Abstract
Eosinophils migrate from the vascular circulation to the inflamed airways during asthma exacerbations. While the mechanism(s) of this process is not known, the expression of urokinase-type plasminogen activator receptor (uPAR) has been found to modulate neutrophil adhesion and migration to inflammatory sites. We hypothesized that increased expression of uPAR and its ligand, uPA, enhance eosinophil adhesion in patients with asthma. Patients with allergic asthma underwent segmental bronchoprovocation with allergen; 48 h later, peripheral blood and airway (from bronchoalveolar lavage fluid) eosinophils were isolated. uPA and uPAR protein expression were measured by flow cytometry and Western blot; mRNA was quantified by real-time PCR. Eosinophil adhesion to intercellular adhesion molecule (ICAM)-1 and vascular cell adhesion molecule (VCAM)-1 was assessed by eosinophil peroxidase activity. Airway eosinophils expressed significantly more uPA and uPAR protein and uPAR mRNA than peripheral blood eosinophils. Removal of cell-bound uPA and/or addition of exogenous uPA had no effect on blood eosinophil adhesion to ICAM-1 or VCAM-1. In contrast, exogenous uPA stimulated ICAM and VCAM adhesion of airway eosinophils. N-formyl-methionyl-leucyl-phenylalanine–activated airway eosinophil adherence to VCAM-1 and ICAM-1 (VCAM-1, 52.8 ± 4.7%; ICAM-1, 49.2 ± 5.3%) was increased over blood eosinophil adhesion (VCAM-1, 38.4 ± 3.6%; ICAM-1, 27.7 ± 4.9%; P < 0.05). Removal of cell-bound uPA from airway eosinophils decreased adhesion to blood cell levels; reintroduction of exogenous uPA completely restored adhesion levels. These data suggest that constitutive uPA primes, and exogenous uPA can activate, airway eosinophil adhesion following segmental allergen challenge and that increased uPA expression may be a mechanism of increased eosinophil infiltration and function in asthma.
Keywords: adhesion, asthma, eosinophil, priming, urokinase-type plasminogen activator
Loss of disease control in asthma results in an initial influx of neutrophils to the airway that is usually followed by a later, selective influx of eosinophils. Both of these granulocyte populations promote airway inflammation through their release of granule proteins and generation of reactive oxygen metabolites. To manifest these functions, however, the cells must first adhere to and traverse the pulmonary vasculature followed by migration into, then through, the interstitial tissue. This movement from the circulation to the airway is dependent on the interaction of granulocyte integrins (such as αMβ2, α4β1) with vascular endothelial cell adhesion molecules (intercellular adhesion moelcule [ICAM]-1 and vascular cell adhesion molecule [VCAM]-1) and extracellular matrix proteins. Moreover, granulocyte functions including degranulation and respiratory burst are dependent on activation of these same integrins (1, 2). Therefore, physiologic agents that modulate integrin expression and activation may regulate granulocyte inflammation in asthma.
Urokinase-type plasminogen activator receptor (uPAR) and its ligand uPA are components of a proteolytic system involving the activation of uPAR-bound pro-uPA and its subsequent conversion of cell-bound plasminogen to plasmin (3). In addition to their proteolytic role, reversible interactions among uPA, uPAR, and integrins promote inflammation through a nonproteolytic mechanism of increased leukocyte adhesion, chemotaxis, respiratory burst, and degranulation (4, 5). uPA and/or uPAR can each interact with β1, β2, β3, and β5 integrins resulting in modulation of each component's activity and expression (6). In vivo and in vitro recruitment of neutrophils and eosinophils in murine models of inflammation is dependent on both αMβ2 integrin and membrane-expressed uPAR (4, 7). uPA and uPAR have been associated with multiple inflammatory diseases and have been proposed as a mechanism of eosinophil recruitment in asthma (8, 9).
We have previously reported that airway eosinophils isolated from bronchoalveolar lavage (BAL) fluid 48 h after segmental bronchoprovocation with allergen (SBP-AG) are primed for increased n-formyl-methionyl-leucyl-phenylalanine (FMLP)-activated adhesion to human umbilical vein endothelial cell monolayers, respiratory burst, and intracellular calcium flux (10). In addition to integrins, uPAR/uPA can form multiprotein complexes with other G protein–linked receptors such as the 7-transmembrane FRPL receptors for FMLP. Sitrin and associates (5) reported that crosslinking uPA-occupied uPAR not only activated the respiratory burst but also primed the FMLP response of human neutrophils. Similarly, Cao and coworkers (11) reported that uPA can increase cytosolic calcium levels and prime human neutrophil FMLP-stimulated respiratory burst in the presence of both uPAR and αMβ2 integrin. Therefore, changes in uPAR and uPA expression may provide a mechanism of enhanced eosinophil participation in airway inflammation in asthma.
Unlike neutrophils, peripheral blood eosinophils express very low levels of uPAR; this expression can be enhanced by in vitro incubation with IL-5 or PAF, modulates in vitro cell chemotaxis, and has been suggested as a mechanism of eosinophil recruitment in asthma (8, 9, 12). We hypothesized that enhanced expression of uPAR and ligation of uPA by airway eosinophils primes these cells for increased cell adhesion. In this study, peripheral blood and airway eosinophils were isolated 48 h after SBP-AG challenge and compared for their expression of uPAR and uPA, and for in vitro adhesion to ICAM-1 and VCAM-1. Although addition of exogenous uPA increased cell-bound uPA on both blood and airway eosinophils, only airway eosinophils demonstrated increased uPA-activated and uPA-primed FMLP-activated adhesion to recombinant human (rh) ICAM-1 and VCAM-1.
MATERIALS AND METHODS
Reagents
Percoll was purchased from Amersham Pharmacia (Piscataway, NJ). PBS, newborn calf serum, FBS, and RPMI were obtained from Invitrogen Life Technologies (Carlsbad, CA). Hanks' balanced salt solution (HBSS) and recombinant human (rh) RANTES, eotaxin, GM-CSF, and IL-8 were purchased from Biosource International (Camarillo, CA). rh VCAM-1, ICAM-1, IL-5 and neutralizing mouse anti-human uPAR monoclonal antibody (mAb; clone: 62,022) were obtained from R&D Systems (Minneapolis, MN). High molecular weight (HMW) tc-uPA and mouse anti-human uPA monoclonal antibodies (mAb) were obtained from American Diagnostica (Stamford, CT). Fluorescent-labeled goat anti-mouse IgG and mouse isotype controls for flow cytometry were obtained from BD Biosciences (San Jose, CA). FMLP, platelet-activating factor (PAF), and phorbol myristate acetate (PMA) were from Sigma Chemical Co. (St. Louis, MO). All other reagents were obtained from Sigma unless otherwise stated.
