Abstract
Mycobacterium avium complex (MAC) causes chronic lung disease in immunocompetent people and disseminated infection in patients with AIDS. MAC is intrinsically resistant to many conventional antimycobacterial agents, it develops drug resistance rapidly to macrolide antibiotics, and patients with MAC infection experience frequent relapses or the inability to completely eradicate the infection with current treatment. Treatment regimens are prolonged and complicated by drug toxicity or intolerances. We sought to identify biochemical pathways in MAC that can serve as targets for novel antimycobacterial treatment. The cytochrome P450 enzyme, CYP51, catalyzes an essential early step in sterol metabolism, removing a methyl group from lanosterol in animals and fungi, or from obtusifoliol in plants. Azoles inhibit CYP51 function, leading to an accumulation of methylated sterol precursors. This perturbation of normal sterol metabolism compromises cell membrane integrity, resulting in growth inhibition or cell death. We have cloned and characterized a CYP51 from MAC that functions as a lanosterol 14α-demethylase. We show the direct interactions of azoles with purified MAC-CYP51 by absorbance and electron paramagnetic resonance spectroscopy, and determine the minimum inhibitory concentrations (MICs) of econazole, ketoconazole, itraconazole, fluconazole, and voriconazole against MAC. Furthermore, we demonstrate that econazole has a MIC of 4 μg/ml and a minimum bacteriocidal concentration of 4 μg/ml, whereas ketoconazole has a MIC of 8 μg/ml and a minimum bacteriocidal concentration of 16 μg/ml. Itraconazole, voriconazole, and fluconazole did not inhibit MAC growth to any significant extent.
Keywords: azoles, CYP51, ERG11, lanosterol
Mycobacterium avium complex (MAC) consists of the nontuberculous mycobacteria M. avium subspecies avium, intracellulare, paratuberculosis, and others (1). MAC is ubiquitous in nature, and is commonly isolated from water or soil. There is a wide spectrum of disease caused by MAC, affecting both immunocompetent and immunocompromised individuals (2, 3). Chronic pulmonary disease is the most common form of localized MAC infection in humans, and manifests as upper lobe infiltrates similar to classic cavitary tuberculosis, or patchy nodular disease in the middle lobe or lingula associated with bronchiectasis (4, 5). Disseminated MAC occurs in immunocompromised patients, such as those with underlying immune defects due to chronic corticosteroid use, hematologic malignancies, organ transplantation, and infection with HIV (6–9). The treatment of patients with chronic pulmonary disease or disseminated infection from MAC is difficult due to the long duration necessary for treatment and the high frequency of drug intolerances or toxicity. Recently, hypersensitivity pneumonitis due to MAC has been described in patients using hot tubs or humidifiers, or exposed to contaminated household water (10, 11). For the majority of these patients, elimination of the exposure typically results in a cure; however, some patients may require a short course of corticosteroids, or specific antimycobacterial therapy.
In comparison with the treatment of tuberculosis, drug therapy for patients with MAC infection has been disappointing (1, 12). Most of the first-line antituberculosis medications have significantly less activity against MAC compared with Mycobacterium tuberculosis. MAC is intrinsically resistant to isoniazid and pyrazinamide, and rapidly develops drug resistance when single agents are used for treatment (13–15). Combination therapy with macrolide antibiotics (such as clarithromycin or azithromycin), rifabutin, or rifampin, and ethambutol is recommended for initial treatment (1). Treatment regimens are usually prolonged, and relapse after treatment is frequent. Pulmonary MAC infection is typically treated for 18 mo, but may require lifelong treatment in cases of disseminated infection. The difficulties of treating chronic and disseminated MAC infections highlight the urgent need to identify novel targets for the development of antimycobacterial medications.
CYP51, called ERG11 in fungi, is a highly conserved cytochrome P450 enzyme required for the early steps of sterol metabolism in the animal, fungus, and plant kingdoms (16–19). CYP51 enzymes have been identified in the genomes of M. tuberculosis and Mycobacterium smegmatis, and the inhibition of mycobacterial sterol biosynthesis may be therapeutically useful for patients with mycobacterial infections (20–22). This essential enzyme catalyzes the removal of a methyl group from lanosterol in animals and fungi, or from obtusifoliol in plants, allowing sterol metabolism to proceed to the end-product of cholesterol, ergosterol, or phytosterol, respectively. Azole medications, which have been the foundation for the treatment of fungal infections, inhibit CYP51 function by binding to the heme cofactor in the active site of the enzyme (23, 24). Sterol metabolism cannot proceed normally due to an accumulation of methylated sterol precursors, which ultimately affect cell membrane integrity, resulting in growth inhibition or cell death. Here, we describe the cloning, expression, purification, and characterization of CYP51 from M. avium subspecies avium. We show the direct interactions of azoles with purified MAC-CYP51. Furthermore, we demonstrate that econazole has a minimum inhibitory concentration (MIC) of 4 μg/ml and a minimum bacteriocidal concentration (MBC) of 4 μg/ml, whereas ketoconazole has a MIC of 8 μg/ml and an MBC of 16 μg/ml. Itraconazole, voriconazole, and fluconazole did not inhibit MAC growth to any significant extent. Our studies suggest that inhibition of CYP51 function in MAC may be of therapeutic benefit for patients with this infection.
MATERIALS AND METHODS
Materials
All reagents were from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. Restriction endonucleases, PCR reagents, and Pfx polymerase were from Invitrogen (Carlsbad, CA).
