INTRODUCTION
Duchenne muscular dystrophy (DMD) is an X-linked recessive disorder characterized by progressive muscle wasting and which occurs in 1 out of 3,500 live male births. DMD is caused by mutations in the gene encoding the cytoskeletal protein dystrophin.[1–3] Absence of dystrophin results in a compromise of sarcolemmal integrity rendering myofibers prone to contraction-induced injury. DMD is not amenable to treatment with existing medical options. Therefore, contemporary medicine is focused on symptom management.[4]
The milder and rarer allelic form of the disease, Becker muscular dystrophy (BMD), is frequently caused by mutations that result in expression of an internally truncated dystrophin protein with reduced functionality. Review of these BMD mutations and the resultant genotype-phenotype relationships has helped to delineate the essential regions and conformations necessary for dystrophin stability and function. These findings have been extended in studies of transgenic mice expressing modified dystrophin constructs to elucidate the importance of the physical links provided at either end of the dystrophin protein.[1, 5] At the cytoplasmic face of the sarcolemma dystrophin plays a pivotal structural role in nucleating assembly of a dynamic multi-protein complex referred to as the dystrophin-glycoprotein complex (DGC). The absence of dystrophin results in a considerable reduction of the DGC components,[5–7] membrane destabilization, and membrane permeability defects leading to increased intracellular calcium, and myofiber degeneration.[8]
Recombinant adeno-associated viral (rAAV) vectors are emerging as a potential gene transfer vehicle for neuromuscular disorders due to their ability to transduce the vast majority of the mouse striated musculature with a single administration of vector, the existence of several muscle-tropic serotypes, and their inherent capacity for persistent transgene expression in post-mitotic cells.[9–11] However, a potential complication of gene therapy in many DMD patients, particularly those with genomic deletions, is a possibility that dystrophin expressed from exogenously administered transgenes could be perceived as a ‘neoantigen’, evoking destruction of transduced myofibers by immune effector cells. Indeed, previous transplantation studies in DMD patients have demonstrated that dystrophin can be recognized as non-self by the immune system with the generation of specific antibodies.[12–14]
Utrophin, an autosomal paralog of dystrophin, was originally named for its ubiquitous expression and high degree of sequence similarity to dystrophin, particularly in regards to critical protein binding domains located near the amino and carboxy termini.[15, 16] This observation raised the possibility of utrophin being used as a therapeutic replacement for dystrophin in DMD. Indeed, investigators have shown that utrophin can compensate for the lack of dystrophin in many circumstances.[11, 17–21] Nonetheless, the functionality of utrophins small enough to be delivered by rAAV vectors has not been investigated. We hypothesized that rAAV6-mediated delivery of a microutrophin (ΔR4-R21/ΔCT) to the mdx:utrn−/− double- knockout mouse model would permit the recruitment of DGC component proteins leading to improved membrane stabilization, enhancement of contractile performance and a halt in progression of the dystrophic phenotype. The mdx:utrn−/− mouse model was chosen because it displays a highly dystrophic phenotype, in contrast with utrn−/− mice that show almost no pathology, or dystrophin-deficient mdx mice where extrasynaptic expression of utrophin contributes to a milder phenotype. Consequently, rAAV6/microutrophin was administered to ~1 month-old mdx:utrn−/− mice by tail vein injection. The most significant finding of this study was that microutrophin is highly functional in the mdx:utrn−/− mouse model, suggesting that it could alter the course of DMD to one that more closely resembles a very mild BMD phenotype.
RESULTS
Generation of a rAAV vector expressing microutrophin
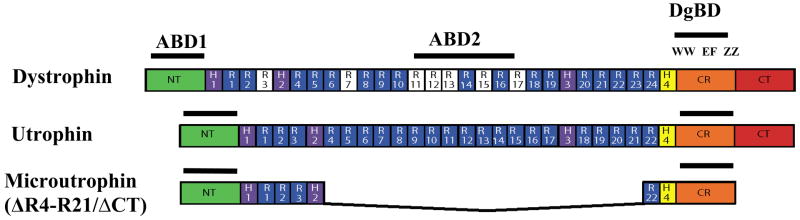
A rAAV vector pseudotyped with serotype 6 capsids was used for gene transfer of a murine codon-optimized microutrophin under the transcriptional control of a minimal cytomegalovirus enhancer plus promoter, and was modeled after the previously described ΔR4-R21/ΔCT microdystrophin construct.[22] This cDNA encodes the amino-terminal actin-binding domain, spectrin-like repeats 1–3, hinge 2, the final (22nd) spectrin-like repeat, and the dystroglycan-binding domain. The resulting microutrophin thus lacks sequences encoding spectrin-like repeats 4 through 21 (Fig. 1). Sequences encoding the carboxy-terminal domain within dystrophin have also been eliminated since those regions have been shown to be non-essential in mice.[22, 23] Production of the microutrophin protein was demonstrated by western analysis of a viral lysate preparation from 293D cells utilizing an antibody specific for the amino-terminus of utrophin (data not shown).
Figure 1. Schematic view of the structural domains in dystrophin, utrophin, and microutrophin.