Human Subjects
Eosinophils were isolated from peripheral blood samples of healthy normal volunteers and subjects with allergic rhinitis or mild allergic asthma. Subject age ranged from 18–55 yr, and sex distribution was equal. Immediate hypersensitivity was demonstrated by at least one positive skin reaction (> 3 mm) by the prick-puncture technique to extracts of common allergens including ragweed, house dust mite, grass pollen, cat dander, and dog dander (Hollister-Stier, Spokane, WA). Determination of asthma was based on a prior physician's diagnosis. Subjects with allergic rhinitis or asthma were not taking any systemic or inhaled medications other than short-acting inhaled β-agonists as needed. A subgroup of subjects with asthma taking long-term (> 1 yr) inhaled corticosteroids (ICS) was only included for group comparisons and did not undergo SBP-AG or BAL. Written informed consent was obtained from each subject before participation in the study, which was approved by the University of Wisconsin Internal Review Board.
Segmental Bronchoprovocation and BAL
At least 1 month before SBP-AG, qualifying subjects with asthma underwent skin testing to determine relevant AG as well as pulmonary function tests to determine percent predicted FEV1, forced vital capacity (FVC), and the dose of AG needed to drop FEV1 by 20% (AGPD20; Madison Scientific Software, Wexford, PA). Airway responsiveness was determined as the methacholine dose needed to drop FEV1 by 20% (MethPC20). Reversibility was defined as the percent increase in FEV1 after two puffs of albuterol. SBP-AG and BAL were performed as previously described (13). Briefly, two bronchopulmonary segments were identified for SBP-AG and a wedge position was achieved for each with the fiberoptic bronchoscope. A baseline BAL was performed, then AG was instilled into each segment through the wedged bronchoscope. One segment received a 10% dose of AGPD20. If this dose was well tolerated, a second segment was challenged with 20% AGPD20. After 48 h, bronchoscopy was repeated and BAL was performed on each of the two previously challenged segments. BAL fluid from the two AG-challenged segments was pooled for analysis. Heparinized venous blood was drawn immediately before the 48 h post-AG bronchoscopy.
Airway Eosinophil Isolation
Airway cells from BAL fluid were washed in HBSS (without Ca+2) + 2% newborn calf serum and fractionated over a 1.085/1.100 g/ml Percoll gradient as previously described (13). The cells at the 1.085/1.100 g/ml interface were collected, washed, and contained > 98% viable eosinophils; contaminating cells were predominantly lymphocytes.
Blood Eosinophil Isolation
Heparinized peripheral blood was density fractionated over 1.090 g/ml Percoll, and highly pure eosinophils were isolated from the resulting granulocyte pellet by negative selection using anti-CD16–, anti-CD3–, and anti-CD14–labeled magnetic beads (Miltenyi Biotec, Auburn, CA) as described previously (14). Eosinophil purity and viability were > 99%; neutrophils were the major contaminating cell.
Blood Neutrophil Isolation
Neutrophils were included as a positive control for uPAR and uPA expression. Neutrophils were isolated during the blood eosinophil separation described above. In subjects with eosinophils comprising less than five percent of the granulocyte pellet, the granulocytes are used as neutrophils without further purification. In subjects with greater than or equal to five percent eosinophils in the granulocyte pellet, neutrophils were obtained by positive selection with anti-CD16-labeled magnetic beads. No differences in uPAR or uPA expression were observed between these two neutrophil populations.
Detection of uPAR Expression
Flow cytometry.
Isolated neutrophil and eosinophil populations were incubated 30 min at 4°C with PE-labeled anti-human uPAR mAb or with unlabeled mouse anti-human uPA followed by FITC-labeled goat anti-mouse Ig secondary polyclonal Ab along with the appropriate isotype controls. Ten thousand events gated for viability and specific cell type were collected and data were recorded as percent positive cells (% positive) with isotype control values subtracted from each measure. Cell events that exceeded the 95 percentile of the isotype control scan were defined as % positive cells. The geometric mean fluorescence was also measured and reflected the same changes as the % positive data.
Western blot.
Purified eosinophils or neutrophils were suspended in RIPA buffer (1% Triton-X-100, 0.25% deoxycholic acid, 0.1% SDS, 20 mM TRIS pH 7.4, 137 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, 20 mM β-glycerophosphate, 10 mM NaF, and a cocktail of protease inhibitors) at a concentration of 25 × 106 cells/ml and frozen. The cell lysates were thawed and sonicated, and the insoluble cellular material was removed by centrifugation at 4°C for 10 min at 14,000 × g. The cell lysates were then diluted with electrophoresis sample buffer (15) and lysate volumes equivalent to 0.3 × 106 cells per lane were added to the wells of a 12.5% SDS-polyacrylamide gel. The proteins were resolved electrophoretically and transferred to PVDF membranes. The membranes were blocked for 16 h at 4°C with 2% BSA in 10 mM Tris-buffered saline (pH 8.0) with 0.1% Tween 20 (TBST). The membrane was incubated for 2 h at ambient temperature with 0.2 μg/ml biotinylated anti-human uPAR (Cat. #BAF807; R&D Systems, Minneapolis, MN) in TBST with 0.2% BSA. The PVDF membrane was then washed and incubated for 1 h with a 1/5,000 dilution of horseradish peroxidase–conjugated streptavidin conjugate (Zymed Laboratories, San Francisco, CA). Antibody labeling of the immobilized proteins was visualized after exposure of the membrane to chemoluminescent substrate reagents (Pierce Biotechnology, Rockford, IL) and the image was digitally photographed. Images were quantified by densitometry using Image J software (version 1.32; National Institutes of Health, Bethesda, MD). The background density was subtracted and immunoblot density values for each uPAR band were normalized to the value of the low molecular weight (LMW) uPAR (35–50 kD) isoforms from neutrophils of the same subject.
Soluble uPAR
Pre– and post–SBP-AG BAL fluids were diluted 1:5 for measurement of suPAR by ELISA (R&D Systems) as per manufacturer's directions. Assay sensitivity was 62.5 pg/ml and data are presented for 1× BAL fluid. Blood eosinophils were incubated in 50% pre– or post–SBP-AG BAL fluid in culture medium (RPMI + 5% FCS + 100 U/ml penicillin and 100 μg/ml streptomycin) for 24 h at 37°C. Cellular expression of uPAR was then determined by flow cytometry.
Detection of uPAR mRNA in Human Eosinophils by Gene Microarray
mRNA from purified human eosinophils was analyzed for the expression of multiple transcripts by hybridization to Affymetrix HuGeneFL oligonucleotide microarrays as previously described (16). Among the transcripts evaluated were human uPAR, as identified by hybridization to probe sets derived from GenBank Accession no. U09937, and uPA (GenBank Accession no. X02419). The hybridization values were expressed as the Model-based Expression Index (MBEI) using dCHip software (Beta test version) (17). Data were summarized as the mean of the MBEI for data on two independent eosinophil sample pools.
Total RNA Extraction and Reverse Transcription
Total RNA was extracted from 4.5 × 106 cells using a one-step phenol/chloroform extraction reagent (Tri Reagent; Sigma) according to manufacturer's instructions. The total RNA was resuspended in 20 μl of nuclease-free water (Promega, Madison, WI). Reverse transcription was then performed by combining 16 μl total RNA solution with 1 μg random hexamer primers (Promega) for 2 min at 70°C, followed by the addition of 400 U reverse transcriptase (SuperScript II RT; Invitrogen, Carlsbad, CA), 8 μl of 5× reaction buffer, 4 μl of 0.01 M dithiothreitol (Invitrogen), 80 U recombinant RNasin (Promega), and 0.02 μmol dNTPs (Promega), in a total volume of 42 μl incubated at 37°C for 1 h, then 94°C for 5 min. Finally, 98 μl of nuclease-free water was added to each tube of cDNA.