Mycobacterial Growth Conditions
M. avium subspecies avium (American Type Culture Collection no. 25,291) was grown in modified Middlebrook 7H9 broth containing oleic albumin dextrose catalase supplement at 37°C in the BACTEC MGIT 960 fluorescent detection instrument (Becton-Dickinson and Co., Franklin Lakes, NJ) until growth was detected.
Cloning of MAC-CYP51
MAC DNA was extracted using standard procedures. We designed degenerate oligonucleotide PCR primers after aligning the amino acid sequences of sterol 14α-demethylase enzymes, as described previously (25). PCR was performed in a 50 μl reaction using 1 μM of each degenerate primer: sense, 5′-AA(A/G)-GA(A/G)-AC(A/T/G/C)-CT(A/T/G/C)-CG(A/T/G/C)-CT(A/T/G/C)-CA(T/C)-CC(A/T/G/C)-CC-3′; and antisense: CA(A/T/G/C)-CG(A/G)-TG(A/T/G/C)-CG(A/T/G/C)-CC(A/T/G/C)-GC(A/T/G/C)-CC(A/G)-AA(A/T/G/C)-GG-3′; with 200 ng of purified MAC DNA. We performed PCR across an annealing temperature gradient from 60 to 72°C in an iCycler thermocycler (Bio-Rad Laboratories, Hercules, CA). A 248-bp PCR amplicon was identified by electrophoresis on agarose after ethidium bromide staining. This fragment was subcloned, sequenced, and analyzed by BlastX (National Center for Biotechnology Information, Bethesda, MD). The PCR amplicon was a novel gene with greatest homology to M. tuberculosis CYP51. This partial gene was then used to search the unannotated MAC genomic database (The Institute for Genomic Research, Rockville, MD; http://www.tigr.org/tdb/mdb/mdbinprogress.html), from which we were able to identify the start and stop codons for the full-length MAC-CYP51. We amplified the complete MAC-CYP51 cDNA by PCR, and subcloned the gene into the bacterial expression plasmid pGEX4T3 (Amersham Biosciences, Piscataway, NJ) to create an N-terminal glutathione S-transferase (GST) fusion.
Expression and Purification of Recombinant MAC-CYP51
The MAC-CYP51 in the pGEX4T3 plasmid was transformed into Escherichia coli BL21 pLysS cells. Transformed colonies were grown overnight at 37°C in 10 ml of Terrific Broth containing 100 μg/ml of carbenicillin and 34 μg/ml of chloramphenicol, and a subculture (1:50 ratio) was expanded in fresh medium and grown to a 0.8 optical density at 600 nm. Protein expression was induced with the addition of 1 mM isopropylthiogalactopyranoside and growth was continued at 25°C with shaking at 200 rpm for approximately 20 h. The cultures were supplemented with 2 mM δ-aminolevulinic acid for heme synthesis. The bacterial pellet was resuspended in 100 mM potassium phosphate (pH 7.4), and protein lysates prepared by French press. The soluble lysate was applied to a glutathione sepharose GSTrap FF 5 ml column (Amersham Biosciences) and washed extensively with PBS. MAC-CYP51 was cleaved from the GST fusion tag by the addition of 80 U of thrombin in PBS applied to the column and incubated at 22°C for 16 h, and purified cleaved MAC-CYP51 was eluted with PBS. Thrombin was removed from the eluted cleaved MAC-CYP51 using a HiTrap Benzamidine FF column (Amersham Biosciences), and buffer exchange was performed using a centricon YM30 (Millipore, Billerica, MA), so that the purified cleaved MAC-CYP51 protein was in 100 mM potassium phosphate (pH 7.4) containing 20% glycerol.
Spectral Analysis
The absolute, dithionate, and carbon monoxide reduced spectra were performed as described by Omura and Sato (26, 27). For the azole binding spectra, MAC-CYP51 was placed in cuvettes in a volume of 1 ml with increasing amounts of azole compounds. The sample was scanned from 500 to 380 nm with a Shimadzu UV-1601 spectrophotometer (Shimadzu Corporation, Kyoto, Japan). The difference spectra were generated in overlay mode by measuring the absorbance in the sample cuvette and subtracting the absorbance of the enzyme mixed with the vehicle control alone. The binding parameters (dissociation constant, Kd) for the interaction between azoles and MAC-CPY51 were determined from the difference spectra by plotting 1/Δabsorbance versus 1/azole concentration. Voriconazole and fluconazole were dissolved in water, whereas econazole, ketoconazole, and itraconazole were dissolved in DMSO.
Electron paramagnetic resonance (EPR) spectra of azole-bound and unbound MAC-CYP51 were measured on an ESP 300 EPR spectrometer (Bruker, Billerica, MA) equipped with an Oxford ESR-910 liquid helium cryostat (28). The microwave frequency was 9.5 GHz (x-band) at 15K, with power and modulation of 10 mW and 1 mT. MAC-CYP51 was at a concentration of 0.1 mM, and each azole concentration was 3 mM.
MAC-CYP51 Catalytic Activity
Lanosterol (Sigma) and obtusifoliol (a generous gift from Dr. Hubert Schaller) were dispersed in Triton WR1339 (29, 30). Purified MAC-CYP51 was incubated with 18 nmol spinach ferredoxin, 2 nmol ferredoxin-NADP reductase, and 2 mM reduced nicotinamide adenine dinucleotide phophate at 37°C for 4 h. Sterols were extracted in hexane, evaporated, followed by silyation with bis(trimethylsilyl)trifluoroacetamide (BSTFA) at 60°C for 1 h. The sterol substrates and metabolites were identified by gas chromatography/mass spectrometry (GC-MS) on a Finnigan Voyager GC-MS (Thermo Inc., San Jose, CA). The column was a DB-1MS 30 m × 0.25 mm ID with 0.25 μm film (J&W Scientific, Folsom, CA). The temperature program was operated at 120–295°C at 10°C/min using helium as the carrier gas.
