Abstract
The myelin sheath insulates axons and allows for rapid salutatory conduction in the nervous system of all vertebrates. The formation of peripheral myelin requires expression of the transcription factor Egr2, which is responsible for inducing such essential myelin-associated genes as Mpz, Mbp, Pmp22, and Mag. Using microarray analysis to compare gene expression patterns in peripheral nerve during development, during remyelination after nerve injury, and in a congenital hypomyelinating mouse model, we identified an evolutionarily conserved novel component of myelin called Mp11 (myelin protein of 11 kDa). The Mp11 genomic locus contains multiple conserved Egr binding sites, and Mp11 induction is regulated by the expression of Egr2. Similar to other Egr2-dependent genes, it is induced during developmental myelination and remyelination after nerve injury. Mp11 is a glycoprotein expressed preferentially in the myelin of the peripheral nervous system versus CNS and is specifically localized to the Schmidt-Lanterman incisures and paranodes of peripheral nerve. The Mp11 protein contains no identifiable similarity to other known protein domains or motifs. However, like other myelin genes, strict Mp11 expression levels are a requirement for the in vitro myelination of DRG neurons, indicating that this previously uncharacterized gene product is a critical component of peripheral nervous system myelin.
Keywords: Mp11, Egr2, myelination, Schmidt-Lanterman incisures, paranode, expression profiling
Introduction
Rapid saltatory conduction of action potentials along the axon in vertebrates is facilitated by myelin in both the CNS and peripheral nervous system (PNS). The myelin sheath is a specialized multilamellar spiraled extension of the plasma membrane of oligodendrocytes in the CNS and Schwann cells in the PNS. The myelin sheath is composed of both compact and noncompact myelin components that each have distinct molecular structures and biological functions. Noncompact myelin contains substantially more cytoplasm than compact myelin and is thought to be critical for signaling, transport of small metabolites, and maintenance of myelin structure and function (Spiegel and Peles, 2002). Noncompact myelin is found in the inner and outer mesaxons, paranodal loops, nodal microvilli, and Schmidt-Lanterman incisures (SLIs). The paranodal regions flank the nodes of Ranvier and are interconnected through adherens, tight and gap junctions, and are linked to the axon through septate-like junctions (Wiley and Ellisman, 1980; Spiegel and Peles, 2002; Salzer, 2003). SLIs are funnel-shaped channels filled with cytoplasm that intermittently interrupt the compact myelin to provide a transverse fast-track route through the myelin sheath from the adaxonal to the abaxonal membrane.
Hereditary motor and sensory neuropathies, such as Charcot-Marie-Tooth (CMT) and congenital hypomyelinating neuropathy (CHN), are among the most common inherited disorders of the nervous system in humans and are caused by abnormalities in myelin and/or axons. There are two main types of CMT subdivided by the primary dysfunction that causes the disease. CMT1 arises primarily because of a dysfunction of the myelin sheath, whereas CMT2 develops primarily as a result of a dysfunction of the neurons or axons. A number of genes involved in myelin development and/or maintenance are mutated in CMT1, including peripheral myelin protein 22 (PMP22), myelin protein zero (MPZ), connexin32 (CX32/GJB1), myotubularin-related protein-2 (MTMR2), and early growth response gene 2 (EGR2/KROX20) (Nave et al., 2007). The association of EGR2 mutations with CMT1 and CHN was entertained because of the peripheral nerve hypomyelination observed in Egr2-deficient mice (Warner et al., 1998). Egr2 mutations are of particular interest in regard to understanding the transcriptional program regulating myelin development and maintenance, because it appears to be the key regulator of myelin gene transcription together with the Nab proteins (Nab1/Nab2) (Nagarajan et al., 2001; Le et al., 2005b).
We sought to identify additional Egr2-regulated genes in hopes of finding novel myelin-associated proteins and additional candidates for mutational analysis in patients with peripheral neuropathy. We examined multiple paradigms in which myelination was altered or dynamically regulated, including development, remyelination after injury, and hypomyelination attributable to Egr2 mutation, to identify genes that are involved in peripheral nerve myelination (Le et al., 2005a). From this analysis, we identified a gene that was coregulated with other myelin proteins in the sciatic nerve during developmental myelination and during remyelination after injury. This novel transcript encodes a highly conserved membrane glycoprotein, which we named Mp11 (myelin protein of 11 kDa), that contains no recognizable functional domains or homologies to other proteins. Mp11 is induced in Schwann cells by Egr2, and its promoter region contains multiple Egr binding sites. Mp11 expression is restricted to the placenta and PNS, where it is expressed by Schwann cells and localized to paranodes and SLIs of noncompact myelin. The functional attributes of Mp11 were examined using in vitro myelination assays in which both decreased and increased levels of Mp11 resulted in defective myelin formation analogous to other myelin proteins such as Pmp22.
Materials and Methods
Computational analysis.
Microarray data were generated and analyzed as described previously (Le et al., 2005a). DNA and protein sequences from multiple species were obtained from searching National Center for Biotechnology Information (NCBI) and Ensembl databases and aligned using Vector NTI 9.0 software (InfoMax). Analysis of the Mp11 protein was performed using the SMART (Simple Modular Architecture Research Tool) program (http://smart.embl-heidelberg.de/), InterPro (http://www.ebi.ac.uk/InterProScan/), TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/), ProtParam software (http://us.expasy.org/tools/protparam.html), and signalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/).
Plasmid construction.
The mouse Mp11 cDNA was obtained from American Type Culture Collection (MGC #11811, GenBank identification number BC005730) and confirmed by DNA sequencing. Epitope tags [hemagglutinin (HA) at the N terminus and 6x His at the C terminus] were linked to the 300 nt Mp11 coding region by PCR and inserted into FUIV, a lentiviral expression vector in which gene expression is driven by the ubiquitin promoter and coexpression of Venus fluorescent protein is enabled by an internal ribosomal entry site (IRES) element (Araki et al., 2004). An Mp11–Venus C-terminal fusion protein was generated by PCR and inserted into FUIV. The small interfering RNA (siRNA) lentiviral vector, FSVsi, was constructed by annealing oligos containing an MP11 target sequence (siRNA #1, AGCCATCGTGGCTGCCAT; siRNA #2, TCTTCTCGCTGAACATGA) and a hairpin loop into the vector. The siRNA sequence was driven by the U6 promoter and followed by an SV40 promoter–Venus fluorescent protein cassette to monitor infection. Nonrecombinant FSVsi vector was used as the control.
RNA preparation and quantitative real-time PCR.
Sciatic nerves were collected from postnatal and adult mice after nerve crush injury as described previously (Le et al., 2005a). Tissues from adult mice were collected, frozen on dry ice, and homogenized in Trizol reagent (Invitrogen). RNA was prepared using Trizol reagent following the manufacturer's protocol and quantified using RiboGreen fluorescence (Invitrogen) on an ND-1000 spectrophotometer (Nanodrop Technologies). Placental cDNA was a kind gift from Y. Sadovsky (Washington University, St. Louis, MO). Quantitative PCR was performed using an SYBR green-based detection system on an ABI 7700 sequence detector instrument as described previously (Nagarajan et al., 2001). Relative gene expression was calculated by normalizing to glyceraldehyde-3-phosphate dehydrogenase expression. cDNA preparations from at least two different animals were pooled to obtain relative expression values.
Transfection and adenoviral infection of cultured cells.
HEK293T and HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum and penicillin/streptomycin at 37°C and 5% CO2. DNA transfection was performed using calcium phosphate or Superfect transfection reagent (Qiagen) following the manufacturer's protocols. Unless stated otherwise, cells were collected for analysis 2 d after transfection. Adenoviruses expressing either Egr2 or green fluorescent protein (GFP) were generated and used to infect Schwann cells as described previously (Le et al., 2005a).
Antibodies and immunohistochemistry.
The primary antibodies used for immunoblotting were rabbit anti-Mp11 (1:1000; see below), mouse anti-polyhistidine (1:500; R&D Systems), mouse anti-α-tubulin clone B-5-1-2 (1:1000; Sigma), mouse anti-MAG (1:1000; Roche Diagnostics), and anti-MBP (Covance Research Products). The secondary antibodies used were anti-mouse and anti-rabbit HRP (1:5000; Jackson ImmunoResearch Laboratories). For immunohistochemistry, primary antibodies used were rabbit anti-Mp11 (1:500; see below), mouse anti-β-tubulin (Tuj1, 1:1000; Covance Research Products), mouse anti-E-cadherin (1:1000; Transduction Laboratories), mouse anti-Caspr #275 (1:1000; kind gift from E. Peles, The Weizmann Institute of Science, Rehovot, Israel), mouse anti-sodium channel (pan, 1:1000; Sigma), anti-HA (1:5000) (Ryu et al., 2007), and mouse anti-MBP (1:200; Covance Research Products). The secondary antibodies used in these analyses, anti-rabbit Cy3 (1:1500) and anti-mouse Alexa 488 (1:1500), were obtained from Jackson ImmunoResearch Laboratories. The signal was captured using a Nikon Eclipse 80i fluorescence microscope and the MetaMorph6.2 program (Molecular Devices).
