Abstract
Despite having identical cystic fibrosis transmembrane conductance regulator genotypes, individuals with ΔF508 homozygous cystic fibrosis (CF) demonstrate significant variability in severity of pulmonary disease. This investigation used high-density oligonucleotide microarray analysis of nasal respiratory epithelium to investigate the molecular basis of phenotypic differences in CF by (1) identifying differences in gene expression between ΔF508 homozygotes in the most severe 20th percentile of lung disease by forced expiratory volume in 1 s and those in the most mild 20th percentile of lung disease and (2) identifying differences in gene expression between ΔF508 homozygotes and age-matched non-CF control subjects. Microarray results from 23 participants (12 CF, 11 non-CF) met the strict quality control guidelines and were used for final data analysis. A total of 652 of the 11,867 genes identified as present in 75% of the samples were significantly differentially expressed in one of the three disease phenotypes: 30 in non-CF, 53 in mild CF, and 569 in severe CF. An analysis of genes differentially expressed by severity of CF lung disease demonstrated significant upregulation in severe CF of genes involved in protein ubiquination (P < 0.04), mitochondrial oxidoreductase activity (P < 0.01), and lipid metabolism (P < 0.03). Analysis of genes with decreased expression in patients with CF compared with control subjects demonstrated significant downregulation of genes involved in airway defense (P < 0.047) and protein metabolism (P < 0.048). This study suggests that differences in CF lung phenotype are associated with differences in expression of genes involving airway defense, protein ubiquination, and mitochondrial oxidoreductase activity and identifies specific new candidate modifiers of the CF phenotype.
Keywords: cystic fibrosis, gene expression, phenotype, respiratory epithelium
Cystic fibrosis (CF) is the most common lethal autosomal recessive disorder in the white population. Although CF affects multiple organ systems, it is the severe lung disease that leads to the shortened life expectancy of 35.1 yr (1). The gene responsible for the disease, the cystic fibrosis transmembrane conductance regulator (CFTR), was first identified in 1989, and since then over 1,000 different mutations have been reported. ΔF508 is the most common mutation, and over 50% of individuals with CF are homozygous for the ΔF508/ΔF508 genotype (1).
Although certain aspects of CF phenotype, such as pancreatic insufficiency, are determined by CFTR genotype, most other aspects are not. Within the group of ΔF508 homozygotes, a full range in severity of pulmonary disease is seen, with some developing severe lung disease at an early age and others reaching adulthood with normal lung function (2). This observed variability has made it clear that the CFTR genotype is not the main determinant of severity of CF lung disease. The reasons for this variation in pulmonary phenotype in patients with identical CFTR mutations remain unclear. Certain infectious exposures, such as mucoid Pseudomonas aeruginosa and Burkholderia cepacia, along with nutritional factors (3, 4), have been demonstrated to contribute to the variability in pulmonary phenotype (3, 4). Other environmental factors, including socioeconomic status, tobacco smoking, and marijuana abuse, have been identified as contributors. There is increasing recognition that genetic modifiers likely play a significant role in influencing pulmonary phenotype in CF (5). The European CF Twin and Sibling Study compared phenotypes for a cohort of 41 sets of twins with identical CFTR genotypes and found that monozygous twins had a significantly higher concordance in nutritional status and severity of lung disease than dizygous twins (suggesting that genetic factors besides the CFTR gene influence phenotype) (6). A more in-depth understanding of the molecular basis of variability in CF lung disease is needed and would have major prognostic and therapeutic implications.
Several obstacles have made investigation into the molecular basis of this variability difficult. First has been the lack of an animal model of CF lung disease, making human studies necessary. Second has been the challenge of accurately identifying the most mild and severe pulmonary phenotypes while controlling for external confounders, such as B. cepacia, resistant P. aeruginosa, allergic bronchopulmonary aspergillus (ABPA), and poor nutrition. Third has been the challenge of identifying molecular differences that lead to a variation in phenotype and are not just a response to chronic infection and inflammation.
The aim of this investigation was to use high-density oligonucleotide microarray studies of in vivo nasal respiratory epithelium to investigate the molecular basis of differences in the CF phenotype by (1) identifying differences in gene expression between ΔF508 homozygotes with very mild lung disease and those with very severe lung disease and (2) identifying differences in gene expression between ΔF508 homozygotes and non-CF control subjects.
We attempted to address each of the previously described obstacles by matching participants with CF for ΔF508 homozygosity and infection status and studying only those representing the most mild and severe 20th percentiles of CF lung disease based on forced expiratory volume in 1 sec (FEV1) criteria. We also chose to study nasal respiratory epithelium to permit analysis of respiratory epithelium from severe phenotype patients, which was less likely to be characterized only by response to chronic infection and inflammation.
The results of this study demonstrate that with the exception of a few potentially interesting genes, there is less difference in nasal respiratory epithelial gene expression profiles between ΔF508 homozygotes with mild CF lung disease and non-CF control subjects than might be anticipated. In contrast, ΔF508 homozygotes with severe lung disease demonstrate upregulation of genes involved in protein ubiquination and mitochondrial oxidoreductase activity. When comparing all ΔF508 homozygotes together with non-CF control subjects, the most significant difference in gene expression is a downregulation in CF of genes involved in airway defense and antigen presentation/immune response. The results also identify a small number of new potential modifiers of CF lung phenotype, including statherin (an antimicrobial peptide present in saliva and the upper airways), adiponectin (a potent anti-inflammatory cytokine and modulator of insulin sensitivity), and dual oxidase 2 (a key producer of hydrogen peroxide in the upper airway).
MATERIALS AND METHODS
Participant Selection
The study was conducted at the Johns Hopkins Cystic Fibrosis Center and was approved by the Johns Hopkins and Western Institutional Review Boards (protocol #1033335). Each participant voluntarily consented to participate. To be eligible for the study, individuals with CF had to be homozygous for the ΔF508 mutation and meet FEV1 criteria that placed them in the top or bottom 20th percentile for FEV1 for their genotype and age (Table 1). These criteria were based on work published by Schlucter and coworkers (7) identifying FEV1 cutoffs that stratify severity of lung disease in ΔF508 homozygotes. Individuals with ABPA, B. cepacia, atypical mycobacteria, methicillin-resistant Staphylococcus aureus, history of significant reactive airway disease, recent viral infection, or active CF exacerbation were excluded. Non-CF control subjects were recruited from age-matched healthy volunteers. If potential participants demonstrated obvious turbinate inflammation or hemorrhage on initial visual inspection, the brushing was rescheduled for a later date.
TABLE 1.
FEV1 PERCENT PREDICTED CRITERIA FOR INCLUSION IN MILD OR SEVERE ΔF508 HOMOZYGOTE PHENOTYPE GROUPS
| Age (yr) | Mild Lung Phenotype (FEV1% predicted) | Severe Lung Phenotype (FEV1% predicted) |
|---|---|---|
| 15 | > 97 | < 65 |
| 16 | > 92 | < 57 |
| 17 | > 90 | < 54 |
| 18 | > 87 | < 53 |
| 19 | > 86 | < 51 |
| 20 | > 84 | < 48 |
| 21 | > 82 | < 46 |
| 22 | > 77 | < 44 |
| 23 | > 70 | < 42 |
| 24 | > 68 | < 40 |
| 25 | > 67 | < 38 |
| 26 | > 62 | < 36 |
Criteria identify the most mild and severe 20th percentiles of FEV1 for each age.