Determination of MICs for Azole Compounds
A 60-μl aliquot of a 1 McFarland unit (1 × 108 organisms/ml) of M. avium subspecies avium American Type Culture Collection no. 25,291 was diluted in 12 ml of modified Middlebrook 7H9 broth containing oleic albumin dextrose catalase growth supplement. This suspension (100 μl; 5 × 104 organisms) was added to 96-well plates containing the various azoles. The plates were sealed and incubated at 37°C in a humidified incubator with 5% CO2 for 8 d. Growth was examined daily in control (no azoles added) and test plates. At 8 d, 15 μl of dimethylthiazoldiphenyltetrazolium bromide stock solution (5 mg/ml) was added to each well for 6 h at 37°C, followed by 100 μl of 10% SDS, and the plate was incubated at 37°C for 16 h. The absorbance at 562 nm of control and treated samples was recorded using a 96-well plate reader. All conditions were performed in triplicate. The MIC was determined as the lowest concentration of azole drug that caused at least a 99.6% reduction of MAC growth by Day 8 (31). Aliquots from control and test wells were plated on modified Middlebrook 7H9 agar and grown at 37°C for 8 d to determine bactericidal or bacteriostatic effects of the azoles.
RESULTS
Cloning, Expression, and Purification of MAC-CYP51
Degenerate PCR was used to amplify a fragment of the MAC-CYP51 gene, which has greatest homology to M. tuberculosis CYP51 (69% identity by BlastX analysis). This partial gene was then used to search the unannotated MAC genomic database at the Institute for Genomic Research, from which we were able to identify the start and stop codons for the full-length MAC-CYP51. We amplified the 1,356-bp complete MAC-CYP51 gene by PCR, sequenced it completely (GenBank Accession no. DQ195502), and subcloned it into the bacterial expression plasmid pGEX4T3 to create an N-terminal GST fusion. The translated MAC-CYP51 open-reading frame encodes a 51.4 kD protein with 451 amino acids, a size typical of other CYP51 proteins. The full-length MAC-CYP51 has greatest homology (BlastX) to M. tuberculosis CYP51 (76% identity), with low homology to human (34% identity) or Saccharomyces (29% identity) CYP51.
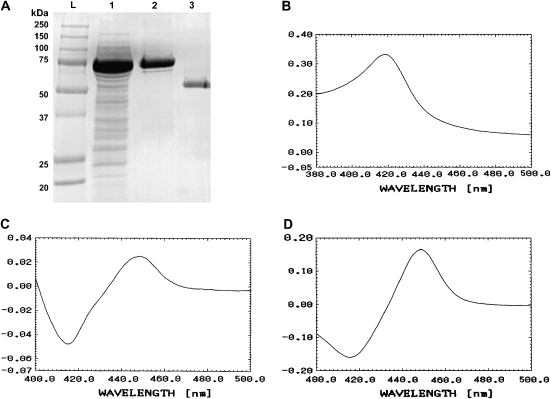
MAC-CYP51 protein was produced in E. coli. The GST fusion protein was column-purified, and MAC-CYP51 was obtained by thrombin cleavage of the GST tag. The migration of the purified MAC-CYP51 corresponded to its predicted molecular weight of 51.4 kD (Figure 1A). The purified MAC-CYP51 has the typical P450 absolute spectrum (Figure 1B), dithionate reduced spectrum (Figure 1C), and carbon monoxide reduced spectrum (Figure 1D) characteristic of this family of cytochrome.
Figure 1.
(A) SDS-PAGE analysis of Mycobacterium avium complex (MAC)-CYP51 expressed in Escherichia coli. Lane L is the molecular weight ladder, lane 1 is bacterial lysate, lane 2 is the glutathione S-transferase (GST)–purified fusion protein, and lane 3 is the thrombin-cleaved MAC-CYP51. (B) Absorption spectrum of absolute oxidized form of MAC-CYP51, (C) the dithionate reduced form, and (D) the carbon monoxide reduced form.
MAC-CYP51 Demethylase Activity
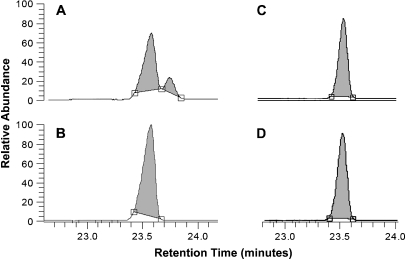
We reconstituted MAC-CYP51 activity in vitro using lanosterol and obtusifoliol as substrates in the presence of reduced nicotinamide adenine dinucleotide phophate. The demethylation activity of MAC-CYP51 on these substrates was analyzed by GC-MS. As shown in Figure 2A, MAC-CYP51 converts lanosterol to its demethylated product. The molecular ion of the silylated lanosterol is m/z 498 with a retention time of 23.5 min, and the molecular ion of the silylated demethylated lanosterol is m/z 482 with a retention time of 23.7 min. We did not observe any demethylation using obtusifoliol as a substrate (Figure 2C). In the control experiments using lanosterol or obtusifoliol without MAC-CYP51 (Figures 2B and 2D), there was no demethylation product. These data demonstrate that the MAC-CYP51 functions as a lanosterol 14α-demethylase.
