Abstract
Objective
T-cell-mediated autoimmunity may be involved in some cases of idiopathic neutropenia. We hypothesized that a precise T-cell receptor repertoire analysis may uncover cytotoxic T-cell (CTL) expansions that are less pronounced than those seen in T large granular lymphocyte leukemia (T-LGL), but are pathophysiologically analogous and thus can serve as markers of a T-cell-mediated process.
Materials and Methods
Using rational algorithms for T-cell receptor analysis and in vivo tracking of CTL responses previously established in our laboratory, we studied patients with unexplained chronic neutropenia (n = 20), T-LGL (n = 15), and healthy controls (n = 12). We further investigated the involvement of soluble inhibitory factors by coculture assays. To determine the level of immune activation, we studied interferon-g expression in CD8+cells using Taqman polymerase chain reaction.
Results
Fifteen expanded (immunodominant) CTL clones were detected in 12 of 20 patients. In comparison to LGL leukemia, these clones were less immunodominant, but clearly discernible from subclinical lymphoproliferations in controls. As a surrogate of cytotoxic activity, we found markedly increased production of interferon-γ in most of the neutropenia patients, irrespective of the presence of immunodominant CTL clones.
Conclusions
These results suggest that, while immunodominant CTL clones are detectable in a proportion of patients only, CTL-mediated pathophysiology may be a general mechanism operating in idiopathic neutropenia. Oligogoclonal CTL expansions in chronic neutropenia may indicate an ongoing autoimmune process, while highly polarized monoclonalities in a subset of neutropenic LGL patients may represent the “extreme” end of the clonal continuum.
Drugs, hereditary factors, infections, and intrinsic bone marrow diseases explain the etiology of most neutropenias. The inability to detect an immune-mediated process or other obvious cause often leads to diagnosis of chronic idiopathic neutropenia (CIN) [1-6]. Similarly, secondary autoimmune neutropenia (AIN), usually associated with collagen vascular diseases, lymphoproliferative disorders and viruses, is typically diagnosed by exclusion and thus relies on the quality of clinical and laboratory workups [6-9]. Lineage-restricted cytopenias, including neutropenia, have been associated with T-cell large granular lymphocyte leukemia (T-LGL) [10], which is diagnosed based on detection of expanded VB+CD3+CD8+CD57+/- and positive T-cell receptor (TCR) rearrangement [11-13].
It is likely that patients with AIN/CIN may belong to a heterogeneous group of diseases combining various autoimmune etiologies. Appearance of the bone marrow may reveal some clues as to the cellular targets of the autoimmune process; left shift could indicate that more mature cells are targets, while pure white cell aplasia suggests that very early myeloid precursors are affected. The similar target spectrum in AIN/CIN and T-LGL suggests that cytotoxic T-cell (CTL)-mediated autoimmunity can also operate in AIN/CIN. Consequently, detection of highly polarized CTL expansions in some cases of neutropenia patients may be consistent with cytotoxicity directed against myeloid precursors. Based on results of previous studies showing that patients with clonal expansions in aplastic anemia or LGL benefit from selective T-cell targeted immunosuppressive therapies, similar treatment might be considered in neutropenia patients who show evidence for LGL-like T-cell expansion.
Clonotypic TCR diagnostics is based on the unique structure of the TCR consisting of variable α- and β-chains, and resulting from complex rearrangement steps. Rearranged T cells express only a single type of variable β-(VB) chain and, therefore, its CDR3 region, that most directly engages the peptide presented within human leukocyte antigen-groove, can serve as a marker of clonal expansion (clonotype) [14,15]. Clonal CTL expansion results in the overrepresentation of the respective TCR VB chain and the molecular analysis of the TCR repertoire can reveal the clonotypic structure of expanded T cell. Previously, based on the study of TCR repertoire in bone marrow failure syndromes, we introduced an efficient approach to detect immunodominant CTL; this methodology can be easily adapted to study CTL clonalities in other diseases [16-18]. The goal of our work was to identify disease-associated CTL clones and characterize cytotoxic responses in patients with unexplained neutropenia as compared to healthy and hematologic controls. This is the first study so far to identify CTL immunodominance and characterize the clonotypic repertoire in patients with unexplained neutropenia.
Materials and methods
Patients and controls
Patients (n = 20) were characterized by a prolonged reduction in absolute neutrophil count below 1500 per mm3 measured on multiple occasions (more than three times) during a period of 3 months (Table 1). Unexplained neutropenia was defined per exclusion of drug effects, hematologic diseases and reactive systemic conditions using routine laboratory tests. A cohort of 12 healthy individuals were studied as normal controls and a previously reported cohort of 15 neutropenic patients associated with T-LGL served as hematologic control [18]. Diagnosis of T-LGL was established by modified clinical and laboratory parameters as suggested by Semenzato et al. [19]. Informed consent for sample collection was given by the individuals according to protocols approved by the Institutional Review Board of the Cleveland Clinic Foundation (Cleveland, OH, USA).
Table 1.