Quantitative Real-Time PCR for Detection of uPAR and β-Actin mRNA
Real-time PCR, used to assay mRNA, was performed on the ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). The thermal cycler protocol consisted of 50°C for 2 min, 95°C for 10 min, then 95°C for 15 s and 60°C for 1 min repeated for 40 cycles. The reaction mixture included 1× TaqMan Universal PCR Master Mix and pre-developed TaqMan Assay Reagents (Applied Biosystems) for [1×] h-β-actin (cat no. 4326315E) and [1×] uPAR (cat no. Hs00182181). Primers and probe were designed to span an exon and will not detect genomic DNA; a DNase treatment, therefore, was not needed. Duplicate 25-μl samples containing 5 μl of cDNA were analyzed in a 96-well plate with an optical cover. A standard curve was developed from bronchial epithelial cells (obtained from lung transplant donors, two to three passages). The relative standard curve was included in each assay to calculate quantity, check the assay for linearity and check primer and probe efficiency. uPAR mRNA was evaluated using two standard curves, one with epithelial cells cultured from lung transplant donors and the other using the SPB21 cell line (a generous gift from J. Jakway, Shering-Plough, Kenilworth, NJ). Results using the two methods were consistent with each other and the epithelial cell values were used in the reported data. Values are reported as the ratio of uPAR mRNA to β-actin mRNA (uPAR:β-actin) and were calculated using the Applied Biosystems software with a cycle threshold selected in the geometric phase of amplification.
Removal of Cell-Bound uPA
Eosinophils were suspended at 10 × 106/ml in cold 50 mM glycine-HCl/100 mM NaCl (pH 3.0) for 3 min, then an equal volume of 0.5 M HEPES/100 mM NACl (pH 7.4) was added to neutralize pH (18). The cells were washed in HBSS and changes in uPA levels were confirmed by flow cytometry.
Eosinophil Adhesion
VCAM-1 (5 μg/ml) or ICAM-1 (10 μg/ml) was incubated in 96-well plates (50 μl/well, Immulon IV; Thermo Lab Systems, Franklin, MA) for 2 h at 37°C (1). The residual fluid was decanted and 200 μl of HBSS + 0.1% gelatin was added for 1 h at 37°C to block nonspecific adhesion. The plate was then washed once with HBSS before use. Eosinophils (1 × 105/ml) were added to the wells, incubated 30 min at 37°C, and rigorously washed 3× with warm HBSS. Adhesion was assessed using eosinophil peroxidase (EPO) activity of adherent cells as previously described (10). Data are presented as percent adhesion based on total EPO activity.
Data Analysis
Data are presented as mean ± SEM unless otherwise indicated. One-way ANOVA was used for multivariate analysis as appropriate and when significant, Student t test was used for paired statistics (Sigma Stat; SPSS Inc., Chicago, IL). Correlations were identified with the Spearman Rank Correlation test. A P value ⩽ 0.05 was considered significant.
RESULTS
Pulmonary Function
Subjects with asthma undergoing SBP-AG had an FEV1/FVC ratio [median (range)] of 84.6 (69–105), a MethPC20 of 1.9 (0.3–7.3) mg/ml, and a reversibility of 5 (0–17)%.
Expression of uPA and uPAR Protein
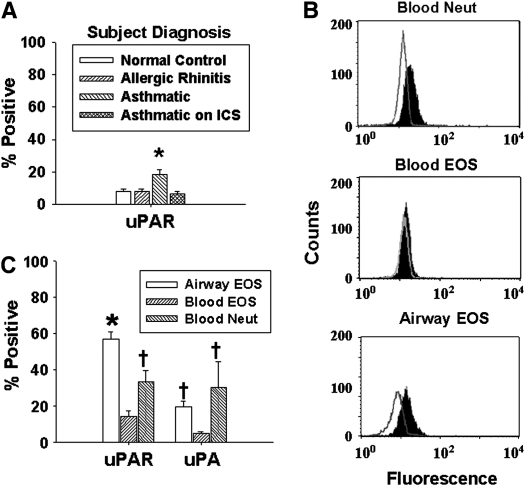
Cell surface expression of uPA and uPAR was evaluated by flow cytometry. Purified peripheral blood eosinophils and neutrophils showed no differences in uPAR or uPA expression (by % positive cells) compared with cells in the mixed granulocyte population (i.e., before negative selection) or in whole blood before purification (data not shown). Even with low expression of uPAR, more blood eosinophils from subjects with asthma expressed membrane uPAR than eosinophils from normal control subjects, subjects with allergic rhinitis, or long-term ICS-treated subjects with asthma (Figure 1A). Significantly more airway eosinophils obtained 48 h after SBP-AG expressed membrane uPAR than the corresponding blood eosinophils and neutrophils (Figures 1B and 1C). Significantly more airway eosinophils expressed uPA than blood eosinophils but were comparable to neutrophil levels. Similar relationships were observed when cell expression was assessed as mean fluorescent units (data not shown). Finally, the percentage of airway eosinophils positive for uPAR in subjects with asthma inversely correlated with MethPC20 and positively correlated with prechallenge reversibility (Table 1), but was not associated with FEV1 or FEV1/FVC (data not shown).
Figure 1.
Expression of uPAR and uPA on neutrophils and eosinophils by flow cytometry. (A) Comparison of eosinophil uPAR expression by subject group. * P < 0.05 versus all other groups (n = 8). (B) Representative histograms of peripheral blood neutrophils, blood eosinophils, and post–SBP-AG airway eosinophils: the unfilled curve represents isotype control and the solid curve is uPAR expression. (C) Comparison of uPAR and uPA expression on blood neutrophils and eosinophils and airway eosinophils. Mean ± SEM. * P < 0.05 versus blood eosinophils and neutrophils by paired t test; † P < 0.005 versus blood eosinophils (n = 20). EOS, eosinophils; Neut, neutrophils; ICS, patients with asthma treated with long-term inhaled corticosteroids.
TABLE 1.
CORRELATION OF AIRWAY FUNCTION WITH uPAR EXPRESSION
| (n = 12) | Membrane uPAR | suPAR | Airway Eosinophils | |
|---|---|---|---|---|
| MethPC20 | rs*= | −0.57 | −0.36 | 0.03 |
| (Baseline) | P = | 0.04 | 0.14 | 0.98 |
| Reversibility | rs = | 0.75 | 0.03 | −0.24 |
| (Pre-AG) | P = | 0.004 | 0.92 | 0.42 |
Definition of abbreviations: MethPC20, methacholine PC20; Pre-AG, before allergen provocation; suPAR, soluble uPAR; uPAR, urokinase-type plasminogen activator.
Spearman Rank Correlation.