The affinity-purified polyclonal Mp11 antibody was generated by immunizing rabbits with the synthetic peptide CNRRMRTRRELEPKSPK (murine residues 53–68) (Pacific Immunology). To obtain affinity-purified Mp11 antibody, the immunizing peptide was immobilized on SulfoLink Coupling Gel (Pierce), and purification was performed according to the manufacturer's instructions.
Preparation of protein lysates and immunoblotting.
Tissues were frozen on dry ice and stored at −80°C. Adult sciatic nerve was immediately desheathed after dissection in cold PBS before freezing. Lysates were prepared by homogenizing the tissue in a buffer containing 2% SDS, 95 mm NaCl, 10 mm EDTA, and Complete Protease Inhibitor Cocktail (Roche Applied Science). For cultured cells, lysis was performed by sonication in a buffer containing 10 mm Tris, pH 7.4, 1% Triton X-100, 0.5% Nonidet P-40, 150 mm NaCl, and Complete Protease Inhibitor Cocktail (Roche Applied Science). The lysates were clarified by centrifugation at 14,000 rpm for 10 min and quantified using the Micro BCA Protein Assay kit (Pierce). For detection of Mp11 in sciatic nerve, 100 μg of protein lysate from tissues was separated on a 15% polyacrylamide gel, and immunoblotting was performed using anti-Mp11 antibody. After incubation with a secondary antibody, the signal was detected using SuperSignal West Dura substrate (Pierce).
Analysis of Mp11 posttranslational modifications.
Lysates were prepared from HEK293T cells transfected with an N-terminal HA-tagged and C-terminal His-tagged Mp11. For PNGase F treatment, His-tagged Mp11 was purified from the lysate using HIS-Select HF Nickel Affinity Gel (Sigma). Purified Mp11 was incubated with PNGase F (New England Biolabs) according to the manufacturer's protocol for 1, 8, or 24 h with the addition of fresh enzyme after 30 min, 4 h, and 8 h, respectively. As a control, the proteins were also incubated at 37°C without enzyme for 8 or 24 h. Samples were analyzed on a 15% SDS polyacrylamide gel.
In vitro myelination assays.
In vitro myelination was performed as described previously (Bermingham et al., 2006). Briefly, dissociated embryonic day 15.5 (E15.5) rat DRG neurons were plated onto four-well Nunc plates at 2.5 × 105 cells per well in DMEM–high glucose with 10% FCS, 2 mm l-glutamine, and 50 ng/ml NGF. To eliminate Schwann cells and fibroblasts, the cultures were cycled between neural basal medium (Invitrogen) supplemented with B-27 (Invitrogen), 2 mm l-glutamine, 50 ng/ml NGF, and 4 g/L d-glucose with or without 5-fluoro-2-deoxyuridine (20 μm; Sigma) plus uridine (20 μm, Sigma) every 2 d. Primary rat Schwann cell cultures were established from newborn rat sciatic nerves using Brockes method. Schwann cells were infected with lentivirus, which resulted in ∼100% of cells infected as monitored by Venus fluorescent protein expression. Five days after rat Schwann cells were infected with lentivirus, they were plated onto DRG neurons at 200,000 cells per well. Five to seven days later, in vitro myelination was initiated by switching the medium to F-12/DMEM–high glucose with 15% FCS, 2 mm l-glutamine, 50 ng/ml NGF, and 50 μg/ml ascorbic acid for 14–21 d. MBP immunostaining was performed as described previously (Bermingham et al., 2006). Percentage of myelination was calculated by counting the number of MBP-positive internodes in five fields at 200× magnification and setting the control to 100%. Each condition was performed in quadruplicate wells, and the data shown are the average of three independent experiments (mean ± SEM).
Membrane localization and permeabilized whole-cell immunohistochemistry.
HeLa cells were plated onto collagen-coated two-well Nunc chamber slides (Nalge Nunc International) and transfected with constructs expressing either Mp11–Venus fusion protein or Venus fluorescent protein alone. Twenty-four hours later, cells were fixed in 4% paraformaldehyde for 10 min, washed three times with PBS, and imaged with a Zeiss LSM 5 PASCAL Vario Two UGB system coupled to a Zeiss Axiovert 200 microscope. HEK293T cells were transfected with an expression vector expressing N-terminally HA-tagged Mp11 and Venus fluorescent protein through an IRES. Cells were washed with PBS and fixed with 4% paraformaldehyde/PBS for 10 min. The fixed cells were incubated for 5 min in either PBS (control) or PBS containing 0.5% Triton X-100 (to permeabilize the membrane). Both permeabilized and nonpermeabilized cells were blocked by incubation in 5% horse serum diluted in PBS and incubated overnight at 4°C in a primary antibody. Slides were washed three times in PBS containing Triton X-100 and incubated at 25°C for 1 h in a secondary antibody (conjugated to either Cy3 or Alexa 488).
Preparation and antibody staining of teased sciatic nerves.
Adult rat sciatic nerves were incubated in Zamboni fixative (2% paraformaldehyde and 15% picric acid in PBS) for 10 min at 25°C. The nerves were washed three times in cold PBS for 5 min and desheathed, and bundles of nerve were dissected with fine needles in cold PBS on Fisherbrand Superfrost/Plus microsope slides (Fisher Scientific). Slides were air dried for 1 h and stored at −20°C. After a postfix in a 1:1 mix of acetone/methanol, the slides were washed in PBS, blocked with 10% fish skin gelatin containing 0.1% Triton for 1 h at 25°C, and incubated with a primary antibody overnight at 4°C. After washing, secondary antibodies were applied for 1 h, and the slides were mounted with Vectashield Mounting Medium with DAPI (Vector Laboratories) for microscopic visualization.
Fractionation of myelin by sucrose gradient centrifugation.
Fresh-frozen adult rat sciatic nerves obtained from Dynal Biotech were homogenized using a glass homogenizer in ice-cold 0.32 m sucrose buffer (0.32 m sucrose, 10 mm Tris-HCl, pH 7.5, and 1 mm EDTA). The homogenates were loaded onto an equal volume of 0.9 m sucrose buffer (0.9 m sucrose, 10 mm Tris-HCl, pH 7.5, and 1 mm EDTA) and centrifuged at 90,000 × g for 16 h. The crude myelin phase, located between the 0.32 and 0.9 m sucrose phases, was collected, mixed with 10× volume of distilled water, and recentrifuged at 90,000 × g for 90 min. The resulting pellet was suspended in 0.32 m sucrose buffer and again recentrifuged at 90,000 × g for 90 min. This final crude myelin pellet was resuspended in PBS containing 1% SDS and stored at −80°C.
Results
Identification of Mp11, a novel myelination-associated transcript
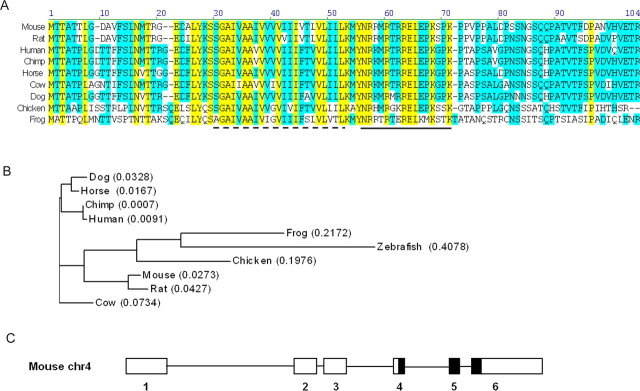
Egr2Lo/Lo mutant mice have severe hypomyelination of peripheral nerves as a consequence of low levels of Egr2, a crucial regulator of the myelination program (Le et al., 2005a). However, as opposed to Egr2 null mice (Topilko et al., 1994), the presence of the hypomorphic allele in Egr2Lo/Lo mice enables them to survive until postnatal day 21 (P21) and allowed us to investigate patterns of gene expression in these abnormally myelinated nerves. This previous analysis revealed that many essential Egr2-regulated genes such as Mpz, Mbp, Pmp22, Periaxin, and Cx32 (Le et al., 2005a) were downregulated in the hypomyelinated sciatic nerves of Egr2Lo/Lo mice. We reasoned that other gene products essential to myelination might also be downregulated in Egr2Lo/Lo nerves, providing us with the opportunity to uncover other Egr2-regulated genes critical to myelination. The expression profile of one particular expressed-sequence tag (EST), Riken A330049M08, did not correspond to any characterized gene product and was clearly downregulated in a pattern similar to that of other bona fide myelin components in hypomyelinated P14 Egr2Lo/Lo sciatic nerve. A search of the NCBI database for mouse RIKEN A330049M08 revealed multiple matching mRNA and EST sequences. Alignment of these sequences by the Vector NTI program produced a single 300 nucleotide open reading frame, encoding a 100 amino acid protein with a calculated molecular weight of 11 kDa (Fig. 1A). We named this novel gene product Mp11 for myelin protein of 11 kDa.