Nasal Respiratory Epithelial Cell Collection
Bilateral nasal mucosal brushing to collect respiratory epithelium was performed on each subject with a Cytosoft cytology brush (Medical Packaging Corp., Camarillo, CA) under direct visualization of the inferior turbinate using a nasal telescope (Karl Storz Endoscopy America, Culver City, CA). The nasal mucosa was anesthetized with two sprays of 4% tetracaine spray. The cytology brush was then gently rotated on the mucosal surface of the inferior turbinate, removed from the nose, and agitated in a DNAase/RNAase-free microcentrifuge tube containing 1 ml of sterile, chilled PBS. After a 40-μl aliquot was set aside for cytologic evaluation, the sample was centrifuged, the supernatant was removed, and 500 μl of TRIzol Reagent (Invitrogen, Carlsbad, CA) was added to the tube. The sample was immediately snap frozen using liquid nitrogen and stored at −80°C.
Cytopatholologic Evaluation
A cytospin centrifuge was used to prepare respiratory epithelial samples for Papanicolaou staining. Slides were evaluated using light microscopy by a blinded cytopathologist, and cells were categorized as ciliated epithelial, squamous epithelial, or inflammatory. Results were expressed as the percentage of total cells.
RNA Extraction and Preparation for Microarray Analysis
Total RNA from each sample was extracted using RNeasy Mini Kits (Qiagen, Valencia, CA). Double-stranded cDNA was synthesized using SuperScript Choice System (Invitrogen) and used for in vitro transcription using ENZO BioArray HighYield RNA transcript labeling kits (Affymetrix, Santa Clara, CA) to produce biotin-labeled cRNA. The cRNA was purified by RNeasy kit (Qiagen) and fragmented randomly to ∼ 50–100 bp (200 mM Tris-acetate [pH 8.2], 500 mM KOAc, and 150 mM MgOAc at 94°C for 35 min). Spike controls were added to fragmented cRNA before hybridization.
Performance of Microarrays
cRNA samples from each subject were separated into two aliquots and hybridized to Affymetrix Human Genome HG-U133 A and B microarrays for 16 h according to the protocols described in the GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA). After hybridization, each microarray was washed, stained, and amplified on the Affymetrix Fluidics Station 400 using a phycoerythrin–streptavidin stain and biotin-labeled, antistreptavidin antibody. Fluorescent images were read using the Hewlett-Packard G2500A Gene Array Scanner (Hewlett-Packard, Palo Alto, CA).
Quality Control Measures
After scanning, array images were assessed by eye to confirm scanner alignment and the absence of significant bubbles or scratches. The 3′/5′ ratio for glyceraldehyde-3-phosphate dehydrogenase was confirmed to be < 4 for all chips used for analysis. The spike controls BioB, BioC, BioD, and CreX were identified as being present in increasing intensity. GeneChip initial expression analysis was performed using GCOS 1.0 software (Affymetrix).
Microarray Data Analysis
GeneChip expression data were exported to GeneSpring where per chip normalization to the 50th percentile expression level and per gene normalization to the median expression intensity in all samples was performed. Only probe sets scored present or marginal in at least 75% of samples were included in the analysis. Data were transformed to log ratio for display and analysis. GeneSpring 7 (Silicon Genetics, Redwood City, CA) and S-Plus 6.2 (MathSoft, Cambridge, MA) software was used in data analysis and visualization. Significantly changing gene expression was set at P ⩽ 0.05 using the GeneSpring t test statistic with cross-gene error model activated using the two-component Rocke-Lorenzato model of normalization (8). The deviation from median option was also used to reduce the effects of outliers. Genes were identified that were significantly differentially expressed in three disease phenotype categories: CF with mild lung disease, CF with severe lung disease, and individuals without CF. A secondary analysis identified genes differentially expressed by sex in the non-CF group to allow exclusion of genes that segregated by sex independent of CF phenotype. The majority of sex-specific genes was already excluded by the requirement that a probe set be present or marginal in at least 75% of the nasal respiratory epithelium samples to be included in the analysis.
Probe sets identified as significantly differentially expressed by disease phenotype underwent an intensive search to identify biologic function. Probe set sequences from the Affymetrix web site were “blasted” against the University of California, Santa Cruz genome database to verify identity and update annotation. For individual genes with multiple probe set sequences specific to different regions of the gene, each probe set was checked separately. The resulting list of genes was submitted to PathwayAssist 3.0 (Stratagene, La Jolla, CA) for automated literature search. Gene Ontology classifications using GOMiner (9), conserved protein family domains, and reference literature were used to construct functional groupings of genes. GOMiner was then used to perform a two-sided Fisher's exact test to determine if a significantly greater number than expected of differentially expressed genes occurred in a category (8). All original array data images and files are available at http://pepr.cnmcresearch.org/browse.do?action=list_prj_expandprojectId=97. This site includes data on all arrays performed. The online supplement identifies which specific arrays were used for analysis and which had unacceptable glyceraldehyde-3-phosphate dehydrogenase ratios (see Tables E1 and E2 in the online supplement).
LightCycler Quantitative RT-PCR Confirmation of Results
Changes in RNA levels of differentially expressed genes of interest were validated with quantitative RT-PCR using LightCycler real-time PCR (Roche, Indianapolis, IN). An aliquot of the nasal epithelium RNA was diluted to a concentration of 1 μg/μl, and cDNA synthesis was performed with 2 μg of RNA using the Avian myoblastoma virus RT enzyme and Oligo-p(dT)15 primers included in the 1 st Strand cDNA Synthesis Kit for RT-PCR (Roche). PCR amplification was performed on the LightCycler using DNA Master SYBR Green I kit and primers. Primers used to amplify STATH were 5′-GAAAAGGCAAGTATCCTGAAACAAA (forward) and 3′-TCCAGAACAACCACTATACCCACAA (reverse). Primers used to amplify DUOX2 were 5′-GCCCCTCTCTGCATCTACTG (forward) and 3′-GGGCAAGAGACTTTCAGTGC (reverse). γ-Actin and Tata Box Protein (TBP) were amplified from the same samples at the same time (separate capillary reaction tubes) as internal controls. Primers used for amplification of the γ-actin and TBP genes were γ-actin: 5′-AAGCCACCGACTTGTCTTCC (forward), 3′-AGATCAAGATCATCGCACCC (reverse) and TBP: 5′-GAATATAATCCCAAGCGGTTTG (forward) and 3′-ACTTCACATCACAGCTCCCC (reverse). PCR product identity was confirmed by agarose gel electrophoresis and product sequencing.
RESULTS
Patients and Samples
A total of 48 individuals (30 with CF, 18 non-CF) underwent nasal brushing to acquire nasal respiratory epithelial cells. Eleven of the brushings (eight CF, three non-CF) did not provide RNA of sufficient quantity or quality after purification to permit microarray analysis. The remaining 37 samples were hybridized to Affymetrix Human Genome HG-U133 A and B microarrays. Twenty-three of the U133A arrays and 21 of the matching U133B arrays passed the stringent quality control guidelines described in Materials and Methods and were used for the final data analysis (12 CF, 11 non-CF). The characteristics of the 23 individuals used in the final data analysis are summarized in Table 2. All of the individuals with severe CF and all but two of the individuals with mild CF were infected with mucoid Pseudomonas.
TABLE 2.
CHARACTERISTICS OF ANALYZED PARTICIPANTS BY PHENOTYPE GROUP
| Severe CF | Mild CF | Non-CF Control Subjects | |
|---|---|---|---|
| Genotype | ΔF508/ΔF508 | ΔF508/ΔF508 | n/a |
| n | 7 | 5 | 11 |
| Age, yr (mean ± SD) | 22.6 ± 4.9 | 22.5 ± 2.5 | 28.2 ± 5.3 |
| FEV1% predicted, mean | 31.3 ± 7.6%* | 100.2 ± 18.5% | n/a |
| FVC % predicted, mean | 49.8 ± 9.0%* | 110.9 ± 18.9% | n/a |
| FEV1, L | 1.2 ± 0.4* | 3.8 ± 0.4 | n/a |
| FVC, L | 2.7 ± 1.1* | 4.7 ± 0.8 | n/a |
| Body mass index, kg/m2 | 21.0 ± 2.1* | 26.2 ± 2.5 | 25.6 ± 1.1 |
| Sex | 6 M/1 F | 3 F/2 M | 6 M/5 F |
P < 0.001 for severe versus mild CF.