Clinical and molecular characteristics of analyzed patients
| Patient no. | ANC (k/uL) | Age (y) | Anti- N-Ab | Colony inhib | VB CTL Clone | Clonotypic | Clonotype exp (%) |
|---|---|---|---|---|---|---|---|
| 1 | 0.50 | 44 | + | No | NIP | VB5- ASTQGNEQFF-JB2-1 | 31 |
| 2 | 0.68 | 58 | NA | No | NA | VB3- ASSPTSAAGELFF-JB2.2 | 22 |
| 3 | 1.7 | 64 | + | No | NIP | VB13-ASSYLTDSDTQYF-JB2.3 | 30 |
| 4 | 0.35 | 61 | + | No | NIP | VB17-ASSIIGNQPQHFGDG-JB1.5 | 100 |
| 5 | 1 | 53 | - | No | VB13 (7%) | VB13-ASSYLLTTGGAVRSPLH-JB1.6 | 76 |
| 6 | 1.07 | 65 | NA | No | NIP | VB4- SVEGSSSYGYTF-JB1-2 VB4- SVWGTGGLYEQYF-JB2-7 VB6- ASSGTGGWNEQFF-JB2-1 |
53 30 50 |
| 7 | 1.3 | 65 | + | NA | NIP | VB14-ASSHQRDYF-JB2.1 VB7- ASSQEGQLSYEQYF-JB2.7 |
40 18 |
| 8 | 1.3 | 41 | NA | No | NA | VB6- ASSPIRDRGAGELFF- JB2.2 | 25 |
| 9 | 0.37 | 57 | - | No | NA | VB13-ASSEGQGADTQYFG-JB2.3 | 54 |
| 10 | 1.3 | 38 | - | No | NA | Polyclonal | - |
| 11 | 1.22 | 42 | - | Yes | NIP | Polyclonal | - |
| 12 | 0.18 | 32 | + | No | NA | Polyclonal | - |
| 13 | 1.8 | 51 | NA | NA | NA | Polyclonal | - |
| 14 | 1.39 | 71 | - | No | NIP | Polyclonal | - |
| 15 | 1.35 | 24 | - | No | NIP | Polyclonal | - |
| 16 | 0.17 | 52 | + | NA | NIP | VB15-ATSEGGNEKLFF-JB1.4 | 81 |
| 17 | 0.73 | 44 | - | Yes | VB2 (8.2%) | VB2-CSAREYAGDQETQYF-JB2.5 | 18 |
| 18 | 0.5 | 41 | + | No | VB13 (9.4%) | Polyclonal | - |
| 19 | 0.51 | 33 | + | No | NIP | Polyclonal | - |
| 20 | 0.26 | 61 | - | No | NIP | VB6-ASSLAVMGNNEQFF- JB2.1 | 50 |
ANC = absolute neutrophil count. ANC at the time of sampling was 0.88 ± 0.52 k/uL and some patients were receiving myeloid growth factors. Anti-N-Ab = antineutrophil antibodies; VB exp = variable β-chain family expansion by flow cytometry; clonotype = VB CDR3 sequence of the expanded clone; clonotypic exp = clonotypic expansion within a given VB family; VB = variable β-chain of the T-cell receptor (TCR) VB; JB = joining region of the TCR VB; polyclonal = polyclonal TCR repertoire; NA = not available at the time of sampling; NIP = not in panel: no VB expansion was detected using the panel of available anti-VB antibodies; Yes/No = inhibition of progenitor growth, expressed as percentage of control colony formation in the presence of normal serum; values lower than mean of all controls +1 × SD of colony formation were defined as positive.
VB flow cytometry
Fresh peripheral blood was stained for VB flow cytometry analysis according to manufacturer’s instructions (Beckman-Coulter, Fullerton, CA, USA) and analyzed as described previously [18,20]. Initially, 12 normal samples were characterized to define the average size and standard deviations for the VB repertoire as detected by the antibodies. A significant expansion was defined as one that was greater than mean + 2 × standard deviation (SD) of the average VB family size obtained in controls.
Antineutrophil antibody testing
Circulating antibodies in patients’ plasma were measured using a standard clinical test performed by ARUP laboratories (http://www.aruplab.com/guides/ug/tests/0055506.jsp)
Plasma colony inhibition assay
Peripheral blood plasma from patients and healthy controls was tested for inhibition of the proliferation of healthy bone marrow (BM) precursor cells. For that purpose, 100 uL of plasma were incubated in duplicates with 100 Tsd. healthy donor BM mononuclear cells in Iscove’s medium containing 15% fetal calf serum for 2 hours. Following this, cultures were plated in 0.8% methylcellulose agar supplemented with a cytokine cocktail containing 10 ng/mL thrombopoietin, 1 U/mL erythropoietin, 10 ng/mL interleukin-3, and 25 ng/mL stem cell factor. After 14 days in a humidified incubator (37°C, 5% CO2 in air), primitive erythroid progenitor bursts and colonies derived from granulocytic/macrophage progenitors were scored using an inverted microscope. Inhibition was expressed as the percent of colony formation by healthy BM incubated with plasma from patients relative to healthy donors. Colony formation lower than mean of 10 controls -1 SD was defined as pathologic.
CD8+ separation, RNA extraction and RT
CD8+ T cells were isolated from Ficoll peripheral blood mononuclear cells by flow cytometric sorting (Epics Altra; Beckman Coulter, Miami, FL, USA) or by magnetic bead separation (Miltenyi, Bergisch Gladbach, Germany). Total RNA was extracted from CD8+ T cells with TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) using phase lock gel tubes (Eppendorf, Germany) and cDNA was reverse transcribed from 6 to 8 μL RNA by first strand cDNA synthesis using SuperScript III RT Kit (Invitrogen).