Figure 2.
MAC-CYP51 demethylase activity. MAC-CYP51 activity was reconstituted in vitro using lanosterol and obtusifoliol and the demethylation activity of MAC-CYP51 was analyzed by GC-MS. As shown in (A), MAC-CYP51 converts lanosterol (m/z 498 with a retention time of 23.5 min) to its demethylated product (m/z 482 with a retention time of 23.7 min). No demethylation of obtusifoliol is observed (C). In the control experiments containing lanosterol (B) or obtusifoliol (D) without MAC-CYP51, there was no observed demethylation product.
Azole Binding Interactions with MAC-CYP51
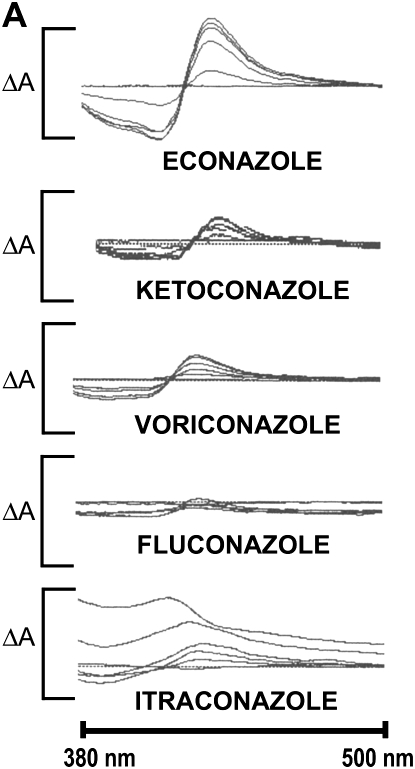
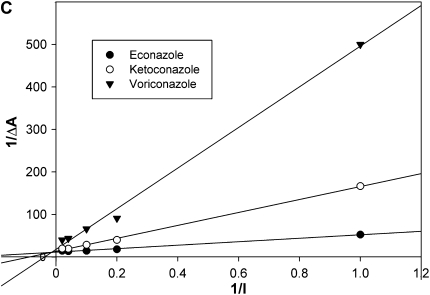
The direct interaction of the azole drugs econazole, ketoconazole, itraconazole, fluconazole, and voriconazole was examined for MAC-CYP51. Absorbance spectroscopy and EPR spectroscopy provides an accurate method of determining the direct binding of azole inhibitors to P450 enzymes such as MAC-CYP51. The nitrogen atom present in the azole ring binds to the heme cofactor present in the active site of MAC-CYP51, resulting in a shift of the absorption spectrum. This spectral shift creates a characteristic type II spectrum, with a peak at 435 nm and a trough at 412 nm. The resulting MAC-CYP51/azole complex can be titrated to provide a measurement of the azole inhibitor dissociation constant (Kd). We tested all of the azoles at concentrations of 0, 1, 5, 10, 25, or 50 μM in the presence of purified MAC-CYP51. As shown in Figure 3A, econazole demonstrated the best interactions with MAC-CYP51, followed by ketoconazole and voriconazole. Minimal binding interactions were detected for itraconazole and fluconazole. We calculated the azole Kd for econazole, ketoconazole, and voriconazole, but were unable to do so for fluconazole and itraconazole due to poor binding interactions with MAC-CYP51 (Figures 3B and 3C). The calculated Kd of MAC-CYP51 for econazole is 3.5 μM, with less affinity for ketoconazole at 11.3 μM, and still less affinity for voriconazole at 26.4 μM.
Figure 3.
MAC-CYP51 azole interactions. The direct interaction of the azole drugs econazole (solid line, circles), ketoconazole (dashed-dotted line, squares), itraconazole (long dashed line, diamonds), fluconazole (dashed line, “x”s), and voriconazole (dotted line, plus symbols) with MAC-CYP51 were examined by absorption spectroscopy (A-B). Econazole demonstrated the best interactions with MAC-CYP51, followed by ketoconazole and voriconazole. Minimal binding interactions were detected for itraconazole and fluconazole. The absorption change depicted on the y axis (ΔA) is 0.13 (0.08 to −0.05). The inhibitor binding constant (Kd) for econazole (3.5 μM), ketoconazole (11.3 μM), and voriconazole (26.4 μM) were calculated, but could not be calculated for fluconazole and itraconazole due to poor binding (C).
