Abstract
TFII-I proteins are ubiquitously expressed transcriptional factors involved in both basal transcription and signal transduction activation or repression. TFII-I proteins are detected as early as at two-cell stage and exhibit distinct and dynamic expression patterns in developing embryos as well as mark regional variation in the adult mouse brain. Analysis of atypical small and rare chromosomal deletions at 7q11.23 points to TFII-I genes (GTF2I and GTF2IRD1) as the prime candidates responsible for craniofacial and cognitive abnormalities in the Williams-Beuren syndrome. TFII-I genes are often subjected to alternative splicing, which generates isoforms that that show different activities and play distinct biological roles. The coding regions of TFII-I genes are composed of more than 30 exons and are well conserved among vertebrates. However, their 5′ untranslated regions are not as well conserved and all poorly characterized. In the present work, we analyzed promoter regions of TFII-I genes and described their additional exons, as well as tested tissue specificity of both previously reported and novel alternatively spliced isoforms. Our comprehensive analysis leads to further elucidation of the functional heterogeneity of TFII-I proteins, provides hints on search for regulatory pathways governing their expression, and opens up possibilities for examining the effect of different haplotypes on their promoter functions.
Keywords: Gtf2i, Gtf2ird1, Alu repeats, Williams-Beuren syndrome
1. Introduction
Molecular analyses during the last decade reveal that alternative splicing often determines the binding properties, intracellular localization, enzymatic activity, and protein stability, and recent estimations indicate that as much as 50–60% of human and mouse genes generate alternatively spliced products (Stamm et al., 2006). In addition, most mouse and human protein-coding genes are associated with more than one promoter region. These alternative promoters and, therefore, alternative exons of 5′ untranslated regions (5′ UTRs) are generally used in different contexts or tissues.
Currently, microarray hybridization is important source of information about gene expression and its regulation. However, a microarray chip frequently contains multiple oligonucleotide probes selected from different regions of the same gene, and different microarray platforms use different probes for the same gene. Without regard for multiple isoforms, averaging of microarray data can result in a paradoxical variation in expression of alternative spliced genes or can lead to misinterpretation of experimental data.
As an attempt to address this problem and with the realization that alternative splicing is an important regulatory mechanism, a number of large-scale efforts are emerging to create resources on splice variants and alternate transcript structures. Specialized databases on alternative splicing are generated by manual collection of experimentally reported alternative exons from peer-reviewed journals or by computational delineation of alternative exons using the transcript resources, such as ESTs and full-length mRNA sequences. Among them, there are Alternative Splice Database of Mammals (Ji et al., 2001), Extended Alternatively Spliced EST Database (Pospisil et al., 2004), Database of Alternatively Spliced Genes (Dralyuk et al., 2000), and Alternative Splicing Database (Stamm et al., 2006).
Direct comparisons of EST and cDNA sequences from the same gene are effective but hampered by protocol differences, transcript end bias, and library coverage limitations. Therefore, microarrays containing about 125,000 different 36-nucleotide junction probes were successfully used to detect the exon-exon connections of 10,000 human multi-exon genes across 52 diverse tissue samples (Johnson et al., 2003), and this data are freely available in the Gene Expression Omnibus database (accession number GSE740). This survey provides unique information on tissue distribution for alternative splicing events, however, it contains junction probes only for limited number of previously reported alternative isoforms.
Using these database and resources, we have refined genomic organization of human and mouse GTF2I and GTF2IRD1 genes, which encode members of the TFII-I family transcription factors. We have discovered new TFII-I splice variants and investigated tissue distribution of both previously reported and novel isoforms. Identification of additional exons in 5′ UTRs of these genes led us to the characterization of alternative promoters and to the description of potential cis-regulator elements governing expression of TFII-I factors.
This work is important in our understanding of the complex expression pattern of TFII-I proteins during mouse pre- and postimplantation development and adult tissues (Bayarsaihan and Ruddle, 2000; Bayarsaihan et al. 2003; Enkhmandakh et al., 2004; Ohazama and Sharpe, 2007; Palmer et al., 2007).