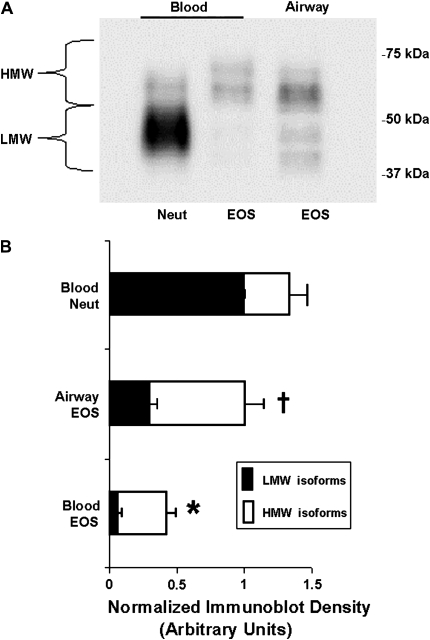
The relative level of immuno-detectable uPAR in lysed airway eosinophils was greater than that observed in blood eosinophils but less than blood neutrophils (Figure 2A). Although neutrophils and eosinophils did not have the same actin content (not shown), the difference in the amount and distribution of uPAR isoforms in blood and airway eosinophils was not the consequence of a systemic difference in protein loading. The protein content of the cell lysates was determined by BCA assay and found to be very similar between neutrophils and eosinophils, with blood eosinophil cell lysates having 86–103% of the protein content of neutrophils. Because blood and airway eosinophils have a very similar repertoire of many cellular proteins, immunoblotting of a “control” protein (actin) was used to evaluate the consistency of protein loading for purposes of this comparison.
Figure 2.
uPAR Western blot. Lysates from purified cells were immunoblotted with anti-uPAR as described in Materials and Methods. (A) Western blot representative of 8 experiments. There was no systematic difference in the total protein loaded on gel. (B) Densitometry analysis normalized to neutrophil LMW uPAR isoforms. * P < 0.004 versus airway eosinophils for both LMW and HMW bands, and neutrophils for LMW band; † P < 0.04 versus neutrophils for both LMW and HMW bands (n = 8).
The size distribution of uPAR isoforms was quite different between eosinophil and neutrophil populations. Immunodetectable uPAR migrated with several different electophoretic mobilities, typical for a protein displaying variable levels of glycosylation (19), post-translational modifications, and/or splice variants. Our quantified data were normalized to the neutrophil LMW species (35–50 kD), which represented 76 ± 4.6% (mean ± SEM) of total uPAR protein (Figure 2B). In contrast, blood eosinophils had much less total cellular immunodetectable uPAR than neutrophils, with the majority above 50 kD (HMW); the LMW isoforms represented only 22 ± 2.5% of the eosinophil total. Airway eosinophils contained significantly more of both HMW and LMW isoforms than blood eosinophils, with the majority of uPAR greater than 50 kD.
Soluble uPAR
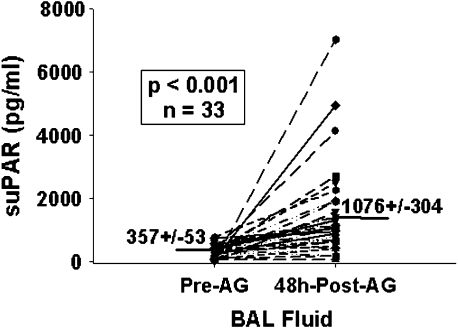
Increased cell surface expression of uPAR on airway eosinophils could be the result of in vivo binding of suPAR released by other cell types (endothelial cells, smooth muscle cells, monocytes/macrophages) (20, 21). suPAR concentrations were significantly increased in BAL fluid 48 h after SBP-AG compared with pre-AG samples (Figure 3). Moreover, after AG challenge, suPAR levels correlated with the percentage of airway eosinophils in the total BAL cell population (rs = 0.720, P < 0.01), but neither of these variables correlated with pulmonary function (Table 1). To determine if exposure to suPAR can increase eosinophil membrane-bound uPAR, blood eosinophils were incubated up to 24 h in culture medium containing 50% BAL fluid from either pre- or post-AG challenge samples and were then evaluated for cell-associated uPAR. Compared with medium, pre–SBP-AG BAL fluid increased eosinophil uPAR expression by 6.5 ± 2.3%, while post-AG BAL fluid increased it by 17.9 ± 2.2%. Although both of these changes were significantly higher than medium alone (P < 0.03), they did not reach those expressed by airway eosinophils.
Figure 3.
Soluble (s)uPAR before and after SBP-AG. suPAR was measured by ELISA in neat, cell-free BAL fluid collected before and 48 h after allergen challenge.
Upregulation of uPAR Expression
Neutrophil stores of intracellular uPAR can be rapidly transported to the cell membrane when the cells are incubated with PAF or IL-8 (22, 23). To determine if the high expression of uPAR by airway eosinophils could be the result of a similar uPAR externalization of blood eosinophils' intracellular stores, blood eosinophils were incubated with individual agonists for 30 min to 24 h in culture medium. uPAR expression was calculated as the increase in membrane-bound uPAR after incubation with agonist minus incubation in medium alone. Incubation with PAF (100 nM, 30 min), IL-5 (1 ng/ml, 3 h), or GM-CSF (1 ng/ml, 3 h) resulted in small, but significant, transient increases (4–10% increase in eosinophils expressing uPAR at 30 min, P < 0.05 versus buffer control) in uPAR expression, which did not reach the levels observed on airway eosinophils. Eotaxin (100 ng/ml), RANTES (100 ng/ml), FMLP (100 nM), and TGF-β (30 ng/ml) had no effect on uPAR expression within this time frame (data not shown). In contrast, incubation with 0.1 ng/ml PMA resulted in significantly increased uPAR expression at 3 h (30% eosinophils expressed uPAR, P < 0.05 versus buffer control) with a maximum response at 20 h (54% positive); these levels of uPAR were comparable with those observed on airway eosinophils (58% positive).
Detection of uPAR mRNA
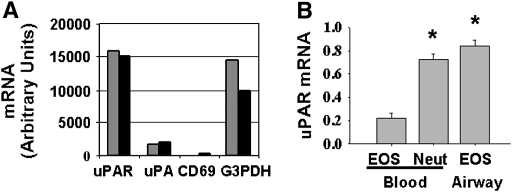
Alternatively, increased expression of uPA and uPAR by airway eosinophils may be the result of increased transcription. To determine if eosinophils expressed uPA and uPAR mRNA, an oligonucleotide microarray was performed on pooled eosinophil lysates from two similar subject groups (the complete data are available online at http://ajrcmb.atsjournals.org/cgi/content/full/2003-0234OC/DC1) (16). The relative levels of mRNA for uPAR and uPA were compared against negative (CD69) and positive (G3PDH) control mRNA. Gene expression analysis identified uPAR mRNA in blood eosinophils (Figure 4A); uPA mRNA was also present, but at a much lower level than uPAR.
Figure 4.
Relative levels of uPAR and uPA mRNA in neutrophils and eosinophils. (A) Gene microarray: blood eosinophils were analyzed for uPAR and uPA gene expression. CD69 and G3PDH were used as negative and positive controls, respectively. The pools consisted of combined eosinophil lysates from three and four subjects. (B) Quantitative real-time PCR of uPAR mRNA in unstimulated cells: values are reported as the ratio of uPAR to β-actin mRNA. * P < 0.003 versus blood eosinophils (n = 4).