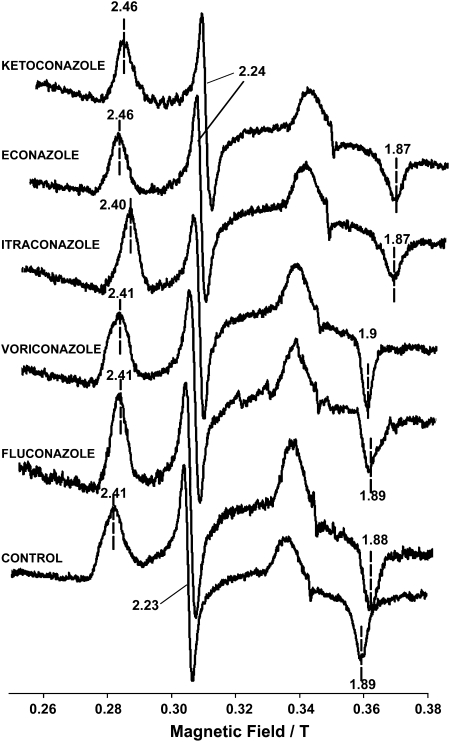
EPR spectroscopy is a powerful technique for demonstrating the direct interaction of MAC-CYP51 with azoles. EPR measures the absorption of microwave radiation by an unpaired electron when it is placed in a strong magnetic field, and spectra are obtained by measuring the absorption of the microwave radiation while scanning the strength of the magnetic field (g values). Azole inhibitors binding to the heme iron of the cytochrome will shift the gz to low field and gx to high field, whereas lack of binding will cause no effect in these values (28). As shown in Figure 4, the set of g values (gz, gy, gx) for the unbound MAC-CYP51 are 2.41, 2.23, and 1.89, respectively. Binding of econazole or ketoconazole resulted in g values (gz, gy, gx) of 2.46, 2.24, 1.87, respectively. There was no significant spectral change detected for fluconazole, voriconazole, or itraconazole. The EPR values for unbound and azole-bound MAC-CYP51 are shown in Table 1.
Figure 4.
Electron paramagnetic resonance (EPR) spectra of azole bound and unbound to MAC-CYP51. EPR spectra of MAC-CYP51 0.1 mM (control) or MAC-CYP51 0.1 mM bound to various azoles (3 mM each of econazole, ketoconazole, itraconazole, voriconazole, fluconazole) were obtained at a microwave frequency of 9.5 GHz (X-band) at 15K, with power and modulation of 10 mW and 1 mT. Direct binding interactions with MAC-CYP51 were noted only with econazole and ketoconazole.
TABLE 1.
ELECTRON PARAMAGNETIC RESONANCE g VALUES FOR MYCOBACTERIUM AVIUM COMPLEX–CYP51 BOUND OR UNBOUND TO AZOLES*
| gz | gy | gx | |
|---|---|---|---|
| Control | 2.41 | 2.23 | 1.89 |
| Unbound MAC-CYP51 | |||
| Econazole | 2.46 | 2.24 | 1.87 |
| Ketoconazole | 2.46 | 2.24 | 1.87 |
| Fluconazole | 2.41 | 2.23 | 1.88 |
| Voriconazole | 2.41 | 2.23 | 1.89 |
| Itraconazole | 2.40 | 2.23 | 1.9 |
Definition of abbreviation: MAC, Mycobacterium avium complex.
Electron paramagnetic resonance (EPR) was performed at a microwave frequency of 95 GHz (X-band) at 15K, with power and modulation of 10 mW and 1 mT. Econazole and ketoconazole binding to MAC-CYP51 shifted the gz to low field and gx to high field, whereas no significant effect was seen for fluconazole, voriconazole, or itraconazole.
Growth Inhibition of M. avium with Azoles
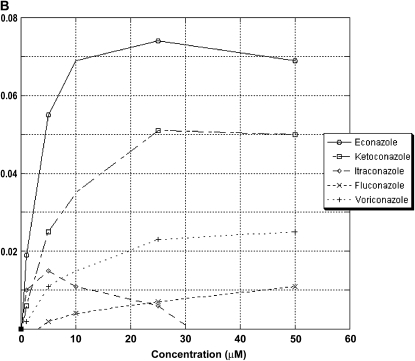
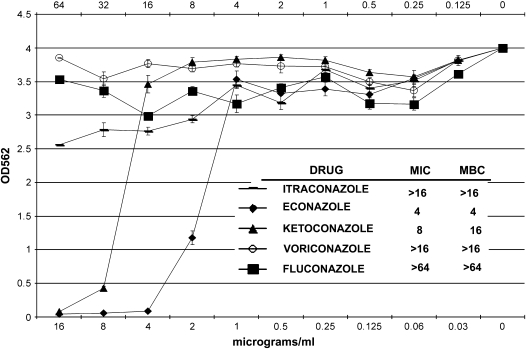
There are no universally accepted guidelines for laboratory susceptibility testing of MAC. The Clinical Laboratory Standards Institute suggests that MAC susceptibility testing be performed with a broth-based method using either macrodilution or microtiter dilution (32). We used a highly sensitive and quantitative microtiter broth-based dilution assay to test the ability of azoles to inhibit MAC growth and to determine MIC and MBC (31). This assay is based on the ability of metabolically active MAC organisms to reduce dimethylthiazoldiphenyltetrazolium bromide to formazan by their mitochondrial dehydrogenases. The MIC for the azoles was determined as the lowest concentration of drug that inhibited mycobacterial growth by at least 99.6% over 8 d of treatment. The percent reductions were taken from azole-treated MAC compared with untreated MAC. The azoles tested at standard doses were econazole, ketoconazole, itraconazole, and voriconazole (0–16 μg/ml) and fluconazole (0–64 μg/ml). We found that econazole had a MIC of 4 μg/ml and an MBC of 4 μg/ml (Figure 5). In contrast, with ketoconazole, we observed an MIC of 8 μg/ml and an MBC of 16 μg/ml. Itraconazole, voriconazole, and fluconazole did not inhibit MAC growth to any significant extent.
Figure 5.
Growth inhibitory effect of azoles on M. avium. Econazole, ketoconazole, itraconazole, and voriconazole were tested from 0 to 16 μg/ml, and fluconazole was tested from 0 to 64 μg/ml. Econazole was found to have a minimum inhibitory concentration (MIC) of 4 μg/ml and a minimum bacteriocidal concentration (MBC) of 4 μg/ml, whereas ketoconazole had an MIC of 8 μg/ml and an MBC of 16 μg/ml. Itraconazole, voriconazole, and fluconazole did not inhibit MAC growth to any significant extent.