2. Materials and methods
2.1. Bioinformatic analysis
List of databases that we used in our analysis: Alternative Splice Database of Mammals (AsMamDB, http://166.111.30.65:100/), Extended Alternatively Spliced EST Database (EASED, http://eased.bioinf.mdc-berlin.de), Database of Alternatively Spliced Genes (ASDB, http://cbcg.nersc.gov/asdb), Alternative Splicing Database (ASD, http://www.ebi.ac.uk/asd/); dbEST (NCBI, www.ncbi.nlm.nih.gov); Cap-Analysis Gene Expression database (CAGE, gerg01.gsc.riken.jp/cage/); Ensemble (www.ensembl.org).
Relative abundance of known alternative isoforms was calculated from the exon junction microarray data (GEO accession GSE740) according to published algorithm (Wang et al., 2003).
Programs that we used for our analysis: RepeatMasker (www.repeatmasker.org); Transcription Element Search System (TESS, www.cbil.upenn.edu/cgi-bin/tess/tess); MacVector version 9.5.4 (Symantec Corporation, www.macvector.com).
2.2. RT-PCR and qRT-PCR analysis
Expression of alternative splice isoforms in different tissues was analyzed using First Choice Human Total RNA Survey and Mouse Total RNA Panels (Ambion). 1 μg of RNA from each sample was reverse-transcribed with gene-specific primers (Tables 1 and 2) or random decamer primers (Ambion) using Omniscript enzyme (Qiagen) according to the manufacturer’s protocol. 2 μl of reverse-transcription reaction was used then for PCR amlification with Taq enzyme (New England Biolabs) and primers listed in Tables 1 and 2. 35 cycles of PCR were performed as follows: denaturation for 30 s at 98°C, annealing for 30 s at 58°C, and extension for 1 min at 72°C. In cases of strong amplifications, numbers of PCR cycles reduced fro more accurate comparisons of relative amounts of isoforms. PCR products resolved in 1.5% agarose gel electrophoresis were purified for sequencing using QIAquick Gel Extraction Kit (Qiagen).
Table 1.
GTF2I primer sequences.
| Name | Location, forward (F) or reverse (R) | Sequence |
|---|---|---|
| Mouse Gtf2i | ||
| RT primers | ||
| MGTF2IRT1 | Exon 5, R | 5′-GGAAATCCCTGCCTTGTTCTCC -3′ |
| MGTF2IRT2 | Exon 15, R | 5′-TCAAGGCGAGGAATGCCG -3′ |
| MGTF2IRT3 | Exon 35, R | 5′-ACACACAGCCCAGCCTGACTC -3′ |
| PCR primers | ||
| MTF2IE01 | Exon 1, F | 5′-TGGCGACCCAGCGTGGAGCG -3′ |
| MTF2IE1A | Exon 1a, F | 5′-TGGGATGGATGGTGAACCTCTGGG -3′ |
| MTF2IE1B | Exon 1b, F | 5′-TGCTCTCAGACCCACTGCCACCTC -3′ |
| MTF2IE5A | Exon 5, R | 5′-AACCCCCTCGGGAAGCCCCTGC -3′ |
| MTF2IE5B | Exon 5, R | 5′-TGAAGGCAACCCCCTCGGGAAG -3′ |
| MTF2IE07 | Exon 5, R | 5′-TGAAGGCAACCCCCTCGGGAAG -3′ |
| MTF2IE07 | Exon 7, F | 5′-GGCTGAGAGGTCCATGCTGTCTCC -3′ |
| MTF2E11F | Exon 11, F | 5′-TCAGAAGGCAACGAGGGAACGG -3′ |
| MTF2E11R | Exon 11, R | 5′-CACTTCCACTTCCGTTCCCTCGTTG -3′ |