Figure 1.
Mp11 is an evolutionary conserved protein. A, Alignment of the protein sequences of Mp11 orthologs in mouse, rat, human, chimp, horse, cow, dog, chicken, and frog. The predicted transmembrane domain in mouse is indicated by a dashed line. Anti-Mp11 antibodies were created using the peptide indicated by the solid line. Yellow represents residues that are identical in all species, whereas blue represents residues that are similar among the orthologs. B, Phylogenetic tree of Mp11 protein sequences reveals the putative evolution of Mp11 orthologs from zebrafish to human. The numbers in parentheses refer to the calculated distance values that represent sequence divergence based on the Neighbor Joining method (Saitou and Nei, 1987). C, Schematic diagram of the mouse Mp11 gene (NC_000070). It comprises six exons: three coding and three noncoding. The thin black line represents introns, white boxes represent noncoding exons, and black boxes represent coding exons.
The coding region of Mp11 was used to search the databases for similar proteins. We found Mp11 orthologs in a number of species, with the most distantly related orthologs occurring in frog and zebrafish (supplemental Table 1, available at www.jneurosci.org as supplemental material). Putative orthologs in mouse, rat, human, chimp, horse, cow, dog, chicken, and frog were aligned to demonstrate the high level of conservation throughout this protein (Fig. 1A). Among these vertebrates, the Mp11 protein sequences were 30% identical and shared 92% similarity. Sequence analysis and a homology search did not reveal significant similarity to any known proteins or functional domains, with the exception of a single hydrophobic domain predicted to represent a transmembrane region (Fig. 1A). Mouse Mp11 has a calculated pI value of 9.69, with most of the positively charged residues located downstream of the predicted transmembrane domain near the C terminus. A phylogenetic analysis provided additional evidence of the high level of conservation of Mp11 among species (Fig. 1B). At the genomic level, the mouse Mp11 gene is located on chromosome 4 and consists of three coding and three noncoding exons (Fig. 1C).
Mp11 is regulated by Egr2 during nerve development and after nerve injury
The goal of our bioinformatics analysis of the microarray data were to identify novel Egr2-regulated genes that are functionally important for myelination. The low levels of Mp11 mRNA in sciatic nerves from Egr2 hypomorphic mice (Egr2Lo/Lo) suggested that the Mp11 gene could be transcriptionally regulated by Egr2. To test this hypothesis, we used the Egr binding profiles curated in the TRANSFAC database to computationally identify Egr binding sites within the Mp11 locus. In this analysis, the entire loci including all introns and exons as well as 50 kb upstream and downstream sequences from all species with Mp11 orthologs were aligned, and conserved regions were scanned for phylogenetically conserved Egr2 binding sites (Fig. 2A). Seven putative Egr2 binding sites that were conserved in human, chimp, rhesus, and at least one rodent were identified distributed throughout the Mp11 locus from ∼14 kb upstream of the first exon to 53 kb downstream of this exon, within the 3′ untranslated region (Fig. 2B). As an example of this analysis, Egr binding site 6, which is an inverted site, is displayed (Fig. 2C). It is conserved in all seven species and obtained an Egr2 binding site score of 6.2 (∼60% match to the Egr binding profile M000807 from TRANSFAC), indicating that there is a very high probability (p < 10−4) that Egr2 will bind to this site (Fig. 2B).
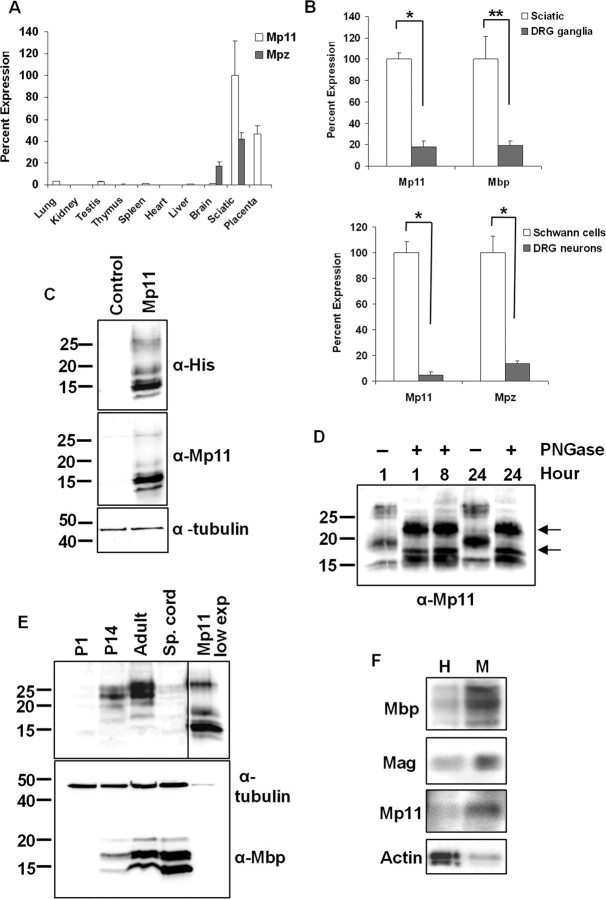
Figure 2.
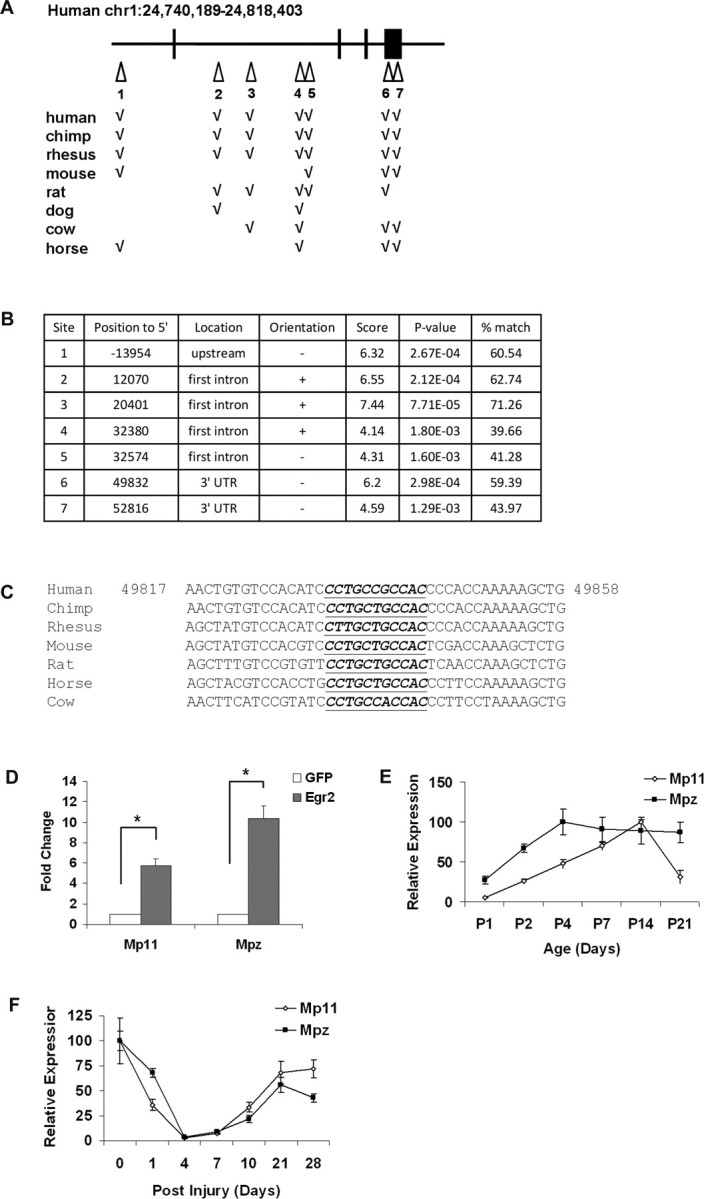
Mp11 expression is regulated by Egr2 during nerve development and after nerve injury. A, A diagram of the human Mp11 gene in which Egr2 binding sites are marked by white triangles. Introns are indicated by black lines, and exons are indicated by black boxes. Genomic loci of MP11 orthologs in multiple species were used to identify conserved sequence regions. These regions were examined for Egr binding sites using matrices from TRANSFAC and JASPAR to identify conserved Egr binding sites. The chart indicates conservation of the human Egr binding sites in the other species examined. B, Positions and probability scores for the seven conserved Egr binding sites. To calculate percentage match, we divided the binding probability score by the maximal Egr binding site score of 10.44 (binding profile M00807 from TRANSFAC). C, Multispecies alignment of the conserved genomic Mp11 region around Egr site 6. The underlined sequence indicates the conserved inverted Egr consensus site. D, Primary cultured rat Schwann cells were infected with adenovirus expressing either Egr2 or GFP (control). Mp11 and Mpz mRNA levels were determined using qRT-PCR. Values shown are means ± SEM of three independent experiments (*p = 0.002). E, RNA was prepared from sciatic nerves isolated from mouse pups of the indicated age. Mp11 and Mpz mRNA levels were determined using qRT-PCR. Values reported are means ± SD. F, RNA was prepared from distal sciatic nerve segments at the indicated time after crush injury (injury = time 0). Mp11 and Mpz mRNA levels were determined using qRT-PCR. Values shown are means ± SD.
