell. 2001;12:1431–1443. doi: 10.1091/mbc.12.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Guo X, Ramirez A, Waddell DS, Li Z, Liu X, Wang X-F. Axin and GSK3-β control Smad3 protein stability and modulate TGF-β signaling. Genes Dev. 2008;22:106–120. doi: 10.1101/gad.1590908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin C-H, Miyazono K, Dijke PT. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Heldin CH, ten Dijke P. SMAD destruction turns off signalling. Nat Cell Biol. 1999;1:E195–E197. doi: 10.1038/70223. [DOI] [PubMed] [Google Scholar]
- Imoto S, Sugiyama K, Sekine Y, Matsuda T. Roles for lysine residues of the MH2 domain of Smad3 in transforming growth factor-beta signaling. FEBS Lett. 2005;579:2853–2862. doi: 10.1016/j.febslet.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- Izzi L, Attisano L. Ubiquitin-dependent regulation of TGF signaling in cancer. Neoplasia. 2006;8:677–688. doi: 10.1593/neo.06472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman L, Massagué J. Distinct oligomeric states of SMAD proteins in the transforming growth factor-beta pathway. J Biol Chem. 2000;275:40710–40717. doi: 10.1074/jbc.M005799200. [DOI] [PubMed] [Google Scholar]
- Kitabayashi AN, Kusuda J, Hirai M, Hashimoto K. Cloning and chromosomal mapping of human casein kinase I gamma 2 (CSNK1G2) Genomics. 1997;46:133–137. doi: 10.1006/geno.1997.4991. [DOI] [PubMed] [Google Scholar]
- Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K, et al. Negative regulation of transforming growth factor-[beta] (TGF-[beta]) signaling by WW domain-containing protein 1 (WWP1) Oncogene. 2004;23:6914–6923. doi: 10.1038/sj.onc.1207885. [DOI] [PubMed] [Google Scholar]
- Kretschmer A, Moepert K, Dames S, Sternberger M, Kaufmann J, Klippel A. Differential regulation of TGF-beta signaling through Smad2, Smad3 and Smad4. Oncogene. 2003;22:6748–6763. doi: 10.1038/sj.onc.1206791. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Datto MB, Frederick JP, Shen X, Wong C, Rougier-Chapman EM, et al. Smads bind directly to the Jun family of AP-1 transcription factors. Proc Natl Acad Sci USA. 1999;96:4844–4849. doi: 10.1073/pnas.96.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati NT, Moniwa M, Borton AJ, Davie JR, Wang XF. An essential role for Mad homology domain 1 in the association of Smad3 with histone deacetylase activity. J Biol Chem. 2001;276:22595–22603. doi: 10.1074/jbc.M010778200. [DOI] [PubMed] [Google Scholar]
- Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, et al. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng X-H. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg G-H, Tan Y, et al. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- Lo RS, Massagué J. Ubiquitin-dependent degradation of TGF-beta-activated Smad2. Nat Cell Biol. 1999;1:472–478. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- Long J, Wang G, Matsuura I, He D, Liu F. Activation of Smad transcriptional activity by protein inhibitor of activated STAT3 (PIAS3) Proc Natl Acad Sci USA. 2004;101:99–104. doi: 10.1073/pnas.0307598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Peters JM, McKay RM, McKay JP, Graff JM. Casein kinase I transduces Wnt signals. Nature. 1999;401:345–350. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- Piek E, Ju WJ, Heyer J, Escalante-Alcalde D, Stewart CL, Weinstein M, et al. Functional characterization of transforming growth factor beta signaling in Smad2- and Smad3-deficient fibroblasts. J Biol Chem. 2001;276:19945–19953. doi: 10.1074/jbc.M102382200. [DOI] [PubMed] [Google Scholar]
- Poncelet A-C, Schnaper HW, Tan R, Liu Y, Runyan CE. Cell phenotype-specific down-regulation of Smad3 involves decreased gene activation as well as protein degradation. J Biol Chem. 2007;282:15534–15540. doi: 10.1074/jbc.M701991200. [DOI] [PubMed] [Google Scholar]
- Price MA. CKI, there’s more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 2006;20:399–410. doi: 10.1101/gad.1394306. [DOI] [PubMed] [Google Scholar]
- Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108:823–835. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- Reguly T, Wrana JL. In or out? The dynamics of Smad nucleocytoplasmic shuttling. Trends Cell Biol. 2003;13:216–220. doi: 10.1016/s0962-8924(03)00075-8. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Tian F, Byfield SD, Stuelten C, Ooshima A, Saika S, et al. Smad3 is key to TGF-[beta]-mediated epithelial-to-mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine Growth Factor Rev. 2006;17:19–27. doi: 10.1016/j.cytogfr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Schilling SH, Datto MB, Wang XF. A phosphatase controls the fate of receptor-regulated Smads. Cell. 2006;125:838–840. doi: 10.1016/j.cell.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Schmierer B, Hill CS. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor beta-dependent nuclear accumulation of Smads. Mol Cell Biol. 2005;25:9845–9858. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielhaber E, Virshup DM. Casein kinase I: from obscurity to center stage. IUBMB Life. 2001;51:73–78. doi: 10.1080/15216540117461. [DOI] [PubMed] [Google Scholar]
- Waddell DS, Liberati NT, Guo X, Frederick JP, Wang X-F. Casein kinase Ie plays a functional role in the transforming growth factor-β signaling pathway. J Biol Chem. 2004;279:29236–29246. doi: 10.1074/jbc.M400880200. [DOI] [PubMed] [Google Scholar]
- Wang W, Koka V, Lan HY. Transforming growth factor-β and Smad signalling in kidney diseases. Review article. Nephrology. 2005;10:48–56. doi: 10.1111/j.1440-1797.2005.00334.x. [DOI] [PubMed] [Google Scholar]
- Weinstein M, Yang X, Deng C. Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev. 2000;11:49–58. doi: 10.1016/s1359-6101(99)00028-3. [DOI] [PubMed] [Google Scholar]
- Xu L, Kang Y, Col S, Massagué J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol Cell. 2002;10:271–282. doi: 10.1016/s1097-2765(02)00586-5. [DOI] [PubMed] [Google Scholar]
- Yagi K, Goto D, Hamamoto T, Takenoshita S, Kato M, Miyazono K. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J Biol Chem. 1999;274:703–709. doi: 10.1074/jbc.274.2.703. [DOI] [PubMed] [Google Scholar]
- Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, et al. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438:873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, Graves PR, Robinson LC, Italiano M, Culbertson MR, Rowles J, et al. Casein kinase I gamma subfamily. Molecular cloning, expression, and characterization of three mammalian isoforms and complementation of defects in the Saccharomyces cerevisiae YCK genes. J Biol Chem. 1995;270:12717–12724. doi: 10.1074/jbc.270.21.12717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.