CDR3 region cloning and sequencing
Quantitative analysis of the clonotypic frequencies was performed as described by us previously [18,21]. Briefly, single VB families were amplified from CD8+cDNA using a combination of a VB family-specific sense primer and a CB antisense primer. Alternatively, multiplex polymerase chain reaction (PCR) was applied to amplify the entire VB TCR repertoire as described previously [22] with the following modifications: only VB and JB primers were used to amplify rearranged VB TCR chains from cDNA instead of genomic DNA. PCR products were gel-purified, cloned into a TA-vector, transformed into Escherichia coli. To avoid unanticipated bacterial cell division, posttransformation phenotypic expression time (37°C, 225 rpm) was reduced from 60 to 40 minutes before plating onto Agar plates. Multiple overnight colonies were subjected to PCR and sequencing. CDR3 sequences were analyzed and translated into amino acids using the online ImMunoGeneTics TCR alignment tool [23]. Pathologic expansion of a VB family or the clonotypic VB CDR3 repertoire was defined in earlier studies [18,24].
Clonotypic Taqman PCR
Clonotype-specific quantitative assay was designed to employ a patient-derived clonotypic sense primer, a JB family specific probe and a CB antisense primer. Based on the sequence of immunodominant CDR3 regions, clonotypic primers were designed to span the terminal VB chain and the entire NDN region, incorporating some bases of the JB chain (for primers and probes see Table 2). This would enhance primer fidelity and reduce the amplification of highly similar clonotypes carrying identical JB chain yet dissimilar variable JB amino acids. FAM-labeled probes specific to all human JB chains were designed with MGB-quencher (Applied Biosystems [ABI], Foster City, CA, USA) or Blackhole quencher 1 (BHQ-1, Eurogentec, San Diego, CA, USA) and CB reverse primer was designed to amplify both human CB families. PCR-mix included 15 μL 2× Taqman universal Mastermix (ABI) with 0.6 μL (50 μM) of each sense clonotypic and antisense CB primers, 0.66 μL (10 μM) JB Taqman probe, 6 μL cDNA (1:3 dilution with water) and 1.34 μL water in a total volume of 25 μL. All samples were run in duplicates after initial 10 minutes at 95°C at 45 cycles of 15 seconds at 95°C and 60 seconds at 60°C.
Table 2.
Sequences of primers and probes used for quantitative polymerase chain reaction assays
| Clonotypic primers | Patient no. 1 (VB5JB2.1) | TGCCAGCACCCAGGGGAAT |
| Patient no. 4 (VB17JB1.5) | GTATTATAGGGAATCAGCCCCAGC | |
| Patient no. 5 (VB13JB1.6) | AGCAGTTACTTACTCACCACAGGG | |
| Patient no. 6 (VB4JB1.2) | TCTGCAGCGTTGAAGGATCCG | |
| Patient no. 7 (VB14JB2.1) | CAGCAGTCACCAACGGGACTAC | |
| JB probes | JB1-2 | FAM-ACCAGGTTAACCGTTGTAG-MGBNFQ |
| JB1-5 | FAM-TTTGGTGATGGGACTCGA-MGBNFQ | |
| JB1-6 | FAM-TCACCCCTCCACTTTGGGAA-BHQ1 | |
| JB2-1 | FAM-AGCAGTTCTTCGGGCCAG-MGBNFQ | |
| CB primer | CB-reverse | CTGCTTCTGATGGCTCAAACAC |
| Taqman IFN-γ primers | IFN-γ forward | TATTCGGTAACTGACTTGAATGTCC |
| IFN-γ reverse | AGGCAGGACAACCATTACTGG | |
| Taqman IFN-γ probe | IFN-γ | FAM-AGGGAAGCGAAAAAGGAGTCAGATGCTG-BHQ1 |
IFN-γ = interferon-γ.
FAM-labeled and minor groove binder nonfluorescent-quencher (MGBNFQ) or black hole quencher1 (BHQ1) probes were used for quantitative polymerase chain reaction. CB reverse primer was designed to span both CB1 and CB2 chains. IFN-γ primer-probe set was designed to span the junction of exon 3 and exon 4 of the target gene. All primers and probes are shown in 5′ -3′order.
Interferon-γ-specific Taqman real-time PCR
Taqman assay spanning the exon-exon junction of the interferon (IFN)-γgene was designed based on the genomic sequence (Table 2). The amplification mix included 15 μL 2× Taqman Universal Mastermix (ABI) 3.5 μL (5 μM) forward and reverse primers, 1.4 μL (5 μM) BHQ-1 probe, 5 μL CD8+cDNA (1:3 dilution) and 1.6 μL water. Samples were run in duplicates on a 7500 real-time PCR system (ABI) at 95°C/10 minutes followed by 50 × 95°C/15 seconds and 60°C/60 seconds. IFN-γ expression was normalized to glyceraldehyde phosphate dehydrogenase expression and calculated relative to normalized expression in CD8+ control samples (n = 10).
Results
Identification of cryptic CTL expansions
We postulated that some cases of otherwise unexplained neutropenia can be mediated by a cytotoxic process, similar to that operating in T-LGL. We studied a cohort of patients with various degrees of neutropenia that was unexplained based on clinical grounds and standard laboratory testing (Table 1). The absolute neutrophil count (ANC) was 0.88 ± 0.52 k/uL (some patients were receiving myeloid growth factors at the time of sampling). Significant inhibition of myeloid colony formation was observed in 2 of 17 patients tested, while antineutrophil antibodies were detected in 8 of 16 patients (Table 1).
