Abstract
Inspired oxygen, an essential therapy for cardiorespiratory disorders, has the potential to generate reactive oxygen species that damage cellular DNA. Although DNA damage is implicated in diverse pulmonary disorders, including neoplasia and acute lung injury, the type and magnitude of DNA lesion caused by oxygen in vivo is unclear. We used single-cell gel electrophoresis (SCGE) to quantitate two distinct forms of DNA damage, base adduction and disruption of the phosphodiester backbone, in the lungs of mice. Both lesions were induced by oxygen, but a marked difference between the two was found. With 40 h of oxygen exposure, oxidized base adducts increased 3- to 4-fold in the entire population of lung cells. This lesion displayed temporal characteristics (a progressive increase over the first 24 h) consistent with a direct effect of reactive oxygen species attack upon DNA. DNA strand breaks, on the other hand, occurred in < 10% of pulmonary cells, which acquired severe levels of the lesion; dividing cells were preferentially affected. Characteristics of these cells suggested that DNA strand breakage was secondary to cell death, rather than a primary effect of reactive oxygen species attack on DNA. By analysis of IL-6– and IL-11–overexpressing transgenic animals, which are resistant to hyperoxia, we found that DNA strand breaks, but not base damage, correlated with acute lung injury. Analysis of purified alveolar type 2 preparations from hyperoxic mice indicated that strand breaks preferentially affected this cell type.
Keywords: 8-oxoguanine, acute lung injury, apoptosis, DNA damage, hyperoxia
Inhaled oxygen is essential in the treatment of respiratory and cardiovascular disorders. Unfortunately, high concentrations of inspired oxygen can be detrimental, because of their propensity to generate reactive oxygen species. One example of this is fatal acute lung injury, which occurs in experimental animals exposed to oxygen concentrations of > 80% (1). Reactive oxygen species can be produced by the interaction of inhaled oxygen with the mitochondrial electron transport chain or membrane-bound NADPH oxidase (2). These reactive species, principally superoxide anion and hydrogen peroxide, damage cellular macromolecules, including lipids, proteins, and nucleic acids (3, 4).
Damage to cellular DNA is of particular interest. Oxidative DNA damage can affect nuclear or mitochondrial DNA and can lead to mutation, gene expression changes, apoptosis, and necrosis (5). Chemical studies indicate that these reactions occur rapidly (6), and in cell culture reactive oxygen species cause DNA strand breaks within minutes of exposure (7, 8). It is also clear that oxidant attack on DNA can be sequence selective, and that the lesions generated can affect gene expression (9). In principle, reactive oxygen species can attack either the phosphodiester backbone of DNA, leading to DNA strand breaks, or the nitrogenous bases, leading to mutagenic base alterations (4). Although qualitative studies suggest the presence of both DNA strand breaks and base lesions in the lungs of oxygen-exposed animals (10–12), their magnitude and relative abundance remains unclear. In addition, controversy exists as to whether DNA strand breaks result from reactive oxygen species attack on DNA, or are a secondary consequence of nucleases activated during programmed cell death (13).
In this report, we present quantitative analysis of DNA strand breaks and DNA base damage in the lungs of mice housed in an elevated oxygen environment. Both lesions were detected. We found, however, a marked dissociation between the two. Base lesions were quantitatively the predominant effect of oxygen, displaying temporal and population characteristics consistent with a primary effect of reactive oxygen species attack on nuclear DNA. DNA strand breaks, on the other hand, occurred in only a small subpopulation of lung cells, and displayed characteristics that suggest they are a secondary effect of nucleases activated during cell death. Using transgenic mice resistant to lung injury, we found that only strand breaks, and not base damage, correlated with lung injury. Strand breaks (likely reflecting cell death) predominantly affected dividing cells in vivo and airway epithelial cells in vitro. Furthermore, by fractionation of lung cell preparations, we found that alveolar type 2 cells undergo DNA strand breakage in oxygen. These results have bearing on the mechanism of acute lung injury and on the ability of inhaled substances to promote mutation.
MATERIALS AND METHODS
Mice and Cells
Animal experiments were performed in compliance with NIH and institutional guidelines for the humane care and use of laboratory animals. C57Bl6 mice were purchased from Charles River Labs (Wilmington, MA). IL-6– and IL-11–overexpressing transgenic mice have been previously described (14, 15); these transgenics are bred in a C57Bl6 background. Genotyping was performed by PCR as previously described (15, 16). Human umbilical vein endothelial cells, human pulmonary microvascular endothelial cells, human lung fibroblasts, and small airway epithelial cells were purchased from Clonetics (Walkersville, MD). Culture conditions were as directed by the manufacturer. A549 and Jurkat cells were purchased from the American Type Culture Collection (Manassas, VA) and cultured under the supplier's recommended conditions.
Chemicals
Unless otherwise specified, chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Oxygen Exposure
Mice were exposed to 95% oxygen at atmospheric pressure in a Lucite chamber. Oxygen concentration inside the chamber was continuously adjusted using an oxygen sensor (Engineering Systems and Designs, Newark, DE) adjusting delivery of 100% oxygen. Oxygen tensions were verified by mass spectrophotometric analysis of chamber gas. Carbon dioxide in the chamber was adsorbed using soda lime (Natronkalk, Fluka; purchased through Sigma). The chamber was maintained at atmospheric pressure. Mice were provided food and water ad libitum. In this system, 2- to 3-mo-old C57Bl mice display signs of illness (lethargy, piloerection) after 60–72 h of oxygen exposure and die after 96–120 h of exposure.
For in vitro experiments, cultured cells were grown to ∼ 80% confluence and transferred to a humidified Lucite container inside a 37°C incubator. At atmospheric pressure, the gas in the Lucite container was maintained at 88–90% oxygen with regulated delivery of 95% O2, 5% CO2 gas via O2 sensor-controlled solenoid (the sensor probe is located within the Lucite chamber; the solenoid and control unit is located outside the incubator; all components from Engineering Systems and Designs). Oxygen tensions were verified daily by mass spectrophotometric analysis. Culture medium was changed every 48 h.
Preparation of Mouse Lung Suspensions for Single-Cell Gel Electrophoresis Analysis
Single-cell gel electrophoresis (SCGE) is a technique to measure DNA damage in which cell populations are embedded in agarose and subjected to electrophoresis; cells without DNA damage migrate as spheres with small tails, while increasing DNA strand breakage causes cells to develop prominent “comet tails” (hence the alternate colloquialism “comet assay”) (17–19). We prepared lung samples for SCGE as follows. Mice were killed with ketamine (100 mg/kg) and xylazine (10–15 mg/kg) and exsanguinated via abdominal aortic transection. The thorax was opened and right and left ventricles were perfused with ice-cold Hanks' balanced salt solution supplemented with 20 mM EDTA and 10% DMSO (“mincing solution”). All lung lobes were removed, placed in 3 cc of mincing solution on ice, and dissected away from mediastinal and bronchial structures. Lung parenchyma was minced and crushed finely with a scalpel blade, agitated briefly by repeated pipetting, and filtered through a 36-μm pore size nylon membrane (Small Parts Inc., Miami Lakes, FL) to yield a single cell suspension. This procedure effectively releases a mixture of cells and nuclei from the distal lung, although the vigorous mechanical dissociation substantially degrades cell morphology. SCGE slides were prepared by mixing 5 μl of cell suspension with 45 μl of 0.9% Seaplaque (FMC Bioproducts, Rockland, ME) low melting agarose (mixed in PBS without calcium or magnesium), applying this to microscope slides precoated with 1% agarose, and covering with a 22-mm square #1 coverslip. A two-layer preparation was used, as previously described (18). Hardened gels were immersed in lysis solution (2.5 M NaCl, 0.1 M EDTA, 10 mM Tris, pH 10, supplemented with freshly added 1% Triton X-100 and 10% DMSO) for 16–24 h at 4°C before SCGE.