The presence of uPAR mRNA was also quantified by real-time PCR; similar to uPAR protein measurements, airway eosinophils and peripheral blood neutrophils expressed significantly more uPAR mRNA than blood eosinophils (Figure 4B). These data were consistent when quantified against two different standard curves: epithelial cells from lung transplant donors and the SPB21 cell line.
Functional Effects of uPAR and uPA on Eosinophil Adhesion
Both uPAR and uPA have been reported to modulate in vivo and in vitro neutrophil adhesion and migration (4, 7, 24). To determine whether uPAR affected eosinophil adhesion to ICAM-1 or VCAM-1, blood and airway eosinophils were incubated with increasing concentrations (1–10 μg/ml) of neutralizing anti-uPAR mAb; no change in adhesion was observed (data not shown).
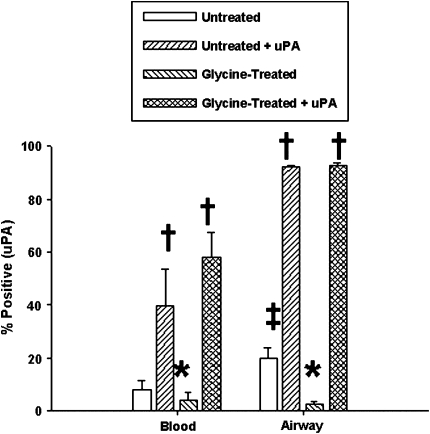
The effects of uPA on eosinophil adhesion were then assessed. A short treatment with acidic glycine has been widely used to remove membrane-bound uPA from a variety of cell types (25). Glycine treatment had no effect on uPAR expression (data not shown). Even though blood eosinophil uPA levels were very low, glycine treatment significantly reduced baseline expression (Figure 5). Similarly, this treatment decreased the elevated uPA levels on airway eosinophils. Addition of exogenous uPA (125 nM) to both untreated and glycine-treated eosinophils increased cell-bound uPA beyond original baseline levels to 40–60% positive for blood cells and almost 100% positive for airway cells, suggesting that maximum uPA expression had not occurred in vivo. This difference in the capacity to bind exogenous uPA was consistent with our earlier finding of distinct eosinophil phenotypes isolated from these two cell compartments (10) and suggests that the ability of airway eosinophils to bind uPA was enhanced during the cell's transition from the circulation to the airway lumen.
Figure 5.
Effects of glycine treatment on eosinophil uPA expression. Peripheral blood and airway eosinophils were treated with buffer ± acidic glycine followed by ± 125 nM exogenous uPA. * P < 0.05 versus untreated cells; † P < 0.02 versus untreated and glycine-treated cells; ‡ P < 0.005 versus untreated blood cells (n = 6).
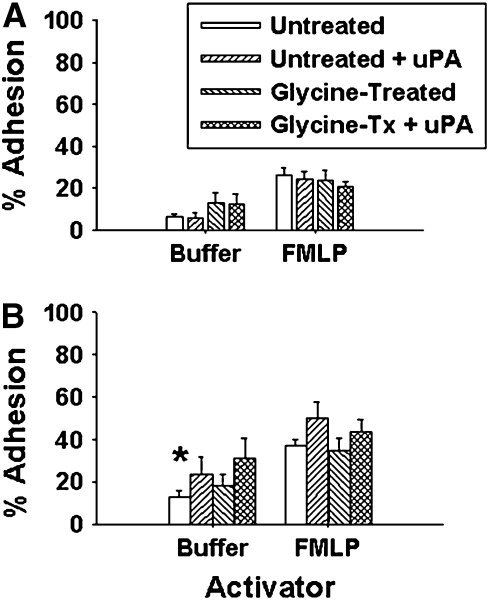
Since uPA could be removed from and added back to eosinophils, its effects on in vitro cell adhesion to ICAM-1 and VCAM-1 were determined. Pretreatment of blood eosinophils (either unchallenged subjects or subjects 48 h after SBP-AG) with glycine and/or exogenous uPA had no significant effect on baseline (unstimulated) or FMLP-activated blood eosinophil adhesion to ICAM-1 (Figure 6). Addition of exogenous uPA to blood eosinophils slightly, but not significantly, increased VCAM-1 adhesion. Therefore, increased cell-bound uPA alone (Figure 5) was not sufficient to modulate blood eosinophil adhesion to these substrates. In contrast, the removal of cell-bound uPA from, and addition of exogenous uPA to, airway eosinophils had very different effects on adhesion.
Figure 6.
Effect of uPA on blood eosinophil adhesion to ICAM-1 (A) and VCAM-1 (B). Peripheral blood eosinophils pretreated with buffer ± glycine, followed by incubation ± 125 nM uPA. * P < 0.005 versus ICAM-1 adhesion (n = 8).
Unstimulated adhesion to ICAM-1 was equivalent for airway and blood eosinophils (Figure 7). Although glycine treatment of airway eosinophils did not significantly affect baseline adhesion, addition of exogenous uPA to both untreated and glycine-treated airway eosinophils resulted in significantly increased ICAM-1 adhesion compared with blood eosinophils. Baseline adhesion of airway eosinophils to VCAM-1 was significantly higher than that of blood eosinophils; however, it decreased to blood eosinophil levels after removal of uPA. Addition of exogenous uPA to both untreated and glycine-treated airway eosinophils increased VCAM-1 adhesion beyond baseline levels similar to its effect on ICAM-1 adhesion. These data indicate that increasing cell-bound uPA on airway, but not blood, eosinophils can stimulate adhesion to both substrates.
Figure 7.
Effect of uPA on airway eosinophil adhesion to ICAM-1 (A) and VCAM-1 (B). Airway eosinophils were pretreated with buffer ± glycine, followed by incubation ± 125 nM uPA. For unstimulated cells: * P < 0.005 versus ICAM-1 adhesion (also versus blood eosinophil VCAM-1 adhesion: Figure 6); † P < 0.05 versus untreated and glycine-treated airway eosinophils; for FMLP-stimulated cells: ‡ P = 0.05 versus glycine-treated airway cells and untreated blood eosinophils (n = 8).
Similar to our previous observation (10), FMLP-activated airway eosinophils were significantly more adherent to both ICAM-1 and VCAM-1 than activated blood eosinophils (Figure 7). Glycine treatment of airway eosinophils to remove cell-bound uPA reduced ICAM-1 and VCAM-1 adhesion to levels comparable with those of blood eosinophils. Subsequent addition of exogenous uPA to the glycine-treated cells restored airway eosinophil adhesion to the original values; uPA addition to untreated, FMLP-activated airway eosinophils had no effect.