To determine whether Mp11 is indeed regulated by Egr2, we infected rat primary Schwann cells with adenovirus expressing either Egr2 or GFP (control) and monitored mRNA levels by quantitative real-time PCR (qRT-PCR). Mp11 mRNA levels were induced approximately sixfold in Egr2-expressing Schwann cells (compared with GFP control) (Fig. 2D). The increased expression of Mp11 in response to Egr2 was closely correlated to the ∼10-fold increase in mRNA levels of Mpz, a well characterized Egr2 target gene that encodes a central constituent of peripheral myelin. Together, these results suggest that Mp11 represents an Egr2-regulated gene that is coregulated with Mpz and that has an expression that is linked to myelination in vivo.
To further explore the potential role of Mp11 in myelination, we examined its expression in sciatic nerve in several paradigms when myelination is dynamically regulated. First, we examined sciatic nerves from early postnatal animals (P1–P21), because this is the most active period of peripheral nerve myelin development in rodents. We compared sciatic nerve Mp11 and Mpz mRNA levels using qRT-PCR and found that, like Mpz, Mp11 expression was virtually undetectable at P0 but steadily increased through P14 (Fig. 2E). Second, we examined Mp11 expression after sciatic nerve crush injury. After nerve injury, there is a characteristic pattern of myelin-associated gene expression (e.g., Egr2, Mpz, Mbp, Pmp22). It begins with a temporary decrease in expression from 0 to 4 d after injury, followed by an increase in expression from days 7–21 that correlates with axonal regeneration and fiber remyelination (Nagarajan et al., 2002; Verheijen et al., 2003; Le et al., 2005a). We examined Mp11 expression in the distal nerve stump after sciatic nerve crush and found that, like other myelination-associated transcripts, Mp11 mRNA levels declined immediately after injury but began to increase 7 d after injury until it plateaus at a level similar to that observed before the injury (Fig. 2F). Again, this pattern of expression paralleled that observed with Mpz (Fig. 2F). Together, these data demonstrate that Mp11 expression is temporally controlled in myelinating Schwann cells in a pattern similar to that of other known myelin genes, suggesting that it may play a role in myelination.
Mp11 is expressed in Schwann cells and is a component of peripheral myelin
In our initial in silico characterization of Mp11, we noted that ESTs corresponding to Mp11 were present in placenta as well as embryonic and extraembryonic tissues. To further explore the tissue distribution of Mp11, we performed qRT-PCR using RNAs from a large number of adult mouse tissues. As expected, Mp11 was highly expressed in sciatic nerve and more moderately in placenta (E15.5) (Fig. 3A). However, Mp11 expression was negligible in most other tissues examined: brain, lung, kidney, testis, thymus, spleen, heart, and liver (Fig. 3A). Notably, unlike Mpz, Mp11 expression appears to be restricted to the PNS, because we did not detect significant levels of Mp11 mRNA in brain.
Figure 3.
Mp11 is a myelin glycoprotein expressed in the PNS. A, RNA was harvested from the indicated adult mouse tissues, and Mp11 mRNA levels relative to glyceraldehyde-3-phosphate dehydrogenase were measured using qRT-PCR. Values reported are means ± SD. B, Mp11 is Schwann cell specific because it showed relatively high expression in rat sciatic nerve compared with whole DRG explants (top graph; *p < 0.0001; **p = 0.003) and high expression in purified rat Schwann cell cultures compared with purified rat DRG neuronal cultures (bottom graph; *p = 0.0003). Results shown are derived from three independent experiments. Values shown are means ± SD. C, Lysates from HEK293T cells transfected with His-tagged Mp11 or GFP (control) expression vector were analyzed by immunoblotting using anti-His or anti-Mp11 antibodies. Several Mp11 species were detected with both antibodies, with the predominant species corresponding to a protein of ∼15 kDa. Two more slowly migrating forms were detected at ∼18 and ∼26 kDa as well as a faster migrating form close to its theoretical weight including tags at 13 kDa. α-Tubulin antibody was used to ensure equivalent loading. D, His-tagged Mp11 was purified from lysates of transfected HEK293T cells and treated with or without PNGase F glycosidase for the indicated time. The proteins were separated by electrophoresis and analyzed by immunoblot with anti-MP11 antibody. An increase in the mobility of the 26 and 18 kDa bands indicates that these Mp11 species are glycosylated. It is unlikely that incomplete deglycosylation accounts for the continued presence of higher molecular weight bands at ∼22 and 17 kDa (arrows) because prolonged incubation (up to 24 h) with PNGase F did not result in additional mobility changes. E, Lysates were prepared using sciatic nerves or spinal cords (Sp. cords) from rats of the indicated ages. Immunoblot analysis was performed using the anti-Mp11 antibody. Mp11 expression is detectable only at later ages, consistent with the postnatal myelination of rodents. The last panel shows lysate from MP11-transfected HEK293T cells exposed for a lesser time (lane MP11 low exp). Note that the 15 kDa Mp11 species that predominates in HEK293T cells is undetectable in vivo. F, Adult rat sciatic nerves were homogenized and fractionated using sucrose gradient centrifugation. The 26 kDa form of Mp11, along with the myelin proteins Mbp and Mag is enriched in the crude myelin fraction (M) compared with the starting homogenate (H). Actin, a nonmyelin protein control, was more abundant in the homogenate.
The high level of Mp11 expression in sciatic nerve strongly suggested that it was expressed in Schwann cells because they are the most predominant cell type in peripheral nerve. However, peripheral nerve does contain additional cell types (perineurial cells, fibroblasts, blood vessel endothelium, some hematopoietic cells), as well as DRG axons that may express Mp11. Therefore, to further examine the site of Mp11 expression, we compared Mp11 mRNA levels in sciatic nerve with those of DRG ganglia, and we compared levels in cultures of purified DRG neurons with those of purified Schwann cell cultures. qRT-PCR analysis demonstrated that Mp11 levels were much higher in sciatic nerve than DRG ganglia and much higher in cultured Schwann cells compared with cultured DRG neurons. These results indicate that Mp11 expression in the PNS is primarily limited to Schwann cells (Fig. 3B).
To examine Mp11 expression in sciatic nerve at the protein level, we developed a polyclonal antibody against an Mp11 peptide (amino acids 53–68 in mouse) that is conserved in both mice and rats. The antibody was affinity purified and used in an immunoblotting assay in which it recognized Mp11 expressed by transfecting the 6xHis-tagged Mp11 vector into HEK293T cells (Fig. 3C). A similar analysis using anti-His antibodies detected similar bands, confirming that the anti-Mp11 antisera we raised recognizes Mp11 (Fig. 3C). Although the transfected Mp11 construct contains a single ORF, multiple distinct Mp11 bands at 26, 18, 15, and 13 kDa were detected by both Mp11 and 6xHis antibodies. Multiple posttranslational modifications, a characteristic of many myelin-associated proteins (e.g., Mag, Mbp, Pmp22), could explain the presence of multiple Mp11 species that appear larger than the predicted 11 kDa. We therefore investigated whether Mp11 is posttranslationally modified by either glycosylation or phosphorylation. His-tagged Mp11 was expressed in HEK293T cells and purified by nickel affinity chromatography. The purified Mp11 was treated with the PNGase F glycosidase to remove carbohydrate groups for 1, 8, or 24 h, and the treated protein was analyzed by immunoblotting with the Mp11 antibody. The 26 and 18 kDa Mp11 species displayed a significant downshift in molecular weight, suggesting that the glycosidase removed carbohydrate moieties (Fig. 3D). However, Mp11 species at ∼22 and 17 kDa remained, suggesting that our attempts to completely remove these groups were unsuccessful (even after 24 h incubation) or that Mp11 undergoes additional posttranslational modification. We also treated the purified Mp11 with lambda phosphatase, a robust enzyme that acts on phosphorylated tyrosine, threonine, and serine residues. However, incubation with this phosphatase (even at very high levels) did not appear to alter the mobility of any of the Mp11 species, suggesting that Mp11 is not phosphorylated under these conditions (data not shown).