All three proteins contain an N-terminal actin-binding domain (ABD1), shown as dotted lines. Within the central region of dystrophin and utrophin there are a number of repeating units with similarity to the coiled-coil repeats of spectrin, and these are interspersed with 4 hinge regions (H1–H4). The central region of dystrophin uniquely contains a second actin binding domain (ABD2, dashed line) not found in utrophin (basically charged repeats are in white). The carboxy-terminal portions of both dystrophin and utrophin contain a cysteine-rich (CR) domain and a carboxy-terminal domain, both of which contain numerous protein interaction motifs for components of the dystrophin-glycoprotein complex. Additonal abbreviations: NT, amino-terminus; R1–24, spectrin-like repeats 1–24; β-DgBD, β-dystroglycan binding domain, which is composed of a WW domain (WW), two EF hand-like domains (EF) and a ZZ zinc finger domain (ZZ). [50]
Intravenous administration of rAAV6/CMV-microutrophin results in persistent expression of microutrophin in adult mdx:utrn−/− mice
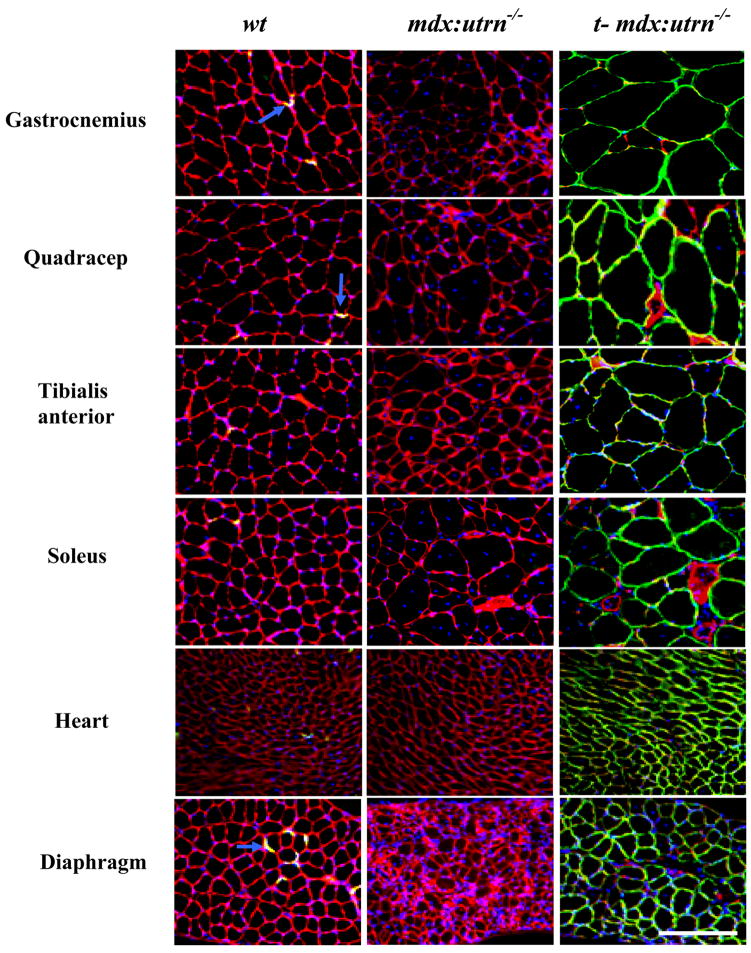
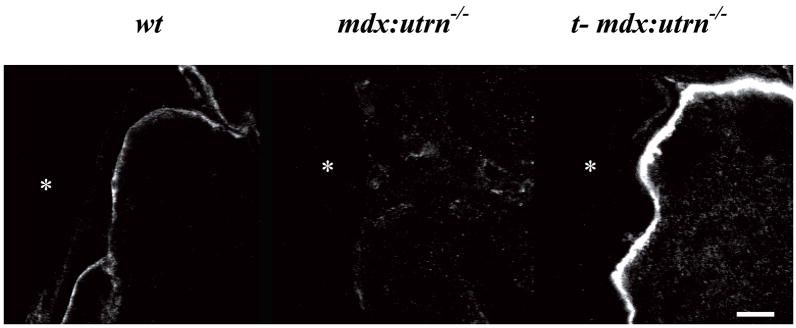
Four week-old mdx:utrn−/− mice were administered 3x1012 vector genomes of rAAV6/CMV-microutrophin by tail vein injection. Cryosections of muscle were stained four months after vector administration with utrophin polyclonal antibodies that recognize the amino-terminus of the protein (Fig. 2). The vast majority of myofibers stained positive for microutrophin at the sarcolemma in all muscles examined (gastrocnemius, quadriceps, tibialis anterior, soleus, heart, and diaphragm). This sarcolemma localization was in sharp contrast to results in age matched wildtype control mice where myofiber utrophin staining was found only at the neuromuscular and myotendinous junctions and in untreated 12 week old mdx:utrn−/− mice that do not express utrophin (Fig. 2 and 3). In accordance with the natural function and localization of utrophin at the myotendinous junction we performed immunostaining followed by confocal microscopy on longitudinal cryosections of gastrocnemius muscles from wildtype, mdx:utrn−/− mice, and treated mdx:utrn−/− mice (Fig. 3). In mice that received rAAV6/CMV-microutrophin there was qualitatively a high degree of enrichment at the MTJ, showing that microutrophin can localize to the myotendinous junction as does full-length utrophin.
Figure 2. Microutrophin properly localizes to the sarcolemma in striated muscle.
TA muscle cryosections from wildtype (wt), mdx:utrn−/− and treated mdx:utrn−/− (t-mdx:utrn−/−) mice were immunofluorescently stained with anti-utrophin polyclonal antibodies. Green fluorescence represents utrophin staining, red represents B2 laminin, blue represents nuclei staining, and blue arrows show utrophin staining (yellow) in wildtype mice. (Scale bar = 100 μm).
Figure 3. Microutrophin is enriched at the myotendinous junction of treated mdx:utrn−/−mice.
Immunofluorescent staining was performed on longitudinally cryosectioned gastrocnemius muscles using a NH2-terminal utrophin antibody followed by confocal microscopy. Representative photos are shown for muscles from wildtype (wt), mdx:utrn−/−, and treated mdx:utrn−/− (t-mdx:utrn−/−) mice. The tendon abutting the myofiber is shown in the left portion of each panel and is denoted by an asterisk (*). (Scale bar = 30 μm).
Microutrophin gene transfer alleviates the dystrophic phenotype in mdx:utrn−/− mice
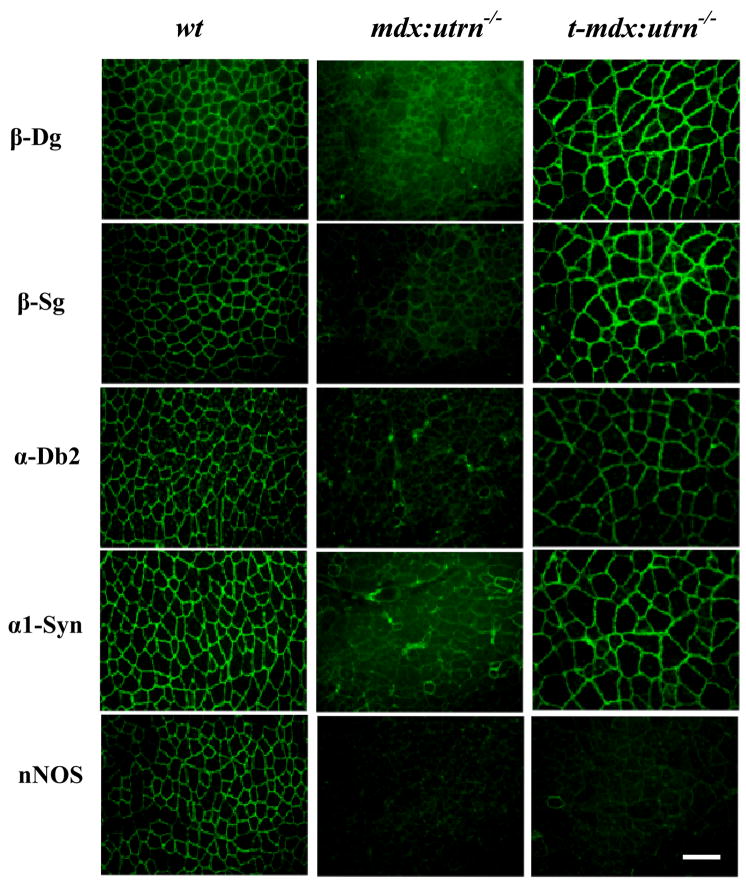
In an attempt to model therapeutic interventions in advanced cases of DMD we assessed the properties of the hind-limb muscles of treated mdx:utrn−/− mice. Analysis of muscle cryosections demonstrated that microutrophin recruited representative DGC components (β-dystroglycan, β-sarcoglycan, α-dystrobrevin-2, and α1-syntrophin) to the sarcolemma, but it did not localize neuronal nitric oxide synthase (nNOS) (Fig. 4). The absence of nNOS recruitment is consistent with results using mini and microdystrophin constructs.[24, 25]
Figure 4. Expression of microutrophin restores the dystrophin-glycoprotein complex, with the exception of nNOS, to the sarcolemma.