DISCUSSION
The results of this study demonstrate that the pulmonary pathogen M. avium contains a gene with homology to CYP51 that, when expressed as a recombinant protein, exhibits spectral properties consistent with cytochrome P450 enzymes, and catalyzes the removal of a methyl group from lanosterol. MAC-CYP51 interacts with the azoles econazole and ketoconazole in vitro, producing typical type II binding absorption spectra and characteristic EPR spectra. We also found that econazole and ketoconazole inhibit MAC growth in culture, whereas the other tested azoles had no effect. As a key enzyme in sterol biosynthesis, CYP51 has been a target for antifungal drug design (24, 33). Azole compounds have proven efficacy for treating localized and systemic fungal infections; however, not all fungi respond to individual azole agents. Although CYP51 proteins isolated from bacteria through mammals have highly conserved regions, significant structural variability occurs in regions of the enzyme associated with the binding of sterol substrates and azole inhibitors (34, 35). For example, CYP51 from human and Candida albicans both catalyze the demethylation of lanosterol, but with significantly different enzymatic activity (36). In contrast, CYP51 from Sorghum bicolor has a strict substrate specificity and selectivity for obtusifoliol, and cannot catalyze the demethylation of lanosterol (37). For MAC-CYP51, we found it could demethylate lanosterol but not the phytosterol obtusifoliol.
The difference in the molecular structure of azole compounds affects their solubility and their ability to bind and inhibit CYP51 enzymes (33). Econazole and ketoconazole are imidazoles, which have five-membered ring structures containing two nitrogen atoms. Fluconazole, itraconazole, and voriconazole are triazoles, which have five-membered ring structures containing three nitrogen atoms. Econazole, ketoconazole, and itraconazole are highly lipophilic compounds, whereas fluconazole and voriconazole are very water soluble. The nitrogen atom in the azole ring coordinates binding of the azole to the heme cofactor present in the active site of CYP51 enzymes. Interestingly, we found that the best in vitro interactions of the azole compounds with MAC-CYP51, as determined by type II spectral analysis and EPR, was for the imidazoles econazole (Kd = 3.5) and ketoconazole (Kd = 11.3). The triazoles demonstrated significantly less binding to MAC-CYP51. Econazole and ketoconazole were also the most effective in inhibiting the growth of MAC in culture.
Three-dimensional computer modeling has been applied to the study of yeast CYP51 in an attempt to design novel compounds to specifically inhibit fungal CYP51 preferentially over mammalian CYP51. The inherent molecular differences in CYP51 enzymes between different organisms make such an approach feasible. Highly specific nonazole compounds were designed to inhibit Candida ERG11 in this manner (38). The crystal structure of M. tuberculosis CYP51 (MTCYP51) reveals several unique structural features, which explains the differences noted between MTCYP51 and human and fungal CYP51s (20, 39). For example, MTCYP51 prefers obtusifoliol as a substrate, yet can also demethylate lanosterol. MTCYP51 was found to have similar affinities for binding ketoconazole and fluconazole, with calculated Kd values of 19 μM and 20 μM, respectively (40). In contrast to MTCYP51, we found that MAC-CYP51 demethylated lanosterol but not obtusifoliol, and had strong affinity for econazole and ketoconazole (Kd = 3.5 μM and 11.3 μM, respectively), but poor affinity for fluconazole. It is conceivable that these differences are due to structural differences between MAC-CYP51 and MTCYP51, as there is only a 76% identity between both proteins on the amino acid level. Similar to our data, however, is that the CYP51 from the nonpathogenic M. smegmatis has high affinity for econazole and ketoconazole, but does not interact at all with fluconazole (21, 41). Econazole was found to be bactericidal for M. smegmatis grown in culture. Additionally, a recent study showed that the biosynthesis of M. smegmatis glycopeptidolipids is inhibited with econazole and clotrimazole treatment, and that both azoles inhibited M. smegmatis growth with MIC values of 2 and 0.5 μg/ml, respectively; the authors speculate that inhibition of the CYP51 from M. smegmatis is involved in the biosynthesis of glycopeptidolipids (42).
In summary, the inhibition of mycobacterial sterol metabolism through the binding of compounds to MAC-CYP51 may provide new treatment options for patients infected with MAC. The application of computer modeling to MAC-CYP51 may aid in the design of novel agents specific for MAC-CYP51 inhibition preferentially over human CYP51, thereby improving efficacy and limiting toxicity.
Acknowledgments
The authors thank Dr. John D. Lipscomb, University of Minnesota, for assistance with performing the EPR analysis. Obtusifoliol was provided as a generous gift from Dr. Hubert Schaller, Institut de Biologie Moléculaire des Plantes du Centre national de la Recherche scientifique, Strasbourg Cedex, France. The authors also appreciate the efforts of David T. Lynch from the Mayo Mycobacteriology Laboratory, and Jerry D. Dewey from the General Clinic Research Centers GC-MS facility for their expert technical assistance.
M.P.P. and P.K.V. contributed equally to this work.
This work was supported by funds from Mayo Foundation and General Clinic Research Centers Program grant M01-RR00585.