Preparation of Alveolar Type 2 Cells
We adapted the previously described method of Corti and coworkers (20). Mice were killed as described above, the trachea was exposed and intubated with an 18-g intravenous cathether (BD Insyte, Becton Dickinson, Sandy, UT), and the right ventricle perfused with 10 cc of room temperature calcium and magnesium PBS. Lungs were then instilled with 1 cc of freshly prepared 1 mg/ml neutral protease (Worthington Biochemical, Lakewood, NJ) in PBS; neutral protease solution was then “chased” into the distal airspaces with a further instillation of 1 cc of 45°C, 1% low melting point agarose. The mouse thorax was immediately covered with ice; after 4 min time to permit agarose hardening, lung lobes were removed and placed in a tube containing a further 1.5 cc neutral protease solution, and incubated another 15 min at room temperature with gentle agitation. Lung tissue was gently teased apart using two fine-tipped forceps; 7 cc of ice-cold Staining Buffer (PBS containing 3% heat-inactivated fetal calf serum [BioWhittaker, Walkersville, MD] and 0.1% sodium azide) added, the mixture gently triturated through a 10-cc pipet, and the suspension progressively filtered through two layers of gauze followed by the aformentioned 36μm mesh nylon membrane. Cells were pelleted at 300 × g for 5 min, resuspended in a further 10 cc of Staining Buffer, counted, pelleted again, and resuspended at a concentration of 2 × 107 cells per cc. To 1 cc of cells, 30 μg of biotin-conjugated rat monoclonal anti-mouse CD45 (clone 30-F11; BD Biosciences, San Jose, CA) and 20 μg of rat monoclonal anti-mouse CD32 (clone 2.4G2; BD Biosciences) were added and incubated on ice for 15–30 min. A quantity of 10 cc of ice-cold Imag buffer (PBS containing 0.5% BSA, 2 mM EDTA, and 0.1% sodium azide) was added and the cells again pelleted at 300 × g for 5 min. Supernatant was carefully removed and 100 μl of streptavidin particles plus-DM (BD Biosciences) added and incubated for 30 min on ice. Positive (i.e., bead-containing) and negative fractions were then separated using a BD Imagnet (BD Biosciences). Beads were washed a second time with Imag buffer, and the wash pooled with the first negative fraction; the combined negative fractions were subjected to a second placement in the magnet to remove any residual beads. Positive fractions were resuspended directly in 100–500 μl of Imag buffer; negative fractions were pelleted (300 × g for 5 min) and resuspended in 50–150 μl. Cells were processed for SCGE as described above.
Modified Papanicolou Staining
Cells were deposited on glass microscope slides via a 1,500-rpm, 5-min centrifugation in a Shandon Cytospin (Thermo Electron Corp., Waltham, MA) and air dried. Modified Papanicolou staining was performed exactly as described (21).
Immunofluorescence Staining
Cytospin preparations were prepared as described above using microscope slides precoated with albumin fixative (EM Science, Gibbstown, NJ). After fixation for 10 min at room temperature in 4% paraformaldehyde, slides were washed twice with PBS and processed immediately or stored at 4°C. Slides were washed in PBS/0.2% Tween 20 and blocked for 1 h with PBS/0.2% Tween 20/5% normal goat serum (Vector Labs, Burlingame, CA). Primary antibody (rabbit anti-human pro–SP-C; Chemicon International, Temecula, CA) was diluted 1:1,000 in blocking buffer and incubated on the slides for 1 h at room temperature. After three washes in PBS/0.2% Tween 20 and another brief incubation with blocking buffer, secondary antibody (fluorescein-labeled goat anti-rabbit IgG; Vector Labs) diluted 1:100 in blocking buffer was incubated on slides for 30 min After three further washes in PBS/0.2% Tween 20, slides were mounted using Vectashield (Vector Labs) and viewed under epifluorescence illumination using XF22 filter set (Omega Optical, Brattleboro, VT) for fluorescein detection and UV2E-C (Nikon USA, Melville, NY) for DAPI counterstain. Slides to be counterstained with DAPI were incubated briefly in 1.5 μg/ml DAPI in PBS before mounting.
Preparation of Cultured Cells for SCGE Analysis
Cells were removed from the culture dish by gentle trypsin treatment (Clonetics trypsin/EDTA solution; Cambrex, Walkersville, MD), trypsin was neutralized (trypsin-neutralizing solution; Cambrex), and cells were resuspended in a small volume of mincing solution (typically 250 μl for a 35-mm culture dish). Gels were prepared as described above.
SCGE
Slides were equilibrated via three changes in 1× FPG buffer (40 mM Hepes, pH 8.0, 0.1 M KCl, 10 mM EDTA, 0.2 mg/ml BSA) at room temperature. Slides to be treated with formamidopyrimidine glycosylase (FPG; Trevigen, Gaithersburg, MD), a DNA repair enzyme that permits quantitation of base lesions by converting them to single-stranded breaks (see Results), were incubated for 30–60 min at 37°C in the same buffer, containing 3 U of Escherichia coli FPG (Trevigen) per 25 cc. Slides were then flooded three times with freshly prepared ice-cold alkaline running buffer (225 mM NaOH, 1 mM EDTA), and immersed in the same solution in a 20-cm gel rig for 10 min before electrophoresis at 350–450 mA (1.5 V/cm) for 15–20 min at 4°C. Slides were neutralized by repeated flooding with 0.2 M Tris pH 7.5, and dehydrated by flooding with 75% and 95% ETOH. Air-dried slides were stored in the dark at room temperature until ready for analysis.
For rehydration and viewing, a 35-μl drop of 0.4 μg/ml ethidium bromide in 10 mM Tris pH7.5, 1 mM EDTA was added and a coverslip applied. Monochromatic photographs of comets were taken under epifluorescence microscopy using a digital CCD camera (SPOT technologies, Sterling Heights, MI) using the following settings: red filter, gain = 1, 2 × 2 binning (to produce ∼ 336-K file size per image), no γ adjustment. Manual exposure was adjusted to assure that the brightest comets on a slide remained below saturable pixel intensity. FPG experiments were quantitated under medium (×200) or high (×400) magnification. Native SCGE experiments were quantitated under low (×100) or medium power, to permit analysis of up to 1,000 comets per sample (see Results).
Statistical Analysis of SCGE
At least 150 comets were analyzed for each mouse or plate of cells. For experiments measuring the mean level of damage, > 150 cells were quantitated. For experiments measuring the population distribution of DNA strand breakage, 500–1,000 cells per mouse were analyzed. Individual comets were quantitated using Cometscore software (Tritek Corp., Sumerduck, VA), which outputs a spreadsheet containing 17 different comet metrics (parameters describing comet head and tail size and intensity). The Olive moment metric was chosen as the measure of DNA damage with the widest dynamic range (Olive moment is calculated as: the summation of tail intensity profile values [tail intensity profile equals the vertical summation of the pixel intensities in the comet tail] by their relative distances to the center of the comet head, divided by total comet intensity). Identical results were obtained with other common comet metrics such as %DNA in Tail (%DNA in Tail is calculated as summation of the pixel intensities in the tail divided by the summation of the total pixel intensities of the entire comet). Both these metrics are advantageous in that they are insensitive to the total brightness of the comet (because each contains total comet intensity in the denominator); therefore they are not affected by changes in overall slide intensity that can occur because of many factors (exposure time, stain freshness, agarose precoating thickness, etc.). The measurement “%DNA in Tail” is also insensitive to the magnification used to photograph the comet, while Olive moment values increase linearly with magnification. For preparation of compact histograms (e.g., Figure 1B), the “% DNA in Tail” metric was preferentially used. Comet metrics were stored using Microsoft Excel spreadsheets and analyzed using Statview (SAS Inc., Cary, NC). In mouse experiments, each mouse was considered to represent a single data point. Therefore the mean olive moment for each mouse was extracted from all the cells counted for that mouse, and the average of the extracted mean olive moments for each treatment condition was then calculated and compared. Error bars reflect the standard deviation of these averaged means. In this way, each experiment was considered to have data points equivalent to the number of mice studied (usually 3–6 per group), rather than the number of comets analyzed (often upward of several thousand). For cell culture experiments, since all cells were considered to be essentially the same, the latter type of population-based comparison was used. In this case, the 95% confidence interval around the mean, which describes the uncertainty regarding the mean value of each treatment condition, was used to present error bars (because SCGE is a single-cell based assay, standard deviations within the raw comet population reflect population variation, rather than assay variability). P values were calculated via ANOVA.