| MTF2E15R | Exon 15, R | 5′-TGGGACTGGAAAAGAGGGTACGGG -3′ |
| MTF2E30 | Exon 30, F | 5′-CACCATCAACCCTGGCTGCGTG -3′ |
| MTF2E35 | Exon 35, R | 5′-GCACTGGCTGCTCACTGGCGG -3′ |
| Human GTF2I | ||
| RT primers | ||
| HTF2IRT1 | Exon 5, R | 5′-GAATGAAATCCCTGCTTTGTTCTCC -3′ |
| HTF2IRT2 | Exon 15, R | 5′-CACAAAACGAATCCTTTCCTTTGC -3′ |
| HTF2IRT3 | Exon 35, R | 5′-CATCCTTAGCCCCCTGACCC -3′ |
| PCR primers | ||
| HTF2IE01 | Exon 1, F | 5′-TCTCGCCTCCCGTCCGCTCG -3′ |
| HTF2IE1A | Exon 1a, F | 5′-GGAGTCTCCCTCTGTCGCACAGGC -3′ |
| HTF2IE1B | Exon 1b, F | 5′-GTGATCCTCTTGCCTCAGGCTCCTG -3′ |
| HTF2IE5 | Exon 5, R | 5′-GCCGACTGGTCTCGCAGCATCTTC -3′ |
| HTF2E11F | Exon 11, F | 5′-GCCACCATTCTTCAGAGGGCAATG -3′ |
| HTF2E11R | Exon 11, R | 5′-CCATTTCTGTGCCTTCATTGCCCTC -3′ |
| HTF2E15R | Exon 15, R | 5′-AAAGAGGGTACGGGATCGTCACCG -3′ |
| HTF2E30 | Exon 30, F | 5′-TGGCTGTGTGGTGGTTGATGGC –3′ |
| HTF2E35 | Exon 35, R | 5′-ACGGCGACCCCTGGAGAGGG –3′ |
Table 2.
GTF2IRD1 primer sequences.
| Name | Location, forward (F) or reverse (R) | Sequence |
|---|---|---|
| Mouse Gtf2ird1 | ||
| RT primers | ||
| MBENRT1 | Exon 6, R | 5′-GTCGCTGGGGGTCAACGG -3′ |
| MBENRT2 | Exon 30, R | 5′-CTCCGTCTATCTTGTGGGCAAAAC -3′ |
| MBENRT3 | Exon 31, R | 5′-TGAGTTGAGGTCCTGATTCGGC -3′ |
| PCR primers | ||
| MBENE01 | Exon 1, F | 5′-TGATCCTGTCCCCCATTCCCG -3′ |
| MBENE1A | Exon 1a, F | 5′-AAGCAGACACCCCTCTCACCCTCG -3′ |
| MBENE5R | Exon 5, R | 5′-GCCCCTGCACCGCCAGTAGC -3′ |
| MBENE17 | Exon 17, F | 5′-AGCGAGCAACAGCATCCAGTTTGTC -3′ |
| MBENE24A | Exon 24, F | 5′-GATGCCAACAGACTGGGGGAGAAG -3′ |
| MBENE24B | Exon 24, F | 5′-GGAGAAGGTGATCCTCCGAGAGCAG -3′ |
| MBENE25 | Exon 25, R | 5′-CCCTTCACCTCCACGGCATCTG -3′ |
| MBENE30 | Exon 30, R | 5′-TCTGGTTGGTGGATGCCACAGACTC -3′ |
| MBENE31 | Exon 31, R | 5′-GTTGAGGTCCTGATTCGGCTCTGAG -3′ |
| Human GTF2IRD1 | ||
| RT primers | ||
| HBENRT1 | Exon 6, R | 5′-CAGTCGGAGAAGTCTTGGGCAC -3′ |
| HBENRT2 | Exon 26, R | 5′-GAAGGTCCCCACTCAAAGCCC -3′ |
| HBENRT3 | Exon 27, R | 5′-TCATTGGAAAAACTAAAAGGCATCG -3′ |
| PCR primers | ||
| HBENE01 | Exon 1, F | 5′-GCCGTCCTCGCCTCCCTCTG -3′ |
| HBENE6R | Exon 6, R | 5′-CCGCCCGCCATCCTCAAGTG -3′ |
| HBENE24A | Exon 24, F | 5′-GTGGAGGTCACGGGTCTGCCTG -3′ |
| HBENE24B | Exon 24, F | 5′-CCGAGAGCATGTCCGCATGGTC -3′ |
| HBENE26 | Exon 26, R | 5′-CGCTTGGGAATGCTGCTGTCTTTG -3′ |
| HBENE27 | Exon 27, R | 5′-CGGGAGCTGCACGTTCAGGC -3′ |
Quantitative PCR was carried out on an ABI PRISM 7300 detection system. We used pre-developed TaqMan gene expression assays (Assay IDs are in the text) and TaqMan Gene Expression Master Mix (Applied Biosystems). Data from triplicates are expressed as normalized expression by using the delta-delta-Ct calculation method (Livak and Schmittgen, 2001) and the Gapdh and GAPDH reference genes (Mm99999915_g1 and Hs99999905_ml, respectively).