To examine Mp11 expressed in vivo, lysates were prepared from rat sciatic nerves, and immunoblotting was performed with the Mp11 antibody. We found that the predominant forms present in sciatic nerve were at ∼25 and 22 kDa, although minor bands at ∼18 and 20 kDa were also visible in P14 and adult rat sciatic nerve (Fig. 3E). We could not detect expression of the 13 and 15 kDa species in vivo, which probably indicates that endogenous Mp11 is efficiently modified posttranslationally in Schwann cells. In fact, the detection of the 13 and 15 kDa species in HEK293T cells may reflect the fact that unmodified forms are prevalent in transfected cells because of the high level of expression and that Mp11 is most likely modified differently in nonglial cell types. Consistent with our previous findings regarding the developmental regulation of Mp11 mRNA, Mp11 protein expression showed a steady increase during development. Mp11 was undetectable at P1 but is present at P14 and is present at slightly higher levels in adulthood (Fig. 3E). This is consistent with the pattern of peripheral nerve myelination in rodents and with the expression of Mbp, another myelin-associated protein (Fig. 3E). Mp11 was undetectable in adult rat spinal cord, confirming that it is not expressed in the CNS (Fig. 3E).
The Mp11 expression pattern strongly suggested that it is associated with the myelination of peripheral nerve. To determine whether Mp11 was indeed localized to peripheral nerve myelin, we prepared crude myelin fractions from adult rat sciatic nerves by sucrose gradient centrifugation and examined the level of Mp11 in this fraction by immunoblotting. We found that Mp11 was enriched in the crude myelin fraction, compared with the nerve homogenate, along with other myelin components such as Mag and Mbp (Fig. 3F). In contrast, actin, a nonmyelin protein, was present at high levels in peripheral nerve homogenate but was essentially excluded from the myelin fraction obtained by centrifugation. Together, these data support the hypothesis that Mp11 is a newly discovered component of peripheral nerve myelin.
Mp11 is a transmembrane protein
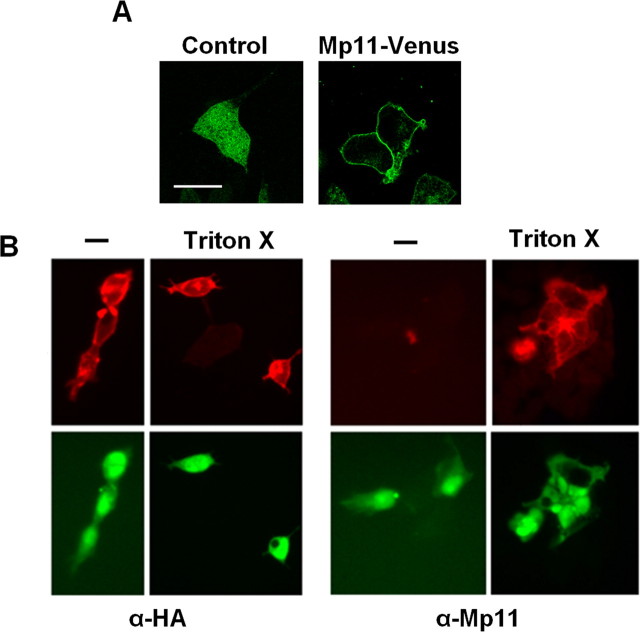
The analysis of the Mp11 protein sequence detected a stretch of amino acids from 27 to 49 with characteristics of a transmembrane domain, indicating that Mp11 could be an integral membrane protein. We also detected a signal anchor sequence, a motif important for membrane protein translocation that differs from a signal sequence by virtue of the fact that it is not cleaved during the translocation process (Bendtsen et al., 2004). To determine the subcellular distribution of Mp11, we fused the Venus fluorescent protein to Mp11 at the C terminus and expressed this fusion protein in HeLa cells. We examined these transfected cells by confocal microscopy to localize the fluorescent Mp11–Venus fusion protein. Consistent with the bioinformatics analysis, we detected the Mp11–Venus fusion protein around the cell surface in a rim-like pattern consistent with association with the cytoplasmic membrane (Fig. 4A). In contrast, the native Venus protein was distributed throughout the cell. These results indicate that Mp11 is localized to the plasma membrane and is likely an integral membrane protein of the type Ib category.
Figure 4.
Mp11 is a type I transmembrane protein. A, HeLa cells were transfected with constructs to express Mp11–Venus fluorescent fusion protein or Venus alone. Fluorescence microscopy detected Mp11–Venus along the outer surface of the cell, consistent with the pattern of a protein associated with the plasma membrane, whereas the signal from the Venus control was detected throughout the cell. Images were taken using a Zeiss laser confocal microscope. Scale bar, 20 μm. B, HEK293T cells were transfected with plasmid expressing N-terminal HA-tagged Mp11. The cells were fixed with 4% paraformaldehyde and visualized with or without treatment with Triton X-100 to permeabilize the cell membrane. Immunocytochemistry was performed with anti-HA antibody to detect the Mp11 N terminus or anti-Mp11, which recognizes an epitope near the C terminus, downstream of the Mp11 transmembrane domain. Note that the N-terminal antibody immunostains the cells with or without permeabilization, whereas a signal with the antibody directed to the C terminus (anti-Mp11) is only detected after permeabilization. The bottom panel demonstrates Venus fluorescent protein that was expressed from an IRES in the construct as a control for transfection and cell visualization.
To further study the membrane topology of Mp11, we generated an N-terminal HA-tagged Mp11 designed such that the predicted transmembrane domain (residues 27–49) is situated between the N-terminal HA epitope and the epitope recognized by the anti-Mp11 antibody (residues 53–68). This construct was transfected into HEK293T cells, and these cells were immunostained with either anti-Mp11 or anti-HA antibodies either before or after permeabilization of the cell membrane with Triton X-100 (0.5%). Cells immunostained with the anti-HA antibody produced a strong signal with or without permeabilization, indicating that the N-terminal HA tag is located extracellularly. However, when cells were immunostained with anti-Mp11 antibody, which recognizes an epitope downstream of the transmembrane domain, we obtained a signal only in permeabilized cells, indicating that this region of Mp11 is located inside the cell (Fig. 4B). To control for cell transfection and visualization, we also imaged the Venus fluorescent protein that was expressed from the plasmid using an IRES (Fig. 4B). These results indicate that the Mp11 N terminus is located extracellularly and its C terminus is located intracellularly, a topology that is characteristic of type I membrane proteins.
Mp11 is required for myelination in vitro
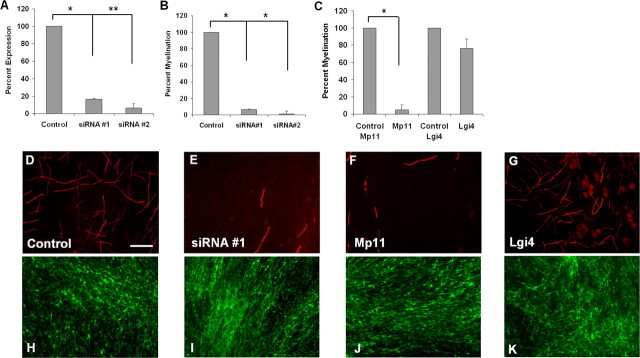
Misexpression of many myelin-associated proteins during development results in improper myelin formation or hypomyelination, and subsequent neuropathy. To determine whether Mp11 function is necessary for myelination, we disrupted Mp11 expression in primary rat Schwann cells and assessed the effects using in vitro myelination assays. To knock down endogenous levels of Mp11 in Schwann cells, we infected them with lentivirus expressing Mp11 targeted siRNAs or no target sequence (control) and Venus fluorescent protein to assess the level of infection. The infected Schwann cells were seeded onto DRG neurons, and the cocultures were allowed to myelinate for 14–21 d. Myelination was quantified by counting the number of MBP-positive internodes that formed. In these assays, we monitored the effects of two different Mp11 siRNAs that both downregulated Mp11 mRNA levels by 80–90% in Schwann cells as measured by qRT-PCR (Fig. 5A). The number of MBP-positive myelin profiles was severely decreased (∼90–95% compared with control) in these cocultures (Fig. 5B,D,E,H,I), indicating that Mp11 is required for myelination in vitro.
Figure 5.