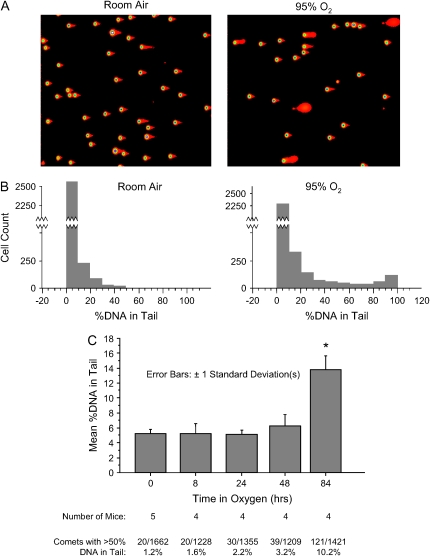
Figure 1.
Native SCGE of lung suspensions from C57Bl6 mice treated with 94% oxygen for 3 d. (A) Representative low-power digital photomicrographs of SCGE gels from a control and an oxygen-exposed mouse. Intensity-map coloration was performed with Cometscore software. (B) Histogram analysis. Discontinuity of the y-axis is used to clarify low-frequency events. (C) Time course. Approximately 300 comets were quantitated for each mouse. Bars depict the mean %DNA in Tail of each group. *P < 0.0001 (by ANOVA) for the difference between the 84-h oxygen exposure group and all other groups. No other comparisons are significant.
5-Bromo-2′-Deoxyuridine Incorporation
The nuceloside analog 5-bromo-2′-deoxyuridine (BrdU; Sigma) was used to specifically label dividing cells. BrdU was dissolved in PBS at a concentration of 10 mM (3.1 mg/ml). For in vitro experiments, BrdU was added directly to tissue culture medium at a final concentration of 100 μm. For in vivo experiments, oxygen-exposed mice were injected intraperitoneally with 1.5 mg BrdU on the day before killing. Care was taken to minimize the time which mice were removed from hyperoxia (2 min or less per cage).
BrdU Detection
A modification of a previously described procedure (22) was used. After SCGE, neutralized gels were rinsed three times in PBS and incubated in 1% blocking solution (Molecular Probes, Eugene, OR). Primary antiserum (biotin-XX–labeled mouse monoclonal anti-BrdU; Molecular Probes) in blocking solution was added to each gel, using a volume of 50 μl per sample; a coverslip was applied and incubation continued for 1 h in a humidified box. Detection was accomplished using horseradish peroxidase–conjugated streptavidin and a tyramide signal amplification kit (both from Molecular Probes), according to manufacturer's directions. Both Alexafluor488 and Alexfluor350 conjugates were used, the former providing a stronger signal. After the detection procedure, slides were rinsed sequentially in TE (10 mM Tris 7.5, 1 mM EDTA), 75% ethanol, and air dried. For viewing and quantitation, slides stained with Alexafluor488 were rehydrated with Hoechst 33342 in TE (6 μg/ml), and those with Alexafluor350 with ethidium bromide (0.4 μg/ ml in TE). Slides were scanned for BrdU-positive cells under epifluorescence illumination using XF22 filter set (Omega Optical) for Alexafluor488 or UV2E-C (Nikon USA) for Alexafluor350. When a positive cell was detected, two monochromatic photographs were taken: one under BrdU-restricted illumination (revealing only the BrdU-positive cell), and another of the same field under Hoechst or ethidium bromide illumination (revealing all the cells by virtue of the nonspecific DNA dye). Matching sets of pictures were then quantitated using Cometscore. Only the Hoechst (or ethidium bromide) stained pictures were quantitated, but the corresponding BrdU-illuminated pictures were viewed simultaneously, allowing assignment of each cell in the field to either BrdU-positive or -negative categories.
RESULTS
DNA Strand Breakage Affects a Minority of Cells in the Oxygen-Exposed Lung
We used the alkaline SCGE to quantitate oxidative DNA damage. Alkaline SCGE (18) detects abasic sites as well as single- and double-strand breaks in the phosphodiester backbone of DNA: these lesions allow greater electrophoretic mobility of the high molecular weight chromatin preparation, producing a “comet tail.” To apply SCGE to the measurement of DNA damage in the lung, we prepared single-cell suspensions of lung tissue from mice exposed to 95% oxygen for 72–96 h. We found that the vast majority of cells did not develop DNA strand breaks in oxygen (Figure 1 and Table 1). In the experiment shown in Figure 1, for example, > 85% of oxygen-exposed cells and > 93% of room air cells fell into the lowest centile of DNA strand breaks. Even with the use of modifications to increase assay sensitivity (prolonged alkali treatment and quantitation using ×400 microscopy), no increase in DNA strand breaks was found in the vast majority of cells (data not shown).
TABLE 1.
FREQUENCY DISTRIBUTION FOR %DNA IN TAIL
| % DNA in Tail
|
Room Air
|
95% Oxygen
|
|||
|---|---|---|---|---|---|
| From | To | Count | Percent | Count | Percent |
| 0 | 10 | 2576 | 84.266 | 2,144 | 72.826 |
| 10 | 20 | 302 | 9.879 | 316 | 10.734 |
| 20 | 30 | 107 | 3.500 | 133 | 4.518 |
| 30 | 40 | 33 | 1.079 | 64 | 2.174 |
| 40 | 50 | 18 | 0.589 | 51 | 1.732 |
| 50 | 60 | 5 | 0.164 | 30 | 1.019 |
| 60 | 70 | 6 | 0.196 | 28 | .951 |
| 70 | 80 | 4 | 0.131 | 26 | .883 |
| 80 | 90 | 2 | 0.065 | 44 | 1.495 |
| 90 | 100 | 4 | 0.131 | 108 | 3.668 |
| Total | 3057 | 100.000 | 2,944 | 100.000 | |
Tabulated results providing raw data from which the histograms shown in Figure 1B were constructed. Lungs from three control (room air) and three oxygen-exposed mice (94% O2 for 3 d) were analyzed by SCGE with low-power quantitation of ∼ 1,000 comets per animal.
There was, however, a change with oxygen exposure: a small proportion of lung cells developed moderate to severe levels of DNA strand breakage (Figure 1). The greatest increase occurred in cells with the most severe damage. For example, only 0.69% (21/3057) of room air–exposed lung cells scored > 50% tail DNA; with oxygen exposure this number increased more than 10-fold, to 8% (236/2944). Significant numbers of these heavily damaged cells were only found with 48 h or more of oxygen exposure (Figure 1C).
DNA Base Damage Affects Most Cells in Oxygen-Exposed Lung
We next investigated oxidative DNA base damage in the lungs by using a modification of SCGE. The bacterial DNA repair enzyme formamidopyrimidine-DNA glycosylase (Fpg) recognizes oxidized purine bases, such as 8-oxoguanine, and nicks the DNA strand adjacent to the damaged base (19); treatment with Fpg thus allows oxidized bases to be scored by alkaline SCGE. Since the number of oxidized bases per cell generally far exceeds the number of alkali-labile sites (see, e.g., Figure 2D), the small contribution of the latter can generally be ignored (17).
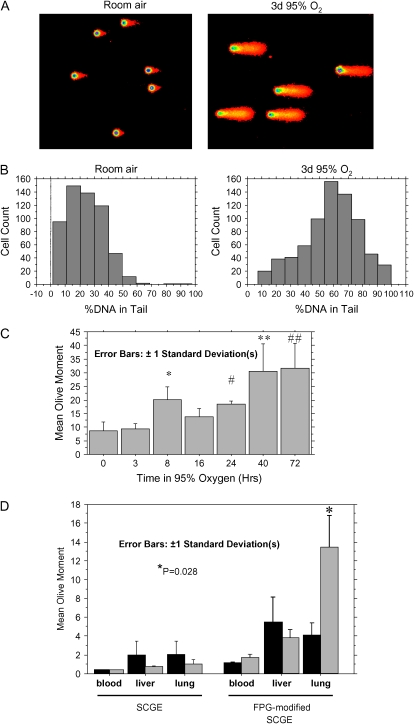
Figure 2.