3. Results
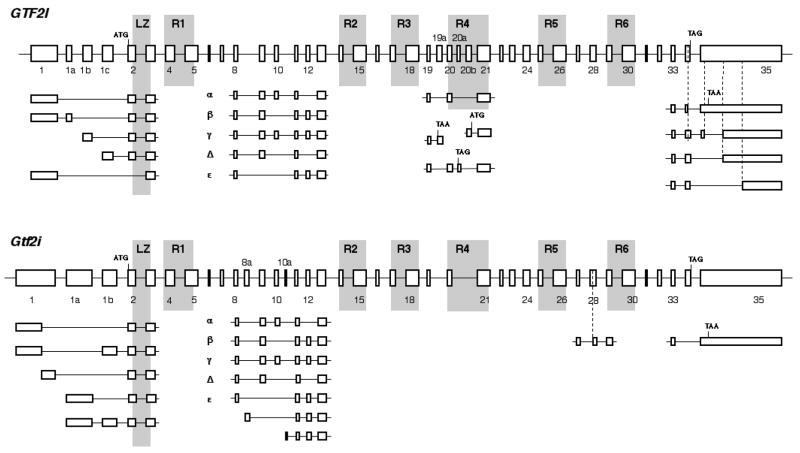
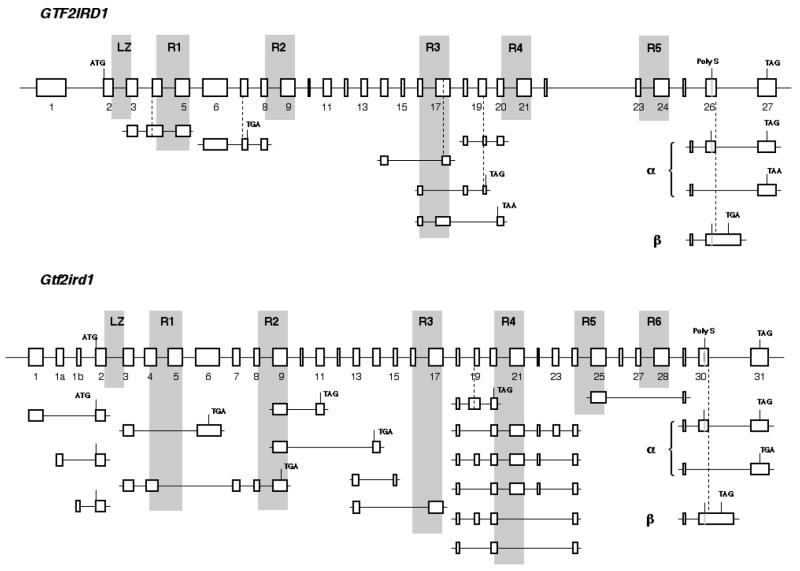
It has been reported that human GTF2I and GTF2IRD1 paralogous genes consist of 35 and 27 exons, respectively (Cheriyath and Roy, 2000; Tassabehji et al,. 1999). Structural analysis revealed that unusual repeated combinations of two exons encode protein domains specific to the TFII-I family and referred as I-repeats. I-repeats have an atypical helix-loop-helix structure that has been resolved by protein crystal analysis (Doi-Katayama et al., 2007) and can serve as independent DNA-binding modules with various DNA-binding efficiency (Vullhorst and Buonanno, 2005). GTF2I and GTF2IRD1 genes contain 6 and 5 I-repeats, respectively (Fig. 1 and 2). Structures of mouse Gtf2i and Gtf2ird1 genes are similar to their human orthologs except duplication of several exons in mouse Gtf2ird1 resulting in the additional I-repeat (Bayarsaihan et al., 2002; Fig. 1 and 2).
