Abstract
Nonsteroidal anti-inflammatory drugs (NSAIDs) are known to prevent colorectal tumorigenesis. Although anti-tumor effects of NSAIDs are mainly due to inhibition of cyclooxgenase (COX) activity, there is increasing evidence that COX-independent mechanisms may also play an important role. The early growth response-1 (EGR-1) gene is a member of the immediate early gene family and has been identified as a tumor suppressor gene. Tolfenamic acid (TA) is an NSAID that exhibits anti-cancer activity in a pancreatic cancer model. In the present study, we investigated the anti-cancer activity of TA in human colorectal cancer cells. TA treatment inhibited cell growth and induced apoptosis as measured by caspase activity and bioelectric impedance. TA induced EGR-1 expression at the transcription level, and analysis of the EGR-1 promoter showed that a putative ETS binding site (EBS), located at −400 and −394 bp, was required for activation by TA. The electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation (ChIP) assay confirmed that this sequence specifically bound to the ETS family protein ESE-1 transcription factor. TA also facilitated translocation of endogenous and exogenous ESE-1 to the nucleus in colorectal cancer cells, and gene silencing using ESE-1 siRNA attenuated TA-induced EGR-1 expression and apoptosis. Overexpression of EGR-1 increased apoptosis and decreased bioelectrical impedance, and silencing of endogenous EGR-1 prevented TA-induced apoptosis. These results demonstrate that activation of ESE-1 via enhanced nuclear translocation mediates TA-induced EGR-1 expression, which plays a critical role in the activation of apoptosis.
Keywords: Tolfenamic acid, EGR-1, ESE-1, NAG-1, NSAID
Introduction
Epidemiological studies have reported an inverse correlation between colon cancer incidence and the use of nonsteroidal anti-inflammatory drugs (NSAIDs) (1). As a result, the regular use of NSAIDs as a chemopreventive strategy for colorectal cancer is now generating a great deal of interest because NSAIDs inhibit cyclooxygenase (COX) and prostaglandin biosynthesis, a pathway strongly associated with colorectal carcinogenesis (2). NSAIDs induce cell cycle arrest and apoptosis during different stages of colorectal tumorigenesis (3–5) and inhibit angiogenesis, invasion, and metastasis in vivo (6, 7).
Tolfenamic acid (TA) has been broadly used for treatment of migraines and has shown fewer upper gastrointestinal side effects than other NSAIDs (8). There is recent evidence showing that TA also affects metastasis and tumorigenesis in pancreatic cancer models (9, 10). TA reduces the expression of vascular endothelial growth factor (VEGF) and its receptor (VEGFR1) (9, 10), which are mediated in part by downregulation of specificity proteins (Sps). Like other NSAIDs, TA acts by inhibiting COX and prostaglandin biosynthesis; however, the molecular basis for induction of apoptosis and the scope of its action in colorectal cancer has not been reported.
One potential molecular target is early growth response-1 (EGR-1), an immediate early gene encoding a zinc finger transcription factor; EGR-1 regulates the expression of genes involved in growth control or survival (11). The EGR-1 gene is stimulated by many extracellular signaling molecules including cytotoxic metabolites, hormones, growth factors, and neurotransmitters. The growth-promoting activity by EGR-1 has been observed in various human cancer models such as prostate (12), skin (13) and kidney (14). Various growth factors target the EGR-1 gene and mediate the mitogenic signaling cascade (15). Despite the discovery of EGR-1 as a growth-promoting protein, a number of reports have described EGR-1 as a pro-apoptotic protein. Indeed, EGR-1 is down-regulated in neoplasia and an array of tumor cell lines (16), and constitutive expression of EGR-1 suppressed growth and transformation in tumor cells (17). Recently, we and others reported that EGR-1 acts as a pro-apoptotic gene in human colorectal cancer cells. EGR-1 is a target of chemopreventive compounds including NSAIDs, LY294002, PPAR ligands (18–21), and dietary compounds such as epicatechin gallate and 1,1-Bis(3'-indolyl)-1-(p-substitutedphenyl) methanes (22–24). In addition, EGR-1 mediates several pro-apoptotic proteins, including p53, phosphatase and tensin homologue (PTEN), activating transcription factor 3 (ATF3), and NSAID-activated gene-1 (NAG-1), which appear to play a critical role in mediating apoptosis in human colorectal cancer cells (18–21, 25, 26). Thus, the pro-apoptotic activity of EGR-1 may depend on the cell type and the nature of the cytotoxic stimulus.
The current study was performed to elucidate whether TA affects human colorectal tumorigenesis. Here, we report, for the first time, that TA suppresses proliferation and induces apoptosis in human colorectal cancer cells through induction of a novel pathway that involves increased nuclear accumulation of epithelial-specific ETS-1 (ESE-1) transcription factor, which in turn activates EGR-1. TA-induced EGR-1 expression results in the induction of apoptosis, which is mediated in part by activation of the pro-apoptotic protein NAG-1.
Material and Methods
Materials
Tolfenamic acid and SC-560 were purchased from Cayman Chemical Company (Ann Arbor, MI), and [5,5-dimethyl-3-(3-fluorophenyl)-4-(4-methylsulphonyl)phenyl-2(5H)-furanone] (DFU) was described previously (20). All other NSAIDs were purchased from Sigma-Aldrich (St. Louis, MO). Antibodies for EGR-1 (sc-110), ESE-1 (sc-17306), actin (sc-1615), control siRNA (sc-37007), and ESE-1 siRNA (sc-37851) were purchased from Santa Cruz (Santa Cruz, CA), and antibody for PARP (#9542) was purchased from Cell Signaling (Beverly, MA). Antibodies for V5 (R960-25) and Sp1 (07-124) were purchased from Invitrogen (Carlsbad, CA) and Upstate Biotechnology (Lake Placid, NY), respectively. TRITC (1031-03) was purchased from Southern Biotech (Birmingham, AL), and antibody for NAG-1 was previously described (27). All chemicals were purchased from Fisher Scientific, unless otherwise specified.
Constructs
Full-length ESE-1 and ELF-1 cDNAs were amplified using ReadyMix Taq polymerase (Sigma, St. Louis, MO) with the following primers: ESE-1, forward 5’-CCAACCTCATCCTCTCTCCCCCTACCC-3’, and reverse 5’-GTTCCGACTCTGGAGA ACCTCTTCCTCC-3’; ELF-1, forward 5’-GGAATTTTTCTCATGTGGATCTAAGGGG-3, and reverse 5’-AAAAGAGTTGGGTTCCAGCAGTTCG-3’. PCR was performed for 30 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The amplified products were subcloned into pcDNA3.1/V5-His-TOPO (Invitrogen) to generate V5-His-tagged proteins. EGR-1 (pcDNA3.1/EGR-1/NEO) expression vector was previously described (19). For deletion analysis of the EGR-1 promoter, pEGR1-1260/+35 (20) was serially deleted using the Erase-a-Base System (Promega, Madison, WI) according to the manufacturer’s protocols. The internal deletion mutant clones for EBS, Sp1, SRF, and C/EBP of the EGR-1 promoter were generated with the pEGR1-1260/+35 wild type clone using the QuickChange II site-directed Mutagenesis Kit (Stratagene, La Jolla, CA).
Cell culture and treatment
Human colorectal carcinoma cell lines HCT-116, SW480, LoVo, and HT-29, were purchased from American Type Culture Collection (Manassas, VA). HCT-116 and HT-29 cells were maintained in McCoy’s 5A, and SW480 and LoVo cells were maintained in RPMI1640 and Ham’s F-12 medium, respectively. All media were supplemented with 10% fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 µg/mL of streptomycin. Cells were incubated at 37°C under a humidified atmosphere of 5% CO2 until cells were 70–80% confluent. The cells were then treated with different concentrations of TA at different time points as indicated in the figure legends.
Cell Proliferation
A cell proliferation assay was performed using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega) as described previously (28). Briefly, cells were seeded at the concentration of 2000 cells/well in 96-well tissue culture plates in four replicates and maintained overnight. The cells were then treated with 0, 1, 5, 10, 20, 30, and 50 µM of TA. At 0, 24, and 48 hours after treatment, 20 µL of CellTiter96 Aqueous One solution was added to each well, and the plate was incubated for 1 h at 37°C. Absorbance at 490 nm was recorded in an ELISA plate reader (Bio-Tek Instruments, Inc. Winooski, VT).
Caspase 3/7 Enzyme Activity
Apoptosis was measured using Apo-ONE Homogeneous Caspase-Glo 3/7 Assay kit (Promega) according to the manufacturer’s protocol. After harvest of the cells using RIPA buffer containing protease inhibitors, the cell lysates (50 µg protein) in 50 µL volume were mixed with 50 µL of Caspase-Glo 3/7 reagent in 96-well plates and incubated at room temperature in the dark for 1 h. The luminescence was measured using a plate-reading luminometer (FLX800, BioTek).