FPG-modified SCGE of lung suspensions from C57Bl6 mice treated with 94% oxygen. (A) Representative medium-power digital photomicrographs from two mice, colored by Autocomet software. (B) Histogram analysis. A 60 h duration of oxygen exposure was used. (C). Time course: for each time point, > 100 comets from each of three mice were quantitated under high-power microscopy (×400) and the mean value for each mouse extracted. Bars depict the average of the extracted means (i.e., each mouse equals one data point). *P = 0.019; #P = 0.039; **P = 0.0006; ##P = 0.0001 (values are provided for comparison to the 0-h group). (D) SCGE and FPG-modified SCGE run in parallel, to compare the relative amounts of DNA strand breakage and base damage. A 40-h duration of oxygen exposure was used. Statistical analysis as in C. P value is shown for comparison of the room air– and oxygen-exposed lung samples.
In contrast to our results with DNA strand breaks, we found that oxygen exposure increased DNA base damage in essentially the entire population of pulmonary cells (Figures 2A and 2B). Increases in base damage occurred with all periods of oxygen exposure greater than 8 h, with the 8- and 24-h time points reaching statistical significance (Figure 2C). Increases in base damage were therefore evident before DNA strand breaks (compare Figure 1C) or clinical toxicity (data not shown) were evident, and increased steadily (Figure 2C). Quantitatively, base damage outweighed DNA strand breaks at all time points measured. The difference was particularly marked at early time points (Figure 2D, and compare Figure 1C and Figure 2C): before 48 h, no increase in the mean level of DNA strand breaks was observed, while base lesions were already increasing 2- to 4-fold between 8 and 48 h. Oxygen-induced increases in base damage were not detected in liver or nucleated blood cells (Figure 2D), consistent with the minimal effect of hyperoxia on systemic oxygen delivery. In summary, these experiments suggested that the predominant form of DNA damage induced by toxic oxygen levels is oxidative base damage, which becomes evident at early time points and affects virtually the entire population of lung cells.
DNA Strand Breakage, but Not Base Damage, Correlates with Lung Injury
Since these two types of oxidative DNA damage, strand breakage and base modification, exhibited markedly different characteristics, we investigated which of the two was more strongly correlated with progression of acute lung injury. Transgenic mice that overexpress the cytokine IL-6 in lung-specific fashion are dramatically protected from hyperoxic lung injury (15). We therefore investigated how IL-6–induced protection affected oxidative DNA damage in the lung. IL-6–expressing animals and transgene-negative littermate control mice were exposed to oxygen for 72–96 h. With this treatment the IL-6 animals appeared well, while control littermates demonstrated piloerection, decreased mobility, and failure to eat or drink. Single-cell lung suspensions were prepared and analyzed by alkaline and Fpg-modified SCGE.
Alkaline SCGE demonstrated that IL-6 expression markedly attenuated the appearance of cells with severe strand break DNA damage: this was evident in both the mean level of DNA damage (Figure 3A) and in a focused analysis of the percentage of cells with severe strand breakage (Figure 3B). In contrast, Fpg-modified SCGE revealed that IL-6 overexpression did not prevent the accumulation of oxidative base damage (Figure 3C). Fpg-modified SCGE was also performed on IL-11–overexpressing transgenic mice, which demonstrate a similar resistance to hyperoxic lung injury (16). IL-11 expression, despite protecting the animals clinically, also failed to prevent the accumulation of oxidized base damage (Figure 3C).
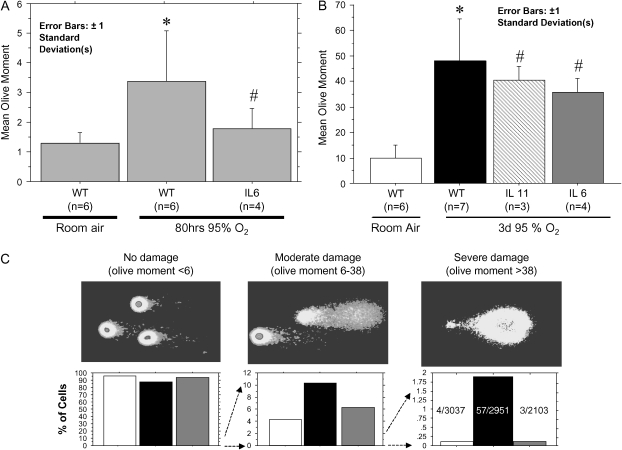
Figure 3.
SCGE analysis of wild-type and IL-6–overexpressing transgenic mice exposed to 94% oxygen for 80 h. (A) Native SCGE analysis quantitated under low-power (×100) microscopy. More than 500 comets from each mouse were quantitated and the mean olive moment for each mouse extracted. Bars depict the average of the extracted mean olive moments for each group. *P < 0.01 versus WT, room air; #P < 0.05 versus WT, oxygen. (B) FPG-modified SCGE. More than 100 comets from each mouse were quantitated under high power with statistical analysis as in A. *P < 0.0001 versus room air; #P < 0.001 versus room air, NS versus WT. (C) Pictorial and graphic depiction of the distribution of cells from experiment shown in A. Lower panels: open bars, WT, room air; black bars, WT, 3 d 95% O2; shaded bars, IL-6, 3 d 95% O2.
In summary, we found that DNA strand breaks, despite only affecting a minority of pulmonary cells, correlated with survival. Oxidized base damage, despite being quantitatively predominant and affecting the majority of cells, did not.
DNA Strand Breakage in the Lung Is Not a Manifestation of DNA Replication
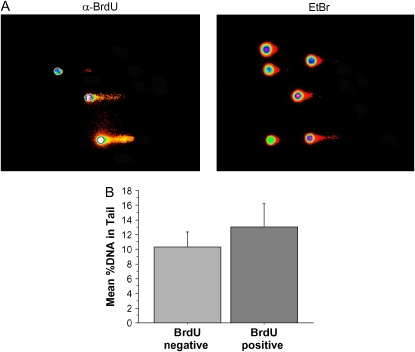
Because of its correlation with lung injury, we wished to further characterize DNA strand breakage. Several characteristics of this lesion suggested that it might be a secondary effect of a cellular process, rather than a direct result of reactive oxygen species attack on DNA. First, we observed increases in DNA strand breakage only after > 48 h of hyperoxia, when animals began to appear ill. Direct, reactive oxygen species attack on DNA would be expected to occur more quickly (as was observed with base damage, Figure 2C). Second, cells developing strand breaks generally had severe levels; intermediate amounts of damage were less common (Figure 1). Sudden appearance of severe levels of strand breakage in a minority of cells did not seem consistent with damage caused by the continuous presence of reactive oxygen species (although we cannot rule out, for example, a delayed oxidant effect occurring in one susceptible cell type). Instead, we thought it might result from either of two cellular processes: DNA replication or programmed cell death. We first investigated DNA replication. Cells undergoing DNA replication inevitably have DNA strand breaks, because of the discontinuous nature of DNA synthesis on one half of the replication fork (18). As oxygen can stimulate cell proliferation, it is possible that cells with DNA strand breakage are undergoing cell division. Determining this required knowledge of how DNA replication affects SCGE under our assay conditions. We used an in vitro strategy to investigate this. Cultured cells were pulsed with the nucleoside analog BrdU for 1 h before being subjected to SCGE. After SCGE, replicating cells, which had incorporated BrdU, were identified using BrdU-specific antisera coupled to a fluorochrome optically separable from the DNA stain. When we looked specifically at newly replicated DNA, using the BrdU-specific stain, we found, as expected, that a considerable proportion was present in the tail (left panel of Figure 4). This is consistent with the known discontinuous nature of eukaryotic lagging strand replication (18). Examining total DNA, however, we found that replicating cells closely resembled their nonreplicating counterparts (Figure 4, right panel), and quantitation revealed only a small increase in tail DNA percentage in the replicating cells (Figure 4C). Similar results were obtained using both transformed cells (A549 lung cancer line) and primary human cultures (human pulmonary microvascular endothelial cells). Thus, DNA replication caused only a small increase in DNA strand breaks. The high levels of strand breaks observed in a minority of oxygen-exposed pulmonary cells (> 50% DNA in tail; Figure 1) were inconsistent with DNA replication, and indicative of much more severe DNA strand breakage.