Originally Published in Press as DOI: 10.1165/rcmb.2005-0398OC on March 16, 2006
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.American Thoracic Society. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. This official statement of the American Thoracic Society was approved by the Board of Directors, March 1997. Medical Section of the American Lung Association. Am J Respir Crit Care Med 1997;156(Suppl. 2 pt 2):S1–S25. [DOI] [PubMed] [Google Scholar]
- 2.Prince DS, Peterson DD, Steiner RM, Gottlieb JE, Scott R, Israel HL, Figueroa WG, Fish JE. Infection with Mycobacterium avium complex in patients without predisposing conditions. N Engl J Med 1989;321:863–868. [DOI] [PubMed] [Google Scholar]
- 3.Aksamit TR. Mycobacterium avium complex pulmonary disease in patients with pre-existing lung disease. Clin Chest Med 2002;23:643–653. [DOI] [PubMed] [Google Scholar]
- 4.Swensen SJ, Hartman TE, Williams DE. Computed tomographic diagnosis of Mycobacterium avium–intracellulare complex in patients with bronchiectasis. Chest 1994;105:49–52. [DOI] [PubMed] [Google Scholar]
- 5.Field SK, Fisher D, Cowie RL. Mycobacterium avium complex pulmonary disease in patients without HIV infection. Chest 2004;126:566–581. [DOI] [PubMed] [Google Scholar]
- 6.Karakousis PC, Moore RD, Chaisson RE. Mycobacterium avium complex in patients with HIV infection in the era of highly active antiretroviral therapy. Lancet Infect Dis 2004;4:557–565. [DOI] [PubMed] [Google Scholar]
- 7.Horsburgh CR Jr, Selik RM. The epidemiology of disseminated nontuberculous mycobacterial infection in the acquired immunodeficiency syndrome (AIDS). Am Rev Respir Dis 1989;139:4–7. [DOI] [PubMed] [Google Scholar]
- 8.Kobashi Y, Matsushima T. Clinical analysis of pulmonary Mycobacterium avium complex disease in association with corticosteroid treatment. J Infect Chemother 2003;9:68–74. [DOI] [PubMed] [Google Scholar]
- 9.Doucette K, Fishman JA. Nontuberculous mycobacterial infection in hematopoietic stem cell and solid organ transplant recipients. Clin Infect Dis 2004;38:1428–1439. [DOI] [PubMed] [Google Scholar]
- 10.Aksamit TR. Hot tub lung: infection, inflammation, or both? Semin Respir Infect 2003;18:33–39. [DOI] [PubMed] [Google Scholar]
- 11.Marras TK, Wallace RJ Jr, Koth LL, Stulbarg MS, Cowl CT, Daley CL. Hypersensitivity pneumonitis reaction to Mycobacterium avium in household water. Chest 2005;127:664–671. [DOI] [PubMed] [Google Scholar]
- 12.Reich JM, Johnson RE. Mycobacterium avium complex pulmonary disease: incidence, presentation, and response to therapy in a community setting. Am Rev Respir Dis 1991;143:1381–1385. [DOI] [PubMed] [Google Scholar]
- 13.Goto M, Katsunuma N. Relationship between porphyrin and isoniazid resistance of Mycobacterium avium. Nature 1958;181:916. [DOI] [PubMed] [Google Scholar]
- 14.Heifets LB, Iseman MD, Crowle AJ, Lindholm-Levy PJ. Pyrazinamide is not active in vitro against Mycobacterium avium complex. Am Rev Respir Dis 1986;134:1287–1288. [DOI] [PubMed] [Google Scholar]
- 15.De Wit S, D'Abbraccio M, De Mol P, Clumeck N. Acquired resistance to clarithromycin as combined therapy in Mycobacterium avium intracellulare infection. Lancet 1993;341:53–54. [PubMed] [Google Scholar]
- 16.Yoshida Y, Aoyama Y, Noshiro M, Gotoh O. Sterol 14-demethylase P450 (CYP51) provides a breakthrough for the discussion on the evolution of cytochrome P450 gene superfamily. Biochem Biophys Res Commun 2000;273:799–804. [DOI] [PubMed] [Google Scholar]
- 17.Aoyama Y, Noshiro M, Gotoh O, Imaoka S, Funae Y, Kurosawa N, Horiuchi T, Yoshida Y. Sterol 14-demethylase P450 (P45014DM*) is one of the most ancient and conserved P450 species. J Biochem (Tokyo) 1996;119:926–933. [DOI] [PubMed] [Google Scholar]
- 18.Fischer RT, Stam SH, Johnson PR, Ko SS, Magolda RL, Gaylor JL, Trzaskos JM. Mechanistic studies of lanosterol 14 α-methyl demethylase: substrate requirements for the component reactions catalyzed by a single cytochrome P-450 isozyme. J Lipid Res 1989;30:1621–1632. [PubMed] [Google Scholar]
- 19.Fischer RT, Trzaskos JM, Magolda RL, Ko SS, Brosz CS, Larsen B. Lanosterol 14 α-methyl demethylase: isolation and characterization of the third metabolically generated oxidative demethylation intermediate. J Biol Chem 1991;266:6124–6132. [PubMed] [Google Scholar]
- 20.Bellamine A, Mangla AT, Nes WD, Waterman MR. Characterization and catalytic properties of the sterol 14α-demethylase from Mycobacterium tuberculosis. Proc Natl Acad Sci USA 1999;96:8937–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson CJ, Lamb DC, Marczylo TH, Parker JE, Manning NL, Kelly DE, Kelly SL. Conservation and cloning of CYP51: a sterol 14 α-demethylase from Mycobacterium smegmatis. Biochem Biophys Res Commun 2003;301:558–563. [DOI] [PubMed] [Google Scholar]