Mp11 is required for myelination in vitro. Schwann cells infected with lentiviruses were seeded onto DRG neurons and allowed to myelinate for 14–21 d. The Schwann cell–neuronal cocultures were immunostained with anti-MBP antibody to detect myelinated axons and quantify the extent of myelination. A, Schwann cells were infected with lentiviruses expressing Mp11 siRNAs (#1 and #2). The efficiency of Mp11 “knockdown” was assessed by measuring Mp11 mRNA levels by qRT-PCR. Values reported are mean ± SEM of two independent experiments (*p = 0.003, **p = 0.002). B, Schwann cells infected with lentiviruses expressing Mp11 siRNAs were used in the in vitro myelination assay. Quantification of MBP-positive internodes in the cocultures showed severe inhibition of myelination by both Mp11 siRNAs but not by the nonrecombinant control siRNA lentivirus. Each condition was performed in quadruplicate wells, and data shown are the average of three independent experiments (mean ± SEM; *p < 0.001). C, Schwann cells infected with lentivirus expressing Mp11 or Venus fluorescent protein (Control Mp11) were tested in the in vitro myelination assay. Overexpression of Mp11 resulted in severe inhibition of myelination as determined by quantification of Mbp-positive internodes. Each condition was performed in quadruplicate wells, and data shown are the average of three independent experiments (mean ± SEM; *p < 0.001). As an overexpression toxicity control, Schwann cells infected with a lentivirus expressing Lgi4 were used in the in vitro myelination assay. There was no significant difference in the ability of Lgi4-overexpressing Schwann cells to myelinate DRG neurons compared with control Venus-infected Schwann cells. Each condition was performed in quadruplicate wells, and data shown are the average of two independent experiments (mean ± SEM; p = 0.13). D–G, Schwann cell–DRG cocultures were stained with anti-MBP, and fluorescence microscopy was used to visualize and quantify the MBP-positive internodes, nonrecombinant siRNA lentivirus, Control (D), Mp11 siRNA #1 (E), Mp11 (F), or Lgi4 (G). H–K, Venus fluorescence of cocultures described above to show that ∼100% of Schwann cells were infected and that similar numbers of Schwann cells were present in each of the cocultures, control (H), Mp11 siRNA #1 (I), Mp11 (J), and Lgi4 (K). Scale bar, 100 μm.
We also assessed the effects of Mp11 overexpression on myelination because it is well recognized that proper myelin formation is exquisitely sensitive to alterations, either increases or decreases, in the levels of myelin proteins. For example, both monosomy and trisomy of PMP22, the most commonly affected gene in CMT1, results in abnormal myelination and neuropathy (Lupski et al., 1991; Raeymaekers et al., 1991). We used lentivirus to overexpress Mp11 in Schwann cells and used these cells to again perform in vitro myelination assays. We found that Mp11 overexpression in Schwann cells also resulted in significant disruption of myelination (∼95% fewer MBP-positive myelin profiles compared with Venus-infected control cells) (Fig. 5C,F,J). As a control to test whether overexpression of glycosylated proteins in Schwann cells leads to myelin inhibition in our assay, we overexpressed Lgi4, a glycoprotein that when mutated in mice gives rise to myelination defects and which results in a decrease in myelin formation when knocked down in Schwann cells in vitro (Bermingham et al., 2006). Lgi4 overexpression in Schwann cells did not significantly inhibit myelin formation, indicating that overexpression of a glycosylated protein in Schwann cells will not necessarily result in myelin inhibition (Fig. 5C,G,K). These results indicate that Mp11 is required for myelination in vitro and that proper levels of Mp11 are critical for myelin formation.
Mp11 is localized to noncompact myelin
Myelin is a highly specialized and complex structure that is composed of distinct domains broadly classified as compact or noncompact myelin. The internode consists of compact myelin, which is tightly spiraled Schwann cell membrane that acts as insulation for the axon. Regions of noncompact myelin, such as the SLIs or paranodes, are thought to be involved in signaling and metabolic duties. To gain further insight into the function of Mp11, we sought to determine where it was located within the myelin structure. We prepared teased adult rat sciatic nerves and coimmunostained them with anti-Mp11 antibody and various markers for nodal, paranodal, and SLI markers. We found that, consistent with our previous results, Mp11 was indeed restricted to the Schwann cell as opposed to the axon because there was no overlap between the axonal marker Tuj1, which recognizes β-tubulin III and Mp11 (Fig. 6A–C). The discrete, periodic pattern of Mp11 staining was reminiscent of that observed for proteins localized in SLIs, the funnel-shaped cytoplasmic structures that traverse the Schwann cell membranes and interrupt the internodal compact myelin. To confirm this idea, we coimmunostained the teased nerve preparations with antibodies against Mp11 and E-cadherin, a well characterized SLI marker. The extensive overlap detected in this analysis confirmed the localization of Mp11 to the SLIs (Fig. 6D–F).
Figure 6.
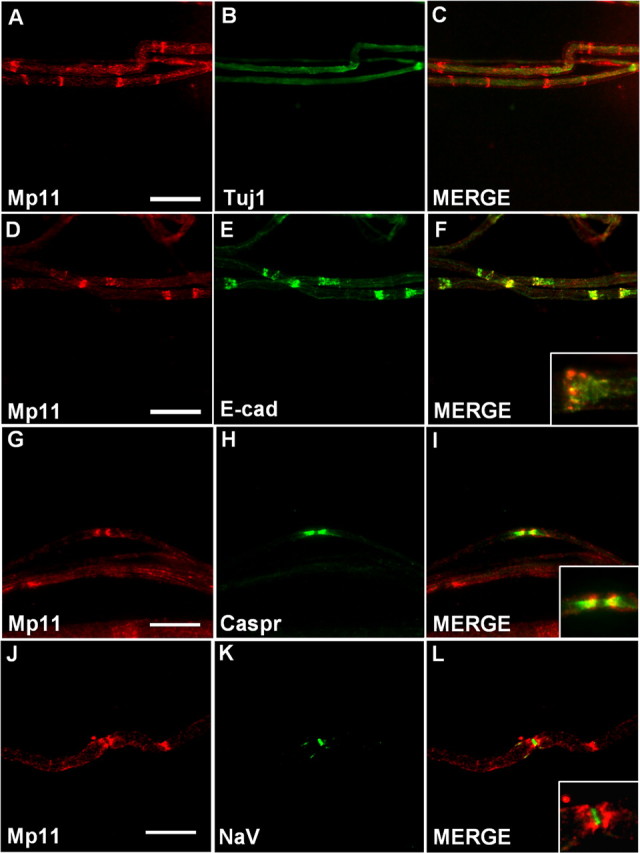
Mp11 is located in SLIs and paranodes. Immunohistochemistry on teased nerve preparations from adult rat sciatic nerve was performed with antibodies that recognize Mp11 or the indicated proteins to identify the location of Mp11. A–C, Double labeling with antibodies against Mp11 and α-tubulin III (Tuj1), an axonal marker, demonstrate that Mp11 expression is restricted to Schwann cells. D–F, Double labeling with antibodies against Mp11 and E-cadherin, a marker for SLIs, demonstrates that Mp11 is present in these structures. The inset in F is a higher-magnification image. G–I, Double labeling with antibodies against Mp11 and Caspr, a paranodal marker, reveals Mp11 is located in paranodes. The inset in I is a higher-magnification image. J–L, Double labeling with antibodies against Mp11 and NaV (pan sodium channel), a marker of the node of Ranvier, demonstrates the close proximity of Mp11 to the node. The inset in L is a higher-magnification image. Scale bars: A–F, 40 μm; G–L, 20 μm.
In addition to Mp11 staining within SLIs, there was also signal detected outside of these structures. We therefore determined whether Mp11 could also be associated with other areas of noncompact myelin, such as paranodes. We again performed colabeling studies, using antibodies to Mp11 and Caspr, a type I axonal membrane protein that is a well characterized marker for the paranodal region (Poliak and Peles, 2003). Mp11 was detected in most, but not all, paranodes, and when detected was localized very close to the node of Ranvier and not seen as far laterally in the paranodal region as Caspr (Fig. 6G–I, inset). Paranodal Mp11 localization was confirmed using a pan anti-sodium channel antibody, which labels nodes of Ranvier. Note that sodium channels at the node are tightly flanked by Mp11 staining on either side (Fig. 6J–L). Together, these data indicate that Mp11 is a novel component of noncompact peripheral nerve myelin.
Discussion
We have used bioinformatics analysis of peripheral nerve gene expression profiles and the principle that coregulated genes often encode proteins involved in similar processes to identify a novel Schwann cell glycoprotein that we have named Mp11 for myelin protein of 11 kDa. This computational method of comparing multiple gene expression profiles has been used previously to link proteins to new cellular processes, not only during myelination but also in cancer, metabolism, and cell cycle, and has been used extensively in yeast to identify protein function (Eisen et al., 1998; Cho et al., 2001; Hansel et al., 2003; Le et al., 2005a; Ma et al., 2005). Mp11 was initially identified as an EST in a microarray data set generated from sciatic nerves of the Egr2Lo/Lo hypomyelination mouse model that expresses low levels of the myelination regulator Egr2. Interestingly, a sequence tag called 18-138 previously identified as downregulated in hypomyelinated sciatic nerves from Pou3f1 (Scip)-deficient and claw paw mutant mice (Bermingham et al., 2002) is contained within the Mp11 cDNA. Egr2 has been shown to directly and indirectly regulate the induction of a number of genes essential for myelin formation including Mpz, Mbp, Cx32, Pmp22, and periaxin (Bondurand et al., 2001; Nagarajan et al., 2002; Verheijen et al., 2003; Le et al., 2005a; LeBlanc et al., 2006). The identification of multiple Egr2 binding sites within the Mp11 locus, along with the induction of Mp11 in Egr2-overexpressing Schwann cells indicates that Mp11 is a novel Egr2-regulated gene. The patterns of expression of Mp11 and other well characterized myelin proteins, such as Mpz in developing sciatic nerve as well as after nerve crush injury, suggest that Mp11 is a myelin component. Indeed, we found that strictly controlled levels of Mp11 are required for myelination in vitro because both overexpression and knockdown of Mp11 resulted in significantly less myelin formation.