Figure 4.
Effect of DNA replication on alkaline SCGE. Primary pulmonary microvascular endothelial cells were pulsed with 100 μm BrdU for 1 h before SCGE. BrdU-containing cells were detected as described in Materials and Methods. (A) Monochromatic images intensity-mapped by the image-analysis program Cometscore; these images are used for quantitation. Right panel, EtBr illumination; left panel, BrdU-specific illumination. (B) Quantitation of 100 BrdU-positive and 200 accompanying BrdU-negative cells.
DNA Strand Breakage in the Lung Is Consistent with Cell Death
Cell death via either apoptosis or necrosis is associated with DNA degradation (18). This seemed a likely explanation of the severe strand breakage seen in some cells in oxygen-exposed lung. We therefore investigated whether apoptosis, necrosis, or both could produce similar comet characteristics under our assay conditions. We first evaluated a well-validated model of apoptosis. After staurosporine treatment, Jurkat cells underwent classical apoptosis, verified by morphologic features and DNA laddering (data not shown). We found that severely damaged comets (> 50% DNA in the tail) appeared relatively early in apoptosis, 6–10 h before cells became nonviable by trypan exclusion (see Figure E1A in the online supplement). Heat-induced necrosis (which caused immediate nonviability by dye exclusion) also, however, resulted in comets with > 50% DNA in tail morphology (Figure E1A); in this case the change occurred within minutes after cells were heated. These results indicated that severely damaged comets were consistent with cell death, but that either apoptosis or necrosis could produce such a comet morphology.
We also noted that the damaged cells from oxygen-exposed lung appeared far dimmer than their undamaged counterparts. We therefore evaluated whether an additional parameter—loss of DNA signal—could distinguish between the different modes of cell death. First we needed to accurately measure how much DNA signal is lost in severely damaged cells from oxygen-exposed lung. Reliance on raw pixel intensity is problematic because numerous technical factors affect how brightly different microscope slides stain (exposure time, bulb alignment, stain freshness, agarose precoating thickness); thus the same cell may have somewhat different intensities on different slides or with different batches of stain. We therefore normalized the total DNA signal from damaged comets to the mean of that measured in 50–100 undamaged comets on the same slide (defining undamaged as containing < 20% DNA in Tail). We searched out cells with strand breakage from slides of oxygen-treated mouse lung: these cells possessed on average only 31% (± 17% SD) of the DNA of surrounding undamaged cells (Figure E1B). Using our in vitro models of necrosis and apoptosis, however, we found that both of these processes could also lead to similar levels of DNA loss (Figure E1B). We therefore could not unambiguously distinguish apoptosis from necrosis using SCGE, and the 30% of DNA remaining in oxygen-damaged lung cells was consistent with either mode of cell death.
Finally, we compared the comet morphology of cell death with that produced by hydrogen peroxide exposure, an oxidant which can produce DNA strand breaks without lethality (at appropriate doses). A549 cells were exposed to a a sublethal dose of hydrogen peroxide (250 μM) before SCGE. Hydrogen peroxide exposure also produced comets with > 50% DNA in the tail, but they possessed different characteristics. First, DNA strand breaks produced by hydrogen peroxide were repaired rapidly (Figure E1A). Second, hydrogen peroxide comets did not lose DNA signal (Figure E1B); that is, despite a major morphology alteration in which the majority of the DNA signal moved to the tail, the hydrogen peroxide comets still had approximately the same total pixel brightness that was present in control cells. This was significantly different (P < 0.001) from the DNA loss seen in apoptosis, necrosis, and in cells from the oxygen-exposed mouse lung. The characteristics of comets found in mouse lung (> 50% DNA in tail and only 30% DNA retention) were therefore most consistent with cell death, via either apoptosis or necrosis.
DNA Strand Breaks Preferentially Affect Dividing Cells
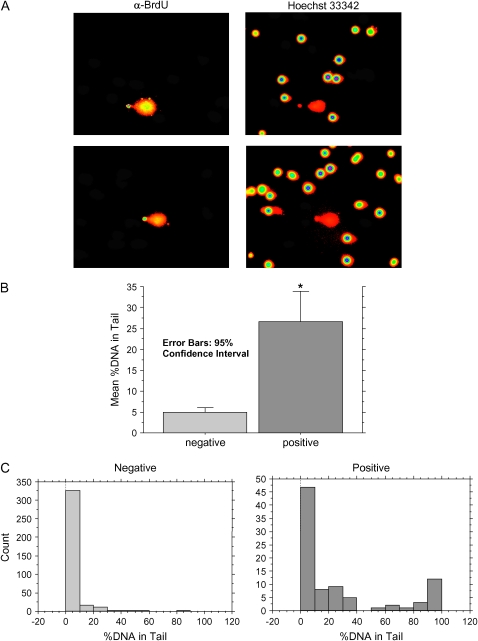
Although the mitotic index of adult lung is very low, oxygen is known to stimulate alveolar epithelial cell proliferation (11). We hypothesized that these dividing cells, as opposed to mitotically quiescent cells, might be more likely to die under the oxidant stress of hyperoxia. We investigated this possibility using in vivo incorporation of BrdU. Oxygen-exposed mice were injected intraperitoneally with BrdU 1 d before lung harvest for SCGE, and dual detection of total DNA and BrdU was performed as described above (Figure 5). BrdU-positive cells showed markedly increased DNA strand breakage compared with BrdU-negative cells (Figure 5B). Histogram analysis revealed that this difference was driven by the appearance of BrdU-positive cells with severe strand breakage (Figure 6C). The majority of all cells, BrdU-negative and -positive alike, displayed little DNA strand breakage; however, of the minority of cells with severe strand breakage, most were BrdU positive. Thus cell death occurred preferentially among dividing cells.
Figure 5.
BrdU incorporation in vivo. Mice exposed to 95% oxygen for 3 d were injected with 1.5 mg of BrdU intraperitoneally 24 h before preparation of lung samples for SCGE followed by BrdU detection as described above. (A) Two representative sets of monochromatic images intensity mapped by Cometscore. Left panels are taken under BrdU-specific illumination; right panels show the same field under total DNA illumination. B and C show, respectively, mean levels of %DNA in Tail and histogram analysis. *P < 0.001 for comparison to BrdU-negative sample.
Figure 6.
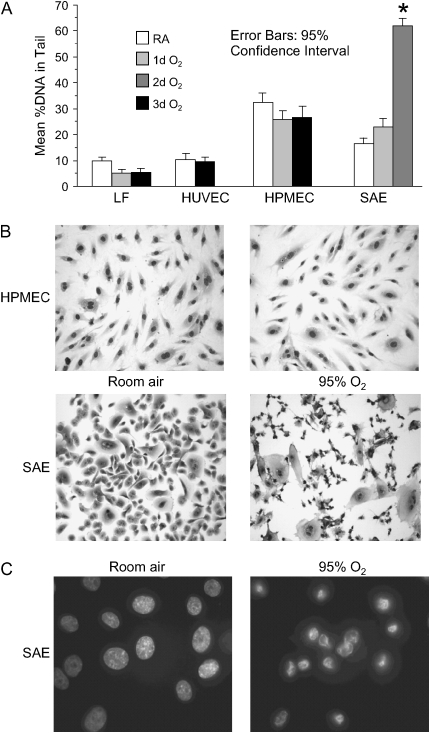
Behavior of primary human cells exposed to 88% oxygen, 5% CO2 atmosphere. (A) Native SCGE analysis: > 150 cells from each treatment condition were quantitated under medium power microscopy. *P < 0.001 for comparison to room air and 1 d O2 samples. LF, lung fibroblasts; HPMEC, pulmonary microvascular endothelial cells; HUVEC, umbilical vein endothelial cells; SAE, small airway epithelial cells. (B) Light microscopy (×400) of Wright-stained cells exposed to humidified 88% oxygen, 5% carbon dioxide atmosphere for 3 d. (C) Fluorescent micrographs (×1,000) of Hoechst 33342–stained SAE cells.