Tibialis anterior muscles from wildtype (wt), mdx:utrn−/−, and treated-mdx:utrn−/− were cryosectioned, and immunoflourescent staining for utrophin was performed. (Abbreviations: β-Dg, β-dystroglycan; β-Sg, β-sarcoglycan; αDb2, alpha-dystrobrevin-2; α1Syn, α-1 syntrophin; nNOS, neuronal nitric oxide synthase) (Scale bar = 100 μm)
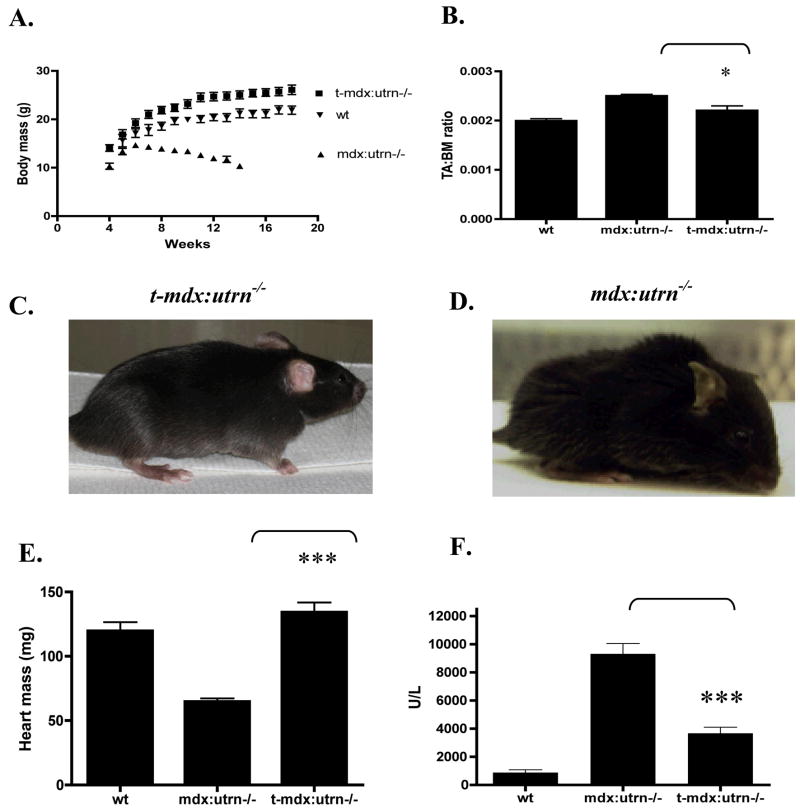
In contrast to mdx mice, mdx:utrn−/− mice experience a considerable reduction in body mass due to progressive muscle wasting that begins at about 6 weeks of age. By 12 weeks of age the body mass of treated mdx:utrn−/− mice was found to be approximately twice that of untreated mdx:utrn−/− mice (Fig. 5a). In addition, untreated mdx:utrn−/− mice display kyphosis from the degenerative process of the disease and decreased ambulation. In contrast, age-matched treated mdx:utrn−/− mice demonstrate minimal kyphosis and sufficient muscularity to participate actively on a voluntary running wheel (Supplement video, S1). We have previously noted that rAAV6 vectors are extremely efficient at transducing cardiac muscle,[9, 26] and treated mdx:utrn−/− mice displayed complete transduction of the myocardium. Heart mass was likewise increased to levels more closely resembling that of wildtype mice (twice that of untreated mdx:utrn−/− mice) (Fig. 5e). In addition, treated mdx:utrn−/− mice experienced an ~60% reduction in serum creatine kinase levels (P < 0.001) compared with untreated mdx:utrn−/− mice (Fig. 5f), consistent with a whole body reduction in muscle degeneration.
Figure 5. Expression of microutrophin compensates for the lack of utrophin and dystrophin.
(A) Increase in body mass over time in mdx:utrn−/− mice (N=5) that received rAAV6/microutrophin relative to untreated control wt and mdx:utrn−/− mice (by week 14 all of the untreated mdx:utrn−/− mice had died). (B) Tibialis anterior muscle mass to body mass ratio. C. Treated mdx:utrn−/− mice show increased musculature and minimal kyphosis and joint contractures as compared with untreated mdx:utrn−/− mice (D) (not to scale.) (E) Treated mdx:utrn−/− mice display increased heart mass compared with control mdx:utrn−/− (12-weeks) and wildtype (5 months). (F) Serum creatine kinase levels are reduced by ~60% in treated mdx:utrn−/− mice. *P < 0.05 and ***P < 0.001 when compared with control mdx:utrn−/− mice; bars represent SEM.