DISCUSSION
Compared with their corresponding blood eosinophils, airway eosinophils isolated from BAL fluid obtained 48 h after SBP-AG demonstrated distinct and significant differences in the expression and functional role of uPA and uPAR. Similar to blood neutrophils, airway eosinophils expressed more uPAR protein and mRNA, as well as increased cell-bound uPA, than blood eosinophils. Levels of suPAR were increased in BAL fluid 48 h after AG challenge, but in vitro incubation of blood eosinophils with these BAL fluids did not substantially increase the percentage of uPAR-positive cells. The number of blood eosinophils expressing uPAR was, however, enhanced by incubation of cells with PAF, IL-5, or GM-CSF; however, these rapid increases were small and transient—only long-term (20 h) incubation with PMA increased the percentage of uPA-positive blood eosinophils to airway eosinophil levels. Although incubation of blood eosinophils with exogenous uPA greatly increased their expression of membrane-bound uPA, it had no affect on cell adhesion to either ICAM-1 or VCAM-1. In contrast, addition of exogenous uPA to airway eosinophils resulted in almost 100% of cells expressing cell-bound uPA as well as stimulated in vitro airway eosinophil adhesion to both VCAM-1 and ICAM-1. Moreover, constitutive cell-bound uPA primed airway cells in vivo for enhanced in vitro FMLP-activated adhesion to these substrates; removal of cell-bound uPA from airway eosinophils reduced their FMLP-stimulated adhesion to the much lower level of blood cells, while subsequent addition of exogenous uPA restored cell adhesion to their original values.
Although the levels of blood eosinophils expressing uPAR were very low, significantly more cells from subjects with asthma expressed uPAR than those from healthy control subjects and patients with allergic rhinitis. Moreover, peripheral eosinophils from ICS-treated patients with asthma expressed the lower “normal” level of uPAR compared with untreated patients with asthma, suggesting that elevated uPAR may be a biomarker of increased disease severity or loss of control. Consistent with this finding, the percentage of airway eosinophils expressing uPAR after SBP-AG correlated with two measures of pulmonary function in the untreated subjects with asthma: airway hyperresponsiveness as measured by MethPC20 and percent airway reversibility. In contrast, correlations of airway or blood eosinophil numbers with airway physiology were weak and not significant. These findings suggest that the phenotype of airway eosinophils is a better predictor of airway physiology than the absolute number of these cells.
There are at least three possible sources of increased cell surface uPAR on airway eosinophils: uptake of suPAR from the BAL fluid, externalization of intracellular uPAR stores, and increased transcription and/or translation. First, cells can increase their membrane expression of uPAR by the uptake of suPAR generated by other cell types including neutrophils (20, 21). In vitro incubation of blood eosinophils with post–SBP-AG BAL fluid containing high levels of suPAR, however, resulted in only small increases in cellular uPAR expression that were well below the observed levels on airway eosinophils. Second, uPAR expression by neutrophils consists of two sources: membrane and intracellular. In neutrophils, intracellular uPAR can be rapidly translocated to the cell surface in response to short-term incubation (30 min) with select chemokines or cytokines (22, 23). We have confirmed reports by others (8, 9) that blood eosinophils can also undergo rapid increases in uPAR expression when incubated in vitro with PAF, IL-5, and GM-CSF. Although significant, these changes were very small, transient, and did not reach the levels of uPAR expression by airway eosinophils. Moreover, eotaxin, RANTES, and FMLP had no effect. These data suggest that intracellular uPAR is an unlikely source of the enhanced uPAR expression observed on airway eosinophils. In contrast to these observations, incubation of blood eosinophils with PMA for 20 h increased uPAR expression to the level of airway eosinophils, suggesting de novo transcription and translation. This is consistent with reports by others (26, 27) that PMA increases both uPAR mRNA and protein in a variety of cell types and lines. Finally, airway eosinophils expressed more uPAR mRNA than blood eosinophils as determined by gene microarray and PCR. These combined data suggest that airway eosinophils express more uPAR by a mechanism of increased transcription and translation.
Most (76%) of the cellular uPAR in neutrophils was between 35 and 50 kD (LMW), whereas the majority of total uPAR protein from airway eosinophils was greater than 50 kD (HMW). uPAR is made up of a 35-kD amino acid backbone that is then glycosylated, resulting in molecular weight variation between and within different cell types (28). Picone and associates (26) reported that PMA treatment of U937 cells for 24 h resulted in a several-fold increase in highly glycosylated 70-kD uPAR, suggesting that this HMW species represents newly generated uPAR. This is consistent with our observations of increased uPAR expression by PMA-activated blood eosinophils, and increased HMW uPAR protein and mRNA in airway cells. These data also suggest that eosinophils and neutrophils are quite different in their expression of uPAR, which may be reflected in their functional responses.
In preliminary experiments, addition of a neutralizing anti-uPAR mAb had no affect on eosinophil adhesion (data not shown), suggesting that increased uPAR expression alone may not be sufficient for enhanced airway eosinophil adhesion and that other factors such as uPA may modulate this function. To assess this possibility, cell-bound uPA expression was decreased or increased and the effect on eosinophil adhesion was determined. Eosinophils were treated with acidic glycine to remove cell-bound uPA (18), significantly reducing even the low levels of uPA observed on circulating eosinophils (Figure 5). Alternatively, addition of exogenous uPA to either untreated or glycine-treated blood eosinophils significantly increased cell-bound uPA expression above baseline levels, suggesting that even untreated blood eosinophils were able to bind more uPA than was typically associated with these cells. Blood eosinophil uPA levels increased to 40–60% positive cells, whereas addition of exogenous uPA to airway eosinophils (untreated or glycine-treated) increased cell-bound uPA expression to almost 100% positive cells, consistent with the observation of phenotypic differences between blood and airway eosinophils (10). This additional uPA binding may be via ligation to newly activated and/or previously empty uPAR sites, to activated integrins such as αMβ2, or to both (29). These data are consistent with Politis and associates' (30) observation that incubation of resting bovine neutrophils with exogenous uPA resulted in a 16-fold increase in cell-bound uPA. It is, of course, possible that the blood and airway eosinophils responded differently to exogenous uPA due to the slight difference in their isolation procedure (airway eosinophils were not exposed to magnetic beads); however, unseparated eosinophils (from blood and BAL fluid) and neutrophils (blood) displayed uPAR and uPA expression similar to purified cells by flow cytometry (data not shown). Moreover, incubating airway eosinophils with the same anti-CD16–labeled magnetic beads used for blood eosinophil isolation had no effect on uPAR expression (data not shown).
Our original observation of enhanced airway eosinophil adhesion after SBP-AG used human umbilical vein endothelial cell monolayers (10). In this study, however, we used ICAM-1 and VCAM-1 proteins to better identify integrin-specific adhesion. Although acid treatment and addition of exogenous uPA significantly decreased and increased, respectively, blood eosinophil expression of cell-bound uPA, it had no significant affect on cell adhesion to either ICAM-1 or VCAM-1. In contrast, addition of exogenous uPA to airway eosinophils increased both cell-bound uPA and adhesion to both of these substrates in the absence of any other agonist. Therefore, it does not appear that exogenous uPA alone is an eosinophil agonist, as it had no effect on blood eosinophil adhesion even though its cell-bound expression was increased. These data suggest that airway eosinophils have been altered in their ability to respond to uPA compared with blood cells, possibly through changes in uPAR and/or integrin activation.