Electrical Impedance Measurements
The cellular electrical impedance method measures the frequency-dependent resistance and reactance of a cell-covered, thin film gold electrode as a function of time. To perform these measurements, a data acquisition and analysis system was implemented using LabVIEW. A reference voltage source, with 50 Ω output impedance, provided an alternating current 1vrms reference signal via a series 1 MΩ resistor to the electrode array. A National Instruments SCXI-1331 switch made successive connections between the various working electrodes and the counter electrode of each array. An SR830 lock-in amplifier (Stanford Research, Sunnyvale, CA) measured the electrode voltage. The input impedance of the lock-in amplifier was equivalent to a parallel resistor and capacitor combination of 10 MΩ and 10 pF, respectively. Direct measurements of the cable parasitic capacitances were made using an LCR meter and incorporated into a circuit model to estimate the impedance based on the lock-in voltage measurements (29). The preliminary naked scan also checked for any debris on electrodes as well as electrode defects. A 1-second naked scan, sampled at a rate of 32 Hz, was then performed for the naked electrodes. The electrodes were then inoculated with 400 µL medium containing HCT-116 cells having a concentration of 105 cells/mL. During the cellular attach scan, data was acquired at a rate of 32 Hz for 2 seconds using a 30 ms filter time constant and a 12 dB/decade roll off. Averages and standard deviation estimates were obtained from the sampled data points. During the experiments, cell-inoculated electrodes were kept in a cell culture incubator that maintained the temperature at 37°C and the CO2 level at 5%.
Transient Transfection and Luciferase Assay
Transient transfection was performed using Lipofectamine (Invitrogen) according to the manufacturer’s instructions. The cells were plated in 12-well plates at a concentration of 2 × 105 cells/well and left overnight. The cells were then transfected with the EGR-1 promoter (0.5 µg DNA) and the pRL-null vector (0.05 µg DNA) for 5 h. For the co-transfection experiment, 0.25 µg of the EGR-1 promoter and 0.25 µg of expression vector were co-transfected with 0.05 µg of the pRL-null vector. The cells were fed fresh medium (McCoy’s 5A medium with 10% FBS) overnight and treated with DMSO or 30 µmol/L of TA for 24 h. After cell harvest using 1X luciferase lysis buffer, luciferase activity was determined and normalized to the pRL-null luciferase activity using a dual luciferase assay kit (Promega).
RNA Interference
HCT-116 cells were transfected with control small interference RNA (siRNA) or ESE-1 siRNA at a concentration of 100 nM, using TransIT-TKO transfection reagent (Mirus Corp., Madison, WI), as described previously (28). After a 24 h transfection, the cells were serum starved overnight and then treated with DMSO or 30 µmol/L of TA for 2 h. For EGR-1 interference, sense EGR-1 oligo (5’-ZECGGGGCGCGGGGAACFOT-3’) and anti-sense EGR-1 oligo (5’-AEZGTTCCCCGCGCCCCGOA-3’) were synthesized by Invitrogen and transfected using TransIT-TKO transfection reagent (Mirus, Madison, WI) at a concentration of 200 nmol/L. After 24 h transfection, the cells were serum starved overnight and treated with DMSO or 30 µmol/L of TA for 2 h.
RNA isolation and RT-PCR
Total RNA was isolated using Trizol Reagent (Invitrogen), according to the manufacturer’s instructions. Total RNA (1 µg) was reverse-transcribed with an iScript cDNA kit (BioRad, Hercules, CA) according to the manufacturer’s instructions. PCR was carried out using ReadyMix Taq polymerase (Sigma) with primers for human EGR-1 and GAPDH as follows: EGR-1, forward, 5’- CTGCGACATCTGTGGAAGAA-3’, and reverse 5’-TGTCCTGGGAGAAAAGGTTG-3’; GAPDH, forward 5’-GGGCTGCTTTTAAC TCTGGT-3’, and reverse 5’- TGGCAGGTTTTTCTAGACGG-3’. PCR was performed at 94°C/30 s, 55°C/30 s, and 72°C/1 min for 25 cycles (EGR-1 and NAG-1) or 19 cycles (GAPDH).
Western Blot Analysis and Immunofluorescence
Western blot and immunofluorescence were performed as described previously (28, 30). For immunofluorescence, the cells were incubated with primary antibody for V5 overnight at 4°C and then with goat anti-mouse TRITC conjugate for 1 h at room temperature in the dark. The cells were stained with 0.5 mg/mL of DAPI for 5 min to counter stain the nucleus.
Electrophoretic Mobility Shift Assay (EMSA)
HCT-116 cells were grown to reach 80% confluence in 100-mm plates. After serum starvation overnight, the cells were treated with DMSO or 30 µmol/L of TA for 2 h. After washing with PBS, nuclear extracts were prepared following the manufacturer’s protocols (Active Motif, Carlsbad, CA). Oligonucleotide probes contained the following sequences: Wild: 5'-AGCAGGAAGGAAGCAGGAAGGAAGCAGGAAGGA-3' and mut: 5'-AGCACGCATGAAGCACGCATGAAGCACGCATGA-3'. EMSA was performed using the LightShift Chemiluminescent EMSA kit according to the manufacturer’s protocols (Pierce, Rockford, IL). Briefly, biotin-labeled oligonucleotide (10 nmol/L) was incubated with nuclear extract (5 µg protein), 1X binding buffer, 2.5% glycerol, 5 mmol/L MgCl2, 50 ng/µL Poly (dI·dC), and 0.05% NP-40 at room temperature for 20 min. For competition assay, nuclear extracts were preincubated with the unlabeled oligonucleotide (X10, X50, and X100) for 10 min. For supershift assay, nuclear extracts were preincubated with 0.6 µg of antibodies (ESE-1, Sp1, and IgG) in 1X binding buffer (Promega) for 15 min prior to binding reactions. DNA-protein complexes were resolved by 5% nondenaturing polyacrylamide gel and transferred to a nylon membrane, followed by chemiluminescent nucleic acid detection according to the manufacturer’s protocols.
Chromatin Immunoprecipitation (ChIP) Assay
HCT-116 cells were plated on a 100-mm culture dish and serum starved overnight. Cells were treated with DMSO or 30 µmol/L of TA for 2 h and fixed with 1% formaldehyde at 37°C for 10 min. The fixed cells were scraped into conical tubes, pelleted, and lysed in lysis buffer (1% SDS, 10 mmol/L EDTA, 50 mmol/L Tris, pH 8.0) containing protease and phosphatase inhibitors as described (28). DNA was sheared to fragments by sonication four times for 10 s at 50% constant maximal power. The sonicated cell supernatant was diluted 10-fold in immunoprecipitate buffer (0.1% SDS, 1% Triton X-100, 0.1% Na-deoxycholate, 140 mmol/L NaCl), and 1% of diluted cell supernatant was kept as a control (input). The chromatin was precleared with protein A/G agarose slurry for 1 h at 4°C. The precleared supernatant was incubated with 50 µL protein A/G agarose, 20 µg heat-denatured salmon sperm DNA, 50 µg BSA, and 10 µg antibodies against ESE-1 or IgG overnight at 4°C. The immunocomplexes were washed five times with wash buffer (20 mmol/L Tris, pH 8.0, 0.1% Triton-X, 150 mmol/L NaCl, 2 mmol/L EDTA,) and eluted with elution buffer (1% SDS, 100 mmol/L NaHCO3). Protein-DNA crosslinks were reversed with 27 µL of 5 mol/L NaCl and 1 µL of 10 mg/mL RNase A at 65°C for 4 h and incubated with 2 µL of 20 mg/mL proteinase K at 45°C for 1 h. The DNA was purified by phenol extraction and ethanol precipitation. The region between −530 and −345 of the human EGR-1 promoter was amplified using the 5’-GCGCCACACGCCACGAGCCCTCCCCGCCT-3’ and 5’-CCACTCCAAATAAGGTGCT GCGTTGTTCCGGCG-3’ primers. The 186 bp products were resolved on a 2% agarose gel and visualized under UV light.