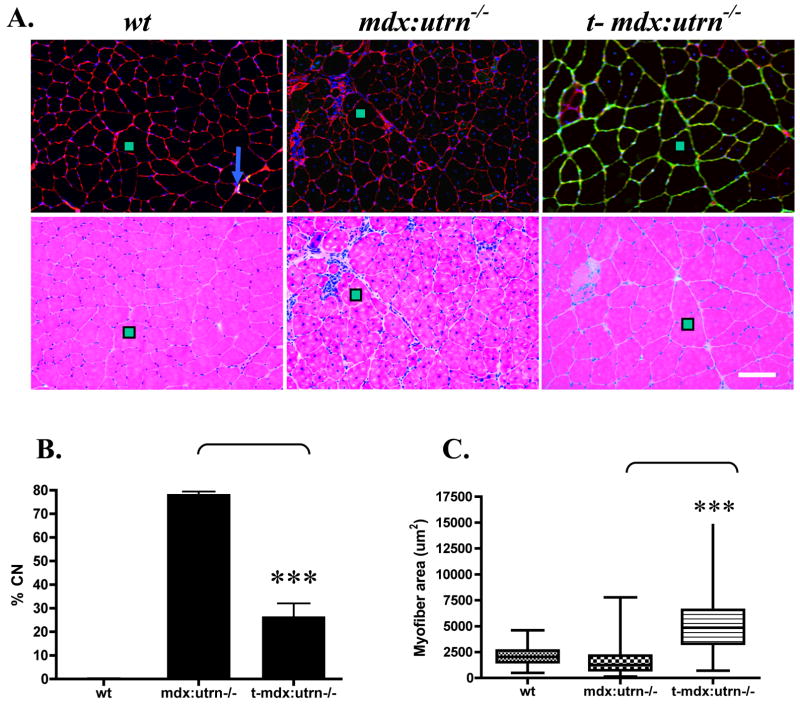
Histologically, mdx:utrn−/− muscles display large inflammatory infiltrates and a high percentage of centrally-nucleated myofibers, which are often quite small in diameter reflecting a continuum of the degeneration-regeneration process. Hematoxylin and eosin staining revealed a considerable reduction in inflammatory infiltrate (Fig. 6a), and a dramatic reduction in the percentage of centrally-nucleated myofibers (26.5%) when compared with untreated gender-matched mice at 14 weeks of age (84.2%) (Fig. 6b). In treated mdx:utrn−/− mice, expression of microutrophin resulted in a shift in myofiber size with considerably larger fibers being present in addition to a reduction in the small fiber population (Fig. 6c).
Figure 6. Microutrophin improves muscle histopathology in mdx:utrn−/− mice.
(A) Upper panel shows immunoflourescent staining for utrophin (green), B2 laminin (red), and nuclei (blue). The lower panel shows adjacent sections stained with hemotoxylin and eosin. The green boxes mark the same muscle fibers in the upper and lower panels. Blue arrows show utrophin staining localized at the neuromuscular junction in wildtype muscles. (Scale bar = 100 μm). (B) Percentage of myofibers with central nucleation (N=1500 myofibers/cohort). (C) Box plots showing variance of the muscle fiber cross-sectional area (N= 800 myofibers/cohort). Boxes represent the middle quartiles from the 25th to the 75th percentiles, bar demonstrates the high and low values. ***P < 0.001 when compared to control mdx:utrn−/− mice, and bars represent SEM.
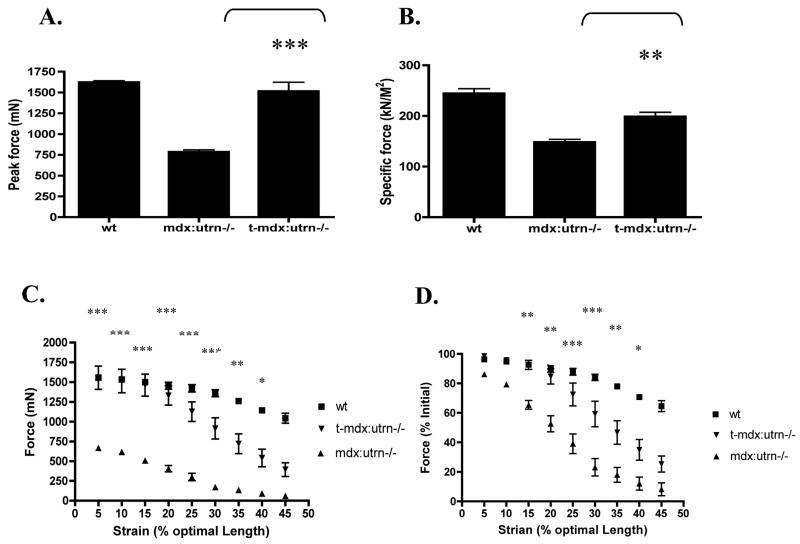
Microutrophin increases force production in limb muscle of mdx:utrn−/− mice
In comparison with untreated mdx:utrn−/− mice there was a significant (N=5; p<0.001) increase in force-producing capacity in tibialis anterior muscles of treated mdx:utrn−/− mice, reaching ~93% of wildtype levels (Fig. 7a). However, when normalizing net force production to the cross-sectional area of the muscle (specific force), the benefit was less dramatic (P<0.01), achieving ~80% of levels in wildtype muscles (Fig. 7a). We also measured the resistance to eccentric-contraction induced injury in the various cohorts. The protocol applied resulted in stressing the muscles with a series of 5% incremental stretches above the optimal length for force generation during in situ stimulated muscle contractions. Microutrophin expressing muscles were protected from contraction-induced injury similar to wildtype muscles up to a 20% strain. For untreated mdx:utrn−/− mice, this same level of strain resulted in a reduction of initial muscle contractile force of ~46% (Fig. 7c,d). Significant differences for treated mdx:utrn−/− compared to untreated mdx:utrn−/− mice represent P < 0.001 at up to a 30% increase in optimal length (Fig. 7c). No significant differences are found with lengthening contractions that are within normal physiological levels of strain (i.e. less than 25%) when compared to wildtype control mice (Fig. 7c,d). These results indicated that expression of microutrophin provided a significant functional benefit to dystrophic muscles even when initiating treatment after the onset of histopathological degeneration in the mdx:utrn−/− mouse model.
Figure 7. Intravenous administration of rAAV6/microutrophin to mdx:utrn−/− mice results in increased muscle function.
The TA muscles of untreated mdx:utrn−/− mice exhibit reduced force producing capacity (A); force generation per cross-sectional area, i.e. specific force (B); and an increased susceptibility to eccentric contraction induced injury (C, D). (C) shows the absolute force developed following strains of a given percentage beyond the optimal length for force development, while (D) shows the percent force development relative to initial values prior to mechanical strain. By comparison, treated mdx:utrn−/− mouse TA muscles exhibited significantly increased absolute force production (A), force per cross-sectional area, (B) (**P < 0.01 and ***P < 0.001 when compared to 3 month old control mdx:utrn−/− mice), and they demonstrated markedly increased resistance to mechanical strain injury (C & D). No significant differences are found at lengthening contractions that are within normal physiological levels of strain (i.e. less than 30%) when compared to wildtype mice in contrast to untreated mdx:utrn−/−mice while treated mdx:utrn−/− mice show a significant difference at up to 40% increase in optimal length when compared to untreated mdx:utrn−/− (Fig.7c,d). (Bars represent the SEM, *P < 0.05, **P < 0.01 and ***P < 0.001).