Nasal Cytology
To assure that the cells collected and analyzed were predominantly respiratory epithelial cells, the nasal brushing smears were blindly read by the Johns Hopkins cytopathology department (Table 3). This analysis demonstrated an overall mean of 86.7 ± 7.1% respiratory epithelial cells (median, 90%; range, 73–98%). Other cell types identified were squamous (5.4 ± 5.6%) and inflammatory cells (7.8 ± 4.1). There was not a significant difference in percentage of inflammatory cells between CF, mild CF, and non-CF samples, although the study was not powered to detect small differences. A mean of 6.6 ± 3.3% of cells were inflammatory in the non-CF group (n = 11), 9.0 ± 6.5% in the mild CF group (n = 5), and 8.9 ± 3.2% in the severe CF group (n = 7) (P = not significant).
TABLE 3.
NASAL BRUSHING CYTOLOGY RESULTS
| Respiratory Epithelial Cells* | Inflammatory Cells† | Squamous Epithelial Cells | |
|---|---|---|---|
| Non-CF (n = 11) | 89.4 ± 5.6% | 6.6 ± 3.3% | 4.0 ± 3.4% |
| Mild CF (n = 5) | 86.0 ± 8.2% | 9.0 ± 6.5% | 5.0 ± 3.5% |
| Severe CF (n = 7) | 82.9 ± 7.6% | 8.9 ± 3.2% | 8.2 ± 8.7% |
| Totals (n = 23) | 86.8 ± 7.1% | 7.8 ± 4.1% | 5.4 ± 5.6% |
Ciliated and columnar, with rare goblet cells.
No significant difference between groups in percentage of inflammatory cells.
Overview of Expression Profiling Results
A total of 11,867 of the 44,760 probe sets on the U133 A and B chips were identified as being present or marginal in at least 75% of the nasal respiratory epithelium samples. Using the GeneSpring t test with cross-gene error modeling to determine significance, 709 of the 11,867 were identified as significantly differentially expressed in one of the three disease phenotypes: 32 in non-CF, 69 in mild CF, and 608 in severe CF (Figure 1, supplemental file 1). Combining multiple probe sets for single genes and eliminating expressed sequence tags reduced the numbers to 30 differentially expressed genes in non-CF, 53 in mild CF, and 569 in severe CF.
Figure 1.
(A) Heat map of differentially expressed genes. Each column represents a separate patient, and each horizontal line represents a separate gene. Only genes differentially expressed by phenotype are included. Red represents increased expression compared with median expression of the gene for all phenotypes; blue represents decreased expression compared with overall median. (B) Probe sets differentially expressed in non-CF. (C) Probe sets differentially expressed in mild CF. (D) Probe sets differentially expressed in severe CF; only 70 of the 608 probe sets are shown for clarity.
K-means clustering analysis divided the differentially expressed probe sets into three main groups: genes with increased expression only in non-CF (Figure 1B), genes with increased expression only in mild CF (Figure 1C), and genes with increased expression only in severe CF (Figure 1D). There were few probe sets that demonstrated significantly decreased expression only in mild or severe CF. One notable exception was STAT1, which demonstrated significantly decreased expression in mild-CF (Figure 1B).
Genes Differentially Expressed between Non-CF and CF
The 30 genes with increased expression only in the non-CF group could alternatively be identified as genes with significantly decreased expression in mild and severe CF. These 30 genes therefore are those differentially expressed in nasal respiratory epithelium of individuals with CF compared with those without (Table 4). The genes most reduced in expression in CF were DUOX2 (2.5-fold) and CD2 (2.3-fold). DUOX2 is a key producer of H2O2 for airway mucosal defense and has previously been identified as potentially playing a role in CF (10, 11). An analysis by gene ontology (GO) categories of the list of all genes with decreased expression in CF demonstrated significant over-representation of those involving response to biotic stimulus (P < 0.047) and protein metabolism (P < 0.048). The biotic stimulus gene classes included those involved in defense response (P < 0.02), particularly the subset of genes involving antigen presentation (HLA-F, HLA-G; P < 0.0001) and lymphocyte differentiation (CD74, CD2; P < 0.003). Of the 11 downregulated genes in the GO protein metabolism category, calreticulin, a multifunctional ER protein chaperone, was of most potential interest. Transcript levels of CFTR were only marginally present on array, but RT-PCR revealed no detectable differences in CFTR transcript levels between ΔF508 homozygotes and non-CF control subjects.
TABLE 4.
GENES WITH SIGNIFICANTLY DECREASED EXPRESSION IN CF COMPARED WITH NON-CF CONTROL SUBJECTS
| Hugo Name | Description | P Value |
|---|---|---|
| ACTB | Beta actin, cell structure | 0.007 |
| ASS | Argininosuccinate synthetase, NO production | 0.038 |
| CALR | Calreticulin, Ca2+ binding and storage | 0.036 |
| CD2 | Immune function | 0.014 |
| CD74 | Immune function, Major histocompatibility complex peptide | 0.029 |
| CLEC2D | C-type lectin, natural killer cell receptor family | 0.029 |
| CPA4 | Carboxypeptidase A4 | 0.038 |
| CSN2 | Casein beta | 0.036 |
| CTSB | Cathepsin B, lysosomal cysteine proteinase | 0.017 |
| DAZAP2 | DAZ-related protein 2, RNA binding | 0.041 |
| DUOX2 | Dual oxidase 2, NADPH oxidase family | 0.021 |
| ENTH | Enthoprotin, transport vesicle associated | 0.034 |
| EPSTI1 | Epithelial stromal interaction | 0.010 |
| FBP1 | Fructose-1,6-bisphosphatase 1 | 0.012 |
| FLJ14281 | Hypothetical protein, contains DnaJ chaperone domain | 0.019 |
| HLA-F | Major histocompatibility complex protein | 0.032 |
| HLA-G | Major histocompatibility complex protein | 0.047 |
| HTFL | Human T-cell leukemia virus enhancer factor, transcription regulation | 0.010 |
| IFITM1 | IFN-induced transmembrane protein | 0.021 |
| IGFBP3 | Insulin-like growth factor binding protein | 0.022 |
| ITM2A | Integral membrane protein | 0.048 |
| LPXN | Leupaxin, may function in cell signaling | 0.018 |
| LRRC1 | Leucine-rich repeat containing 1 | 0.046 |
| MFTC | Mitochondrial folate transporter | 0.023 |
| MYH9 | Myosin, nonmuscle | 0.048 |
| PADI1 | Peptidyl arginine deiminase | 0.032 |
| PRKACB | Protein kinase, cAMP dependent | 0.048 |
| RPS2 | Ribosomal protein | 0.023 |
| S100A16 | Activator of gene transcription | 0.007 |
| SP110 | Nuclear body protein | 0.019 |
| ZNF198 | Transcription regulation | 0.036 |
Genes Differentially Expressed in Mild CF
There were 69 probe sets representing 53 genes that were significantly differentially expressed in individuals with mild CF lung disease compared with those with severe CF and non-CF control subjects. Fifty-two of the 53 genes demonstrated increased expression in mild CF. STAT1, an inducible transcription factor mediating response to IFN (12) and represented by two probe sets (Figure 1C), was the only gene significantly decreased in expression in mild CF. An analysis of the upregulated list demonstrated significant over-representation in several GO categories: lipid metabolism (P < 0.032), G-coupled protein receptors (P < 0.024) and ion transport (P < 0.03) (Table 5). In addition to the genes in these categories, two other genes of interest were found to be upregulated in individuals with mild CF lung disease: statherin (STATH) and adiponectin (ADIPOQ). STATH is a calcium-binding protein found in saliva and is primarily known for its regulation of calcium deposition (13). STATH is also well documented to have significant antibacterial properties (14) and is produced in submucosal glands of the nasal cavity and upper airway (15). ADIPOQ is a potent anti-inflammatory cytokine and inducer of IL-10 and is a modulator of insulin sensitivity (16, 17).