Statistical Analysis
Statistical analysis was performed using a Student unpaired t test, with statistical significance set at *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Results
Identification of TA as a Strong Inducer of EGR-1 and NAG-1
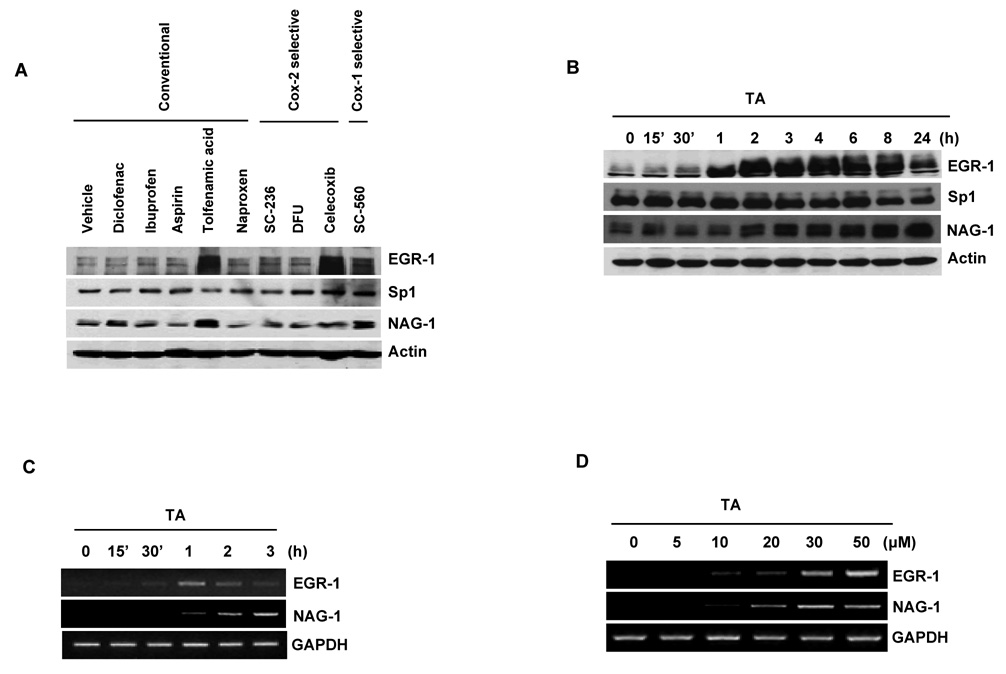
A number of studies indicate that EGR-1 plays a role as a tumor suppressor gene (16, 17, 25, 26), and EGR-1 mediates pro-apoptotic activity of the NSAID-activated gene-1 (NAG-1) by NSAIDs (20). In order to identify a potent NSAID that induces the two pro-apoptotic EGR-1 and NAG-1 genes in human colorectal cancer cells, we screened induction responses in HCT-116 cells by conventional (diclofenac, ibuprofen, aspirin, tolfenamic acid, naproxen) and COX-2 (SC-236, DFU, celecoxib) or COX-1 (SC-560) selective inhibitors. Since EGR-1 is an early induced gene (20, 21), we initially treated cells with 30 µmol/L of NSAIDs for 6 h. As shown in Fig. 1A, the expression of EGR-1 and NAG-1 was obviously increased in the cells treated with TA and SC-560. Celecoxib also highly increased EGR-1, but not NAG-1 expression. Because induction of EGR-1 was more dramatic in TA-treated cells than in SC-560 treated cells, TA was selected for further study. EGR-1 protein began to increase at 1 h, reached a plateau at 2 h, and gradually decreased after 8 h, whereas after 30 µmol/L of TA treatment, NAG-1 expression started to increase at 2 h and continued to increase until 24 h (Fig. 1B). Sp1 expression was decreased after 8 h, consistent with the data seen in pancreatic cancer cells (9, 10). We observed a significant increase in EGR-1 transcripts after 30 min and subsequent induction of NAG-1 transcripts in the cells treated with 30 µmol/L TA for 1 h (Fig. 1C). EGR-1 and NAG-1 mRNA levels were increased in a dose-dependent manner, reaching a plateau in the cells treated with 30 µmol/L of TA (Fig. 1D).
Figure 1. Tolfenamic acid (TA) increases the expression of EGR-1 and NAG-1 in human colorectal cancer cells.
A Screening of various NSAIDs based on EGR-1 and NAG-1 expression. HCT-116 cells were incubated with the indicated NSAIDs at 30 µmol/L for 6 h. Total cell lysates were harvested and subsequently Western blot analysis was performed for EGR-1, Sp1, NAG-1, and actin as described in Materials and Methods. B Western blot analysis. HCT-116 cells were serum-starved overnight and treated with 30 µmol/L of TA for 0, 15’, 30’, 1, 2, 3, 4, 6, 8, and 24 h. Total cell lysates were harvested, and subsequently Western blot analysis was performed. C Time-dependent expression of EGR-1 and NAG-1. After serum starvation overnight, HCT-116 cells were treated with 30 µmol/L of TA for 0, 15’, 30’, 1, 2, and 3 h. RT-PCR was performed for EGR-1 and NAG-1 as described in Materials and Methods. GAPDH represents a loading control. D Dose-dependent expression of EGR-1 and NAG-1. After serum starvation, HCT-116 cells were treated with 0, 5, 10, 20, 30, and 50 µmol/L of TA for 2 h. RT-PCR was performed.
TA Suppresses Cell Growth and Increases Apoptosis in HCT-116 Cells
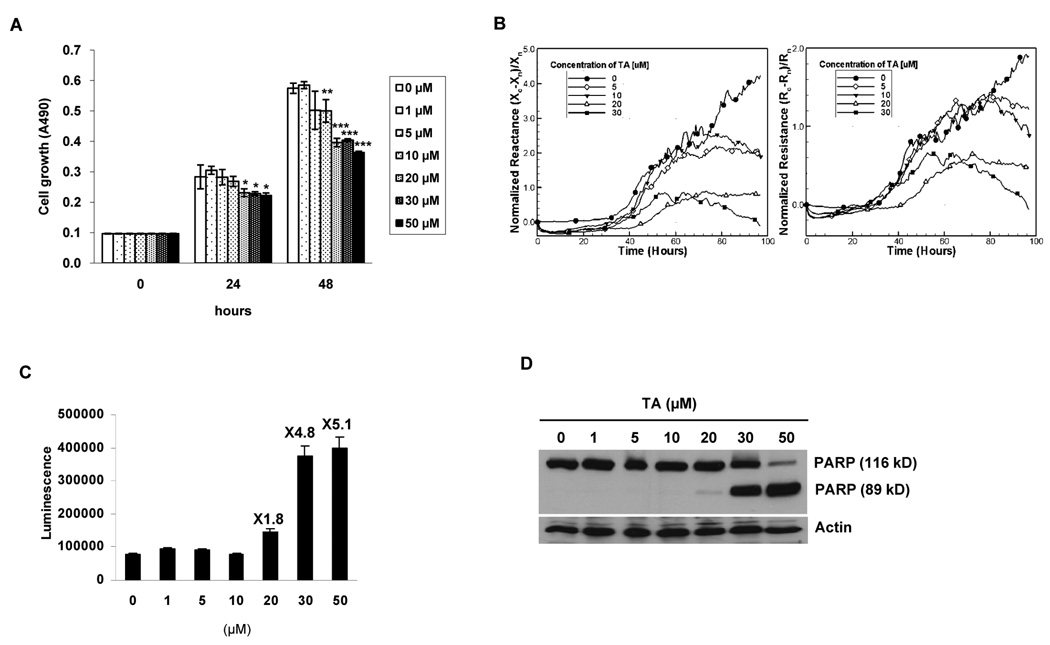
Since TA exhibited anti-tumorigenic activity in pancreatic cancer cells (9, 10), HCT-116 human colorectal cancer cells were incubated with 0, 1, 5, 10, 20, 30, and 50 µmol/L of TA for 24 and 48 h, and cell proliferation was measured. As shown in Fig. 2A, HCT-116 cells treated with 20, 30 and 50 µmol/L of TA significantly decreased cell growth after 24 and 48 h. Longer exposure of TA (48 h) also significantly inhibited cell growth at a lower concentration of TA (10 µmol/L). To further examine TA-induced apoptosis, dynamic and noninvasive cellular electrical micro-impedance measurements were performed. This method measures the frequency- and time-dependent resistance and reactance of a cell-covered thin film gold electrode and is very sensitive to apoptosis. As shown in Fig. 2B, 20 and 30 µmol/L TA decreased the normalized impedance components consistent with increasing numbers of apoptotic cells. Both resistance and reactance using vehicle-treated cells increased in cell-cell and cell-substrate adhesion. TA-treated cells, however, produced normalized resistances and reactances that decreased after they reached their maximum. These changes are, therefore, represented as the induction of apoptosis (31, 32). To investigate the effects of TA on caspase-dependent apoptosis, HCT-116 cells were treated with 0, 1, 5, 10, 20, 30, and 50 µmol/L of TA for 24 h, and caspase 3/7 activity and apoptosis-related cleaved poly (ADP-ribose) polymerase (PARP) were measured by Western blot analysis. As shown in Fig. 2C, caspase 3/7 activity was slightly increased in the cells treated with 20 µmol/L of TA and dramatically increased by 4.8- and 5.1-fold in cells treated with 30 and 50 µmol/L of TA, respectively. In addition, PARP cleavage was dramatically increased in HCT-116 cells treated with 30 and 50 µmol/L of TA with a slight increase in 20 µmol/L TA-treated cells (Fig. 2D). Therefore, we decided to use 30 µmol/L TA for further experiments because this concentration induced both apoptosis and EGR-1/NAG-1 gene expression.
Figure 2. TA suppresses cell growth and increases apoptosis.
A Cell growth. HCT-116 cells were treated with 0, 1, 5, 10, 20, 30, and 50 µmol/L of TA for 0, 24 and 48 h. Cell growth was measured using CellTiter96 Aqueous One Solution Cell Proliferation Assay as described in Materials and Methods. Values are expressed as mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus DMSO-treated cells at each time point. B Cellular micro-impedance. Normalized resistance, (Rc-Rn)/Rn, and normalized reactance, (Xc-Xn)/Xn, of HCT116 cells following treatment with 0, 5, 10, 20, and 30 µmol/L of TA were obtained using electrical impedance measurement technique as described in Materials and Methods for indicated time points. The subscripts c and n indicate cell covered and naked scans, respectively. Measurements were performed simultaneously using the same batch of HCT-116 cells. The representative time-dependent normalized resistances and reactances shown here were scanned at 5.62 kHz and 100 kHz, respectively. For the sake of clarity, symbols are selectively marked. C Apoptosis detection (caspase 3/7 activity). HCT-116 cells were treated with 0, 1, 5, 10, 20, 30, and 50 µmol/L of TA for 24 h. Caspase 3/7 activity was measured as described in Materials and Methods. The data represent mean ± SD from three independent experiments. D Apoptosis detection (PARP cleavage). HCT-116 cells were treated with 0, 1, 5, 10, 20, 30, and 50 µmol/L of TA for 24 h. Poly ADP ribose polymerase (PARP) cleavage was measured using Western blot analysis.