Fig. 1. Diagram of genomic organization and alternatively spliced forms of human GTF2I gene and its mouse ortholog Gtf2i.
Exons are shown as boxes and drawn to scale, and the introns, which vary greatly in size, are represented in a uniform manner. LZ, leucine zipper; R1–R6, I-repeats. Alternative variants of exon junctions are shown beneath the diagrams of genes.
Fig. 2. Diagram of genomic organization and alternatively spliced forms of human GTF2IRD1 gene and its mouse ortholog Gtf2ird1.
Notation and labeling as in Fig. 1.
In contrast to the coding regions of TFII-I genes, the exons encoding their 5′ UTRs had not been fully defined. To identify alternatively started TFII-I transcripts, we searched specialized databases on alternative splicing listed in Materials and methods. However, none of these database provides exhaustive data, therefore, we have complemented this analysis with our search in the NCBI EST database. We restricted our search to correctly spliced transcripts utilizing for splicing only canonic donor and acceptor sequences.
3.1. Identification of new alternative splicing in human GTF2I and mouse Gtf2i transcripts
Four isoforms of human and mouse GTF2I were described previously that are resulted from skipping cassette exons 10 and/or 12 between the first and the second I-repeats (Cheriyath and Roy, 2000). Exon 12 is absent in isoform α (transcript variant 3, human RefSeq accession NM_033001, mouse RefSeq accession NM_001080747), whereas exon 10 is missing in isoform β (transcript variant 2, human RefSeq accession NM_033000, mouse RefSeq accession NM_010365). Skipping of these exons is not mutually exclusive, and isoform Δ lacks both of them (transcript variant 4, human RefSeq accession NM_033003, mouse RefSeq accession NM_001080748). Exons 9 – 13 have the same phase and, therefore, their skipping does not change the open reading frame (ORF) of the full-length transcript (isoform γ, transcript variant 1, human RefSeq accession NM_032999, mouse RefSeq accession NM_001080746).
In addition to these four isoforms, we have found isoform ε, transcripts of which lack exons 9 and 10, but retain exon 12 (Fig. 1). Such cDNAs occur in both human (EST accession DA448103) and mouse (EST accession CX562749, CD348277, and CN527965). Isoform ε is very close in size to isoform Δ and, therefore, not easily can be identified in experiments, and its transcripts appear to be rare (see below).
One more region of extensive alternative splicing of GTF2I has been found at its 3′ end. We have identified an optional intron I35 within non-coding exon 35, which obeys classical AG-GT splicing rules on both sides, and some human transcripts (EST accession BG202932 and AA393465) possess 299 bp shorter 3′ UTR than most GTF2I mRNAs retaining this sequence (Fig. 1). Some human transcripts in stomach lymph node and epithelial cells (EST accession BM761842 and BM760328) can skip 5′ portion of exon 35 and join exon 34 directly to 3′ part of exon 35 using its internal acceptor site (Fig. 1).
In addition, exon 34 contains a cryptic splice donor site and its harnessing for splicing in metastatic chondrosarcoma (EST accession BM994169) results in missing TAG stop codon. The next stop codon TAA is located in exon 35 and, therefore, such transcripts can be translated into proteins that are 23 amino acid longer than the major GTF2I sequence (Fig. 1). Similarly, 3′ extension of the ORF in mouse Gtf2i mRNAs has been observed as a result of skipping the entire exon 34 (EST accessions CA579745, CK619025, and AA197421). However, there is no obvious homology between human and mouse novel carboxyl termini.
Comprehensive analysis of EST sequences has also revealed additional genomic sequences that can be included as exons in rare human and mouse GTF2I transcripts (Fig. 1). After exon 19, some human GTF2I cDNAs obtained from testis, lung, and placenta are unexpectedly being continued with the sequence identified as a part of intron 19 (EST accessions CD244641, BG485093, and H87691). This sequence is correctly spliced through the canonic acceptor site and referred as exon 19a. There is TAA stop codon in the very beginning of exon 19a and, therefore, exon 19a encodes an alternative 3′ UTR of GTF2I. A putative protein product of such transcripts contains 560 amino acids and has size of 63.3 kDa.