TABLE 5.
GENE ONTOLOGY CATEGORIES AND MEMBER GENES WITH SIGNIFICANTLY INCREASED EXPRESSION IN INDIVIDUALS WITH MILD CF LUNG DISEASE
| Name | Function | P Value |
|---|---|---|
| GO:0006629 | Lipid metabolism | 0.041 |
| ADIPOQ | Adiponectin | 0.026 |
| C21orf106 | DIP2-like protein | 0.024 |
| PLA2G5 | Phospholipase A2, group 5 | 0.046 |
| PPP2CA | Protein phosphatase 2 catalytic subunit | 0.049 |
| SCD4 | Stearyl-CoA desaturase | 0.031 |
| SFTPB | Surfactant, pulmonary protein B | 0.047 |
| GO:0006811 | Ion transport | 0.045 |
| SLC9A2 | Na/H exchanger | 0.045 |
| MINK | Potassium channel | 0.041 |
| FXYD2 | Na/H exchanger | 0.024 |
| COL7A1 | Collagen, phosphate transport regulation | 0.007 |
| ADIPOQ | Adiponectin | 0.026 |
| GO:0007186 | G-protein coupled receptor | 0.045 |
| PGF | Placental growth factor | 0.048 |
| HTR2A | Serotonin receptor 2A | 0.035 |
| GPR12 | G protein-coupled receptor 12 | 0.023 |
| C21orf106 | DIP2-like protein | 0.024 |
| GO:0030135 | Coated vesicle | 0.017 |
| SCD4 | Stearyl-CoA desaturase | 0.031 |
| SCAMP1 | Secretory carrier, post Golgi recycling | 0.045 |
| ADIPOQ | Adiponectin | 0.026 |
| GO:0005576 | Extracellular | 0.027 |
| STATH | Statherin | 0.020 |
| SFTPB | Surfactant, pulmonary protein B | 0.047 |
| PLA2G5 | Phospholipase A2, group 5 | 0.046 |
| PGF | Placental growth factor | 0.048 |
| LRRN1 | Leucine-rich repeat neuronal 1 | 0.032 |
| KLK13 | Kallikrein serine protease | 0.036 |
| COL7A1 | Collagen, phosphate transport regulation | 0.007 |
| ADIPOQ | Adiponectin | 0.026 |
Genes Differentially Expressed in Severe CF
A total of 569 genes demonstrated significant upregulation in individuals with severe CF lung disease compared with those with mild disease and non-CF control subjects. Analysis of these genes by gene ontology categories revealed a striking over-representation of the upregulated genes involved oxidoreductase activity (P = 0.01), the ubiquitin cycle (P = 0.04), and lipid metabolism (P = 0.04) (Table 6). One particular cluster of upregulated oxidoreductase genes were those involved in NADH dehydrogenase:ubiquinone complex I, a mitochondrial subunit essential for electron transfer (NDUFS1, NDUFS7, NDUFB3, NDUFB5, NDUFAB1, NDUFA3). Numerous ubiquitin-conjugating enzymes were also significantly upregulated in individuals with severe CF (UBE2A, UBE2B, UBE2E1, UBE2E3, FBXW2, HIP2, and NEDD8) along with two ubiquitin-activating enzymes (UBA2 and UBE1C). Other upregulated genes of interest in severe CF included glutamate-cysteine ligase, the rate-limiting enzyme of glutathione synthesis (18), and activating transcription factor 1, a transcription factor involved in increasing IL-8 inflammatory response (19). A full list of all genes differentially expressed in individuals with severe CF is available in the online supplement (Table E3).
TABLE 6.
GENE ONTOLOGY CATEGORIES WITH SIGNIFICANTLY INCREASED EXPRESSION IN INDIVIDUALS WITH SEVERE CF LUNG DISEASE
| GO ID | Name* | n Total† | n Changed | P Value‡ |
|---|---|---|---|---|
| GO:0016491 | Oxidoreductase activity | 322 | 39 | 0.011 |
| GO:0005489 | Electron transporter activity | 138 | 21 | 0.005 |
| GO:008137 | NADH dehydrogenase (ubiquinone) | 32 | 6 | 0.047 |
| GO:0005783 | Endoplasmic reticulum | 265 | 31 | 0.034 |
| GO:0006629 | Lipid metabolism | 269 | 31 | 0.041 |
| GO:0019752 | Carboxylic acid metabolism | 216 | 27 | 0.022 |
| GO:0006082 | Organic acid metabolism | 217 | 27 | 0.032 |
| GO:0009308 | Amine metabolism | 153 | 21 | 0.016 |
| GO:0006066 | Alcohol metabolism | 129 | 18 | 0.021 |
| GO:0006629 | Lipid biosynthesis | 105 | 17 | 0.006 |
| GO:0006631 | Fatty acid metabolism | 79 | 13 | 0.013 |
| GO:0016125 | Sterol metabolism | 39 | 9 | 0.004 |
| GO:0006399 | tRNA metabolism | 40 | 9 | 0.005 |
| GO:0006512 | Ubiquitin cycle | 228 | 27 | 0.040 |
| GO:4840 | Ubiquitin conjugating enzyme activity | 38 | 7 | 0.036 |
The indented names are subsets or largely overlapping sets of genes in the broader categories listed above.
n total is the number of genes in a category, and n changed is the number of those genes significantly altered in expression.
P value is the Fisher's two-sided exact test that the number of genes altered in that category is greater than expected by chance.
IL-8
The inflammatory chemokine IL-8 has previously been identified as being characteristically elevated in CF (20). IL-8 transcript levels were noted to be significantly elevated in the nasal epithelium of the patients with severe CF compared with non-CF control subjects (Figure 2). In contrast, a wide range of IL-8 transcript expression was exhibited in the patients with mild CF; although the two highest expression levels of IL-8 occurred in individuals with mild CF, the remainder had IL-8 levels similar to non-CF control subjects. Overall, the median IL-8 level in individuals with mild CF lung disease was not significantly different from that seen in non-CF (Figure 2).
Figure 2.
Box plot of microarray IL-8 mRNA expression in nasal epithelial cells by disease status. IL-8 expression in individuals with severe CF was significantly greater than in non-CF control subjects. Although the highest levels of IL-8 expression occurred in individuals with mild CF, there was a much greater range, with some individuals with mild CF not demonstrating IL-8 levels greater than non-CF control subjects. Open square is mean; horizontal line is median. Upper and lower box bounds mark the 75th and 25th percentiles, respectively. Expression is normalized by making median expression across all phenotypes equal 1.0.
STATH and DUOX2 rt-PCR Results
Because of its potential importance, we sought to verify the significantly increased expression of STATH in mild CF by quantitative RT-PCR in the samples used for microarray analysis and in an additional 12 mild and severe patient samples collected after the microarray experiments were complete. This separate analysis confirmed a significant difference in STATH expression between those with mild CF lung disease (n = 12) and those with severe CF lung disease (n = 11) (Figure 3; Kruskal-Wallis rank sum test, P = 0.042).
Figure 3.