Airway Epithelial Cells Develop DNA Strand Breaks In Vitro
In an attempt to determine which cell types of the lung are more susceptible to oxygen toxicity, we measured oxidative DNA damage in a variety of primary human cells exposed to elevated oxygen levels. Cultures were exposed to humidified 88% O2, 5% CO2 for 1–3 d and analyzed by native and FPG-modified SCGE. Neither human pulmonary microvascular endothelial cells, human umbilical vein endothelial cells, or human lung fibroblasts accumulated strand break DNA damage with oxygen exposure (Figure 6A); these cells did, however, show an increase in oxidized base lesions (Figure E2). These three cell types exhibited little morphologic change with oxygen exposure (Figure 6B and data not shown).
Small airway epithelial cells, on the other hand, demonstrated a dramatic response to hyperoxia. A large increase in strand break DNA damage was seen with 2–3 d of oxygen exposure (Figure 6A). Morphologically, marked cytotoxic change was evident, with cells losing a plump, ovoid outline and developing an elongated, tethered shape with condensed, pyknotic nuclei (Figure 6B). Hoechst 33342 staining revealed condensed, polygonal nuclei with rimming of chromatin (Figure 6C). In small airway cells exposed to oxygen, DNA strand breakage did not develop gradually, but appeared suddenly along with the occurrence of morphologic change, suggesting that DNA strand breakage was a secondary to cytotoxicity.
DNA Strand Breaks Preferentially Affect Type 2 Alveolar Cells In Vivo
These in vitro results prompted us to examine epithelial cells in vivo. The dividing cells that repopulate alveolar epithelium are type 2 alveolar cells. We therefore adapted a previously published method of type 2 cell preparation (20, 23) for use with SCGE. This method uses neutral protease digestion of distal airways to produce a crude, epithelial-enriched cell suspension, followed by immunomagnetic bead–based depletion of contaminating hematopoetic cells. We made the following changes. To minimize cell metabolic activity, including DNA repair, we maintained suspensions on ice for all steps after the initial protease digestion; limited this step to 15 min at room temperature; and also omitted a several-hour, differential adherence to plastic step. DNase treatment of the cell suspensions was also omitted, to prevent artifactual DNA damage. Modified Papanicolou staining permitted unambiguous identification of type 2 cells by virtue of their dark blue cytoplasmic granules (Figure 7A, left and middle panels of upper row; larger versions of these pictures are also provided as Figures E3A–E3C). Detection of surfactant protein C by indirect immunofluorescence confirmed this identification (Figure E4). Analysis of crude cell fractions using a Wright stain revealed the following composition: ∼ 24% epitheloid-appearing cells, 37% lymphocytes, 15% monocytes, 15% granulocytes, 1% eosinophils, and 10% cells that defied certain identification.
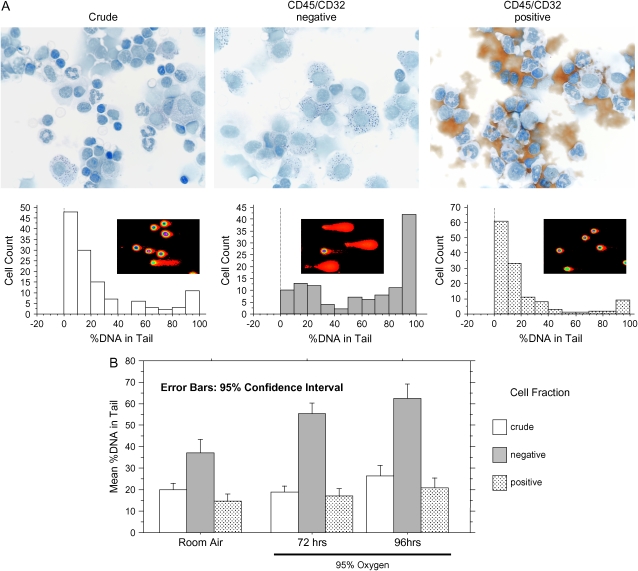
Figure 7.
DNA strand breaks in alveolar cell preparations. (A) Top panels: modified Papanicolou staining. Brown material in positive fraction is the magnetic beads. Bottom panels: histogram analysis of SCGE results for > 100 cells from each fraction, quantitated under ×400 magnification. (B) Mean values from SCGE quantitation of the indicated fractions from three mice. More than 100 cells were quantitated under high power for each sample. Concordant results were obtained in three independent experiments.
Our protocol did not yield the 90% pure type 2 preparation of published reports (20, 23), likely because of the changes made. However, substantial purification was achieved: CD45/CD32-negative fractions contained 50% (± 4% SD) type 2 cells, compared with 6% (± 3% SD) present in crude suspensions (Figures 7A and E3A–E3B). CD45/CD32-positive fractions contained primarily lymphocytes and granulocytes, with lesser amounts of macrophage, monocytoid, and epitheloid appearing cells (Figures 7A and E3C).
SCGE was performed on alveolar cell preparations from oxygen-exposed mice (Figure 7A, bottom panels). Dramatically more cells with DNA strand breakage were found in the type 2 cell–enriched fractions (Figure 7A, middle panel) than in the crude and the CD45/CD32-positive fractions (Figure 7A, left and right panels). Quantitation of the mean amount of DNA damage revealed significantly higher levels in the alveolar fraction (Figure 7B), and oxygen-exposed alveolar cells displayed more damage compared with control preparations (Figure 7B).
DISCUSSION
Based on the in vitro properties of oxygen and other oxidants such as hydrogen peroxide, inhaled oxygen has been assumed to cause oxidative damage to nucleic acids in vivo (13). In this report, we used alkaline SCGE to directly measure DNA damage at the single-cell level in the lungs of mice exposed to 95% oxygen. We quantitated two chemically distinct forms of oxidative DNA damage: breaks in the phosphodiester backbone, and oxidative alterations to nitrogenous bases. Although both types of damage were detected, we found a marked dichotomy between the two.
Quantitatively, oxidized bases were far and away the predominant lesion, in vivo and in vitro. Oxidized bases displayed characteristics (affecting the vast majority of cells in the lung but not other tissues, occurring within hours of oxygen exposure, and gradually increasing over time) consistent with a primary effect of reactive oxygen species attack on DNA. DNA strand breaks, on the other hand, displayed features which suggest that they are a secondary consequence of nucleases activated during cell death, rather than a primary effect of oxygen-generated reactive species. These features included the following: (1) strand breaks affected only a small minority of cells within the lung; (2) strand breaks appeared only with prolonged (> 48 h) oxygen exposure; (3) cells possessing strand breaks were severely affected, intermediate levels of damage being less common; and (4) cells with strand breaks displayed a comet morphology similar to that observed with apoptosis and necrosis in vitro, including marked loss of DNA.
These results are significant for several reasons. First, the finding of DNA base damage with even short exposure to oxygen highlights the important effect of inhaled substances on the pulmonary genome. This has implications for a number of lung diseases associated with oxidative injury, including lung cancer, emphysema, and pulmonary fibrosis. The lack of correlation between base damage and acute lung injury is not unexpected, because 8-oxoguanine is predominantly a mutagenic, rather than directly toxic lesion; by mispairing with adenine it can cause guanine to thymine transversions (24). Considerable evidence implicates 8-oxoguanine in lung cancer (25–27), and our findings directly link this cancer promoting lesion to inhaled oxidants. Although we used oxygen as a model, many other environmental pollutants, including ozone and particulate matter, have oxidizing properties (28, 29).
Second, our results with DNA strand breaks have implications for the mechanism of acute lung injury. We found that progressive lung injury was accompanied by death of a small number of dividing cells within the lung, that primary respiratory epithelial cells developed DNA strand breaks and cytotoxicity when exposed to oxygen in vitro, and that alveolar type 2 cells from oxygen-exposed mice display severe DNA strand breakage suggestive of cell death. Taken together, these findings suggest that acute lung injury involves death of dividing epithelial progenitors, type 2 cells being the epithelial progenitor cells of the alveolar compartment (30). Considerable prior work supports this conclusion. First, alveolar type 2 cells proliferate in response to hyperoxia (11). Second, numerous studies have found evidence of alveolar epithelial necrosis and apoptosis in oxygen-exposed lung (12, 31–33), as well as in cultured respiratory epithelial cells in vitro (34); oxygen-stimulated apoptosis of primary alveolar type 2 cells has also been reported (35). Finally, the concept that proliferating cells are more susceptible to programmed cell death under oxidant stress than mitotically quiescent cells is consistent with the known linkage between cell cycle checkpoints and apoptosis (5). We hypothesize that repair of alveolar damage in hyperoxia, which requires type 2 cell proliferation, may be crippled by death of these dividing progenitors.