We observed several interesting uPA-dependent effects on airway, but not blood, eosinophil adhesion. First, as we have previously reported (31), blood eosinophils were more adhesive to VCAM-1 than ICAM-1; this is consistent with the partial activation of α4β1 integrin on these cells (32). Airway eosinophil adhesion to VCAM-1 was not only significantly higher than the corresponding ICAM-1 adhesion, but was also increased compared with blood eosinophil VCAM-1 adhesion. This difference in VCAM-1 adhesion was completely eliminated when cell-bound uPA was removed from airway eosinophils; similar treatment of blood eosinophils had no effect, suggesting that this small increase in VCAM-1 adhesion was not dependent on uPA expression. Second, airway eosinophil VCAM-1 and ICAM-1 adhesion were significantly enhanced, beyond even the high baseline VCAM-1 levels, with the addition of exogenous uPA. Under these conditions, exogenous uPA was a stimulant of airway eosinophil adhesion even though it had no effect on blood eosinophil adhesion. Since eosinophil adhesion to ICAM-1 and VCAM-1 is dependent on cell expression of αMβ2 and α4β1 integrins, respectively (33), the effect of exogenous uPA on airway eosinophil adhesion may again be due to increased uPA binding to activated uPAR and/or activated integrins. Third, eosinophils express the low-affinity receptor for FMLP (FPRL) (34), which can interact directly with cell integrins and uPAR (35). Therefore, it is possible that a multiprotein complex of integrin, uPAR, uPA, and FMLP promotes optimal cell adhesion to select substrates. FMLP stimulated airway eosinophil adhesion to ICAM-1 and VCAM-1 above blood cell levels. This effect could not be further enhanced by the addition of exogenous uPA, but could be completely eliminated by removal of cell-bound uPA. Addition of exogenous uPA to the glycine-treated airway cells completely restored FMLP-activated adhesion to its original level. It therefore appears that the higher constitutive levels of cell-bound uPA on airway eosinophils after AG challenge primes these cells for enhanced in vitro FMLP-activated ICAM-1 and VCAM-1 adhesion.
Where airway eosinophils acquired this difference in uPAR and uPA expression and function is unknown, but there are several possibilities. It may begin while the cells are still in the circulation, because subjects with asthma expressed more uPAR on their blood eosinophils than healthy control subjects and patients with allergic rhinitis, a difference that was lost upon ICS treatment. Although circulating eosinophils could take up exogenous uPA, this had no effect on their in vitro adhesion to ICAM-1 or VCAM-1. It is possible that a modification of circulating eosinophil adhesion occurs only in close proximity to the airway vasculature where inflammatory mediators would be maximal after AG challenge. This may then be followed by rapid eosinophil adhesion and diapedesis into the airway tissue. Alternatively, eosinophils may acquire higher uPAR and/or uPA expression as they transverse the vascular endothelium or during migration through the interstitial tissue and airway epithelium. It may even occur within the airway lumen itself with its high levels of suPAR. In any case, it appears that these cells undergo a transformation during their transition from the circulation to the airway lumen in which cell-bound PA primes FMLP-stimulated airway eosinophil adhesion to ICAM-1 and VCAM-1. Finally, additional exposure of airway eosinophils to uPA in vitro can activate adhesion to these substrates. The observed effects of uPA on airway eosinophil adhesion may be dependent on direct uPA binding to activated integrins or activated uPAR, or it may form a tricomplex with both. Studies to define these uPA interactions and their effects on other enhanced airway eosinophil functions are underway.
uPA is a primer/activator of airway eosinophil adhesion and may have similar effects on in vivo eosinophil recruitment during asthma exacerbations. Moreover, membrane levels of airway eosinophil uPAR expression correlated with airway hyperresponsiveness and reversibility far better than absolute eosinophil levels and, therefore, may be a biomarker of disease severity. Identification of these mechanisms of airway eosinophil activation after AG challenge may provide new therapeutic tools for the treatment of asthma.
Acknowledgments
The authors thank Dr. Keith Meyer, Dr. Richard Cornwell, and the Pulmonary Research nurses for pre– and post–SBP-AG BAL fluid and blood samples from subjects with asthma.
This study was supported by NIH grants HL05696 and RR03186.
Originally Published in Press as DOI: 10.1165/rcmb.2006-0113OC on May 25, 2006
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Nagata M, Sedgwick JB, Bates ME, Kita H, Busse WW. Eosinophil adhesion to VCAM-1 activates superoxide anion generation. J Immunol 1995;155:2194–2202. [PubMed] [Google Scholar]
- 2.Fujiu T, Kato M, Kimura H, Tachibana A, Suzuki M, Nako Y, Morikawa A. Cellular adhesion is required for effector functions of human eosinophils via G-protein coupled receptors. Ann Allergy Asthma Immunol 2002;89:90–98. [DOI] [PubMed] [Google Scholar]
- 3.Mondino A, Resnati M, Blasi F. Structure and function of the urokinase receptor. Thromb Haemost 1999;82:19–22. [PubMed] [Google Scholar]
- 4.Waltz DA, Fujita RM, Yang X, Natkin L, Zhuo S, Gerard CJ, Rosenberg S, Chapman HA. Nonproteolytic role for the urokinase receptor in cellular migration in vivo. Am J Respir Cell Mol Biol 2000;22:316–322. [DOI] [PubMed] [Google Scholar]
- 5.Sitrin RG, Pan PM, Harper HA, Todd RF III, Harsh DM, Blackwood RA. Clustering of urokinase receptors (uPAR; CD87) induces proinflammatory signaling in human polymorphonuclear neutrophils. J Immunol 2000;165:3341–3349. [DOI] [PubMed] [Google Scholar]
- 6.Reuning U, Magdolen V, Hapke S, Schmitt M. Molecular and functional interdependence of the urokinase-type plasminogen activator system with integrins. Biol Chem 2003;384:1119–1131. [DOI] [PubMed] [Google Scholar]
- 7.Gyetko MR, Sud S, Kendall T, Fuller JA, Newstead MW, Standiford TJ. Urokinase receptor-deficient mice have impaired neutrophil recruitment in response to pulmonary Pseudomonas aeruginosa infection. J Immunol 2000;165:1513–1519. [DOI] [PubMed] [Google Scholar]
- 8.Guilbert M, Ferland C, Bosse M, Flamand N, Lavigne S, Laviolette M. 5-Oxo-6,8,11,14-eicosatetraenoic acid induces important eosinophil transmigration through basement membrane components: comparison of normal and asthmatic eosinophils. Am J Respir Cell Mol Biol 1999; 21:97–104. [DOI] [PubMed] [Google Scholar]