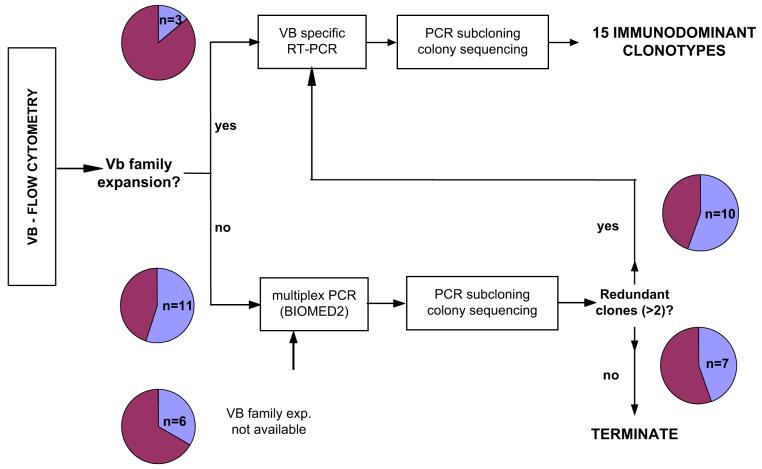
Clonal CTL expansions were detected using a diagnostic algorithm shown in Figure 1: Initially VB family expansions were detected in 3 patients, while subsequent TCR single colony sequencing identified immunodominant clonotypes (identical clones occurring repetitively) in two patients. In the remaining cases, the entire TCR VB repertoire was sequenced (generally ≥15 per VB family or 40 per entire VB repertoire). If redundant clonotypes were encountered, clonotypic repertoire within respective VB was sequenced and allowed for the identification of clonotype expansions in 10 additional patients. Despite overall CTL polyclonality, 15 immunodominant clonotypes were found in a total of 12 patients (Table 1; Fig. 1).
Figure 1.
Experimental strategy for the detection and molecular characterization of clonal immunodominant cytotoxic T-cell (CTL) populations. Organizational chart illustrates the rational diagnostic approach for the detection of cryptic clonal CTL expansions in neutropenia patients. For details see text.
Detection of immunodominant clonotypes
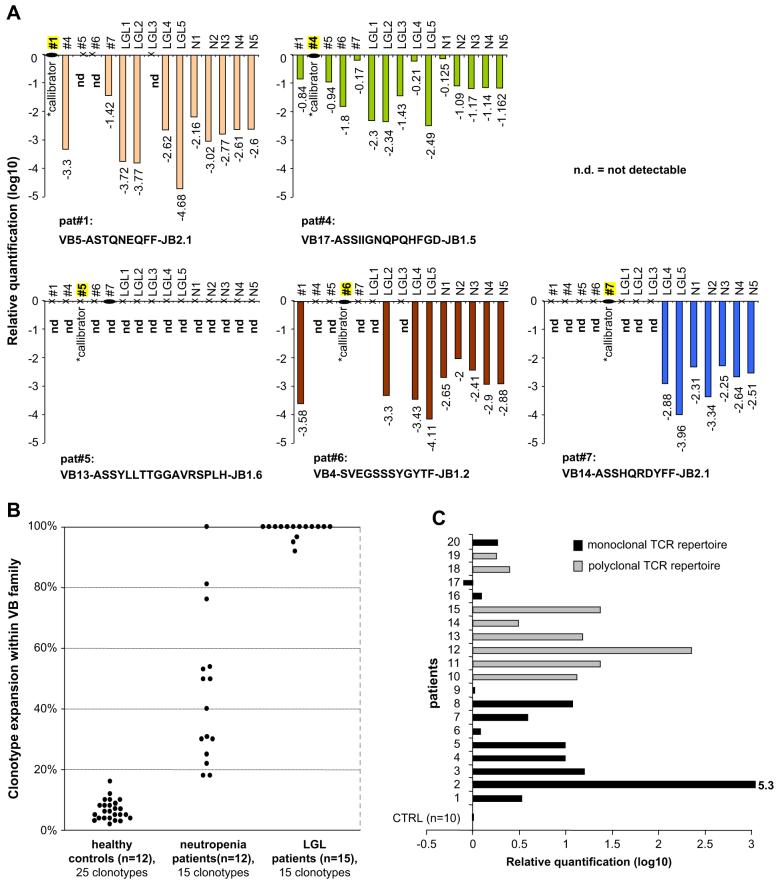
When compared to healthy controls, expansion of CTL clones was significantly higher in the patient group 45.2% ± 24.7% (Fig. 2B). As expected, CTL expansions in typical T-LGL patients (n = 15) comprised 96% of a given VB family. Conversely, in healthy controls the average rate of identical sequences corresponding to “expanded” clones can account for up to 10.3% of a VB family and is encountered at a similar rate among individual VB populations. Interestingly, one patient initially diagnosed with CIN showed an expanded clone and in the subsequent course developed features consistent with T-LGL leukemia, including positive TCR-γ rearrangement by PCR; this patient was excluded from the analysis in our study.
Figure 2.
Clonotypic analysis of patients with neutropenia. (A) Quantitative detection of disease-associated immunodominant clonotypes. Clonotypic size was measured using Taqman polymerase chain reaction (PCR) in five neutropenia patients, five large granular lymphocyte leukemia (LGL) patients and five healthy controls. Each patient-specific clonotypic assay was also performed on other patients and controls. For relative quantification the original patient sample harboring the expanded clonotype as initially identified by sequencing was used as a calibrator (*). Values are shown in log10 scale. ND = not detectable. (B) Clonotypic T-cell receptor (TCR) CDR3 expansions in neutropenia, typical LGL leukemia and healthy controls. Expansions of 15 disease-associated immunodominant clonotypes in neutropenia patients (n = 12) are shown in relation to the 15 highly expanded monoclonal clonotypes in typical LGL leukemia (n = 15) and 25 redundant clonotypes in healthy controls (n = 12). (C) Levels of interferon-γ (IFN-γ) expression in patients with neutropenia. Expression of IFN-γ in sorted CD8+ cells of neutropenia patients and healthy controls was measured using Taqman PCR. Mean expression values of sorted CD8+ cells from 10 healthy controls were used as a baseline (calibrator). Gray bars represent patients without detectable immunodominant expansions (log10-fold increase vs controls: 1.07 ± 0.69), black bars correspond to patients in whom expanded clones were identified (log10 fold increase vs controls: 0.84 ± 1.42).