TA induces EGR-1 at the transcription level
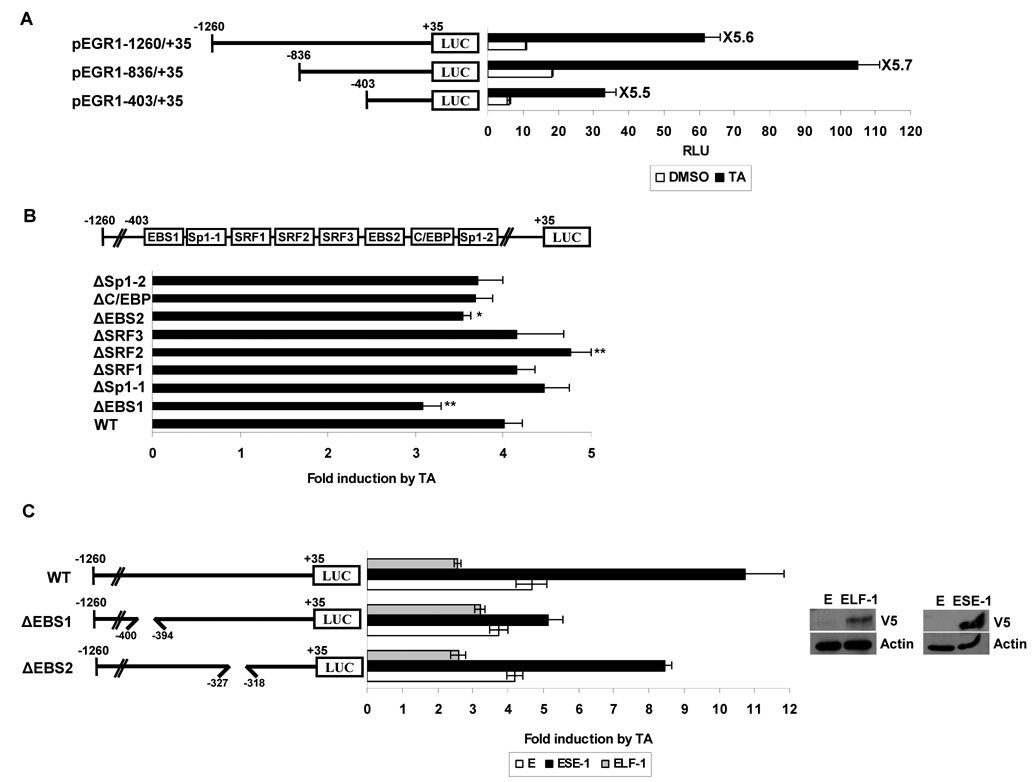
The effects of TA on EGR-1 expression at the transcriptional level were investigated using an EGR-1 promoter in transient transfection studies. HCT-116 cells were transfected with different promoter constructs spanning the −1260 to +35 promoter regions as shown in Fig. 3A. TA treatment increased luciferase activity by 5.6-, 5.7-, and 5.5-fold in cells transfected with pEGR1-1260/+35, pEGR1-836/+35, and pEGR1-403/+35, respectively, indicating that the −403 and +35 region of the promoter was sufficient for TA-induced EGR-1 transactivation. To further identify potential regulatory cis-acting elements that mediate the stimulatory effects of TA, the Transcription Element Search System (http://www.cbil.upenn.edu/cgi-bin/tess/tess) was used to search for conserved transcription factor binding sites within the −403 and +35 region. The EGR-1 promoter (pEGR1-403/+35) contains multiple transcription factor binding sites including EBS, Sp1, SRF, and C/EBP (Fig. 3B, top). We constructed internal deletion clones lacking specific binding sites as indicated in Fig. 3B. HCT-116 cells were transfected with these deletion constructs, treated with DMSO or 30 µmol/L of TA for 24 h, and luciferase activity was measured. As shown in Fig. 3B, in cells transfected with wild type pEGR1-1260/+35 promoter, TA increased luciferase activity by 4-fold, whereas TA-induced activity was significantly decreased in cells transfected with a promoter construct lacking an EBS1 (pEGR1-1260/+35ΔEBS1). Moreover, deletion of another EBS (pEGR1-1260/+35ΔEBS2) also showed a slight decrease of luciferase activity. Interestingly, deletion of an SRF site (pEGR1-1260/+35ΔSRF2) resulted in increased luciferase activity induced by TA. Other deletion constructs did not influence TA-induced luciferase activities. These data suggest that the EBS region of the EGR-1 promoter may play a role in TA-induced EGR-1 transactivation.
Figure 3. ETS binding site (EBS) located at −400 to −394 of the EGR-1 promoter is necessary for EGR-1 transcription induced by TA.
A Structures of EGR-1 sequential deletion constructs and deletion promoter assay. The EGR-1 promoter fragments of a different length but with the same 3'-end were cloned into pGL3-Basic. HCT-116 cells were co-transfected with 0.5 µg of each reporter construct containing the EGR-1 promoter and 0.05 µg of pRL-null vector using Lipofectamine. After growth overnight with fresh media, the cells were treated with 30 µmol/L of TA for 24 h. Luciferase activity was measured as a ratio of firefly luciferase signal/renilla luciferase signal and was shown as mean ± S.D. of three independent transfections. B The putative transcription binding sites within the −403 to +35 region in the EGR-1 promoter and luciferase assay with internal deletion clones. The boxes in the promoter represent the binding site of indicated transcription factors and are used for construction of internal deletion clones. HCT-116 cells were co-transfected with 0.5 µg of each internal deletion construct of the EGR-1 promoter and 0.05 µg of pRL-null vector using Lipofectamine and treated with 30 µmol/L of TA for 24 h. The X axis shows fold induction over vehicle as 1.0. The results are presented as means ± S.D. of three independent transfections. *, P < 0.05; **, P < 0.01 versus pEGR1-1260/+35 WT transfected cells. C Effect of ESE-1 overexpression on EGR-1 transactivation. HCT-116 cells were co-transfected with wild type EGR-1 promoter (pEGR1-1260/+35 WT or internal deletion clone pEGR1-1260/+35ΔEBS) in the presence of empty (E), ESE-1, or ELF-1 expression vector using Lipofectamine. After growth overnight with fresh media, the cells were treated with 30 µmol/L of TA for 24 h. The X axis shows fold induction over vehicle as 1.0. The results are presented as the means ± S.D. of three independent transfections. The overexpression of ELF-1 and ESE-1 was confirmed by Western analysis using V5 tag (GKPIPNPLLGLDST) antibody.
EBS is characterized by a 5’-GGA(A/T)-3’ DNA core motif, and all ETS proteins contain a highly conserved DNA-binding domain (ETS domain) and bind to this site as a monomer (33). To obtain further evidence that TA induces EGR-1 through EBS, we cotransfected the EGR-1 promoter constructs and expression vectors for ESE-1, ELF-1, and ETS-1 into HCT-116 cells and determined luciferase activity. Expression of transfected vectors was confirmed by Western blot analysis (Fig. 3C, right). ESE-1 expression caused a dramatic increase of TA-induced luciferase activity, compared with empty vector-transfected cells. Interestingly, ESE-1 expression attenuated the increase of EGR-1 promoter activity in cells transfected with a construct lacking the EBS1 (−400 to −394), whereas the deletion clone lacking the EBS2 (−372 to −318) did not show dramatic attenuation of TA-induced activation (Fig. 3C), suggesting that EBS1 is most likely required for activation of the EGR-1 promoter induced by TA. ELF-1 expression decreased TA-induced EGR-1 promoter activity. We excluded the contribution of ELK-1 on TA-induced EGR-1 expression because our RT-PCR and Western blot data showed that HCT-116 cells do not express ELK-1 (data not shown).