Analysis of STATH expression by RT-PCR in a separate patient population confirming microarray findings. STATH expression was significantly elevated in those with mild CF lung disease (n = 12) compared with those with severe CF lung disease (n = 11) (Kruskal-Wallis rank sum test, P = 0.042). Open square is mean; horizontal line is median. Upper and lower box bounds mark the 75th and 25th percentiles, respectively. RT-PCR data were normalized to expression level in the shared control sample.
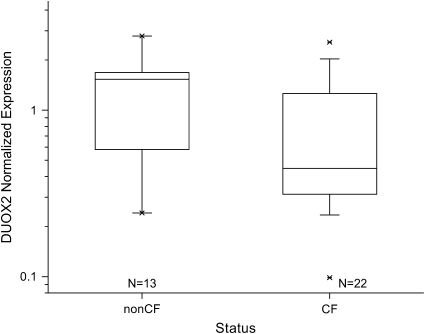
Decreased expression of DUOX2 in individuals with CF was also confirmed by RT-PCR in an independent larger group. DUOX2 expression was significantly lower in individuals with CF (n = 22) compared with those without CF (n = 13) (Figure 4; Kruskal-Wallis rank sum test, P = 0.047).
Figure 4.
Analysis of DUOX2 expression by RT-PCR in a separate patient population confirming microarray findings. DUOX2 expression was significantly lower in individuals with CF (n = 22) compared with those without CF (n = 13) (Kruskal-Wallis rank sum test, P = 0.047). Horizontal line is median. Upper and lower box bounds mark the 75th and 25th percentiles, respectively. RT-PCR data were normalized to expression level in the shared control sample.
DISCUSSION
Despite having identical CFTR genotypes, ΔF508 homozygous CF individuals demonstrate a full range of pulmonary disease. Although several environmental factors influencing severity of lung disease have been identified, there is growing evidence that genetic and molecular differences contribute to the significant variability seen in the CF phenotype. This study used microarray analysis of nasal respiratory epithelium to investigate the molecular basis of variability in CF phenotype by identifying differences in gene expression between ΔF508 homozygotes in the most severe 20th percentile of lung disease and those in the most mild 20th percentile and identifying differences in gene expression between ΔF508 homozygotes and age-matched non-CF control subjects. The results suggest that the most significant differences in gene expression between those with CF and those without include those involved with airway defense, antigen presentation, and protein metabolism. There are also differences in gene expression between those with mild CF lung disease and those with severe lung disease, even in nasal respiratory epithelium without evidence of significant differences in inflammation. These include differential expression of genes involving the ubiquitin cycle, oxidoreductase activity, and lipid metabolism.
A previous comparison of gene expression in CF and non-CF respiratory epithelium has been performed in mice and identified differential expression of multiple gene classes, including those involved in transcription, inflammation, intracellular trafficking, signal transduction, and ion transport (21). Because of the lack of pulmonary pathology in these CF mice, no analysis could be performed to evaluate differences in gene expression associated with severity of lung disease. Another recent study of ΔF508 and non-CF primary respiratory cell cultures grown in sterile conditions for 60 d suggested minimal differences in gene expression (22). To design our study evaluating differences in gene expression by severity of lung disease, we first needed to select samples from ΔF508 homozygotes in distinct contrasting phenotypic groups. A recent study by Schlucter and coworkers aided in selecting these groups by determining FEV1 criteria identifying those with the most mild 20% and most severe 20% for lung disease among ΔF508 homozygotes between the ages of 15 and 26 (7). Our investigation used these criteria as the basis for identifying “mild” and “severe” CF phenotypes after controlling for other known environmental influences, including ABPA, B. cepacia, and atypical mycobacteria.
One challenge to this study was identified from the onset: Could we identify differences in mild and severe CF respiratory epithelium that were not due just to response to chronic infection? This challenge was made evident in initial attempts to study lower airway cells from patients with severe CF by the amount of purulence in cell samples obtained by bronchoscopy in patients with CF just before lung transplantation. To address this concern, we studied gene expression in nasal ciliated respiratory epithelial cells, a commonly used surrogate in CF for lower airway respiratory cells. Just as in CF lower airway respiratory epithelium, a markedly decreased amount of CFTR reaches the apical surface membrane of nasal epithelial cells (23). Electrolyte transport characteristics have also been shown to be nearly identical (24). We tried to minimize the potential influence of active local infection by excluding individuals with symptoms of sinus or pulmonary exacerbation, with the hope that collected cells would be more likely to reflect intrinsic differences in gene expression not due solely to response to local infection.
CF versus Non-CF Differential Gene Expression
Perhaps the most striking of this group is DUOX2, which had an average 2.5-fold lower expression in individuals with CF. DUOX2 is an NADPH oxidase that has recently been identified as the key producer of H2O2 in the airway and oral cavity and is an essential component of the lactoperoxidase (LPO) airway epithelium defense system (10). LPO uses H2O2 to oxidize the anion thiocyanate to hypothiocyanite, a strong antimicrobial agent that prevents the growth of bacteria and fungi in the airways (25). Individuals with chronic granulomatous disease known to have deficiency in the production of airway reactive oxygen species have been noted to be susceptible to the gram-negative infections often characteristic of CF, such as B. cepacia (11). Although authors have previously noted these similarities and have commented on the need for further investigation (11, 26), these array findings provide the first in vivo support for a potential link between CF and abnormalities in the DUOX-LPO airway defense system.
One gene ontology category demonstrating clear decreased expression in CF was that involving genes responding to biotic stimulus. These genes included β-casein (CSN2), which demonstrated on average a 1.5-fold lower expression in individuals with CF, and a group of genes involving lymphocyte differentiation (CD2, CD74) and antigen presentation (HLA-F, HLA-G). CSN2 has been identified as inhibiting the growth of viruses and bacteria, particularly S. aureus, through its strong inhibition of cysteine proteases (27). CD2 is a cell surface adhesion protein expressed on nearly all T cells that mediates STAT involvement in the regulation of IFN-γ expression, particularly in mucosal T cells (28). Detection of differential expression of CD2 suggests that the 6–9% inflammatory cells present in the nasal samples made a contribution to the expression profiles. CD2 has previously been shown to be cleaved by polymorphonuclear leukocyte elastase and cathepsin G in patients with CF (29). IFN-γ dysregulation has repeatedly been proposed to play a role in CF pathophysiology, and these array findings suggest that decreased expression of CD2 in mucosal lymphocytes might contribute to IFN-γ dysregulation.
Also significantly decreased in CF was insulin-like growth factor binding protein-3 (IGFBP3), a protein known to be a key modulator of the effects of insulin-like growth factor. Serum levels of IGFBP3 have been repeatedly demonstrated to be decreased in individuals with CF and in some cases correlate with lung function and nutritional status (30). Microarray analysis of CFTR null mice lung tissue has also previously demonstrated a significant decrease in insulin-like growth factor binding proteins (21). Although decreases in IGFBP3 can be related to chronic malnutrition, the significant decrease even in well nourished individuals with mild CF suggests an association between the loss of CFTR function and the decrease in insulin-like growth factor binding proteins.
Six of the 30 genes downregulated in CF were involved in lipid metabolism (PIGB, PIGF, PITPNB, SC4MOL, SLC27A2, and UGCG) and are located in the endoplasmic reticulum according to GO. None of these genes has been identified as potentially involved in CF, although abnormalities in lipid metabolism in CF are well known. Despite the large number of differentially expressed genes located in the endoplasmic reticulum, there was no indication of an ER overload response.