Tracking of patient-specific clonotypes
In order to confirm the clonotypic size and characterize behavior of immunodominant clonotypes, we applied Taqman PCR cross-sectionally to five patients, five healthy controls, and five T-LGL patients (Fig. 2A). Clonotypic primers were derived from patients’ expanded clonotypes; therefore, the initial sample (used for TCR sequencing) served as calibrator for relative quantification of samples derived from other patients and controls. The individual clonotype was detected by PCR in each respective neutropenia patient. In addition, patient-specific primers also amplified a product in some patients and healthy controls, however, always at greatly lower magnitudes. For one clonotypic assay (patient no. 5), the amplification was limited to the corresponding patient only.
Intraclonotypic diversity in patients with CIN
To gain insight into the intraclonotypic diversity, TCR repertoires were analyzed within individual patients (Table 3). While several clonotypes showed considerable similarity within private repertoires, we could not reproduce this finding in TCR repertoires of healthy controls. For example, a high degree of similarity of major or minor clonotypes in patient nos. 3 and 6 within four VB families (VB3, VB4, VB6, VB13) was encountered, and the majority of sequenced VB3 clonotypes in patient no. 3 show JB2.7-restriction.
Table 3.
Examples of T-cell receptor repertoire restriction and selection of homologous clonotypes in individual patients
| Patient no. | VB | CDR3 | JB | Exp (%) |
|---|---|---|---|---|
| C S V E G S G S Y -G Y T F | JB1-2 | 53 | ||
| C S V W G T G G L -Y E Q Y F | JB2-7 | 30 | ||
| 6 | VB4 | C S V W T G G G -T E A F F | JB1-1 | 6 |
| C S V D H R A G -E Q Y F | JB2-7 | 6 | ||
| C A S S G T G G W -N E Q F | JB2-1 | 50 | ||
| C A S S L G G P F -Y E Q Y F | JB2-7 | 15 | ||
| 6 | VB6 | C A S S L G G A V D -E Q Y F | JB2-7 | 5 |
| C A S S R Y R S -E A F F | JB1-1 | 20 | ||
| C A S S S R E E S S -Y E Q Y F | JB2-7 | 10 | ||
| 3 | VB13 | C A S S Y L T D S D -T Q Y F | JB2-3 | 30 |
| C A S S L T L D S K A -Y E Q Y F | 6.5 | |||
| C A S S L T G L A -Y E Q Y F | 13 | |||
| C A S S L Y G Q S -Y E Q Y F | 13 | |||
| 3 | VB3 | C A S S L L G G D L -Y E Q Y F | JB2-7 | 13 |
| C A S R M R G S S -Y E Q Y F | 13 | |||
| C A S N S G E -Y E Q Y F | 6.5 | |||
| C A S S S G P S P -Y E Q Y F | 6.5 |
Clonotypes derived from T-cell receptor repertoire of VB4/VB6 in patient no. 6 and VB13/VB3 in patient no. 3 are shown. Immunodominant clonotypes defined by major expansion are in italics; all other clonotypes were categorized as minor.
Expression of IFN-γ in CD8+ lymphocytes of CIN patients
IFN-γ production is a tightly regulated marker of cytotoxic activity and was previously reported to be overexpressed in T-LGL and neutropenia [25,26]. We initially hypothesized that spontaneous IFN-γ overexpression would be found in patients with highly immunodominant CTL, indicative of strong cytotoxic activity. However, we detected markedly increased IFN-γ production in most of the patients (16 of 20, Fig. 2C) suggesting that CTL activation may be present in patients with both highly skewed and polyclonal TCR repertoires.
Discussion
Some cases of chronic neutropenia appear idiopathic despite intense clinical testing. Isolated neutropenia of various degrees often accompanies T-LGL and it is possible that semiautonomous CTL clones characteristic of this disease are intrinsically responsible for neutropenia via TCR-directed killing of myeloid precursors and mature neutrophils.
In the current study, we postulated that certain cases of otherwise unexplained neutropenia are mediated by a cytotoxic process, but unlike in T-LGL, these CTL responses are less polarized and unlike in a typical LGL, significant expansion of CD8+CD57+TCRab+ cells cannot be detected and more sensitive methods for identification of clonal CTLs need to be employed.
Despite overall CTL polyclonality, immunodominance was detected in a significant proportion of patients, suggesting a continuum from highly skewed CTL expansions in T-LGL to entirely polyclonal responses. The detection of increased IFN-γ expression in most of the patients with or without CTL expansions may imply that CTL-mediated pathophysiology is operative in most of the studied patients and perhaps constitutes a better marker of T-cell-mediated inhibition of hematopoiesis. In general, our results are in agreement with previous reports dealing with increased Fas-L and IFN-γ production in both CIN and LGL patients [26-28].