Effect of TA on ESE-1 DNA Binding Activity
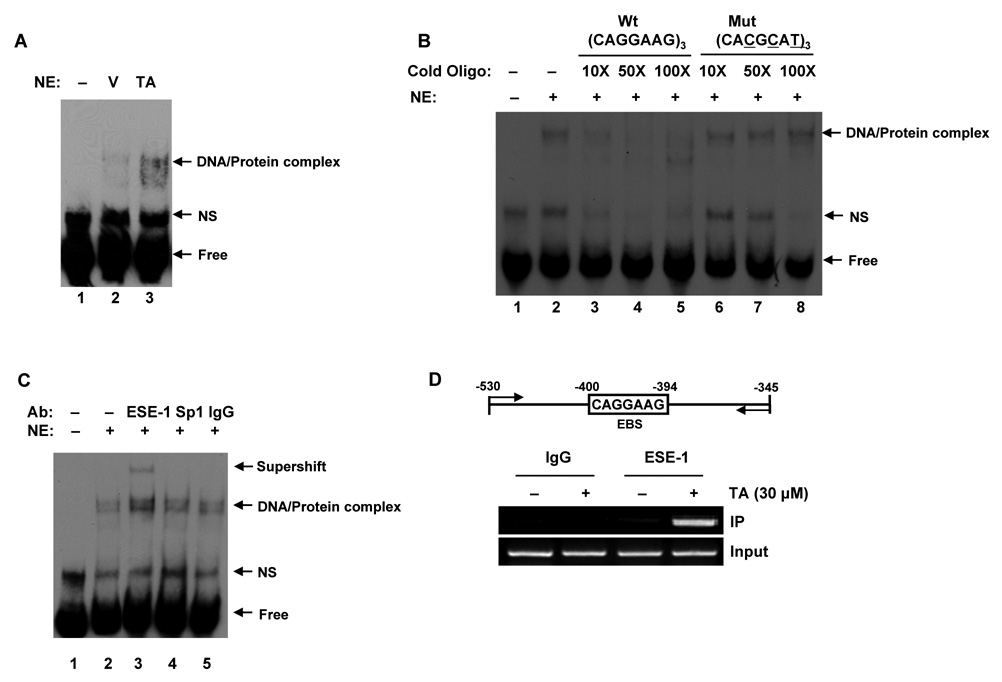
To examine whether the transactivation of the EGR-1 gene promoter is mediated by ESE-1, an electrophoretic mobility shift assay (EMSA) was performed using oligonucleotide probes derived from the EGR-1 promoter (−400 to −394) and nuclear extracts prepared from HCT-116 cells treated with DMSO or TA for 2 h. As shown in Fig. 4A, TA treatment caused an induction of DNA-protein complex formation. Preincubation of nuclear extracts with 10X, 50X, and 100X excess unlabeled EBS oligonucleotide abolished the binding activity (Fig. 4B, Lane 3–5), whereas preincubation of nuclear extracts with unlabeled mutant EBS oligonucleotide did not compete with labeled oligonucleotides for binding (Fig. 4B, Lane 6–8), suggesting that the binding protein is specific for the EBS1 sequence. A supershift assay was performed to confirm that ESE-1 protein binds to this site. Preincubation of nuclear extracts with ESE-1 antibody resulted in a supershift of the DNA-protein complexes, whereas antibodies for Sp1 and IgG did not affect the mobility of the DNA-protein complex, suggesting that ESE-1 is specifically bound to this site (Fig. 4C). Finally, a ChIP assay was performed to confirm that ESE-1 binds to EBS1 in the EGR-1 promoter. As shown in Fig. 4D, immunoprecipitation of the chromatin-protein complex with ESE-1 amplified 186 bp of the PCR products, and TA treatment increased the binding affinity of ESE-1 to DNA. An aliquot (1%) of the total chromatin DNA was used for input. The ChIP assay was also performed at 0, 30’, 1, and 2 h time points, and we observed that the binding affinity increased as early as 30 min after treatment (Data not shown). These results confirm that endogenous ESE-1 strongly binds to EBS on the EGR-1 promoter after treatment of HCT-116 cells with TA.
Figure 4. Identification of TA-responsive DNA binding activity of ESE-1 in the EGR-1 promoter.
A Electrophoretic mobility shift assay (EMSA). After serum starvation overnight, HCT-116 cells were treated with DMSO (V) or 30 µmol/L of TA (TA) for 2 h and EMSA was performed using nuclear extracts (5 µg), as described in Materials and Methods. The specific DNA-protein complexes are indicated by arrows. NE, nuclear extract; NS, non specific. B Competition assay. The competition of the DNA binding was obtained using a 10, 50, and 100-time excess of the unlabeled oligonucleotide (lane 3–5). The specificity of the complex for EBS was confirmed by the absence of competition with an excess of cold EBS mutated oligonucleotide (lane 6–8). C Supershift experiment. A gel supershift experiment was performed to detect binding activity of ESE-1 to EBS after exposure to 30 µmol/L of TA. The indicated antibodies were added to the incubation mixture. An arrow indicates the antibody-induced supershift band. The EMSA conditions and the sequence of the probes used in the experiment are described in Materials and Methods. D Chromatin immunoprecipitation (ChIP) assay. After serum starvation overnight, HCT-116 cells were treated with 30 µmol/L of TA for 2 h. The in vivo DNA-protein complexes were cross-linked by formaldehyde treatment, and chromatin pellets were extracted and sonicated. The associated EGR-1 DNA was isolated as described in Materials and Methods. The sequence of the human EGR-1 promoter region (−530/−345) was amplified by PCR primer pairs as indicated by the arrows. The input represents PCR products obtained from 1% aliquots of chromatin pellets prior to immunoprecipitation.
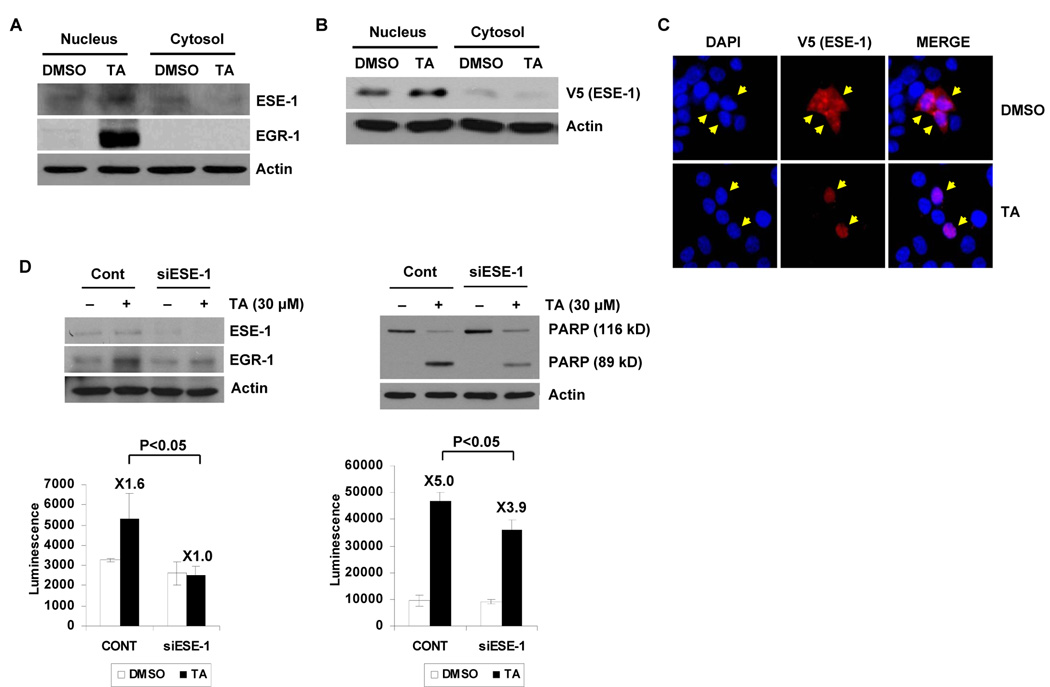
TA Mediates Translocation of ESE-1 and EGR-1-Induced Apoptosis
EGR-1 protein was strongly induced by TA at 2 h; however, ESE-1 protein levels were not changed by TA treatment for up to 24 h (data not shown). The activity of ESE- 1 can be regulated by subcellular localization (34), and therefore, effects of TA on nuclear translocation of ESE-1 were investigated. Fig. 5A shows depleted cytosolic ESE-1 and increased expression of this protein in the nuclear fraction after TA treatment for 2 h. We also observed that TA induced nuclear translocation of ectopically expressed ESE-1 in a manner similar to that shown for endogenous ESE-1 (Fig. 5B). To verify the Western blot data, we used immunofluorescence imaging to directly visualize localization of ESE-1 after TA treatment in cells transfected with an expression vector encoding V5-His epitope-tagged ESE-1. The result shows that the nuclear staining of ESE-1 was significantly increased in HCT-116 cells after treatment with 30 µmol/L TA for 1 h, confirming data obtained from the fractionation experiments (Fig. 5C). To determine the requirement of ESE-1 for EGR-1 expression, HCT-116 cells were transfected with control or ESE-1 siRNA, and then treated with 30 µmol/L TA for 2 h. Immunoblotting of the lysates confirmed efficient knockdown of ESE-1 expression (Fig. 5D). TA induced EGR-1 expression in control siRNA-transfected-cells; however, ESE-1 siRNA transfection attenuated TA-induced EGR-1 expression (Fig. 5D, left). The same result was obtained from RT-PCR (data not shown). Two hours after TA treatment, a 1.6-fold higher caspase 3/7 enzyme activity was observed in control siRNA-transfected cells, whereas the activity was not changed in ESE-1 siRNA transfected cells. To demonstrate the specific involvement of ESE-1 in TA-induced apoptosis, control or ESE-1 siRNA transfected cells were treated with 30 µmol/L TA for 24 h, and caspase 3/7 enzyme activity and PARP cleavage were determined. As shown in Fig. 5D (right), 24 h after treatment with TA, high levels (5-fold) of caspase 3/7 enzyme activity were observed, indicative of cell death, whereas induction of caspase 3/7 enzyme activity (3.9-fold) was decreased in ESE-1 siRNA-treated cells (Fig. 5D, right). Similar results were obtained for PARP cleavage, where knockdown of ESE-1 decreased TA-induced PARP cleavage compared to cells transfected with control siRNA. Together, these results demonstrate that ESE-1 may contribute to TA-induced EGR-1 expression and cell death.