DISCUSSION
Somatic gene transfer technology is one of a number of approaches being developed for the treatment of DMD. Numerous studies have demonstrated the potential of rAAV as a vector for gene transfer. rAAV has been shown to be not only a valuable research tool for the delivery of transgenes, as well as gene-targeting cassettes such as those encoding shRNA[27] or zinc finger[28] technologies, but also an important reagent in numerous human clinical trials. Undoubtedly, there are issues of risk that should be considered not only for individual vectors but also for the inherent characteristics of specific disease processes. Understanding the factors involved in immune activation following gene transfer is of great clinical significance and much progress has been made in this area.[29] In many DMD patients there is little or no dystrophin expression and in others the residual truncated protein produced in revertant myofibers lacks significant portions of the full amino acid sequence. Exogenous dystrophin production resulting from gene therapy protocols could in theory elicit a destructive cellular immune response against transduced muscle fibers. Using a therapeutic transgene based on a protein normally expressed in muscles of DMD patients, such as utrophin, could be important for improving the efficiency and efficacy of gene therapy for DMD.
Utrophin is structurally similar to dystrophin, with expression normally being restricted to the neuromuscular and myotendinous junctions within adult skeletal muscle. As a strategy to overcome the potential risk of an immune response to dystrophin, a microutrophin cassette was developed that can be packaged within rAAV vectors. The goal of this study was to characterize whether microutrophin could display functional efficacy in the mdx:utrn−/− mouse model. In contrast to dystrophin-deficient mdx mice, whose mild skeletal muscle pathology is attributed, at least in part, to compensatory overexpression of utrophin at the sarcolemma,[30] mdx:utrn−/− mice display an earlier onset of dystrophy, more extensive muscle wasting, significant weight loss, joint contractures, and a considerably shortened lifespan.[20, 31] A variety of investigators have previously shown that utrophin can compensate for the lack of dystrophin in many circumstances, including via adenoviral delivery or in transgenic mice.[11, 17–21] Further evidence for a compensatory role for utrophin derives from studies in the distal toe and extraocular muscles, which are largely spared from dystrophic degeneration, and where utrophin expression is normally high.[32–34] Thus, a large body of data supports the idea that utrophin can functionally compensate for the absence of dystrophin in striated muscles. Indeed, upregulation of the utrophin gene has been actively pursued as a pharmacological target as a potential therapy for DMD.[35, 36] Here we demonstrated that intravenous injection of a rAAV6/microutrophin transgene could improve morphology and function of muscles within the severely dystrophic mdx:utrn−/− mouse model.
The DGC is distributed across the sarcolemma of striated muscle, with biochemical studies indicating that it provides a structural link between the extracellular matrix and the intracellular actin cytoskeleton.[37] Structurally, the DGC is thought to assemble in a dynamic manner in response to stress or contractile perturbation.[38] In the absence of dystrophin there is extensive loss of sarcolemmal localization of DGC component proteins in both DMD patients and mdx:utrn−/− mice. As with dystrophin, utrophin interacts with this large oligomeric complex.[39] Previous experiments have shown the binding of dystrophin and utrophin to actin filaments to be non-competitive, though with similar affinities.[40] Similarly, mutagenesis experiments have demonstrated that dystrophin and utrophin exhibit different modes of contact with the transmembrane protein β-dystroglycan.[41] Despite these differences we observed that microutrophin readily recruited and localized transmembrane and cytoplasmic components of the DGC (Fig. 4). The level of staining for the transmembrane components qualitatively appeared to be higher when compared with wildtype staining. This observation is consistent with transgenic studies using the larger mini-utrophin, where it was proposed that dystrophin (or utrophin) glycoprotein complexes are limited by the amount of dystrophin or utrophin present in the myofibers.[21] However, nNOS localization to the sarcolemma was not restored by expression of microutrophin, consistent with previous reports showing that neither micro- nor minidystrophins could localize nNOS to the sarcolemma.[24, 42]
During muscle contraction, the integrity of the myotendinous junction is physiologically vital for the transmission of force from muscle to tendon. As the muscle transitions into tendon, the increasing proportion of connective tissue establishes a stiffness gradient, thus preventing injury at the myotendinous junction. The high transverse stiffness of the muscle at the myotendinous junction and inextensibility of the central tendon combine to eliminate stress concentrations at the muscle-tendon interface, and may play an important role in preventing injury.[43] Consistent with the natural distribution and function of utrophin at the myotendinous junction we found that microutrophin was highly-enriched at this anatomical location (Fig. 3). However, in contrast to the neuromuscular and myotendinous junction restricted localization of endogenous utrophin found in wildtype mice, the microutrophin delivered by rAAV was readily targeted to the sarcolemma. The uniform expression pattern of microutrophin likely reflects the use of the CMV promoter, which lacks N-boxes that can restrict transcription to nuclei adjacent to neuromuscular junctions, and the lack of most of the untranslated regions that also can influence mRNA trafficking and/or stability.[44–46] The extrasynaptic localization of microutrophin is vital in the consideration of a possible treatment for DMD.
To evaluate the benefit of treatment on a representative limb-muscle, we chose to assess the morphological and functional properties of tibialis anterior (TA) muscles in treated mdx:utrn−/− mice. Delivery of microutrophin was found to result in vast reduction in mononuclear cell infiltrates, and the presence of myofibers of a more consistent, albeit larger, size in addition to a reduction in the number of small caliber fibers. This change in myofiber population size could be the result of an architectural specialization that may be influenced by activity patterns over the course of treatment. In all muscles tested we found a similar and significant reduction in the presence of regenerating myofibers in the treated mdx:utrn−/− mice when compared with untreated mdx:utrn−/− mice. A further encouraging finding from our study was a highly significant improvement in the contractile properties of the treated TA muscles (Fig. 7). Previous studies of dystrophin-deficient mdx mice have shown that muscles from those animals display numerous centrally-nucleated myofibers, ongoing degeneration/regeneration, a striking loss of force generating capacity and a susceptibility to contraction-induced injury.[22] Our results show that delivery of microutrophin to muscles of mdx:utrn−/− mice leads to a striking normalization of all these parameters, showing that treated muscles are physiologically closer to wildtype muscles than those of mdx mice.[22] Mice lacking only utrophin but not dystrophin display no signs of dystrophy or weakness, and therefore our results with microutrophin are in agreement with previous studies using mini- and full-length utrophin that utrophin constructs are able to compensate for the lack of both dystrophin and utrophin.[17, 21]
Taken together our results show that delivery of a highly truncated microutrophin cassette using rAAV6 vectors is able to achieve widespread and persistent transduction of the striated musculature in mdx:utrn−/− mice. Microutrophin expression resulted in a significant amelioration of the histopathological features in muscles of mdx:utrn−/− mice and led to a nearly complete recovery of contractile properties. While we did not perform a detailed lifespan analysis, the data showed a significant extension of lifespan in treated mdx:utrn−/− mice (Fig. 5a and data not shown). Overall, the phenotypic consequences of microutrophin delivery were broadly similar to those we previously observed following rAAV-mediated delivery of a structurally similar microdystrophin.[47] Microutrophin and microdystrophin each resulted in significant and similar reductions in centrally nucleated fibers, increased resistance to contraction-induced injury, decreased serum levels of creatine kinase and a halt to ongoing myofiber necrosis.[47] In moving towards clinical applications for rAAV gene transfer, potential immune responses of the host against both the viral capsid and the therapeutic protein encoded by the vector will need to be carefully considered. Since utrophin is normally expressed in nearly all tissues, including muscle, the results reported herein suggest that gene therapy protocols for DMD could potentially benefit from the use of utrophin based transgenes.