The Mp11 protein is well conserved in higher vertebrates from human to chicken and is moderately conserved in lower vertebrates such as frog and zebrafish, suggesting that it possesses a critically conserved evolutionary function. Coincidentally, the lower vertebrates in which Mp11 is expressed, such as zebrafish and frog, also possess well developed myelin sheaths, including myelination of peripheral nerves (Avila et al., 2007; Luo et al., 2007). Furthermore, zebrafish and frog express other critically important myelin proteins such as Mpz, Mbp, and Pmp22, lending more evidence that Mp11 may be an evolutionarily conserved essential component of myelin. Besides peripheral nerve myelin, Mp11 is expressed in placenta indicating that it may also play a critical role during embryonic development.
The lack of recognizable protein motifs, functional domains, or structural homology of Mp11 to other proteins makes it difficult to surmise functional attributes based on protein similarity. The relatively small size of Mp11 as well as our finding that it is a positively charged (pI = 9.69) transmembrane glycoprotein expressed in Schwann cells provides some clues, because these attributes are similar to other myelin proteins such as Mbp and Mpz. Interestingly, Mbp and Mpz can interact with negatively charged phospholipid bilayers of the cell membrane to influence formation of the major dense lines of compact myelin (Ding and Brunden, 1994; Martini et al., 1995). Whether Mp11 also interacts with lipid bilayers in regions of noncompact myelin is intriguing, because it is currently unknown how regions of noncompact myelin maintain their structural integrity.
Mp11 expression is restricted to the PNS versus CNS. One notable difference of PNS versus CNS myelin is the node of Ranvier. In peripheral myelin, the nodes are covered by Schwann cell microvilli, whereas in the CNS the nodes are associated with perinodal astrocytes (Poliak and Peles, 2003). Another difference is that PNS myelin requires a complex extracellular matrix called the basal lamina to surround the Schwann cell–axonal unit in order for myelination to occur. The basal lamina is secreted by the differentiating Schwann cell and plays multiple critical roles during myelination such as in radial sorting, adhesion, differentiation, and proliferation of Schwann cells (Court et al., 2006). The limited expression of Mp11 to the Schwann cell membrane and PNS suggests that Mp11 could interact with the basal lamina.
Mp11 is localized to noncompact myelin, specifically to SLIs and paranodes. Noncompact myelin contains multiple channels and a variety of specialized junctions including gap, adherens, and tight junctions by which the Schwann cell accomplishes intracellular signaling and extracellular communication with the axon and basal lamina (Spiegel and Peles, 2002). One of these cytoplasmic channels is the SLI, which is intermittently dispersed throughout the compact myelin and traverses the myelin sheath. The maintenance of the SLI is thought to be dependent on adherens junctions (Fannon et al., 1995; Tricaud et al., 2005), which consist of small, membrane-spanning proteins; thus it is possible that Mp11 is a component of these junctions. The paranodes, located at the ends of the internode and flanking the nodes of Ranvier, are the regions where compact myelin ends and opens up into cytoplasm-filled loops that adhere to the axon and form the axoglial junction. Septate-like junctions connect the Schwann cell paranodal loops to the axon, with the loops closest to the node forming the initial junctions (Tao-Cheng and Rosenbluth, 1983). Mp11 expression is most prominent in the paranodal region directly flanking the node, suggesting that it plays a specialized function at the axoglial junction, possibly as part of more mature septate junctions, or as a barrier to ion diffusion. Finally, tight junctions are a prominent feature of myelinating Schwann cells and provide a physical barrier to the diffusion of ions and molecules and to the movement of proteins and lipids in the plasma membrane (Mitic and Anderson, 1998). The junctional adhesion molecule is a constituent of tight junctions and is similar to Mp11 in that it is also a type I membrane protein with a single transmembrane domain (Dejana et al., 2000), suggesting that Mp11 may be involved in the formation of tight junctions. The identification of Mp11-interacting partners will undoubtedly provide important insights into the functions of this novel protein.
Both overexpression and inhibition of Mp11 expression in Schwann cells leads to a significant decrease in myelin formation in vitro. A similar sensitivity to overexpression or inhibition of gene expression in myelinating Schwann cell–DRG cocultures was observed with the polarity protein Par-3 (Chan et al., 2006). In this case, mislocalization of Par-3 was most likely responsible for the inhibition of myelination when either overexpressed or inhibited. As for Mp11, overexpression could result in an accumulation of the protein in the endoplasmic reticulum (ER) where it is processed for glycosylation as well as membrane insertion, thus interfering with myelin formation. Lgi4 is another protein that is glycosylated and processed in the ER and when mutated is responsible for the hypomyelinating claw paw phenotype (Bermingham et al., 2006). This mutant form of Lgi4 is retained within the ER and presumably leads to the myelination defects. However, simple overexpression of glycosylated proteins is not the root cause of a decrease in myelin formation in vitro because we found that overexpression of the glycoprotein Lgi4 did not lead to an inhibition of myelin formation.
The gene dosage effect observed with Mp11 also occurs with mutations in the PMP22 gene, in that both deletion and overexpression of PMP22 leads to a demyelinating neuropathy in humans and in animal models (Meyer Zu Hörste and Nave, 2006). Pmp22 is an integral membrane glycoprotein found in compact peripheral nerve myelin, and its exact function is unknown but is believed to be involved in the structure and maintenance of myelin (Amici et al., 2006). Primary Schwann cells isolated from Pmp22-overexpressing mice contain aggregates of Pmp22 and exhibit reduced proteasome activity that leads to an accumulation of ubiquitinated substrates (Fortun et al., 2006). It has been hypothesized that overloading of the protein degradation machinery is carried over to other essential cellular proteins and results in inhibition of myelination (Nave et al., 2007). A similar scenario could occur with Mp11, because it is also a membrane protein that inhibits myelination when it is either overexpressed or underexpressed.
Footnotes
This work was supported by National Institutes of Health (NIH) Neuroscience Blueprint Center Core Grant P30 NS057105 to Washington University, the HOPE Center for Neurological Disorders, and NIH Grant NS040745 (J.M.). We thank Carmen Melendez-Vasquez, Carla Taveggia, and Jim Salzer for help with the in vitro myelination and members of the Milbrandt laboratory for helpful discussions.