Figure 5. TA mediates translocation of ESE-1 into the nucleus, and siRNA-mediated inhibition of ESE-1 expression suppressed TA-induced EGR-1 expression and apoptosis.
A Nuclear translocation of endogenous ESE-1. HCT-116 cells were serumstarved overnight and then treated with 30 µmol/L of TA for 2 h. Nuclear and cytosol fractions were isolated, and Western blot analysis was performed for ESE-1, EGR-1, and actin antibodies. B Nuclear translocation of exogenous ESE-1. HCT-116 cells were transiently transfected with pcDNA3.1/V5-His/ESE-1 expression vector using Lipofectamine as described in Materials and Methods. After serum-starvation overnight, the cells were treated with 30 µmol/L of TA for 1 h. Nuclear and cytosol fractions were isolated, and Western blot was performed for V5 and actin antibodies. C Immunohistochemistry. After transfection as described in (B), the cells were serum-starved overnight and treated with 30 µmol/L of TA for 1 h. The cells were fixed and stained with anti-V5 antibody overnight and subsequently, secondary anti-mouse TRITC conjugate (red). DAPI staining was used to visualize the nucleus of the cells (blue). Magnifications correspond to 400X. The arrows indicate ESE-1 localization. D Effect of ESE-1 knockdown on TA-induced EGR-1 expression and apoptosis. HCT-116 cells were transfected with control siRNA (100 nmol/L) or ESE-1 siRNA (100 nmol/L) for 24 h using a TransIT-TKO transfection reagent. After serum starvation overnight, the cells were treated with 30 µmol/L of TA for 2 h (left panel) or for 24 h (right panel). Western blot analysis was performed for ESE-1, EGR-1, PARP, and actin antibodies, and caspase 3/7 activity was measured as described in Materials and Methods. The same cell lysates were used to measure caspase 3/7 activity as described in Materials and Methods (bottom panels). The data represent mean ± SD from three independent experiments.
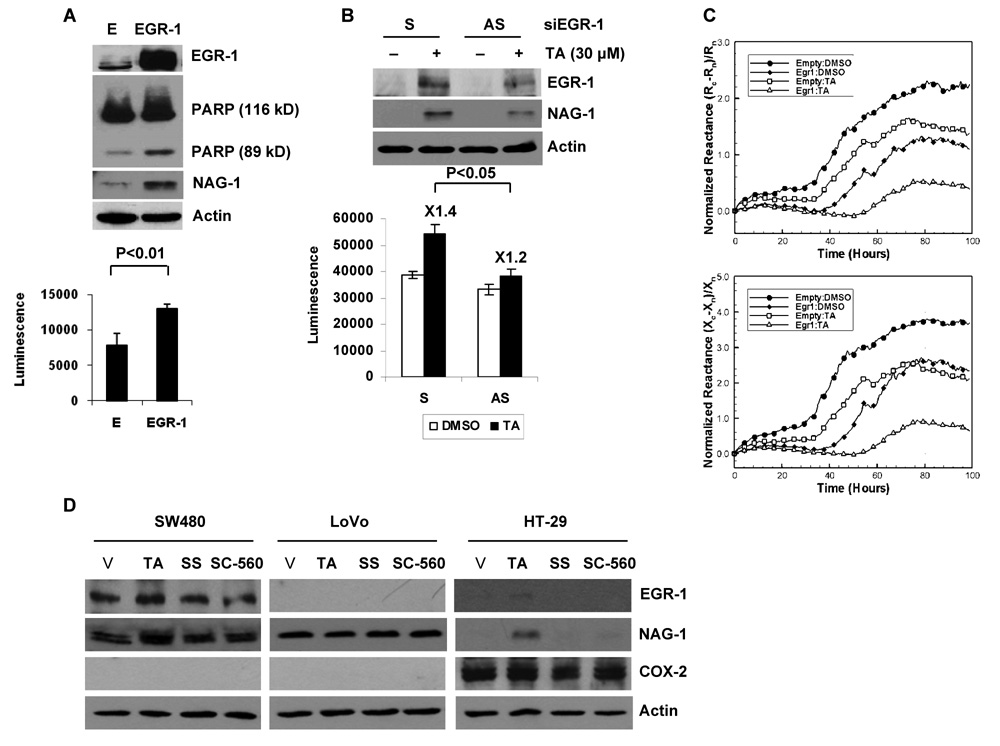
EGR-1 Contributes at Least in Part to Apoptosis by NAG-1 Expression in the Presence of TA
The involvement of EGR-1 in apoptosis was investigated in cells that overexpressed EGR-1. As shown in Fig. 6A, expression of EGR-1 increased PARP cleavage and caspapse 3/7 enzyme activity. Interestingly, the pro-apoptotic protein NAG-1 was induced in EGR-1 overexpressing cells. In addition, suppression of endogenous expression of the EGR-1 gene using antisense oligonucleotide blunted TA-induced caspase 3/7 activity and NAG-1 expression, suggesting a linkage between EGR-1 expression, NAG-1 expression, and TA-induced apoptosis (Fig. 6B). We also measured electrical impedance after EGR-1 overexpression. Fig. 6C shows that overexpression of EGR-1 dramatically reduced the normalized resistance and reactance. In addition, in the presence of EGR-1, TA decreased impedance components more dramatically, which is consistent with increasing numbers of apoptotic cells. These results demonstrate that EGR-1 mediates TA-induced apoptosis in HCT-116 cells.
Figure 6. EGR-1 induces apoptosis and mediates TA-induced apoptosis.
A Apoptosis detection after EGR-1 overexpression. HCT-116 cells were transfected with the empty or EGR-1 expression vector. PARP cleavage was measured by Western blot analysis (top panel) and caspase 3/7 activity (bottom panel) was determined as described in Materials and Methods. The data represent mean ± SD from three independent experiments. B Effect of EGR-1 knockdown on TA-induced apoptosis. HCT-116 cells were transfected with sense (S) or anti-sense (AS) oligo for human EGR-1 as described in Materials and Methods. After serum starvation overnight, the cells were treated with 30 µmol/L of TA for 2 h. Western analysis was performed for NAG-1 and actin antibodies (top panel), and caspase 3/7 activity (bottom panel) was measured. The data represent mean ± SD from three independent experiments. C Cellular micro-impedance to detect the effect of EGR-1 transfection. Normalized resistance, (Rc-Rn)/Rn, and normalized reactance, (Xc-Xn)/Xn, of HCT116 cells transiently transfected with empty (E) or EGR-1 expression vector following treatment with TA were measured using the cellular micro-impedance measurement technique as described in Materials and Methods. The representative time-dependent normalized resistances and reactances shown here were scanned at 5.62 kHz and 100 kHz, respectively. Filled symbols represent cell measurements with DMSO, and blank symbols represent cells treated with 30 µmol/L of TA. For the sake of clarity, symbols are selectively marked. D Expression of EGR-1, NAG-1 and COX-2 in other colorectal cancer cells. SW480, LoVo and HT-29 cells were grown, serum starved overnight, and treated with 30 µmol/L of TA, sulindac sulfide (SS) and SC-560 for 2 h. Western blot analysis was performed for EGR-1, NAG-1, COX-2, and actin antibodies.
Finally, to investigate whether TA modulates EGR-1-dependent NAG-1 expression in other human colorectal cancer cells, SW480, LoVo, and HT-29 cells were treated with TA, sulindac sulfide (SS), and SC-560 for 2 h, and induction of EGR-1 and NAG-1 were determined. TA increased EGR-1 and NAG-1 expression in SW480 (COX-2 null cells) and HT-29 (COX-2 expressing cells), but not in LoVo cells (COX-2 null cells). However, expression of EGR-1 and NAG-1 were not changed at 2 h after treatment of sulindac sulfide and SC-560 in any of the tested cells. These data indicate that NAG-1 induction by NSAIDs may be mediated in a cell context-dependent and COX-2 independent manner (Fig. 6D).
Discussion
Recent studies showed TA suppressed cell growth and angiogenesis in in vitro and in vivo models of pancreatic cancer (10). This response was mediated by decreased expression of Sp1, Sp3, and Sp4 proteins and subsequent downregulation of Sp-dependent genes such as VEGF and VEGFR1 (9, 10). In this study, expression of Sp1 was also decreased after treatment with TA for 8 h, indicating that Sp1 down-regulation occurred and may contribute to TA-dependent anti-tumorigenic activity in colorectal cancer cells. However, Sp1 (Fig. 1A) and VEGF (data not shown) expression did not change at earlier time points (6 h) in TA-treated cells. This suggests that TA affects anti-tumorigenic activity through Sp1 downregulation at later time points (8–48 h), whereas TA induces apoptosis through EGR-1 induction at early time points (0–6 h).