Another premature truncated ORF of human GTF2I has been detected as a result of insertion of exon 20a between exons 20 and 21 (Fig. 1). mRNAs containing exon 20a were found among GTF2I transcripts in testis, thymus, amygdala, and caudate nucleus (EST accessions DB459813, BX437343, DA196174, and DA270594) and frequently in human embryonic stem cells (EST accessions CD643599, CN368579, and CN368612). Exon 20a is spliced correctly in the content of GTF2I mRNAs but contains TAG stop codon and results in ORF shortening to 580 aa. It is interesting that some GTF2I transcripts found in human hippocampus, keratoconus cornea, and large cell carcinoma (EST accessions AV725148, CV572902, and BQ223449) start from exon 20b, which is located within intron 20 just after exon 20a (Fig. 1). Exon 20b contains ATG start codon, which is in frame with the rest of amino sequence, and therefore, transcripts starting from exon 20b would be translated into N-terminus truncated TFII-I proteins lacking the leucine zipper and first three I-repeats.
A novel exons 8a and 10a were discovered within mouse Gtf2i introns 8 and 10, respectively (Fig. 1). mRNAs starting from exon 8a were found only in mouse blastocysts (EST accessions CJ089319 and BY031968) and sequences starting from exon 10a were found only among transcripts in 17 days pregnant adult female amnion (EST accessions BY074213, BY070145, and BY068783). Existence of alternative transcriptional starts within mouse intron 8 and 10 has been also confirmed by CAGE analysis (tag clusters T05R07DE4682 and T05R07DE28AF, respectively). In contrast to human exon 20b, mouse exons 8a and 10a do not contain in frame ATG and the next translational start codon is located in the beginning of the third I-repeat. Therefore, mRNAs starting from exon 8a and exon 10a differ in their 5′ UTRs only and can be translated into a N-terminus truncated TFII-I protein similar to product of human transcripts starting from exon 20b.
Finally, we will mention an exon sliding in the Gtf2i gene: in contrast to human, mouse exon 28 has an additional splice acceptor site. Alternative splicing at 5′ end of exon 28 does not shift ORF and just results in replacement of Arg-826 with IFLSG amino sequence (Fig. 1).
3.2. Identification of new alternative splicing in human GTF2IRD1 and mouse Gtf2i rd1 transcripts
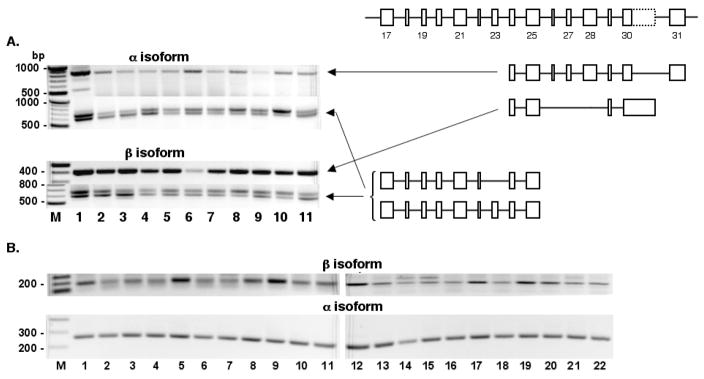
Previously it has been shown that combinations of five splicing events in the 3′ region of Gtf2ird1 gives rise to 11 mRNA isoforms in mouse skeletal muscles (Tay et al., 2003): 1) mutually exclusive exons 30 and 31 encode two alternative carboxyl termini and 3′ UTRs of α (exon 31) and β (exon 30) groups of transcripts and 2) four alternatively spliced cassettes can be removed independently in each of these two groups. We have identified additional splicing events in the Gtf2ird1 gene. Some of them are common between human and mouse but most are unique for mouse (Fig. 2).