In addition, our results are significant in that they suggest that the progression of acute lung injury does not depend simply on the total amount of oxidative stress incurred, but rather on the reaction to such stress. We found that IL-6–overexpressing mice, which are resistant to oxygen toxicity, accumulated levels of oxidative base damage similar to that of susceptible mice, indicative of similar oxidative stress. This is consistent with prior work failing to find changes in antioxidant pathways with IL-6 overexpression (15). However, the protected mice did not develop the DNA strand breaks that mark dying cells, suggesting that the cytokine alters the cell death response in the face of oxidative stress. This may occur by upregulation of the anti-apoptotic regulatory protein BCL-2, which has been reported in the lungs of these mice (15). Overall, these data suggest that when oxidative stress cannot be avoided, modulation of cell death regulatory pathways may prevent short-term organ damage.
Our data do not shed light on the question of whether apoptosis or necrosis is the primary mode of cell death in the alveolar epithelium. We were unable to find properties of SCGE specific for either mode of cell death. Cell death that we observed in oxygen-exposed small airway epithelial cells morphologically resembled apoptosis (see Figure 7), but we have not found evidence of DNA laddering or caspase 3 activation (data not shown). Features of both apoptosis and necrosis are present in the intact lung (12), and a strict demarcation between these pathways has been questioned (36).
Our results with base damage agree with those of two prior studies, which examined 8-oxoguanine levels in oxygen-exposed rodent lungs using immunohistochemical detection. Roper and coworkers reported that 48 h of hyperoxia increased 8-oxoguanine in > 60% of mouse lung cells co-expressing alveolar type 2 cell markers (10), and Auten and colleagues reported extensive 8-oxoguanine staining in a newborn rat lung (37). We note that Roper and coworkers found intense 8-oxoguanine staining localized to mitochondria (10). As mitochondria are lost in the SCGE, we cannot exclude the possibility that the damage we observed in nuclear DNA represents only the lower limit of oxidative base damage present. We also cannot exclude the possibility that oxygen might cause DNA strand breaks in mitochondrial DNA. As oxygen can stimulate production of reactive oxygen species from mitochondria (2), damage to mitochondrial DNA may even be more prominent than damage to nuclear DNA, and such mitochondrial DNA damage may initiate apoptosis (38).
Our finding that base damage did not correlate with toxicity conflicts with the results of Wu and colleagues, who reported that overexpression of enzymes specific for the repair of 8-oxoguanine increased A549 cell survival in hyperoxia (39). The discrepancy is likely due to the fact that although A549 survival correlated with decreased DNA damage in the study by Wu and coworkers, only DNA strand breaks and not base damage was measured. Our findings with small airway cells also differ from those reported by Jyonouchi and colleagues (40), who reported a more modest effect of enriched oxygen on morphologically determined apoptotic index using the same primary cell cultures. As primary small airway cells are prepared from human cadavers, genetic and environmental differences between lots may be responsible for such differences.
Our finding that oxygen exposure caused predominant base modification rather than DNA strand breakage may seem surprising, as it is generally assumed that reactive oxygen species induce the latter (4, 11). However, surprisingly few studies have quantitated DNA damage induced in cells or animals by oxygen. Cacciuttolo and coworkers (8) found transient DNA strand breakage in response to very high dissolved oxygen concentrations in cells cultured in vitro using an alkaline unwinding assay; however, the assay methodology did not distinguish between moderate strand breakage occurring in most cells from severe fragmentation occurring in a minority, and the atmosphere was hyperbaric. Evidence for DNA strand breakage in the oxygen-exposed lung relies on TUNEL staining; despite great utility as a marker of programmed cell death, TUNEL is not quantitative and can be difficult to adjust to an appropriate level of sensitivity. We suspect that TUNEL identifies the small proportion of cells that score as having severe DNA damage in SCGE. In fact, our SCGE estimate of the percentage of cells with severe DNA damage agrees well with the TUNEL results of Barazzone and colleagues (12). After 80 h of oxygen exposure, we found that 5–8% of cells possessed severe DNA damage (see Figure 1B and Table 1); Barazzone and coworkers estimated the TUNEL positivity rate as 6% at 48 h, increasing to 15% at 90 h (12). Our results are hard to reconcile, however, with studies that show very extensive TUNEL staining involving the majority of bronchial and alveolar epithelium even at early time points after exposure (41). A possible explanation could be that apoptotic cells are so adherent that they stick to debris present after disaggregation, and thus fail to be liberated into our cell suspension (adhering instead to the filtered material).
In summary, these results demonstrate that even brief oxygen exposure caused base damage in the lung, which appeared early and demonstrated characteristics consistent with a primary effect of reactive oxygen species attack on DNA. This finding has implications for the ability of inhaled oxidants to alter the pulmonary genome. Prolonged oxygen exposure also led to DNA strand breaks in a small proportion of pulmonary cells; the distribution and characteristics of this lesion suggested that it is likely a secondary effect of nucleases activated during cell death. DNA strand breaks affected primarily type 2 alveolar epithelial cells and correlated with the progression of lung disease, suggesting that type 2 cell injury plays a key role in pulmonary oxygen toxicity. Finally, the effects of IL-6 expression on DNA damage suggest that modulation of cell death pathways, even in the face of continued oxidative stress, may prevent short-term tissue damage.
Supplementary Material
Acknowledgments
The authors are grateful to Po-Shun Lee, M.D., for helpful discussions.