Inflammatory chemokine IL-8 has been identified as being characteristically elevated in CF (20). Although IL-8 expression was significantly elevated in the severe CF group compared with non-CF control subjects, there was much more variability in IL-8 expression in individuals with the mild CF group. The highest absolute IL-8 values in the study group were found in two individuals with mild CF lung disease and minimal sinus disease. This suggests that although IL-8 levels are usually elevated in CF, marked elevation is not necessarily associated with more aggressive disease. Alternatively, CF nasal respiratory epithelial cells may demonstrate IL-8 characteristics independent of lower airway respiratory epithelium.
There were no differences in CFTR transcript levels between ΔF508 homozygotes and non-CF control subjects. This is not surprising given that it is thought that ΔF508 homozygotes develop CF pathology based on abnormalities at the protein trafficking and function level. This finding is also consistent with the recent results of Zabner and colleagues (22).
Mild versus Severe CF Differential Gene Expression
Some of the most striking differences in gene expression were seen when we analyzed the results of specific phenotypes within CF. There were 53 genes that were significantly differentially expressed in individuals with mild CF lung disease compared with individuals with severe CF and non-CF control subjects. The GO categories most represented included lipid metabolism, airway defense, G-coupled protein receptors, and ion transport. Among these genes, several strong candidate modifiers of CF phenotype were apparent.
STATH, a calcium-binding, 43-amino-acid phosphopeptide known to have antibacterial properties is found in saliva, nasal secretions, and the upper airway. STATH was clearly upregulated in individuals with mild CF lung disease. This increased expression was confirmed in an additional 12 mild and severe patient samples collected after the microarray experiments were complete. STATH plays a key role in the development of the oral cavity biofilm by mediating adhesion of bacteria and was recently identified as being the most prominent protein in the saliva–air interface (31). It is known to have bacterial binding epitopes that promote the growth and adhesion in the oral cavity of some organisms (Porphyromonas gingivalis and Fusobacterium nucleatum) while inhibiting the growth of others (Peptostreptococci and S. aureus) (14, 32, 33). Its antimicrobial effect on P. aeruginosa has not been investigated. Given that colonization with mucoid Pseudomonas is known to accelerate a decline in lung function in CF (3), a protein acting as a key determinant of bacterial adhesion in the oral cavity and upper airway is of significant interest.
ADIPOQ, a protein usually produced in adipocytes that potently inhibits inflammation and modulates insulin sensitivity, was also significantly upregulated in ΔF508 homozygotes with mild disease. ADIPOQ induces in leukocytes the production of the anti-inflammatory mediators IL-10 and IL-1 receptor antagonist (16). It also stimulates the release of IL-1β, IL-6, and TNF-α from adipocytes (34) and modulates energy metabolism and glucose sensitivity (17). Functional polymorphisms have been identified that influence circulating ADIPOQ levels (35). If the increased expression of ADIPOQ was due to the amount of adipose tissue alone, it would be expected that the well nourished, non-CF control subjects would have the highest levels, not individuals with mild CF. Functional polymorphisms leading to increased levels of ADIPOQ in CF would likely lead to more mild disease by suppression of CF-related inflammation and improved utilization of nutritional intake. Alternatively, low levels of ADIPOQ in poorly nourished individuals with CF might contribute to the known relationship between poor nutritional status and decline in lung function.
ADIPOQ is classically identified as being produced only by adipocytes, although array studies have identified ADIPOQ expression in trachea, skin, adrenal gland, thymus, and thyroid (36). Separate RT-PCR studies confirmed the presence of ADIPOQ transcripts in CF and control nasal brushings, although further studies are needed to determine whether this expression is from respiratory epithelial cells or other cell types present in the sample.
Signal transducer and activator of transcription 1 (STAT1), represented by two probe sets, was the only gene identified to be significantly decreased in expression in mild CF. In mucosal T cells, STAT1 is activated by CD2 receptors (37), which are also identified by the microarray data as being decreased in CF. In epithelial cells, STAT1 is essential for cellular antiviral defense and is central in activating the transcription of IFN-induced genes, particularly nitric oxide synthase-2 (38). STAT1 activates transcription by binding directly to regulatory DNA elements (38). STAT1-deficient mice display an absence of responsiveness to IFN and are highly sensitive to infection by virus (39). It has previously been shown that STAT1 induction and activation are impaired in CF, and STAT1 has been proposed as a potential modifier of the CF phenotype (40). Although the microarray data corroborate the decrease of STAT1 in CF, it is unclear why this would be more apparent in individuals with mild CF because low levels might be expected to result in more susceptibility to infection. One possibility is the recently identified increased antiapoptotic effect of IFN in the absence of STAT1 (41). An alternative explanation is that STAT1 is usually low in all CF, but the studied severe group had more active inflammation leading to STAT1 induction.
Individuals with severe CF demonstrated the largest number of differentially expressed genes. A total of 569 genes demonstrated significant upregulation in individuals with severe CF lung disease compared with those with mild CF lung disease and non-CF control subjects. Although the respiratory epithelial cells sampled were not acutely infected or exposed to chronic purulence, the increased number of differentially expressed genes may not only reflect intrinsic differences but also cellular exposure to elevated serum levels of circulating inflammatory mediators present in individuals with severe CF lung disease.
Genes involved in the ubiquitin cycle, oxidoreductase activity, and lipid metabolism were those most strongly upregulated in severe CF. Of the 569 upregulated genes identified, nine were ubiquitin-activating and ubiquitin-conjugating enzymes. This strongly suggests a significant increase in the activity of the ubiquitin system in individuals with severe CF. Among the numerous upregulated ubiquitin cycle genes were the specific ubiquitin-activating enzyme UBA2 and its ubiquitin-like protein target NEDD8 (42). Also upregulated was ubiquitin-conjugating enzyme HIP2 (E2–25K), which by its covalent attachment of ubiquitin identifies proteins for intracellular proteolysis by the 26S proteasome (43). ΔF508-mutated CFTR is known to be targeted for degradation by the proteasome, and increased degradation would result in less partially functional CFTR reaching the epithelial cell apical membrane. Although functional polymorphisms in the identified ubiquitin-conjugating enzymes are not known, those leading to increased activity of the ubiquination-proteasome pathway would almost certainly be detrimental to CFTR expression in CF and potentially modify the CF phenotype.
Six of the 569 upregulated genes in severe CF were subunits of the NADH:ubiquinone oxidoreductase complex I, the initial enzyme complex in the electron transport chain of mitochondria. Complex I catalyses the first step in the respiratory electron transport chain in mitochondria, the reduction of ubiquinone by NADH (44). It also produces superoxide in the mitochondrial matrix, which is converted by superoxide dismutase into hydrogen peroxide (45). Abnormalities in NADH dehydrogenase have been identified in CF (46). Further investigation is required to determine if the upregulation of complex I seen in individuals with severe CF is due to increased oxidative stress or is a primary contributor to pathology.
One challenge that we had to address in this study was the risk of type I error (false-positive differentially expressed genes) due to the multiple comparisons present in microarray analysis. There are several classic approaches to controlling for multiple comparisons; however, the normal physiologic variability and overlap between CF and non-CF gene expression in vivo, even in genes known to be differentially expressed, such as IL-8 (47), makes the stringent P values required for significance by these classic methods nearly impossible to obtain. This was noted recently in a microarray study of CF versus non-CF epithelium grown in cell culture by Zabner and coworkers (22). Even when they analyzed numerous CF and non-CF epithelial cell samples grown under tightly controlled conditions, their initial correction for multiple comparisons resulted in 0 of 22,238 tested genes being identified as significantly differentially expressed.