Nominal expansions of immunodominant clones were compared to those occasionally encountered in healthy controls. Previously, we studied a large number of clonotypes of several VB families in healthy controls of various ages and we defined clonotypic frequency of greater than, 12.5% (mean + 2 × SD) as pathologic [17,18]. While studying TCR repertoire in CIN patients, the size of clones encountered was significantly higher than that seen in controls. We concluded that immunodominant CTL clones encountered in CIN are clearly distinctive from the previously postulated monoclonal clonopathy of unclear significance and from clonal polarity seen in the elderly [29,30]. Previously, CIN has been associated with humoral antineutrophilic responses. Antineutrophil-antibodies have unclear clinical significance and the routinely available tests identify a relatively low proportion of cases attributable to these antibodies [31,32]. Nevertheless, antineutrophilic humoral responses were present in some our patients, however, in general, they did not correlate with the presence of immunodominant CTL clones. Similarly, antineutrophil-antibodies did not cause significant inhibition of myeloid colony formation questioning the role of soluble inhibitors as the sole factor in the pathogenesis of defective neutrophil production. In this respect, the increased IFN-γ production in CIN may not translate into the “inhibitory humoral activity” as the protein serum concentration of this cytokine is low and most of the effects are exerted locally. Both humoral and cellular responses may also be concurrently present in CIN/AIN, analogous to immune reactions that occur in autoimmune or infectious setting.
If immunodominant CTL clones are specific for the pathophysiologic process, corresponding clonotypes can be utilized as surrogate markers for the inciting antigens and also for characterization of disease activity or response to therapy [18,33]. When we analyzed the amino acid sequence of neutropenia-associated clonotypes, several of the identified clonotypes showed considerable similarity. It is possible that the homology results from the presence of common antigens recognized by these expanded clones. Structural analysis of T-LGL-specific clonotypes delivered similar results [18]. In contrast, comparable degree of homology was not found between the clonotypes of healthy TCR VB repertoires.
The knowledge of clonotypic sequences can be exploited for the design of quantitative clonotypic assays. We were able to precisely quantify the levels of immunodominant clonotypes in patients using Taqman-PCR. Some of the patient-specific clonotypic tests were also positive in several other patients and controls. However, as expected, the clonotypic expression was at all times several magnitudes lower in other subjects tested than in patients from whom the clonotypes were derived. The amplification of these clonotypes in control subjects is not surprising, but rather anticipated, considering the vast diversity of human TCR CDR3 repertoire, which may harbor highly homologous TCRs, often differing only in one nucleotide. The amplification of these TCR sequences suggests that highly homologous or identical, yet not pathologically expanded clonotypes may be present in repertoires of other patients and controls. Their presence in controls may be consistent with the fact that the antigen recognized is an autoantigen rather than foreign antigen/neoantigen.
The spectrum of antigens implicated in antibody responses in autoimmune neutropenia include NA1, NA2, CD11a and CD11b, CD16, thyrotropin receptor-like molecules and membrane actin-like molecules [31,34-39]. Due to the nature of serological assays, the specificity of the autoantibodies can be more or less precisely established. However, determination of antigenic specificity is difficult to discern for TCR and their specificity can be inferred indirectly by analyzing the clinical context. In this regard, single-lineage cytopenias may be a result of cytotoxicity directed toward differentiated progenitors.
In our study, we did not address functional properties of the expanded CTL clones; however, this question was previously investigated: T-cell-mediated inhibition of progenitor growth was demonstrated in coculture experiments in patients with pure red cell aplasia, Felty’s syndrome, and aplastic anemia [40-43]. While direct cell-mediated toxicity of CTL in AIN was recently disputed, it does not rule out the theory of antigen-specific CTL-mediated pathogenic response in neutropenia. Presence of autoantibodies does not exclude T-cell-mediated cytotoxic mechanisms. For example, in autoimmune hepatitis, despite occurrence of autoantibodies, liver necrosis is thought to be caused by T cells [44]. The role of direct T-cell-mediated cytotoxicity has also been established in autoantibody-mediated diseases, such as idiopathic thrombocytopenic purpura [45].
Strict lineage-specific inhibition of myelopoiesis in CIN/AIN is difficult to explain only with effects of cytokines such as IFN-γ and Fas-L, which would be expected to have a broader range of suppression affecting more than one cell lineage. In our study, we used IFN-γ expression, which under normal circumstances is a very tightly regulated process, as a surrogate marker of CTL activation. The finding that in some patients the CTL process may remain polyclonal despite increased IFN-γ expression may be explained in different ways. Firstly, common epitope spread occurring in the process of an antigen-driven immune reaction may result in a diversification of the autoreactive TCR repertoire. Secondly, clonal CTL may simply remain undetected, perhaps due to their small size or the inability to detect them systemically.
In summary, based on our findings we propose that a clonal continuum exists between unexplained neutropenia and LGL leukemia. Furthermore, we believe that cases of seemingly idiopathic neutropenia may in fact correspond to variants of LGL leukemia. Therefore, it is likely that similar pathophysiologic processes operate in both disease entities and the antigens that trigger clonal expansions may likely be shared. The question regarding antigenic specificity of the expanded CTL clones is very interesting and requires additional investigations.
Clinically, detection of increased IFN-γ production and immunodominant CTL clones in CIN/AIN indicates that some of those patients could potentially benefit from T-cell-targeted therapies. This intriguing conclusion should be tested in a clinical study of severely neutropenic patients refractory to traditional therapy.
Acknowledgments
This work was supported in part by National Institutes of Health RO1 HL73429 (awarded to J.P.M.), CA11397201 (J.M.P.), U54 RR 019391 (J.P.M.) and an award from AA & MDS Foundation (J.P.M.). The authors thank Stacy Gilfillan for critical reading and correcting the manuscript.