Epithelial-specific ETS-1 (ESE-1) protein belongs to the ETS family of transcription factors and is also identified as ERT (35), ELF3 (36), and ESX (37). ESE-1 proteins contain a highly conserved ETS domain that recognizes the core motif GGA(A/T). ESE-1 proteins are constitutively expressed in many types of epithelia, including lung and intestine (38), and regulate terminal differentiation of the epidermis (38, 39). ESE-1 have multiple functions in the transcriptional regulation of genes involved in epithelial differentiation and development of cancer (40), depending on cell context, and exact molecular mechanisms and their transcriptional targets need to be defined in cancer cells. The present study supports the anti-tumorigenic role of ESE-1 by facilitating cell growth arrest and inducing apoptosis in HCT-116 cells. We have also shown that knockdown of ESE-1 by RNA interference (RNAi) inhibited TA-induced cell death as assessed by caspase 3/7 enzyme activity and PARP cleavage, which is associated with decreased expression of EGR-1 (Fig. 5D). Furthermore, inhibition of EGR-1 attenuated cell death as well as NAG-1 expression by TA treatment (Fig. 6B). The main finding of the present study is that TA resulted in translocation of ESE-1 protein to the nucleus. This temporal pattern of nuclear ESE-1 translocation and binding of ESE-1 in the EGR-1 promoter nicely corresponded to TA-induced activation of EGR-1 gene expression at the transcriptional level. Under these experimental conditions, nuclear ESE-1 translocation was almost exclusively observed in cells 1 h after administration of TA. It is likely that ESE-1 has two distinct but separable biological functions, namely apoptosis and transformation, which depend on the subcellular localization of this protein. Indeed, Prescott et al. reported that endogenous ESE-1 localize in the cytoplasm of human breast cancer cells; however, ectopically expressed ESE-1 in the nucleus resulted in the induction of apoptosis (34). These findings support our presumption that cytoplasmic accumulation of ESE-1 may be involved in the development of colon cancer, while nuclear translocation of ESE-1 enhances both EGR-1 transactivation and apoptosis. Interestingly, a recent study showed that p21-activated kinase 1 targets ESE-1 for phosphorylation, resulting in increased protein stability and higher accumulation in cells (41). Therefore, it is likely that TA may also trigger an upstream pathway followed by phosphorylation of ESE-1, which results in ESE-1 nuclear translocation. Future studies are needed to determine the molecular mechanism by which TA modifies ESE-1 by phosphorylation.
A combination of EMSA and ChIP assay on the binding of ESE-1 protein to the EGR-1 promoter has shown that the GGAA core sequence located in −400 to −394 is essential for the specific binding of ESE-1, indicating ESE-1 specificity in TA-induced EGR-1 expression. However, we could not exclude the possibility that this EBS is not sufficient to fully activate the EGR-1 promoter, since the EGR-1 promoter assay using internal deletion clones is not enough to decrease the promoter activity to basal levels. Human EGR-1 promoter also contains the second EBS at −327 to 318 (EBS2). As shown in Fig. 3B, deletion on this site results in the marginal decrease of TA-induced luciferase activity, indicating that this site also contributes to the TA-induced EGR-1 expression. However, co-transfection experiments suggest that the EBS2 site is less significant than the EBS1 site in TA-induced EGR-1 expression (Fig. 3C).
The NAG-1, one of the transforming growth factor-beta (TGF-β) superfamily genes, has been reported as pro-apoptotic and anti-tumorigenic by our group and others (27, 42). Recently, we generated NAG-1 transgenic mice that overexpress human NAG-1, and have demonstrated that these mice (NAG-Tg) are much less sensitive to carcinogens or genetic toxicity (43). The results support our previous study that EGR-1 mediates activation of NAG-1 gene expression by sulindac sulfide in human colorectal cancer cells (20). Thus, it appears that EGR-1 and NAG-1 play a significant role in the chemopreventive effect of NSAIDs in human colon or other cancers.
On the other hand, TA induced both EGR-1 and NAG-1 expression in COX-2 wild type (HT-29) and COX-2 mutant (SW480) cell lines, supporting that TA may affect tumorigenesis in a COX-2 independent manner. Although we did not observe that TA increased EGR-1 and NAG-1 expression in LoVo cells, another group has shown that treatment with NCX4040 (nitric oxide-releasing aspirin) induced NAG-1 expression in LoVo cells (44). These conflicting results may be explained by cell / NSAID specificity or duration of NSAID treatment.
Here we report that TA inhibited proliferation and induced apoptosis in human colorectal cancer cells through a COX-independent pathway. This new pathway involved 1) activation of ESE-1 via nuclear translocation, 2) EGR-1 transactivation and apoptosis induction by ESE-1, and 3) NAG-1 transactivation and apoptosis induction by EGR-1.
ACKNOWLEDGEMENTS
We thank Ms. Misty R. Bailey for her critical reading of the manuscript.
This work was supported by grants from the American Cancer Society (CNE-111611), National Institutes of Health (RO1CA108975), and the University of Tennessee Center of Excellence in Livestock Diseases and Human Health to Seung Joon Baek. This work was also supported in part by an NSF CAREER Award (BES-0238905) to Anthony E. English.
Abbreviations List
- CRC
colorectal carcinoma
- ETS
E26 transformed-specific
- EBS
ETS binding site
- EGR-1
early growth response factor 1
- ESE
epithelial-specific ETS protein
- NSAIDs
nonsteroidal anti-inflammatory drugs
- NAG-1
NSAID-activated gene-1
- NE
nuclear extract
- PARP
poly (ADP-ribose) polymerase
- TA
tolfenamic acid
REFERENCES
- 1.Huls G, Koornstra JJ, Kleibeuker JH. Non-steroidal anti-inflammatory drugs and molecular carcinogenesis of colorectal carcinomas. Lancet. 2003;362:230–232. doi: 10.1016/s0140-6736(03)13915-3. [DOI] [PubMed] [Google Scholar]
- 2.Gupta RA, Dubois RN. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer. 2001;1:11–21. doi: 10.1038/35094017. [DOI] [PubMed] [Google Scholar]
- 3.Grosch S, Tegeder I, Niederberger E, Brautigam L, Geisslinger G. COX-2 independent induction of cell cycle arrest and apoptosis in colon cancer cells by the selective COX-2 inhibitor celecoxib. Faseb J. 2001;15:2742–2744. doi: 10.1096/fj.01-0299fje. [DOI] [PubMed] [Google Scholar]
- 4.Sheng H, Shao J, Kirkland SC, et al. Inhibition of human colon cancer cell growth by selective inhibition of cyclooxygenase-2. J Clin Invest. 1997;99:2254–2259. doi: 10.1172/JCI119400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leahy KM, Ornberg RL, Wang Y, Zweifel BS, Koki AT, Masferrer JL. Cyclooxygenase-2 inhibition by celecoxib reduces proliferation and induces apoptosis in angiogenic endothelial cells in vivo. Cancer Res. 2002;62:625–631. [PubMed] [Google Scholar]
- 6.Tsujii M, Kawano S, Tsuji S, Sawaoka H, Hori M, DuBois RN. Cyclooxygenase regulates angiogenesis induced by colon cancer cells. Cell. 1998;93:705–716. doi: 10.1016/s0092-8674(00)81433-6. [DOI] [PubMed] [Google Scholar]
- 7.Yao M, Kargman S, Lam EC, et al. Inhibition of cyclooxygenase-2 by rofecoxib attenuates the growth and metastatic potential of colorectal carcinoma in mice. Cancer Res. 2003;63:586–592. [PubMed] [Google Scholar]
- 8.Hansen PE. Tolfenamic acid in acute and prophylactic treatment of migraine: a review. Pharmacol Toxicol. 1994;75 Suppl 2:81–82. doi: 10.1111/j.1600-0773.1994.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 9.Abdelrahim MB, Abbruzzese CH, Sheikh-Hamad JL, et al. Regulation of vascular endothelial growth factor receptor-1 expression by specificity proteins 1, 3, and 4 in pancreatic cancer cells. Cancer Res. 2007;67:3286–3294. doi: 10.1158/0008-5472.CAN-06-3831. [DOI] [PubMed] [Google Scholar]
- 10.Abdelrahim M, Baker CH, Abbruzzese JL, Safe S. Tolfenamic acid and pancreatic cancer growth, angiogenesis, and Sp protein degradation. J Natl Cancer Inst. 2006;98:855–868. doi: 10.1093/jnci/djj232. [DOI] [PubMed] [Google Scholar]