- 9.Mabilat-Pragnon C, Janin A, Michel L, Thomaidis A, Legrand Y, Soria C, Lu H. Urokinase localization and activity in isolated eosinophils. Thromb Res 1997;88:373–379. [DOI] [PubMed] [Google Scholar]
- 10.Sedgwick JB, Calhoun WJ, Vrtis RF, Bates ME, McAllister PK, Busse WW. Comparison of airway and blood eosinophil function after in vivo antigen challenge. J Immunol 1992;149:3710–3718. [PubMed] [Google Scholar]
- 11.Cao D, Mizukami IF, Garni-Wagner BA, Kindzelskii AL, Todd RF III, Boxer LA, Petty HR. Human urokinase-type plasminogen activator primes neutrophils for superoxide anion release: possible roles of complement receptor type 3 and calcium. J Immunol 1995;154:1817–1829. [PubMed] [Google Scholar]
- 12.Ferland C, Guilbert M, Davoine F, Flamand N, Chakir J, Laviolette M. Eotaxin promotes eosinophil transmigration via the activation of the plasminogen-plasmin system. J Leukoc Biol 2001;69:772–778. [PubMed] [Google Scholar]
- 13.Liu LY, Sedgwick JB, Bates ME, Vrtis RF, Gern JE, Kita H, Jarjour NN, Busse WW, Kelly EAB. Decreased expression of membrane IL-5Rα on human eosinophils: I. Loss of membrane IL-5alpha on eosinophils and increased soluble IL-5R alpha in the airway after antigen challenge. J Immunol 2002;169:6452–6458. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto H, Sedgwick JB, Busse WW. Differential regulation of eosinophil adhesion and transmigration by pulmonary microvascular endothelial cells. J Immunol 1998;161:971–977. [PubMed] [Google Scholar]
- 15.Bates ME, Green VL, Bertics PJ. ERK1 and ERK2 activation by chemotactic factors in human eosinophils is interleukin 5-dependent and contributes to leukotriene C(4) biosynthesis. J Biol Chem 2000;275: 10968–10975. [DOI] [PubMed] [Google Scholar]
- 16.Bates ME, Liu LY, Esnault S, Stout BA, Fonkem E, Kung V, Sedgwick JB, Kelly EA, Bates DM, Malter JS, et al. Expression of IL-5- and GM-CSF-responsive genes in blood and airway eosinophils. Am J Respir Cell Mol Biol 2004;30:736–743. [DOI] [PubMed] [Google Scholar]
- 17.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 2001;98:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoppelli MP, Tacchetti C, Cubellis MV, Corti A, Hearing VJ, Cassani G, Appella E, Blasi F. Autocrine saturation of pro-urokinase receptors on human A431 cells. Cell 1986;45:675–684. [DOI] [PubMed] [Google Scholar]
- 19.Degryse B. Is uPAR the centre of a sensing system involved in the regulation of inflammation? Curr Med Chem 2003;2:237–259. [Google Scholar]
- 20.Mustjoki S, Sidenius N, Vaheri A. Enhanced release of soluble urokinase receptor by endothelial cells in contact with peripheral blood cells. FEBS Lett 2000;486:237–242. [DOI] [PubMed] [Google Scholar]
- 21.Mizukami IF, Todd RF III. A soluble form of the urokinase plasminogen activator receptor (suPAR) can bind to hematopoietic cells. J Leukoc Biol 1998;64:203–213. [DOI] [PubMed] [Google Scholar]
- 22.Jardi M, Ingles-Esteve J, Burgal M, Azqueta C, Velasco F, Lopez-Pedrera C, Miles LA, Felez J. Distinct patterns of urokinase receptor (uPAR) expression by leukemic cells and peripheral blood cells. Thromb Haemost 1996;76:1009–1019. [PubMed] [Google Scholar]
- 23.Plesner T, Ploug M, Ellis V, Ronne E, Hoyer-Hansen G, Wittrup M, Pedersen TL, Tscherning T, Dano K, Hansen NE. The receptor for urokinase-type plasminogen activator and urokinase is translocated from two distinct intracellular compartments to the plasma membrane on stimulation of human neutrophils. Blood 1994;83:808–815. [PubMed] [Google Scholar]
- 24.Pluskota E, Soloviev DA, Plow EF. Convergence of the adhesive and fibrinolytic systems: recognition of urokinase by integrin alpha Mbeta 2 as well as by the urokinase receptor regulates cell adhesion and migration. Blood 2003;101:1582–1590. [DOI] [PubMed] [Google Scholar]
- 25.Ragno P, Montuori N, Vassalli JD, Rossi G. Processing of complex between urokinase and its type-2 inhibitor on the cell surface: a possible regulatory mechanism of urokinase activity. FEBS Lett 1993;323: 279–284. [DOI] [PubMed] [Google Scholar]
- 26.Picone R, Kajtaniak EL, Nielsen LS, Behrendt N, Mastronicola MR, Cubellis MV, Stoppelli MP, Pedersen S, Dano K, Blasi F. Regulation of urokinase receptors in monocytelike U937 cells by phorbol ester phorbol myristate acetate. J Cell Biol 1989;108:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lund LR, Ellis V, Ronne E, Pyke C, Dano K. Transcriptional and post-transcriptional regulation of the receptor for urokinase-type plasminogen activator by cytokines and tumour promoters in the human lung carcinoma cell line A549. Biochem J 1995;310:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behrendt N, Ronne E, Ploug M, Petri T, Lober D, Nielsen LS, Schleuning WD, Blasi F, Appella E, Dano K. The human receptor for urokinase plasminogen activator. NH2-terminal amino acid sequence and glycosylation variants. J Biol Chem 1990;265:6453–6460. [PubMed] [Google Scholar]
- 29.Pluskota E, Soloviev DA, Bdeir K, Cines DB, Plow EF. Integrin alpha(M)beta(2) orchestrates and accelerates plasminogen activation and fibrinolysis by neutrophils. J Biol Chem 2004;279:18063–18072. [DOI] [PubMed] [Google Scholar]
- 30.Politis I, Zavizion B, Cheli F, Baldi A. Expression of urokinase plasminogen activator receptor in resting and activated bovine neutrophils. J Dairy Res 2002;69:195–204. [DOI] [PubMed] [Google Scholar]
- 31.Sedgwick JB, Jansen KJ, Kennedy JD, Kita H, Busse WW. Effects of VLA-4 antagonist WAY103 on human peripheral blood eosinophil VCAM-1 dependent functions. J Allergy Clin Immunol 2005;116:812. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto K, Sterbinsky SA, Bickel CA, Zhou DM, Kovach NL, Bochner BS. Regulation of α4 integrin-mediated adhesion of human eosinophils to fibronectin and vascular cell adhesion molecule-1. J Allergy Clin Immunol 1997;99:648–656. [DOI] [PubMed] [Google Scholar]
- 33.Nagata M, Sedgwick JB, Kita H, Busse WW. Granulocyte-macrophage colony-stimulating factor augments ICAM-1 and VCAM-1 activation of eosinophil function. Am J Respir Cell Mol Biol 1998;19:158–166. [DOI] [PubMed] [Google Scholar]
- 34.Svensson L, Dahlgren C, Wenneras C. The chemoattractant Trp-Lys-Tyr-Met-Val-D-Met activates eosinophils through the formyl peptide receptor and one of its homologues, formyl peptide receptor-like 1. J Leukoc Biol 2002;72:810–818. [PubMed] [Google Scholar]
- 35.Montuori N, Carriero MV, Salzano S, Rossi G, Ragno P. The cleavage of the urokinase receptor regulates its multiple functions. J Biol Chem 2002;277:46932–46939. [DOI] [PubMed] [Google Scholar]