References
- 1.Starkebaum G. Chronic neutropenia associated with autoimmune disease. Semin Hematol. 2002;39:121–127. doi: 10.1053/shem.2002.31918. [DOI] [PubMed] [Google Scholar]
- 2.Berliner N, Horwitz M, Loughran TP., Jr. Congenital and acquired neutropenia. Hematology. 2004 Jan;:63–79. doi: 10.1182/asheducation-2004.1.63. [DOI] [PubMed] [Google Scholar]
- 3.Papadaki HA, Palmblad J, Eliopoulos GD. Non-immune chronic idiopathic neutropenia of adult: an overview. Eur J Haematol. 2001;67:35–44. doi: 10.1034/j.1600-0609.2001.00473.x. [DOI] [PubMed] [Google Scholar]
- 4.Palmblad J, Papadaki HA, Eliopoulos G. Acute and chronic neutropenias. What is new? J Intern Med. 2001;250:476–491. doi: 10.1046/j.1365-2796.2001.00915.x. [DOI] [PubMed] [Google Scholar]
- 5.Palmblad JE, von dem Borne AE. Idiopathic, immune, infectious, and idiosyncratic neutropenias. Semin Hematol. 2002;39:113–120. doi: 10.1053/shem.2002.31919. [DOI] [PubMed] [Google Scholar]
- 6.Boxer L, Dale DC. Neutropenia: causes and consequences. Semin Hematol. 2002;39:75–81. doi: 10.1053/shem.2002.31911. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann HW, Von LP, Modrow S. Parvovirus B19 infection and autoimmune disease. Autoimmun Rev. 2003;2:218–223. doi: 10.1016/s1568-9972(03)00014-4. [DOI] [PubMed] [Google Scholar]
- 8.Boxer LA, Greenberg MS, Boxer GJ, Stossel TP. Autoimmune neutropenia. N Engl J Med. 1975;293:748–753. doi: 10.1056/NEJM197510092931505. [DOI] [PubMed] [Google Scholar]
- 9.Bux J, Behrens G, Jaeger G, Welte K. Diagnosis and clinical course of autoimmune neutropenia in infancy: analysis of 240 cases. Blood. 1998;91:181–186. [PubMed] [Google Scholar]
- 10.Loughran TP, Jr, Hammond WP. Adult-onset cyclic neutropenia is a benign neoplasm associated with clonal proliferation of large granular lymphocytes. J Exp Med. 1986;164:2089–2094. doi: 10.1084/jem.164.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loughran TP, Jr, Starkebaum G. Large granular lymphocyte leukemia. Report of 38 cases and review of the literature. Medicine (Baltimore) 1987;66:397–405. [PubMed] [Google Scholar]
- 12.Lima M, Almeida J, Santos AH, et al. Immunophenotypic analysis of the TCR-Vbeta repertoire in 98 persistent expansions of CD3(+)/TCR-alphabeta(+) large granular lymphocytes: utility in assessing clonality and insights into the pathogenesis of the disease. Am J Pathol. 2001;159:1861–1868. doi: 10.1016/s0002-9440(10)63032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loughran TP, Jr, Starkebaum G, Aprile JA. Rearrangement and expression of T-cell receptor genes in large granular lymphocyte leukemia. Blood. 1988;71:822–824. [PubMed] [Google Scholar]
- 14.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 15.Padovan E, Casorati G, Dellabona P, et al. Expression of two T cell receptor alpha chains: dual receptor T cells. Science. 1993;262:422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 16.Risitano AM, Maciejewski JP, Green S, et al. In-vivo dominant immune responses in aplastic anaemia: molecular tracking of putatively pathogenetic T-cell clones by TCR beta-CDR3 sequencing. Lancet. 2004;364:355–364. doi: 10.1016/S0140-6736(04)16724-X. [DOI] [PubMed] [Google Scholar]
- 17.Beck RC, Wlodarski M, Gondek L, et al. Efficient identification of T-cell clones associated with graft-versus-host disease in target tissue allows for subsequent detection in peripheral blood. Br J Haematol. 2005;129:411–419. doi: 10.1111/j.1365-2141.2005.05472.x. [DOI] [PubMed] [Google Scholar]
- 18.Wlodarski MW, O’Keefe C, Howe EC, et al. Pathologic clonal cytotoxic T-cell responses: nonrandom nature of the T-cell-receptor restriction in large granular lymphocyte leukemia. Blood. 2005;106:2769–2780. doi: 10.1182/blood-2004-10-4045. [DOI] [PubMed] [Google Scholar]
- 19.Semenzato G, Zambello R, Starkebaum G, Oshimi K, Loughran TP., Jr The lymphoproliferative disease of granular lymphocytes: updated criteria for diagnosis 13. Blood. 1997;89:256–260. [PubMed] [Google Scholar]
- 20.Langerak AW, van den BR, Wolvers-Tettero IL, et al. Molecular and flow cytometric analysis of the Vbeta repertoire for clonality assessment in mature TCRalphabeta T-cell proliferations. Blood. 2001;98:165–173. doi: 10.1182/blood.v98.1.165. [DOI] [PubMed] [Google Scholar]
- 21.Wlodarski MW, Gondek LP, Nearman ZP, et al. Molecular strategies for detection and quantitation of clonal cytotoxic T cell responses in aplastic anemia and myelodysplastic syndrome. Blood. 2006:2632–2641. doi: 10.1182/blood-2005-09-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Dongen JJ, Langerak AW, Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936 2. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 23.Lefranc MP, Giudicelli V, Ginestoux C, et al. IMGT-ONTOLOGY for immunogenetics and immunoinformatics. In Silico Biol. 2004;4:17–29. [PubMed] [Google Scholar]