References
- Amici et al., 2006.Amici SA, Dunn WA, Jr, Murphy AJ, Adams NC, Gale NW, Valenzuela DM, Yancopoulos GD, Notterpek L. Peripheral myelin protein 22 is in complex with α6β4 integrin, and its absence alters the Schwann cell basal lamina. J Neurosci. 2006;26:1179–1189. doi: 10.1523/JNEUROSCI.2618-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki et al., 2004.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Avila et al., 2007.Avila RL, Tevlin BR, Lees JP, Inouye H, Kirschner DA. Myelin structure and composition in zebrafish. Neurochem Res. 2007;32:197–209. doi: 10.1007/s11064-006-9136-5. [DOI] [PubMed] [Google Scholar]
- Bendtsen et al., 2004.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bermingham et al., 2002.Bermingham JR, Jr, Shumas S, Whisenhunt T, Sirkowski EE, O'Connell S, Scherer SS, Rosenfeld MG. Identification of genes that are downregulated in the absence of the POU domain transcription factor pou3f1 (Oct-6, Tst-1, SCIP) in sciatic nerve. J Neurosci. 2002;22:10217–10231. doi: 10.1523/JNEUROSCI.22-23-10217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham et al., 2006.Bermingham JR, Jr, Shearin H, Pennington J, O'Moore J, Jaegle M, Driegen S, van Zon A, Darbas A, Ozkaynak E, Ryu EJ, Milbrandt J, Meijer D. The claw paw mutation reveals a role for Lgi4 in peripheral nerve development. Nat Neurosci. 2006;9:76–84. doi: 10.1038/nn1598. [DOI] [PubMed] [Google Scholar]
- Bondurand et al., 2001.Bondurand N, Girard M, Pingault V, Lemort N, Dubourg O, Goossens M. Human Connexin 32, a gap junction protein altered in the X-linked form of Charcot-Marie-Tooth disease, is directly regulated by the transcription factor SOX10. Hum Mol Genet. 2001;10:2783–2795. doi: 10.1093/hmg/10.24.2783. [DOI] [PubMed] [Google Scholar]
- Chan et al., 2006.Chan JR, Jolicoeur C, Yamauchi J, Elliott J, Fawcett JP, Ng BK, Cayouette M. The polarity protein Par-3 directly interacts with p75NTR to regulate myelination. Science. 2006;314:832–836. doi: 10.1126/science.1134069. [DOI] [PubMed] [Google Scholar]
- Cho et al., 2001.Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge SJ, Davis RW, Lockhart DJ. Transcriptional regulation and function during the human cell cycle. Nat Genet. 2001;27:48–54. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- Court et al., 2006.Court FA, Wrabetz L, Feltri ML. Basal lamina: Schwann cells wrap to the rhythm of space-time. Curr Opin Neurobiol. 2006;16:501–507. doi: 10.1016/j.conb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Dejana et al., 2000.Dejana E, Lampugnani MG, Martinez-Estrada O, Bazzoni G. The molecular organization of endothelial junctions and their functional role in vascular morphogenesis and permeability. Int J Dev Biol. 2000;44:743–748. [PubMed] [Google Scholar]
- Ding and Brunden, 1994.Ding Y, Brunden KR. The cytoplasmic domain of myelin glycoprotein P0 interacts with negatively charged phospholipid bilayers. J Biol Chem. 1994;269:10764–10770. [PubMed] [Google Scholar]
- Eisen et al., 1998.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannon et al., 1995.Fannon AM, Sherman DL, Ilyina-Gragerova G, Brophy PJ, Friedrich VL, Jr, Colman DR. Novel E-cadherin-mediated adhesion in peripheral nerve: Schwann cell architecture is stabilized by autotypic adherens junctions. J Cell Biol. 1995;129:189–202. doi: 10.1083/jcb.129.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortun et al., 2006.Fortun J, Go JC, Li J, Amici SA, Dunn WA, Jr, Notterpek L. Alterations in degradative pathways and protein aggregation in a neuropathy model based on PMP22 overexpression. Neurobiol Dis. 2006;22:153–164. doi: 10.1016/j.nbd.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Hansel et al., 2003.Hansel DE, Rahman A, Hidalgo M, Thuluvath PJ, Lillemoe KD, Shulick R, Ku JL, Park JG, Miyazaki K, Ashfaq R, Wistuba II, Varma R, Hawthorne L, Geradts J, Argani P, Maitra A. Identification of novel cellular targets in biliary tract cancers using global gene expression technology. Am J Pathol. 2003;163:217–229. doi: 10.1016/S0002-9440(10)63645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le et al., 2005a.Le N, Nagarajan R, Wang JY, Araki T, Schmidt RE, Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc Natl Acad Sci U S A. 2005a;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le et al., 2005b.Le N, Nagarajan R, Wang JY, Svaren J, LaPash C, Araki T, Schmidt RE, Milbrandt J. Nab proteins are essential for peripheral nervous system myelination. Nat Neurosci. 2005b;8:932–940. doi: 10.1038/nn1490. [DOI] [PubMed] [Google Scholar]
- LeBlanc et al., 2006.LeBlanc SE, Jang SW, Ward RM, Wrabetz L, Svaren J. Direct regulation of myelin protein zero expression by the Egr2 transactivator. J Biol Chem. 2006;281:5453–5460. doi: 10.1074/jbc.M512159200. [DOI] [PubMed] [Google Scholar]
- Luo et al., 2007.Luo X, Cerullo J, Dawli T, Priest C, Haddadin Z, Kim A, Inouye H, Suffoletto BP, Avila RL, Lees JP, Sharma D, Xie B, Costello CE, Kirschner DA. Peripheral myelin of Xenopus laevis: role of electrostatic and hydrophobic interactions in membrane compaction. J Struct Biol. 2007;162:170–183. doi: 10.1016/j.jsb.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski et al., 1991.Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, Saucedo-Cardenas O, Barker DF, Killian JM, Garcia CA, Chakravarti A, Patel PI. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- Ma et al., 2005.Ma PC, Chan KC, Chiu DK. Clustering and re-clustering for pattern discovery in gene expression data. J Bioinform Comput Biol. 2005;3:281–301. doi: 10.1142/s0219720005001053. [DOI] [PubMed] [Google Scholar]
- Martini et al., 1995.Martini R, Mohajeri MH, Kasper S, Giese KP, Schachner M. Mice doubly deficient in the genes for P0 and myelin basic protein show that both proteins contribute to the formation of the major dense line in peripheral nerve myelin. J Neurosci. 1995;15:4488–4495. doi: 10.1523/JNEUROSCI.15-06-04488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Zu Hörste and Nave, 2006.Meyer Zu Hörste G, Nave KA. Animal models of inherited neuropathies. Curr Opin Neurol. 2006;19:464–473. doi: 10.1097/01.wco.0000245369.44199.27. [DOI] [PubMed] [Google Scholar]
- Mitic and Anderson, 1998.Mitic LL, Anderson JM. Molecular architecture of tight junctions. Annu Rev Physiol. 1998;60:121–142. doi: 10.1146/annurev.physiol.60.1.121. [DOI] [PubMed] [Google Scholar]
- Nagarajan et al., 2001.Nagarajan R, Svaren J, Le N, Araki T, Watson M, Milbrandt J. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron. 2001;30:355–368. doi: 10.1016/s0896-6273(01)00282-3. [DOI] [PubMed] [Google Scholar]
- Nagarajan et al., 2002.Nagarajan R, Le N, Mahoney H, Araki T, Milbrandt J. Deciphering peripheral nerve myelination by using Schwann cell expression profiling. Proc Natl Acad Sci U S A. 2002;99:8998–9003. doi: 10.1073/pnas.132080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave et al., 2007.Nave KA, Sereda MW, Ehrenreich H. Mechanisms of disease: inherited demyelinating neuropathies–from basic to clinical research. Nat Clin Pract Neurol. 2007;3:453–464. doi: 10.1038/ncpneuro0583. [DOI] [PubMed] [Google Scholar]
- Poliak and Peles, 2003.Poliak S, Peles E. The local differentiation of myelinated axons at nodes of Ranvier. Nat Rev Neurosci. 2003;4:968–980. doi: 10.1038/nrn1253. [DOI] [PubMed] [Google Scholar]
- Raeymaekers et al., 1991.Raeymaekers P, Timmerman V, Nelis E, De Jonghe P, Hoogendijk JE, Baas F, Barker DF, Martin JJ, De Visser M, Bolhuis PA, et al. Duplication in chromosome 17p11.2 in Charcot-Marie-Tooth neuropathy type 1a (CMT 1a). The HMSN Collaborative Research Group. Neuromuscul Disord. 1991;1:93–97. doi: 10.1016/0960-8966(91)90055-w. [DOI] [PubMed] [Google Scholar]
- Ryu et al., 2007.Ryu EJ, Wang JY, Le N, Baloh RH, Gustin JA, Schmidt RE, Milbrandt J. Misexpression of Pou3fl results in peripheral nerve hypomyelination and axonal loss. J Neurosci. 2007;27:11552–11559. doi: 10.1523/JNEUROSCI.5497-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou and Nei, 1987.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salzer, 2003.Salzer JL. Polarized domains of myelinated axons. Neuron. 2003;40:297–318. doi: 10.1016/s0896-6273(03)00628-7. [DOI] [PubMed] [Google Scholar]
- Spiegel and Peles, 2002.Spiegel I, Peles E. Cellular junctions of myelinated nerves. Mol Membr Biol. 2002;19:95–101. doi: 10.1080/09687680210130009. [DOI] [PubMed] [Google Scholar]
- Tao-Cheng and Rosenbluth, 1983.Tao-Cheng JH, Rosenbluth J. Axolemmal differentiation in myelinated fibers of rat peripheral nerves. Brain Res. 1983;285:251–263. doi: 10.1016/0165-3806(83)90023-8. [DOI] [PubMed] [Google Scholar]
- Topilko et al., 1994.Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Chennoufi AB, Seitanidou T, Babinet C, Charnay P. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- Tricaud et al., 2005.Tricaud N, Perrin-Tricaud C, Brusés JL, Rutishauser U. Adherens junctions in myelinating Schwann cells stabilize Schmidt-Lanterman incisures via recruitment of p120 catenin to E-cadherin. J Neurosci. 2005;25:3259–3269. doi: 10.1523/JNEUROSCI.5168-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen et al., 2003.Verheijen MH, Chrast R, Burrola P, Lemke G. Local regulation of fat metabolism in peripheral nerves. Genes Dev. 2003;17:2450–2464. doi: 10.1101/gad.1116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner et al., 1998.Warner LE, Mancias P, Butler IJ, McDonald CM, Keppen L, Koob KG, Lupski JR. Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet. 1998;18:382–384. doi: 10.1038/ng0498-382. [DOI] [PubMed] [Google Scholar]
- Wiley and Ellisman, 1980.Wiley CA, Ellisman MH. Rows of dimeric-particles within the axolemma and juxtaposed particles within glia, incorporated into a new model for the paranodal glial-axonal junction at the node of Ranvier. J Cell Biol. 1980;84:261–280. doi: 10.1083/jcb.84.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]