MATERIALS AND METHODS
Microutrophin cloning and virus production
The microutrophin transgene (ΔR4–R21/ΔCT) was engineered to contain the amino-terminal actin binding domain, the first 3 spectrin-like repeats followed by the second hinge, spectrin-like repeat 22, hinge 4, and the cysteine-rich domain. Twenty-five nucleotides of the adjacent 5’ and 3’ untranslated region were included, followed by the polyadenylation signal from the rabbit beta-globin gene at the carboxy terminus. The coding sequence of microutrophin was codon optimized and synthesized (Blueheron Biotech, Bothell WA) (GenBank accession EU293093). This synthesized microutrophin cDNA was excised from pENTR (Invitrogen, Carlsbad, CA) using Pme I, and subcloned into SnaB I-digested pARAP4.[48] The CMV promoter obtained from pAAV-LacZ (Stratagene, La Jolla, CA) was excised by digestion with SpeI/BstBI digestion, and ligated into the Xho I/BstB I site of pAAV/microutrophin to create pAAV/CMV-microutrophin. Production of the rAAV6/microutrophin vector was as described.[49]
Animal experiments
Male and female wildtype C57Bl/6J mice (The Jackson Laboratory, Bar Harbor, ME) and dystrophin/utrophin deficient (mdx:utrn−/−) mice (N=5) were derived as previously described.[31] All animals were experimentally manipulated in accordance with the Institutional Animal Care and Use Committee (IACUC) of the University of Washington. Four week old dystrophic mdx:utrn−/− mice received 3x1012 vector genomes of rAAV6 vector in a single 300 μl bolus intravenous injection via the tail vein. Mice were weighed on a weekly basis and sacrificed at 5 months of age for further evaluation.
Histological analyses
Following functional analysis, mice were sacrificed and a necropsy was performed. Muscles were frozen in liquid nitrogen-cooled isopentane embedded in Tissue-Tek OCT medium (Sakura Finetek USA, Torrance, CA), and sectioned transversely in a cryostat at 10 μm. For brightfield microscopy, sections were stained with hematoxylin and eosin and mounted with Permount (Fisher Scientific, Fairlong, NJ). For immunofluorescence studies, sections were blocked in 4% bovine serum albumin and 0.05% Tween-20 in 1X Dulbecco’s phosphate buffered saline (PBS; Invitrogen GIBCO, Grand Island, NY). Sections were then washed three times in 1X PBS for 5 minutes. The slides were then followed with an incubation of 60 minutes using rabbit anti-utrophin polyclonal antibody (1:400), (kindly provided by Stan Froehner, University of Washington) and rat anti-B2 laminin (1:800) (Sigma, St. Louis, MO) in PBS containing 2% goat serum. Sections were then rinsed three times in PBS and incubated for 45 minutes with goat anti-rabbit-alexa-488 (1:1200) and goat anti-rat-alexa-540 (1:800) (Molecular Probes Inc., Eugene, OR). Slides were then mounted in anti-fade mounting media containing DAPI (Vector Labs, Burlingame, CA). Fluorescent sections were imaged using a Nikon eclipse E1000 fluorescent microscope (Nikon, NY). Images were captured using a QIcam digital camera and processed using Qcapture Pro (Qimaging Corp.; BC Canada). Immunoflourescent detection of representative components of the DGC were performed on TA muscles with β-Sg and β-Dg monoclonal antibodies (1:100) (Novocastra, Newcastle, UK) utilizing a mouse on mouse blocking kit (Novacastra, Newcastle UK ). Primary rabbit polyclonal antibodies used were α-dystrobrevin-2, α1-syntrophin (1:200) (kindly provided by Stan Froehner, University of Washington) and nNOS (Sigma, St. Louis MO). For localization of microutrophin at myotendinous junctions confocal microscopy was performed on longitudinal gastrocneminus cryosections with immunoflourescent staining of utrophin as described above with the exception of utilizing 50% glycerol in PBS as mounting media. Visualization was performed on a Zeiss 510 Meta using 60X magnification. Images were taken with the same settings and were processed in an identical way. Limits were placed and maintained throughout image processing on the image ‘gain’ to ensure avoidance of immunofluorescence saturation of the images.
Myofibers with centrally located nuclei were quantified from utrophin positive myofibers in TA muscles by counting fibers in a minimum of 4 random fields (~800 myofibers/cohort). The mean number of centrally-nucleated fibers was compared between treated and untreated mice using an unpaired Students t-test. The myofiber cross-sectional area within TA muscle (~1500 myofibers/cohort) was measured using Image J computer software (National Institutes of Health). Two days prior to sacrifice animals were bled retro-orbitally for determination of serum creatine kinase levels using a commercial kinetic kit (CK LiquiUV, Stanbio Laboratory, Boerne, TX).
Contractility assays
Sixteen weeks following vector administration, mice were anesthetized with 2,2,2-tribromoethanol (Sigma, St. Louis MO) and assayed in situ (tibialis anterior) for force generation and susceptibility to eccentric contraction-induced injury using equipment and protocols described previously.[9, 49] Control wildtype mice at 5 months of age and untreated mdx:utrn−/− mice at an age of 3 months, where they were not yet approaching death, were used as controls. The conditions established with this assay occur over a broad range of physiological operating conditions with the potential to produce injurious overload of the contractile apparatus.[19] Briefly, via nerve stimulation we determined the maximum isometric force-producing capacity at optimal muscle fiber length. The TA muscle was then subjected to a series of progressively greater (5%) length changes under maximum stimulation (lengthening contractions) at twenty second intervals. The impact of each lengthening contraction upon force production was recorded from the peak isometric force generated just prior to the initiation of the subsequent lengthening contraction. Cohorts from the lengthing contraction assay were analyzed using two-way ANOVA with the incorporation of a Bonferroni post-test using the PRISM software.