Just as mouse mRNAs, human GTF2IRD1 transcripts possess α or β variants of 3′ termini (Fig. 2). Both α and β human variants of protein tails show a clear homology to the corresponding mouse amino acid sequences, suggesting specific functional roles for alternative carboxyl termini. In addition, exon 30 in mouse and its homologous exon 26 in human encode sequence containing polyserine tract and we have found that these exons can be omitted in both mouse (EST accession DV658314) and human (EST accession BU629129) providing the third variant of C-terminal end of GTF2IRD1 (Fig. 2).
Four alternatively spliced cassettes in the 3′ region of mouse Gtf2ird1 gene include: 1) exon 19, 2) exon 23, 3) exons 21 + 22 + 23, and 4) exons 26 + 27 + 28. The first three cassettes combine with each other independently (Fig. 2) and examples of all six possible combinations can be found in the EST database. The fourth cassette has been found in combination with the absence of exon 23 (RefSeq accession NM_001081470). Although these isoforms were described for the first time in mouse skeletal muscles (Tay et al., 2003), EST database shows that they are present also in many other tissues.
Because the structure of the human gene has diverged from its mouse ortholog in this region, human GTF2IRD1 mRNAs show lesser diversity of isoforms (Fig. 2). Other alternatively spliced cassettes include exon 14 solely (EST accession DT918981) or in combination with exons 15 + 16 (EST accession DT930126) in mouse and exons 15 + 16 in combination with 5′ part of exon 17 (EST accessions DY655747 and DY655748) in human.
Mouse exon 19 not only can be skipped in Gtf2ird1 transcripts but also it can be 5′ extended as a result of presence of alternative splice acceptor within intron 18 (EST accession BM950194 and BQ960475). This splicing event shifts the ORF and results in premature translational termination within the next exon (Fig. 2). In contrast, human exon 19 contains internal alternative splice acceptor corresponding to the conventional 5′ border of mouse exon 19 and its use does not change the ORF (EST accessions CU454871 and CU445164). However, human variants, which are similar to mouse transcripts containing premature terminated ORFs, can be produced as a combination of the 5′-truncated exon 19 with skipping exons 15 + 16 (EST accession BX104539) or by skipping exons 18 + 19 (EST accession AW749392) (Fig. 2). mRNAs encoding severely truncated polypeptides can be also generated by skipping of exons 4 + 5 (EST accession BB656497), by skipping of exons 5 + 6 (EST accession BI689851), by skipping of exon 10 (EST accession DT911408), or by skipping of exons 10 + 11 + 12 + 13 (EST accession DT916347) in mouse or by sliding of acceptor splice site of exon 7 (EST accession BQ639001) in human (Fig. 2).
3.3. Expression profiles of TFII-I transcripts in human and mouse tissues
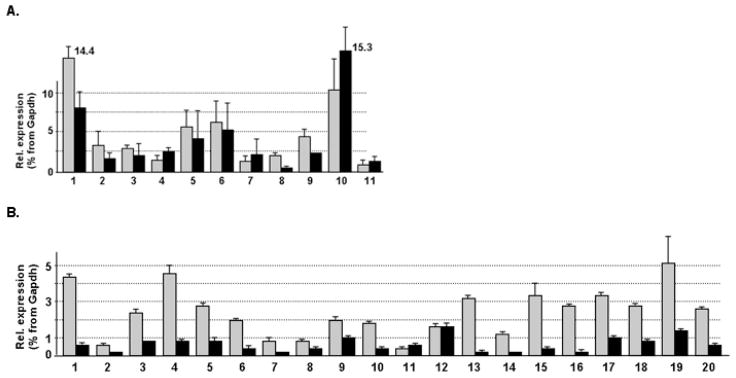
To compare the expression levels of TFII-I genes in different tissues, we took advantage of commercial panels of total RNA preparations from Ambion and used pre-developed TaqMan gene expression assays for quantitative detection of Gtf2i (Mm00494841_ml), Gtf2ird1 (Mm00465654_ml), GTF2I (Hs00263393_ml), and GTF2IRD1 (Hs00249456_ml). Each of these assays comprises a large group of oligos representing most of exons in the analyzing genes and, therefore, allows detection of all isoforms. Expression levels of TFII-I genes were normalized using Gapdh and GAPDH genes and presented as percent of these housekeeping gene expression (Fig. 3). TFII-I transcripts were detected in all tissues analyzed. Although levels of GTF2I expression were higher than expression of their corresponding GTF2IRD1 paralogs in many tissues, the expression profiles of Gtf2ird and GTF2IRD1 were similar to that of Gtf2i and GTF2I. The highest levels of TFII transcripts were registered in embryonic fibroblasts and testicle in mouse (Fig. 3A) and in brain, adipose, thymus, and thyroid in human (Fig. 3B). Similar tissue profiles were calculated from exon junction microarrays for constitutively expressed exons (data not shown).