Our goal was to use available statistical tools to minimize the possibility of type I error as much as possible without being so stringent that we would eliminate all leads to potentially important differentially expressed genes. We did this by first minimizing variability before conducting ANOVA analysis by using a two-component Rocke-Lorenzato model normalization, which corrects for the absolute error that dominates at low expression and the relative errors present at high expression levels (8). Second, we used a deviation from median correction to minimize the effect of outliers. These adjustments resulted in a reduction of 70.1% in the number of genes identified as significantly differentially expressed compared with ANOVA alone. Next, we used gene ontology group analysis to further reduce type I error because false–positive, differentially expressed genes should be randomly distributed across ontology groups. Finally, we assured that our key findings in individual genes were not due to false discovery by confirming the Statherin and Duox2 data by RT-PCR in a larger, separate CF population.
There are other potential limitations to using an array approach to identify candidate modifiers of CF phenotype. First is the lack of tight correlation between transcript levels and functional protein expression. All of the candidate genes identified here require further study at the protein level. Second is an inability to detect meaningful modifiers, which occur in a small percentage of the population. An example of this is the nonfunctional variant of the mannose-binding lectin gene that is present in 5–10% of the population and has been suggested as being associated with severe CF lung disease (48). The statistics of microarray analysis do not identify as significant a marked decrease in expression in only 5–10% of the samples. Finally, although using nasal respiratory epithelial cells for analysis is well accepted and provides some significant advantages, these cells may not fully represent the characteristics of lower airway cells.
Overall, the number of genes differentially expressed in the nasal respiratory epithelial cells of individuals with CF compared with non-CF control subjects is less than might be expected. This is particularly true for cells from individuals with mild CF lung disease, with only 69 of the 44,670 assessed probe sets being clearly differentially expressed. This finding is consistent with the recent study by Zabner and colleagues of ΔF508 and non-CF respiratory primary cell cultures grown in sterile conditions for 60 d (22). They identified only 24 of 22,000 probed genes to be differentially expressed. When in vivo nasal respiratory epithelial cells from individuals with severe CF lung disease are compared separately with non-CF control subjects, a larger number of differentially expressed genes are identified. Whether this is due to intrinsic cellular differences or to chronic exposure to circulating cytokines requires further investigation.
In summary, this study provides the first in vivo comparison of respiratory epithelial cell gene expression profiles in ΔF508 homozygotes with mild and severe CF lung disease and non-CF control subjects. Although the number of differentially expressed genes might be considered low, particularly in ΔF508 homozygotes with mild disease, specific candidate modifiers of interest involved in airway defense and inflammation are identified. ΔF508 homozygotes with severe disease demonstrate a unique expression profile, with significant upregulation of genes involved in protein ubiquination and mitochondrial oxidoreductase activity. Further investigation of genes identified in this study may aid in the greater understanding of the molecular basis of variability in severity of CF lung disease.
Supplementary Material
Acknowledgments
The authors acknowledge the work of the Children's National Medical Microarray Center in processing the samples.
This work was supported by grants K23-HL071847, U01-HL66618, R025-CR02, CFFMerlo00Q0, R01-HL68927, and BAA HL 02–04 from the NHLBI and the Cystic Fibrosis Foundation.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2005-0359OC on April 13, 2006
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Cystic Fibrosis Foundation Patient Registry. 2004 annual data report to the center directors. Bethesda, MD: Cystic Fibrosis Foundation; 2005.
- 2.Kerem E, Corey M, Kerem BS, Rommens J, Markiewicz D, Levison H, Tsui LC, Durie P. The relation between genotype and phenotype in cystic fibrosis: analysis of the most common mutation (delta F508). N Engl J Med 1990;323:1517–1522. [DOI] [PubMed] [Google Scholar]
- 3.Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, Grimwood K. Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J Pediatr 2001;138:699–704. [DOI] [PubMed] [Google Scholar]
- 4.Corey M, McLaughlin FJ, Williams M, Levison H. A comparison of survival, growth, and pulmonary function in patients with cystic fibrosis in Boston and Toronto. J Clin Epidemiol 1988;41:583–591. [DOI] [PubMed] [Google Scholar]
- 5.Merlo CA, Boyle MP. Modifier genes in cystic fibrosis lung disease. J Lab Clin Med 2003;141:237–241. [DOI] [PubMed] [Google Scholar]
- 6.Mekus N, Ballmann M, Bronsveld I, Bijman J, Veeze H, Tummler B. Categories of deltaF508 homozygous cystic fibrosis twin and sibling pairs with distinct phenotypic characteristics. Twin Res 2000;3:277–293. [DOI] [PubMed] [Google Scholar]
- 7.Schluchter MD, Konstan MW, Davis PB. Jointly modelling the relationship between survival and pulmonary function in cystic fibrosis patients. Stat Med 2002;21:1271–1287. [DOI] [PubMed] [Google Scholar]
- 8.Rocke DM, Lorenzato S. A two-compenent model for measurement error in analytical chemistry. Technometrics 1995;37:176–184. [Google Scholar]
- 9.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol 2003;4:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forteza R, Salathe M, Miot F, Forteza R, Conner GE. Regulated hydrogen peroxide production by Duox in human airway epithelial cells. Am J Respir Cell Mol Biol 2005;32:462–469. [DOI] [PubMed] [Google Scholar]
- 11.Geiszt M, Witta J, Baffi J, Lekstrom K, Leto TL. Dual oxidases represent novel hydrogen peroxide sources supporting mucosal surface host defense. FASEB J 2003;17:1502–1504. [DOI] [PubMed] [Google Scholar]
- 12.Kristof AS, Marks-Konczalik J, Billings E, Moss J. Stimulation of signal transducer and activator of transcription-1 (STAT1)-dependent gene transcription by lipopolysaccharide and interferon-gamma is regulated by mammalian target of rapamycin. J Biol Chem 2003;278:33637–33644. [DOI] [PubMed] [Google Scholar]
- 13.Raj PA, Johnsson M, Levine MJ, Nancollas GH. Salivary statherin: dependence on sequence, charge, hydrogen bonding potency, and helical conformation for adsorption to hydroxyapatite and inhibition of mineralization. J Biol Chem 1992;267:5968–5976. [PubMed] [Google Scholar]
- 14.Kochanska B, Kedzia A, Kamysz W, Mackiewicz Z, Kupryszewski G. The effect of statherin and its shortened analogues on anaerobic bacteria isolated from the oral cavity. Acta Microbiol Pol 2000;49:243–251. [PubMed] [Google Scholar]
- 15.Sabatini LM, Warner TF, Saitoh E, Azen EA. Tissue distribution of RNAs for cystatins, histatins, statherin, and proline-rich salivary proteins in humans and macaques. J Dent Res 1989;68:1138–1145. [DOI] [PubMed] [Google Scholar]
- 16.Wolf AM, Wolf D, Rumpold H, Enrich B, Tilg H. Adiponectin induces the anti-inflammatory cytokines IL-10 and IL-1RA in human leukocytes. Biochem Biophys Res Commun 2004;323:630–635. [DOI] [PubMed] [Google Scholar]