- 22.Lamb DC, Kelly DE, Manning NJ, Kelly SL. A sterol biosynthetic pathway in Mycobacterium. FEBS Lett 1998;437:142–144. [DOI] [PubMed] [Google Scholar]
- 23.Hitchcock CA, Dickinson K, Brown SB, Evans EG, Adams DJ. Interaction of azole antifungal antibiotics with cytochrome P-450–dependent 14 α-sterol demethylase purified from Candida albicans. Biochem J 1990;266:475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev 1999;12:501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales IJ, Vohra PK, Puri V, Kottom TJ, Limper AH, Thomas CF Jr. Characterization of a lanosterol 14 α-demethylase from Pneumocystis carinii. Am J Respir Cell Mol Biol 2003;29:232–238. [DOI] [PubMed] [Google Scholar]
- 26.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I: evidence for its hemoprotein nature. J Biol Chem 1964;239:2370–2378. [PubMed] [Google Scholar]
- 27.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. II: solubilization, purification, and properties. J Biol Chem 1964;239:2379–2385. [PubMed] [Google Scholar]
- 28.Lipscomb JD. Electron paramagnetic resonance detectable states of cytochrome P-450cam. Biochemistry 1980;19:3590–3599. [DOI] [PubMed] [Google Scholar]
- 29.Schaller H. The role of sterols in plant growth and development. Prog Lipid Res 2003;42:163–175. [DOI] [PubMed] [Google Scholar]
- 30.Xu SH, Norton RA, Crumley FG, Nes WD. Comparison of the chromatographic properties of sterols, select additional steroids and triterpenoids: gravity-flow column liquid chromatography, thin-layer chromatography, gas–liquid chromatography and high-performance liquid chromatography. J Chromatogr 1988;452:377–398. [DOI] [PubMed] [Google Scholar]
- 31.Gomez-Flores R, Gupta S, Tamez-Guerra R, Mehta RT. Determination of MICs for Mycobacterium avium–M. intracellulare complex in liquid medium by a colorimetric method. J Clin Microbiol 1995;33:1842–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clinical Laboratory Standards Institute. Susceptibility testing of Mycobacteria, Nocardiae, and other aerobic actinomycetes; approved standard. Wayne, PA: CLSI; 2003. CLSI document M24-A. [PubMed]
- 33.Lamb D, Kelly D, Kelly S. Molecular aspects of azole antifungal action and resistance. Drug Resist Updat 1999;2:390–402. [DOI] [PubMed] [Google Scholar]
- 34.Aoyama Y, Yoshida Y. Different substrate specificities of lanosterol 14a-demethylase (P- 45014DM) of Saccharomyces cerevisiae and rat liver for 24-methylene-24,25-dihydrolanosterol and 24,25-dihydrolanosterol. Biochem Biophys Res Commun 1991;178:1064–1071. [DOI] [PubMed] [Google Scholar]
- 35.Aoyama Y, Yoshida Y, Sonoda Y, Sato Y. Structural analysis of the interaction between the side-chain of substrates and the active site of lanosterol 14 α-demethylase (P- 450(14)DM) of yeast. Biochim Biophys Acta 1992;1122:251–255. [DOI] [PubMed] [Google Scholar]
- 36.Lamb DC, Kelly DE, Kelly SL. Molecular diversity of sterol 14α-demethylase substrates in plants, fungi and humans. FEBS Lett 1998;425:263–265. [DOI] [PubMed] [Google Scholar]
- 37.Cabello-Hurtado F, Taton M, Forthoffer N, Kahn R, Bak S, Rahier A, Werck-Reichhart D. Optimized expression and catalytic properties of a wheat obtusifoliol 14α-demethylase (CYP51) expressed in yeast: complementation of erg11Delta yeast mutants by plant CYP51. Eur J Biochem 1999;262:435–446. [DOI] [PubMed] [Google Scholar]
- 38.Ji H, Zhang W, Zhang M, Kudo M, Aoyama Y, Yoshida Y, Sheng C, Song Y, Yang S, Zhou Y, et al. Structure-based de novo design, synthesis, and biological evaluation of non-azole inhibitors specific for lanosterol 14α-demethylase of fungi. J Med Chem 2003;46:474–485. [DOI] [PubMed] [Google Scholar]
- 39.Podust LM, Poulos TL, Waterman MR. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc Natl Acad Sci USA 2001;98:3068–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuura K, Yoshioka S, Tosha T, Hori H, Ishimori K, Kitagawa T, Morishima I, Kagawa N, Waterman MR. Structural diversities of active site in clinical azole-bound forms between sterol 14α-demethylases (CYP51s) from human and Mycobacterium tuberculosis. J Biol Chem 2005;280:9088–9096. [DOI] [PubMed] [Google Scholar]
- 41.Jackson CJ, Lamb DC, Kelly DE, Kelly SL. Bactericidal and inhibitory effects of azole antifungal compounds on Mycobacterium smegmatis. FEMS Microbiol Lett 2000;192:159–162. [DOI] [PubMed] [Google Scholar]
- 42.Burguiere A, Hitchen PG, Dover LG, Dell A, Besra GS. Altered expression profile of mycobacterial surface glycopeptidolipids following treatment with the antifungal azole inhibitors econazole and clotrimazole. Microbiol 2005;151:2087–2095. [DOI] [PubMed] [Google Scholar]