- 11.Sukhatme VP, Cao XM, Chang LC, et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 12.Abdulkadir SA, Qu Z, Garabedian E, et al. Impaired prostate tumorigenesis in Egr1-deficient mice. Nat Med. 2001;7:101–107. doi: 10.1038/83231. [DOI] [PubMed] [Google Scholar]
- 13.Riggs PK, Rho O, DiGiovanni J. Alteration of Egr-1 mRNA during multistage carcinogenesis in mouse skin. Mol Carcinog. 2000;27:247–251. doi: 10.1002/(sici)1098-2744(200004)27:4<247::aid-mc1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Scharnhorst V, Menke AL, Attema J, et al. EGR-1 enhances tumor growth and modulates the effect of the Wilms' tumor 1 gene products on tumorigenicity. Oncogene. 2000;19:791–800. doi: 10.1038/sj.onc.1203390. [DOI] [PubMed] [Google Scholar]
- 15.Svaren J, Ehrig T, Abdulkadir SA, Ehrengruber MU, Watson MA, Milbrandt J. EGR1 target genes in prostate carcinoma cells identified by microarray analysis. J Biol Chem. 2000;275:38524–38531. doi: 10.1074/jbc.M005220200. [DOI] [PubMed] [Google Scholar]
- 16.Huang RP, Fan Y, de Belle I, et al. Decreased Egr-1 expression in human, mouse and rat mammary cells and tissues correlates with tumor formation. Int J Cancer. 1997;72:102–109. doi: 10.1002/(sici)1097-0215(19970703)72:1<102::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 17.Huang RP, Liu C, Fan Y, Mercola D, Adamson ED. Egr-1 negatively regulates human tumor cell growth via the DNA-binding domain. Cancer Res. 1995;55:5054–5062. [PubMed] [Google Scholar]
- 18.Yamaguchi K, Lee SH, Kim JS, Wimalasena J, Kitajima S, Baek SJ. Activating transcription factor 3 and early growth response 1 are the novel targets of LY294002 in a phosphatidylinositol 3-kinase-independent pathway. Cancer Res. 2006;66:2376–2384. doi: 10.1158/0008-5472.CAN-05-1987. [DOI] [PubMed] [Google Scholar]
- 19.Baek SJ, Wilson LC, Hsi LC, Eling TE. Troglitazone, a peroxisome proliferator-activated receptor gamma (PPAR gamma ) ligand, selectively induces the early growth response-1 gene independently of PPAR gamma. A novel mechanism for its anti-tumorigenic activity. J Biol Chem. 2003;278:5845–5853. doi: 10.1074/jbc.M208394200. [DOI] [PubMed] [Google Scholar]
- 20.Baek SJ, Kim JS, Moore SM, Lee SH, Martinez J, Eling TE. Cyclooxygenase inhibitors induce the expression of the tumor suppressor gene EGR-1, which results in the up-regulation of NAG-1, an antitumorigenic protein. Mol Pharmacol. 2005;67:356–364. doi: 10.1124/mol.104.005108. [DOI] [PubMed] [Google Scholar]
- 21.Baek SJ, Kim JS, Nixon JB, DiAugustine RP, Eling TE. Expression of NAG-1, a transforming growth factor-beta superfamily member, by troglitazone requires the early growth response gene EGR-1. J Biol Chem. 2004;279:6883–6892. doi: 10.1074/jbc.M305295200. [DOI] [PubMed] [Google Scholar]
- 22.Moon Y, Lee M, Yang H. Involvement of early growth response gene 1 in the modulation of microsomal prostaglandin E synthase 1 by epigallocatechin gallate in A549 human pulmonary epithelial cells. Biochem Pharmacol. 2007;73:125–135. doi: 10.1016/j.bcp.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Chintharlapalli S, Papineni S, Baek SJ, Liu S, Safe S. 1,1-Bis(3'-indolyl)-1-(p-substitutedphenyl) methanes are peroxisome proliferator-activated receptor gamma agonists but decrease HCT-116 colon cancer cell survival through receptor-independent activation of early growth response-1 and nonsteroidal anti-inflammatory drug-activated gene-1. Mol Pharmacol. 2005;68:1782–1792. doi: 10.1124/mol.105.017046. [DOI] [PubMed] [Google Scholar]
- 24.Cho KN, Sukhthankar M, Lee SH, Yoon JH, Baek SJ. Green tea catechin (−)-epicatechin gallate induces tumour suppressor protein ATF3 via EGR-1 activation. Eur J Cancer. 2007;43:2404–2412. doi: 10.1016/j.ejca.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair P, Muthukkumar S, Sells SF, Han SS, Sukhatme VP, Rangnekar VM. Early growth response-1-dependent apoptosis is mediated by p53. J Biol Chem. 1997;272:20131–20138. doi: 10.1074/jbc.272.32.20131. [DOI] [PubMed] [Google Scholar]
- 26.Virolle T, Adamson ED, Baron V, et al. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol. 2001;3:1124–1128. doi: 10.1038/ncb1201-1124. [DOI] [PubMed] [Google Scholar]
- 27.Baek SJ, Kim KS, Nixon JB, Wilson LC, Eling TE. Cyclooxygenase inhibitors regulate the expression of a TGF-beta superfamily member that has proapoptotic and antitumorigenic activities. Mol Pharmacol. 2001;59:901–908. [PubMed] [Google Scholar]
- 28.Lee SH, Yamaguchi K, Kim JS, et al. Conjugated linoleic acid stimulates an anti-tumorigenic protein NAG-1 in an isomer specific manner. Carcinogenesis. 2006;27:972–981. doi: 10.1093/carcin/bgi268. [DOI] [PubMed] [Google Scholar]
- 29.Choi CK, English AE, Jun SI, Kihm KD, Rack PD. An endothelial cell compatible biosensor fabricated using optically thin indium tin oxide silicon nitride electrodes. Biosens Bioelectron. 2007;22:2585–2590. doi: 10.1016/j.bios.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Lee SH, Cekanova M, Baek SJ. Multiple mechanisms are involved in 6-gingerol-induced cell growth arrest and apoptosis in human colorectal cancer cells. Mol Carcinog. 2008;47:197–208. doi: 10.1002/mc.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arndt S, Seebach J, Psathaki K, Galla HJ, Wegener J. Bioelectrical impedance assay to monitor changes in cell shape during apoptosis. Biosens Bioelectron. 2004;19:583–594. doi: 10.1016/s0956-5663(03)00269-0. [DOI] [PubMed] [Google Scholar]
- 32.Yin HW, Wang FL, Wang AL, Cheng J, Zhou Y. Bioelectrical Impedance Assay to Monitor Changes in Aspirin-Treated Human Colon Cancer HT-29 Cell Shape during Apoptosis. Anal Lett. 2007;40:85–94. [Google Scholar]
- 33.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 34.Prescott JD, Koto KS, Singh M, Gutierrez-Hartmann A. The ETS transcription factor ESE-1 transforms MCF-12A human mammary epithelial cells via a novel cytoplasmic mechanism. Mol Cell Biol. 2004;24:5548–5564. doi: 10.1128/MCB.24.12.5548-5564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi SG, Yi Y, Kim YS, et al. A novel ETS-related transcription factor, ERT/ESX/ESE-1, regulates expression of the transforming growth factor-beta type II receptor. J Biol Chem. 1998;273:110–117. doi: 10.1074/jbc.273.1.110. [DOI] [PubMed] [Google Scholar]
- 36.Tymms MJ, Ng AY, Thomas RS, et al. A novel epithelial-expressed ETS gene, ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene. 1997;15:2449–2462. doi: 10.1038/sj.onc.1201427. [DOI] [PubMed] [Google Scholar]
- 37.Chang CH, Scott GK, Kuo WL, et al. ESX: a structurally unique ETS overexpressed early during human breast tumorigenesis. Oncogene. 1997;14:1617–1622. doi: 10.1038/sj.onc.1200978. [DOI] [PubMed] [Google Scholar]
- 38.Oettgen P, Alani RM, Barcinski MA, et al. Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family. Mol Cell Biol. 1997;17:4419–4433. doi: 10.1128/mcb.17.8.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng AY, Waring P, Ristevski S, et al. Inactivation of the transcription factor ELF3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology. 2002;122:1455–1466. doi: 10.1053/gast.2002.32990. [DOI] [PubMed] [Google Scholar]
- 40.Brembeck FH, Opitz OG, Libermann TA, Rustgi AK. Dual function of the epithelial specific ets transcription factor, ELF3, in modulating differentiation. Oncogene. 2000;19:1941–1949. doi: 10.1038/sj.onc.1203441. [DOI] [PubMed] [Google Scholar]
- 41.Manavathi B, Rayala SK, Kumar R. Phosphorylation-dependent regulation of stability and transforming potential of ETS transcriptional factor ESE-1 by p21-activated kinase 1. J Biol Chem. 2007;282:19820–19830. doi: 10.1074/jbc.M702309200. [DOI] [PubMed] [Google Scholar]
- 42.Baek SJ, Eling TE. Changes in gene expression contribute to cancer prevention by COX inhibitors. Prog Lipid Res. 2006;45:1–16. doi: 10.1016/j.plipres.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Baek SJ, Okazaki R, Lee SH, et al. Nonsteroidal anti-inflammatory drug-activated gene-1 over expression in transgenic mice suppresses intestinal neoplasia. Gastroenterology. 2006;131:1553–1560. doi: 10.1053/j.gastro.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 44.Tesei A, Rosetti M, Ulivi P, et al. Study of molecular mechanisms of pro-apoptotic activity of NCX 4040, a novel nitric oxide-releasing aspirin, in colon cancer cell lines. J Transl Med. 2007;5:52. doi: 10.1186/1479-5876-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]