This work was supported by National Institutes of Health grants HL074859 and HL03888–01 to A.B.W. and 2004 GlaxoSmithKline Pulmonary Fellowship Award to G.F.B.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2005-0340OC on March 30, 2006
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Smith LJ. Hyperoxic lung injury: biochemical, cellular, and morphologic characterization in the mouse. J Lab Clin Med 1985;106:269–278. [PubMed] [Google Scholar]
- 2.Chandel NS, Schumacker PT. Cellular oxygen sensing by mitochondria: old questions, new insight. J Appl Physiol 2000;88:1880–1889. [DOI] [PubMed] [Google Scholar]
- 3.Cochrane CG. Cellular injury by oxidants. Am J Med 1991;91:23S–30S. [DOI] [PubMed] [Google Scholar]
- 4.Halliwell B, Aruoma OI. DNA damage by oxygen-derived species: its mechanism and measurement in mammalian systems. FEBS Lett 1991; 281:9–19. [DOI] [PubMed] [Google Scholar]
- 5.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature 2000;408:433–439. [DOI] [PubMed] [Google Scholar]
- 6.Termini J. Hydroperoxide-induced DNA damage and mutations. Mutat Res 2000;450:107–124. [DOI] [PubMed] [Google Scholar]
- 7.Cantoni O, Cattabeni F, Stocchi V, Meyn RE, Cerutti P, Murray D. Hydrogen peroxide insult in cultured mammalian cells: relationships between DNA single-strand breakage, poly(ADP-ribose) metabolism and cell killing. Biochim Biophys Acta 1989;1014:1–7. [DOI] [PubMed] [Google Scholar]
- 8.Cacciuttolo MA, Trinh L, Lumpkin JA, Rao G. Hyperoxia induces DNA damage in mammalian cells. Free Radic Biol Med 1993;14:267–276. [DOI] [PubMed] [Google Scholar]
- 9.Ziel KA, Grishko V, Campbell CC, Breit JF, Wilson GL, Gillespie MN. Oxidants in signal transduction: impact on DNA integrity and gene expression. FASEB J 2005;19:387–394. [DOI] [PubMed] [Google Scholar]
- 10.Roper JM, Mazzatti DJ, Watkins RH, Maniscalco WM, Keng PC, O'Reilly MA. In vivo exposure to hyperoxia induces DNA damage in a population of alveolar type II epithelial cells. Am J Physiol Lung Cell Mol Physiol 2004;286:L1045–L1054. [DOI] [PubMed] [Google Scholar]
- 11.O'Reilly MA. DNA damage and cell cycle checkpoints in hyperoxic lung injury: braking to facilitate repair. Am J Physiol Lung Cell Mol Physiol 2001;281:L291–L305. [DOI] [PubMed] [Google Scholar]
- 12.Barazzone C, Horowitz S, Donati YR, Rodriguez I, Piguet PF. Oxygen toxicity in mouse lung: pathways to cell death. Am J Respir Cell Mol Biol 1998;19:573–581. [DOI] [PubMed] [Google Scholar]
- 13.Albertine KH, Plopper CG. DNA oxidation or apoptosis: will the real culprit of dna damage in hyperoxic lung injury please stand up? Am J Respir Cell Mol Biol 2002;26:381–383. [DOI] [PubMed] [Google Scholar]
- 14.Waxman AB, Einarsson O, Seres T, Knickelbein RG, Homer R, Warshaw JB, Johnston R, Elias JA. Targeted lung expression of interleukin-11 enhances murine tolerance of 100% oxygen and diminishes hyperoxia-induced DNA fragmentation. Chest 1999;116:8S–9S. [DOI] [PubMed] [Google Scholar]
- 15.Ward NS, Waxman AB, Homer RJ, Mantell LL, Einarsson O, Du Y, Elias JA. Interleukin-6-induced protection in hyperoxic acute lung injury. Am J Respir Cell Mol Biol 2000;22:535–542. [DOI] [PubMed] [Google Scholar]
- 16.Waxman AB, Einarsson O, Seres T, Knickelbein RG, Warshaw JB, Johnston R, Homer RJ, Elias JA. Targeted lung expression of interleukin-11 enhances murine tolerance of 100% oxygen and diminishes hyperoxia-induced DNA fragmentation. J Clin Invest 1998;101:1970–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins AR. The comet assay: principles, applications, and limitations. Methods Mol Biol 2002;203:163–177. [DOI] [PubMed] [Google Scholar]
- 18.Olive PL. The comet assay: an overview of techniques. Methods Mol Biol 2002;203:179–194. [DOI] [PubMed] [Google Scholar]
- 19.Piperakis SM, Visvardis EE, Tassiou AM. Comet assay for nuclear DNA damage. Methods Enzymol 1999;300:184–194. [DOI] [PubMed] [Google Scholar]
- 20.Corti M, Brody AR, Harrison JH. Isolation and primary culture of murine alveolar type II cells. Am J Respir Cell Mol Biol 1996;14:309–315. [DOI] [PubMed] [Google Scholar]
- 21.Dobbs LG. Isolation and culture of alveolar type II cells. Am J Physiol 1990;258:L134–L147. [DOI] [PubMed] [Google Scholar]
- 22.McGlynn AP, Wasson G, O'Connor J, McKerr G, McKelvey-Martin VJ, Downes CS. The bromodeoxyuridine comet assay: detection of maturation of recently replicated DNA in individual cells. Cancer Res 1999;59:5912–5916. [PubMed] [Google Scholar]
- 23.Rice WR, Conkright JJ, Na CL, Ikegami M, Shannon JM, Weaver TE. Maintenance of the mouse type II cell phenotype in vitro. Am J Physiol Lung Cell Mol Physiol 2002;283:L256–L264. [DOI] [PubMed] [Google Scholar]
- 24.Culp SJ, Cho BP, Kadlubar FF, Evans FE. Structural and conformational analyses of 8-hydroxy-2'-deoxyguanosine. Chem Res Toxicol 1989;2: 416–422. [DOI] [PubMed] [Google Scholar]
- 25.Asami S, Manabe H, Miyake J, Tsurudome Y, Hirano T, Yamaguchi R, Itoh H, Kasai H. Cigarette smoking induces an increase in oxidative DNA damage, 8-hydroxydeoxyguanosine, in a central site of the human lung. Carcinogenesis 1997;18:1763–1766. [DOI] [PubMed] [Google Scholar]
- 26.Lu R, Nash HM, Verdine GL. A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr Biol 1997;7:397–407. [DOI] [PubMed] [Google Scholar]
- 27.Xie Y, Yang H, Cunanan C, Okamoto K, Shibata D, Pan J, Barnes DE, Lindahl T, McIlhatton M, Fishel R, et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res 2004;64: 3096–3102. [DOI] [PubMed] [Google Scholar]
- 28.Knaapen AM, Borm PJ, Albrecht C, Schins RP. Inhaled particles and lung cancer. Part A: Mechanisms. Int J Cancer 2004;109:799–809. [DOI] [PubMed] [Google Scholar]
- 29.Donaldson K, Stone V, Borm PJ, Jimenez LA, Gilmour PS, Schins RP, Knaapen AM, Rahman I, Faux SP, Brown DM, et al. Oxidative stress and calcium signaling in the adverse effects of environmental particles (PM10). Free Radic Biol Med 2003;34:1369–1382. [DOI] [PubMed] [Google Scholar]
- 30.Kotton DN, Summer R, Fine A. Lung stem cells: new paradigms. Exp Hematol 2004;32:340–343. [DOI] [PubMed] [Google Scholar]
- 31.Lee PJ, Choi AM. Pathways of cell signaling in hyperoxia. Free Radic Biol Med 2003;35:341–350. [DOI] [PubMed] [Google Scholar]
- 32.Budinger GR, Chandel NS. The role of cell suicide or apoptosis in the pathophysiology of acute lung injury. Intensive Care Med 2001;27: 1091–1093. [DOI] [PubMed] [Google Scholar]
- 33.Otterbein LE, Kolls JK, Mantell LL, Cook JL, Alam J, Choi AM. Exogenous administration of heme oxygenase-1 by gene transfer provides protection against hyperoxia-induced lung injury. J Clin Invest 1999; 103:1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buccellato LJ, Tso M, Akinci OI, Chandel NS, Budinger GR. Reactive oxygen species are required for hyperoxia-induced Bax activation and cell death in alveolar epithelial cells. J Biol Chem 2004;279:6753–6760. [DOI] [PubMed] [Google Scholar]
- 35.Buckley S, Barsky L, Driscoll B, Weinberg K, Anderson KD, Warburton D. Apoptosis and DNA damage in type 2 alveolar epithelial cells cultured from hyperoxic rats. Am J Physiol 1998;274:L714–L720. [DOI] [PubMed] [Google Scholar]
- 36.Proskuryakov SY, Konoplyannikov AG, Gabai VL. Necrosis: a specific form of programmed cell death? Exp Cell Res 2003;283:1–16. [DOI] [PubMed] [Google Scholar]
- 37.Auten RL, Whorton MH, Nicholas Mason S. Blocking neutrophil influx reduces DNA damage in hyperoxia-exposed newborn rat lung. Am J Respir Cell Mol Biol 2002;26:391–397. [DOI] [PubMed] [Google Scholar]
- 38.Ruchko M, Gorodnya O, LeDoux SP, Alexeyev MF, Al-Mehdi AB, Gillespie MN. Mitochondrial DNA damage triggers mitochondrial dysfunction and apoptosis in oxidant-challenged lung endothelial cells. Am J Physiol Lung Cell Mol Physiol 2005;288:L530–L535. [DOI] [PubMed] [Google Scholar]
- 39.Wu M, He YH, Kobune M, Xu Y, Kelley MR, Martin WJ II. Protection of human lung cells against hyperoxia using the DNA base excision repair genes hOgg1 and Fpg. Am J Respir Crit Care Med 2002;166:192–199. [DOI] [PubMed] [Google Scholar]
- 40.Jyonouchi H, Sun S, Abiru T, Chareancholvanich S, Ingbar DH. The effects of hyperoxic injury and antioxidant vitamins on death and proliferation of human small airway epithelial cells. Am J Respir Cell Mol Biol 1998;19:426–436. [DOI] [PubMed] [Google Scholar]
- 41.O'Reilly MA, Staversky RJ, Stripp BR, Finkelstein JN. Exposure to hyperoxia induces p53 expression in mouse lung epithelium. Am J Respir Cell Mol Biol 1998;18:43–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.