- 24.O’Keefe CL, Rodriguez A, Sobecks RM, Bolwell B, Maciejewski JP. Molecular TCR Diagnostics can be used to identify shared clonotypes after allogeneic hematopoietic stem cell transplantation. Exp Hematol. 2004;32:1010–1022. doi: 10.1016/j.exphem.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Pistoia V, Prasthofer EF, Tilden AB, et al. Large granular lymphocytes from patients with expanded LGL populations acquire cytotoxic functions and release lymphokines upon in vitro activation. Blood. 1986;68:1095–1100. [PubMed] [Google Scholar]
- 26.Papadaki HA, Stamatopoulos K, Damianaki A, et al. Activated T-lymphocytes with myelosuppressive properties in patients with chronic idiopathic neutropenia. Br J Haematol. 2005;128:863–876. doi: 10.1111/j.1365-2141.2005.05380.x. [DOI] [PubMed] [Google Scholar]
- 27.Perzova R, Loughran TP., Jr Constitutive expression of Fas ligand in large granular lymphocyte leukaemia. Br J Haematol. 1997;97:123–126. doi: 10.1046/j.1365-2141.1997.d01-2113.x. [DOI] [PubMed] [Google Scholar]
- 28.Loughran TP., Jr Clonal diseases of large granular lymphocytes. Blood. 1993;82:1–14. [PubMed] [Google Scholar]
- 29.Dhodapkar MV, Li CY, Lust JA, Tefferi A, Phyliky RL. Clinical spectrum of clonal proliferations of T-large granular lymphocytes: a T-cell clonopathy of undetermined significance? 2. Blood. 1994;84:1620–1627. [PubMed] [Google Scholar]
- 30.Schwab R, Szabo P, Manavalan JS, et al. Expanded CD4+ and CD8+ T cell clones in elderly humans. J Immunol. 1997;158:4493–4499. [PubMed] [Google Scholar]
- 31.Hartman KR, LaRussa VF, Rothwell SW, et al. Antibodies to myeloid precursor cells in autoimmune neutropenia. Blood. 1994;84:625–631. [PubMed] [Google Scholar]
- 32.Bruin MC, von dem Borne AE, Tamminga RY, et al. Neutrophil antibody specificity in different types of childhood autoimmune neutropenia. Blood. 1999;94:1797–1802. [PubMed] [Google Scholar]
- 33.O’Keefe CL, Plasilova M, Wlodarski M, et al. Molecular analysis of TCR clonotypes in LGL: a clonal model for polyclonal responses. J Immunol. 2004;172:1960–1969. doi: 10.4049/jimmunol.172.3.1960. [DOI] [PubMed] [Google Scholar]
- 34.Hartman KR, Mallet MK, Nath J, Wright DG. Antibodies to actin in autoimmune neutropenia. Blood. 1990;75:736–743. [PubMed] [Google Scholar]
- 35.Hartman KR, Wright DG. Identification of autoantibodies specific for the neutrophil adhesion glycoproteins CD11b/CD18 in patients with autoimmune neutropenia. Blood. 1991;78:1096–1104. [PubMed] [Google Scholar]
- 36.Huizinga TW, Kleijer M, Tetteroo PA, Roos D, von dem Borne AE. Biallelic neutrophil Na-antigen system is associated with a polymorphism on the phospho-inositol-linked Fc gamma receptor III (CD16) Blood. 1990;75:213–217. [PubMed] [Google Scholar]
- 37.Ory PA, Clark MR, Kwoh EE, Clarkson SB, Goldstein IM. Sequences of complementary DNAs that encode the NA1 and NA2 forms of Fc receptor III on human neutrophils. J Clin Invest. 1989;84:1688–1691. doi: 10.1172/JCI114350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ory PA, Goldstein IM, Kwoh EE, Clarkson SB. Characterization of polymorphic forms of Fc receptor III on human neutrophils. J Clin Invest. 1989;83:1676–1681. doi: 10.1172/JCI114067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weitzman SA, Stossel TP, Harmon DC, et al. Antineutrophil autoantibodies in Graves’ disease. Implications of thyrotropin binding to neutrophils. J Clin Invest. 1985;75:119–123. doi: 10.1172/JCI111663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starkebaum G, Singer JW, Arend WP. Humoral and cellular immune mechanisms of neutropenia in patients with Felty’s syndrome. Clin Exp Immunol. 1980;39:307–314. [PMC free article] [PubMed] [Google Scholar]
- 41.Abkowitz JL, Kadin ME, Powell JS, Adamson JW. Pure red cell aplasia: lymphocyte inhibition of erythropoiesis. Br J Haematol. 1986;63:59–67. doi: 10.1111/j.1365-2141.1986.tb07495.x. [DOI] [PubMed] [Google Scholar]
- 42.Zeng W, Maciejewski JP, Chen G, Young NS. Limited heterogeneity of T cell receptor BV usage in aplastic anemia. J Clin Invest. 2001;108:765–773. doi: 10.1172/JCI12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handgretinger R, Geiselhart A, Moris A, et al. Pure red-cell aplasia associated with clonal expansion of granular lymphocytes expressing killer-cell inhibitory receptors 43. N Engl J Med. 1999;340:278–284. doi: 10.1056/NEJM199901283400405. [DOI] [PubMed] [Google Scholar]
- 44.Ichiki Y, Aoki CA, Bowlus CL, et al. T cell immunity in autoimmune hepatitis. Autoimmun Rev. 2005;4:315–321. doi: 10.1016/j.autrev.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 45.Olsson B, Andersson PO, Jernas M, et al. T-cell-mediated cytotoxicity toward platelets in chronic idiopathic thrombocytopenic purpura. Nat Med. 2003;9:1123–1124. doi: 10.1038/nm921. [DOI] [PubMed] [Google Scholar]