- 17.Yamauchi T, Kamon J, Minokoshi Y, Ito Y, Waki H, Uchida S, Yamashita S, Noda M, Kita S, Ueki K, et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat Med 2002;8:1288–1295. [DOI] [PubMed] [Google Scholar]
- 18.Krzywanski DM, Dickinson DA, Iles KE, Wigley AF, Franklin CC, Liu RM, Kavanagh TJ, Forman HJ. Variable regulation of glutamate cysteine ligase subunit proteins affects glutathione biosynthesis in response to oxidative stress. Arch Biochem Biophys 2004;423:116–125. [DOI] [PubMed] [Google Scholar]
- 19.Zhu YM, Bradbury DA, Pang L, Knox AJ. Transcriptional regulation of interleukin (IL)-8 by bradykinin in human airway smooth muscle cells involves prostanoid-dependent activation of AP-1 and nuclear factor (NF)-IL-6 and prostanoid-independent activation of NF-kappaB. J Biol Chem 2003;278:29366–29375. [DOI] [PubMed] [Google Scholar]
- 20.Tabary O, Zahm JM, Hinnrasky J, Couetil JP, Cornillet P, Guenounou M, Gaillard D, Puchelle E, Jacquot J. Selective up-regulation of chemokine IL-8 expression in cystic fibrosis bronchial gland cells in vivo and in vitro. Am J Pathol 1998;153:921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Clark JC, Aronow BJ, Dey CR, Liu C, Wooldridge JL, Whitsett JA. Transcriptional adaptation to cystic fibrosis transmembrane conductance regulator deficiency. J Biol Chem 2003;278:7674–7682. [DOI] [PubMed] [Google Scholar]
- 22.Zabner J, Scheetz T, Abdulkawy H, Casavant T, Huang J, Keshavjee S, McCray PB Jr. The CFTR DeltaF508 mutation has minimal effect on the gene expression profile of differentiated human airway epithelia. Am J Physiol Lung Cell Mol Physiol 2005;289:L545–L553. [DOI] [PubMed] [Google Scholar]
- 23.Puchelle E, Gaillard D, Ploton D, Hinnrasky J, Fuchey C, Boutterin MC, Jacquot J, Dreyer D, Pavirani A, Dalemans W. Differential localization of the cystic fibrosis transmembrane conductance regulator in normal and cystic fibrosis airway epithelium. Am J Respir Cell Mol Biol 1992;7:485–491. [DOI] [PubMed] [Google Scholar]
- 24.Knowles M, Gatzy J, Boucher R. Increased bioelectric potential difference across respiratory epithelia in cystic fibrosis. N Engl J Med 1981; 305:1489–1495. [DOI] [PubMed] [Google Scholar]
- 25.Wijkstrom-Frei C, El Chemaly S, Ali-Rachedi R, Gerson C, Cobas MA, Forteza R, Salathe M, Conner GE. Lactoperoxidase and human airway host defense. Am J Respir Cell Mol Biol 2003;29:206–212. [DOI] [PubMed] [Google Scholar]
- 26.Ratner AJ, Prince A. Lactoperoxidase: new recognition of an “old” enzyme in airway defenses. Am J Respir Cell Mol Biol 2000;22:642–644. [DOI] [PubMed] [Google Scholar]
- 27.Ohashi A, Murata E, Yamamoto K, Majima E, Sano E, Le QT, Katunuma N. New functions of lactoferrin and beta-casein in mammalian milk as cysteine protease inhibitors. Biochem Biophys Res Commun 2003; 306:98–103. [DOI] [PubMed] [Google Scholar]
- 28.Gonsky R, Deem RL, Young HA, Targan SR. CD2 mediates activation of the IFN-gamma intronic STAT binding region in mucosal T cells. Eur J Immunol 2003;33:1152–1162. [DOI] [PubMed] [Google Scholar]
- 29.Doring G, Frank F, Boudier C, Herbert S, Fleischer B, Bellon G. Cleavage of lymphocyte surface antigens CD2, CD4, and CD8 by polymorphonuclear leukocyte elastase and cathepsin G in patients with cystic fibrosis. J Immunol 1995;154:4842–4850. [PubMed] [Google Scholar]
- 30.Ozen M, Cokugras H, Ozen N, Camcioglu Y, Akcakaya N. Relation between serum Insulin-like growth factor-I and insulin-like growth factor-binding protein-3 levels, clinical status and growth parameters in prepubertal cystic fibrosis patients. Pediatr Int 2004;46:429–435. [DOI] [PubMed] [Google Scholar]
- 31.Proctor GB, Hamdan S, Carpenter GH, Wilde P. A statherin and calcium enriched layer at the air interface of human parotid saliva. Biochem J 2005;389:111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niemi LD, Johansson I. Salivary statherin peptide-binding epitopes of commensal and potentially infectious Actinomyces spp. delineated by a hybrid peptide construct. Infect Immun 2004;72:782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole AM, Dewan P, Ganz T. Innate antimicrobial activity of nasal secretions. Infect Immun 1999;67:3267–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lappas M, Permezel M, Rice GE. Leptin and adiponectin stimulate the release of proinflammatory cytokines and prostaglandins from human placenta and maternal adipose tissue via nuclear factor-kappaB, peroxisomal proliferator-activated receptor-gamma and extracellularly regulated kinase 1/2. Endocrinology 2005;146:3334–3342. [DOI] [PubMed] [Google Scholar]
- 35.Vasseur F, Helbecque N, Lobbens S, Vasseur-Delannoy V, Dina C, Clement K, Boutin P, Kadowaki T, Scherer PE, Froguel P. Hypoadiponectinaemia and high risk of type 2 diabetes are associated with adiponectin-encoding (ACDC) gene promoter variants in morbid obesity: evidence for a role of ACDC in diabesity. Diabetologia 2005;48:892–899. [DOI] [PubMed] [Google Scholar]
- 36.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA 2002;99:4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahajan S, Gollob JA, Ritz J, Frank DA. CD2 stimulation leads to the delayed and prolonged activation of STAT1 in T cells but not NK cells. Exp Hematol 2001;29:209–220. [DOI] [PubMed] [Google Scholar]
- 38.Zheng S, Xu W, Bose S, Banerjee AK, Haque SJ, Erzurum SC. Impaired nitric oxide synthase-2 signaling pathway in cystic fibrosis airway epithelium. Am J Physiol Lung Cell Mol Physiol 2004;287:L374–L381. [DOI] [PubMed] [Google Scholar]
- 39.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 1996;84:431–442. [DOI] [PubMed] [Google Scholar]
- 40.Zheng S, De BP, Choudhary S, Comhair SA, Goggans T, Slee R, Williams BR, Pilewski J, Haque SJ, Erzurum SC. Impaired innate host defense causes susceptibility to respiratory virus infections in cystic fibrosis. Immunity 2003;18:619–630. [DOI] [PubMed] [Google Scholar]
- 41.Tanabe Y, Nishibori T, Su L, Arduini RM, Baker DP, David M. Cutting edge: role of STAT1, STAT3, and STAT5 in IFN-alpha beta responses in T lymphocytes. J Immunol 2005;174:609–613. [DOI] [PubMed] [Google Scholar]
- 42.Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature 2003;422:330–334. [DOI] [PubMed] [Google Scholar]
- 43.Yao T, Cohen RE. Cyclization of polyubiquitin by the E2–25K ubiquitin conjugating enzyme. J Biol Chem 2000;275:36862–36868. [DOI] [PubMed] [Google Scholar]
- 44.Hirst J. Energy transduction by respiratory complex I: an evaluation of current knowledge. Biochem Soc Trans 2005;33:525–529. [DOI] [PubMed] [Google Scholar]
- 45.Lambert AJ, Brand MD. Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (complex I). J Biol Chem 2004;279:39414–39420. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro BL. Evidence for a mitochondrial lesion in cystic fibrosis. Life Sci 1989;44:1327–1334. [DOI] [PubMed] [Google Scholar]
- 47.McGarvey LP, Dunbar K, Martin SL, Brown V, Macmahon J, Ennis M, Elborn JS. Cytokine concentrations and neutrophil elastase activity in bronchoalveolar lavage and induced sputum from patients with cystic fibrosis, mild asthma and healthy volunteers. J Cyst Fibros 2002; 1:269–275. [DOI] [PubMed] [Google Scholar]
- 48.Yarden J, Radojkovic D, De Boeck K, Macek M Jr, Zemkova D, Vavrova V, Vlietinck R, Cassiman JJ, Cuppens H. Polymorphisms in the mannose binding lectin gene affect the cystic fibrosis pulmonary phenotype. J Med Genet 2004;41:629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.