Statistics
All results are expressed as mean ± s.e.m. unless otherwise stated. Differences identified between cohorts were determined using one-way ANOVA with a Student’s t-test with the exception of the contractility assays as stated above. All data analyses were performed using the PRISM software.
Supplementary Material
Mice treated with rAAV6/microutrophin retained sufficient muscularity and mobility to readily use a voluntary running wheel whereas untreated mice could not. Treated and untreated mdx:utrn−/− littermates were evaluated in terms of mobility at 12 weeks of age. At the start of the video the untreated mouse is shown entering the running wheel initially but is quickly displaced by the treated mouse that demonstrates considerable enhanced mobility.
Acknowledgments
We are grateful to Leonard Meuse for animal husbandry, Caitlyn Doremus for rAAV vector production assistance and Miki Haraguchi for histopathological support. We thank Stan Froehner and Marvin Adams of the University of Washington for providing antibodies and critical reading of this manuscript, respectively. We also thank Greg Martin at the Keck Imaging Center University of Washington for assistance with confocal microscopy and Chamberlain lab members for critically reviewing the manuscript. This work was supported by grants from the National Institutes of Health (R37AR40864) and the Muscular Dystrophy Association (to J.S.C.). G.L.O. was supported by a National Institutes of Health National Research Service Award (T32 HL07828). P.G. was supported by a Development Grant from the Muscular Dystrophy Association.
References
- 1.Dalkilic I, Kunkel LM. Muscular dystrophies: genes to pathogenesis. Curr Opin Genet Dev. 2003;13:231–238. doi: 10.1016/s0959-437x(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 4.Angelini C. The role of corticosteroids in muscular dystrophy: a critical appraisal. Muscle Nerve. 2007;36:424–435. doi: 10.1002/mus.20812. [DOI] [PubMed] [Google Scholar]
- 5.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida M, Ozawa E. Glycoprotein complex anchoring dystrophin to sarcolemma. J Biochem (Tokyo) 1990;108:748–752. doi: 10.1093/oxfordjournals.jbchem.a123276. [DOI] [PubMed] [Google Scholar]
- 8.Watchko JF, O'Day TL, Hoffman EP. Functional characteristics of dystrophic skeletal muscle: insights from animal models. J Appl Physiol. 2002;93:407–417. doi: 10.1152/japplphysiol.01242.2001. [DOI] [PubMed] [Google Scholar]
- 9.Gregorevic P, Blankinship MJ, Allen JM, Crawford RW, Meuse L, Miller DG, et al. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat Med. 2004;10:828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Podsakoff G, Wong KK, Jr, Chatterjee S. Efficient gene transfer into nondividing cells by adeno-associated virus-based vectors. J Virol. 1994;68:5656–5666. doi: 10.1128/jvi.68.9.5656-5666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakefield PM, Tinsley JM, Wood MJ, Gilbert R, Karpati G, Davies KE. Prevention of the dystrophic phenotype in dystrophin/utrophin-deficient muscle following adenovirus-mediated transfer of a utrophin minigene. Gene Ther. 2000;7:201–204. doi: 10.1038/sj.gt.3301066. [DOI] [PubMed] [Google Scholar]
- 12.Huard J, Roy R, Bouchard JP, Malouin F, Richards CL, Tremblay JP. Human myoblast transplantation between immunohistocompatible donors and recipients produces immune reactions. Transplant Proc. 1992;24:3049–3051. [PubMed] [Google Scholar]
- 13.Karpati G, Ajdukovic D, Arnold D, Gledhill RB, Guttmann R, Holland P, et al. Myoblast transfer in Duchenne muscular dystrophy. Ann Neurol. 1993;34:8–17. doi: 10.1002/ana.410340105. [DOI] [PubMed] [Google Scholar]
- 14.Ohtsuka Y, Udaka K, Yamashiro Y, Yagita H, Okumura K. Dystrophin acts as a transplantation rejection antigen in dystrophin-deficient mice: implication for gene therapy. J Immunol. 1998;160:4635–4640. [PubMed] [Google Scholar]
- 15.Love DR, Hill DF, Dickson G, Spurr NK, Byth BC, Marsden RF, et al. An autosomal transcript in skeletal muscle with homology to dystrophin. Nature. 1989;339:55–58. doi: 10.1038/339055a0. [DOI] [PubMed] [Google Scholar]
- 16.Tinsley JM, Blake DJ, Roche A, Fairbrother U, Riss J, Byth BC, et al. Primary structure of dystrophin-related protein. Nature. 1992;360:591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- 17.Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis JM, et al. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 18.Squire S, Raymackers JM, Vandebrouck C, Potter A, Tinsley J, Fisher R, et al. Prevention of pathology in mdx mice by expression of utrophin: analysis using an inducible transgenic expression system. Hum Mol Genet. 2002;11:3333–3344. doi: 10.1093/hmg/11.26.3333. [DOI] [PubMed] [Google Scholar]
- 19.Rafael JA, Tinsley JM, Potter AC, Deconinck AE, Davies KE. Skeletal muscle-specific expression of a utrophin transgene rescues utrophin-dystrophin deficient mice. Nat Genet. 1998;19:79–82. doi: 10.1038/ng0598-79. [DOI] [PubMed] [Google Scholar]
- 20.Deconinck AE, Rafael JA, Skinner JA, Brown SC, Potter AC, Metzinger L, et al. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90:717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- 21.Tinsley JM, Potter AC, Phelps SR, Fisher R, Trickett JI, Davies KE. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature. 1996;384:349–353. doi: 10.1038/384349a0. [DOI] [PubMed] [Google Scholar]
- 22.Harper SQ, Hauser MA, DelloRusso C, Duan D, Crawford RW, Phelps SF, et al. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 23.Crawford GE, Faulkner JA, Crosbie RH, Campbell KP, Froehner SC, Chamberlain JS. Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J Cell Biol. 2000;150:1399–1410. doi: 10.1083/jcb.150.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Judge LM, Haraguchiln M, Chamberlain JS. Dissecting the signaling and mechanical functions of the dystrophin-glycoprotein complex. J Cell Sci. 2006;119:1537–1546. doi: 10.1242/jcs.02857. [DOI] [PubMed] [Google Scholar]
- 25.Yue Y, Liu M, Duan D. C-terminal-truncated microdystrophin recruits dystrobrevin and syntrophin to the dystrophin-associated glycoprotein complex and reduces muscular dystrophy in symptomatic utrophin/dystrophin double-knockout mice. Mol Ther. 2006;14:79–87. doi: 10.1016/j.ymthe.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Townsend D, Blankinship MJ, Allen JM, Gregorevic P, Chamberlain JS, Metzger JM. Systemic administration of micro-dystrophin restores cardiac geometry and prevents dobutamine-induced cardiac pump failure. Mol Ther. 2007;15:1086–1092. doi: 10.1038/sj.mt.6300144. [DOI] [PubMed] [Google Scholar]
- 27.Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 28.Yokoi K, Zhang HS, Kachi S, Balaggan KS, Yu Q, Guschin D, et al. Gene Transfer of An Engineered Zinc Finger Protein Enhances the Anti-angiogenic Defense System. Mol Ther. 2007;15:1917–1923. doi: 10.1038/sj.mt.6300280. [DOI] [PubMed] [Google Scholar]
- 29.Mingozzi F, Maus MV, Hui DJ, Sabatino DE, Murphy SL, Rasko JE, et al. CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat Med. 2007;13:419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- 30.Hirst RC, McCullagh KJ, Davies KE. Utrophin upregulation in Duchenne muscular dystrophy. Acta Myol. 2005;24:209–216. [PubMed] [Google Scholar]
- 31.Grady RM, Teng H, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell. 1997;90:729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 32.Dowling P, Culligan K, Ohlendieck K. Distal mdx muscle groups exhibiting up-regulation of utrophin and rescue of dystrophin-associated glycoproteins exemplify a protected phenotype in muscular dystrophy. Naturwissenschaften. 2002;89:75–78. doi: 10.1007/s00114-001-0289-4. [DOI] [PubMed] [Google Scholar]
- 33.Porter JD. Commentary: extraocular muscle sparing in muscular dystrophy: a critical evaluation of potential protective mechanisms. Neuromuscul Disord. 1998;8:198–203. doi: 10.1016/s0960-8966(98)00015-7. [DOI] [PubMed] [Google Scholar]
- 34.Porter JD, Merriam AP, Hack AA, Andrade FH, McNally EM. Extraocular muscle is spared despite the absence of an intact sarcoglycan complex in gamma- or delta-sarcoglycan-deficient mice. Neuromuscul Disord. 2001;11:197–207. doi: 10.1016/s0960-8966(00)00171-1. [DOI] [PubMed] [Google Scholar]
- 35.Khurana TS, Davies KE. Pharmacological strategies for muscular dystrophy. Nat Rev Drug Discov. 2003;2:379–390. doi: 10.1038/nrd1085. [DOI] [PubMed] [Google Scholar]
- 36.Stocksley MA, Chakkalakal JV, Bradford A, Miura P, De Repentigny Y, Kothary R, et al. A 1.3 kb promoter fragment confers spatial and temporal expression of utrophin A mRNA in mouse skeletal muscle fibers. Neuromuscul Disord. 2005;15:437–449. doi: 10.1016/j.nmd.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Campbell KP. Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell. 1995;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 38.Rybakova IN, Patel JR, Ervasti JM. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol. 2000;150:1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumura K, Ervasti JM, Ohlendieck K, Kahl SD, Campbell KP. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360:588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- 40.Rybakova IN, Humston JL, Sonnemann KJ, Ervasti JM. Dystrophin and utrophin bind actin through distinct modes of contact. J Biol Chem. 2006;281:9996–10001. doi: 10.1074/jbc.M513121200. [DOI] [PubMed] [Google Scholar]
- 41.Ishikawa-Sakurai M, Yoshida M, Imamura M, Davies KE, Ozawa E. ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to beta-dystroglycan. Hum Mol Genet. 2004;13:693–702. doi: 10.1093/hmg/ddh087. [DOI] [PubMed] [Google Scholar]
- 42.Chao DS, Gorospe JR, Brenman JE, Rafael JA, Peters MF, Froehner SC, et al. Selective loss of sarcolemmal nitric oxide synthase in Becker muscular dystrophy. J Exp Med. 1996;184:609–618. doi: 10.1084/jem.184.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang W, Kelly NG, Boriek AM. Passive mechanics of muscle tendinous junction of canine diaphragm. J Appl Physiol. 2005;98:1328–1333. doi: 10.1152/japplphysiol.00816.2004. [DOI] [PubMed] [Google Scholar]
- 44.Chakkalakal JV, Miura P, Belanger G, Michel RN, Jasmin BJ. Modulation of utrophin A mRNA stability in fast versus slow muscles via an AU-rich element and calcineurin signaling. Nucleic Acids Res. 2008;36:826–838. doi: 10.1093/nar/gkm1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miura P, Thompson J, Chakkalakal JV, Holcik M, Jasmin BJ. The utrophin A 5′-untranslated region confers internal ribosome entry site-mediated translational control during regeneration of skeletal muscle fibers. J Biol Chem. 2005;280:32997–33005. doi: 10.1074/jbc.M503994200. [DOI] [PubMed] [Google Scholar]
- 46.Perkins KJ, Basu U, Budak MT, Ketterer C, Baby SM, Lozynska O, et al. Ets-2 repressor factor silences extrasynaptic utrophin by N-box mediated repression in skeletal muscle. Mol Biol Cell. 2007;18:2864–2872. doi: 10.1091/mbc.E06-12-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregorevic P, Allen JM, Minami E, Blankinship MJ, Haraguchi M, Meuse L, et al. rAAV6-microdystrophin preserves muscle function and extends lifespan in severely dystrophic mice. Nat Med. 2006;12:787–789. doi: 10.1038/nm1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen JM, Halbert CL, Miller AD. Improved adeno-associated virus vector production with transfection of a single helper adenovirus gene, E4orf6. Mol Ther. 2000;1:88–95. doi: 10.1006/mthe.1999.0010. [DOI] [PubMed] [Google Scholar]
- 49.Blankinship MJ, Gregorevic P, Allen JM, Harper SQ, Harper H, Halbert CL, et al. Efficient transduction of skeletal muscle using vectors based on adeno-associated virus serotype 6. Mol Ther. 2004;10:671–678. doi: 10.1016/j.ymthe.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 50.Abmayr S. The Structure and Function of Dystrophin. In: Winder S, editor. Molecular Mechanisms of Muscular Dystrophies. Landes Bioscience; Georgetown: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mice treated with rAAV6/microutrophin retained sufficient muscularity and mobility to readily use a voluntary running wheel whereas untreated mice could not. Treated and untreated mdx:utrn−/− littermates were evaluated in terms of mobility at 12 weeks of age. At the start of the video the untreated mouse is shown entering the running wheel initially but is quickly displaced by the treated mouse that demonstrates considerable enhanced mobility.