Fig. 3. Expression profiles of mouse (A) and human (B) transcripts of GTF2I (grey bars) and GTF2IRD1 (black bars).
Quantitative RT-PCR was carried out with pre-developed TaqMan gene assays and TFII-I isoforms were detected on gel after amplification with primers listed in Tables 1 and 2. Error bars represent standard deviation calculated from three independent PCR reactions. Tissues assayed are:
A: 1 - mouse embryonic fibroblasts, 2 - total embryo E11–12, 3 - brain, 4 - heart, 5 - lung, 6 - thymus, 7 - spleen, 8 - liver, 9 - kidney, 10 - testicle, 11 – ovary;
B: 1 - brain, 2 - heart, 3 - lung, 4 - adipose, 5 - kidney, 6 - bladder, 7 - liver, 8 - cervix, 9 -esophagus, 10 - small intestine, 11 - colon, 12 - skeletal muscle, 13 - trachea, 14 - spleen, 15 -thymus, 16 - placenta, 17 - ovary, 18 - testes, 19 - thyroid, and 20 – prostate.
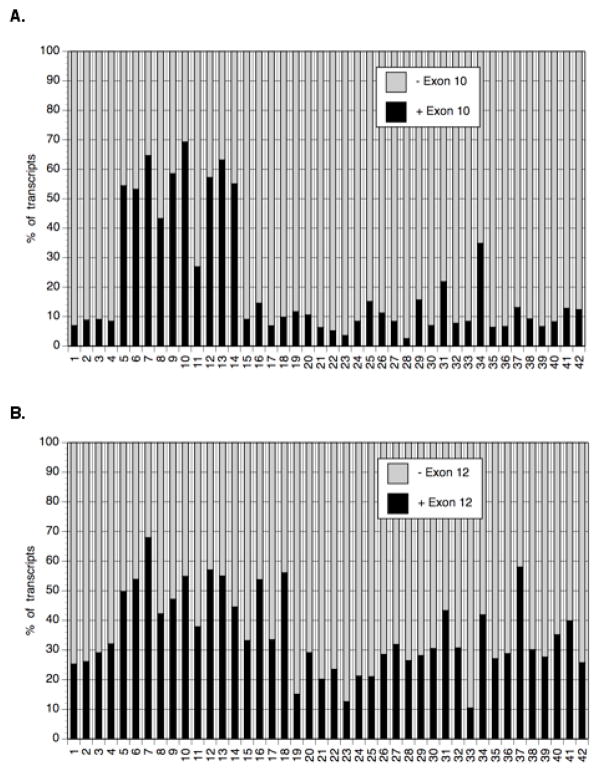
3.4. Neuronal tissues are enriched by GTF2I transcripts retaining exons 10 and 12
Splicing-specific microarrays (GEO accession GSE740) contain oligonucleotide probes not only for neighbor exons, but also for junction between exons 9 and 11 and between exons 11 and 13 of human GTF2I gene. Unfortunately, these data cannot allow to calculate an individual contribution of each isoform, however, results demonstrate clearly that isoforms α and γ retaining exon 10 are significantly in neuronal tissues (including whole brain, its subdivisions, and spinal cord) and in retina (Fig. 4A). Similar though less prominent specificity has been obtained for retention of exon 10 (isoforms β, γ, and Δ) (Fig. 4B).
Fig. 4. Relative distribution in human tissues of GTF2I splice isoform retaining exon 10 (A) and exon 12 (B).
Ratios of alternative isoforms were calculated from the relative hybridization intensity of total transcripts with 36-nucleotide probes representing junctions between exons 9 and 10, exons 9 and 11, exons 10 and 11, exon 11 and 12, exons 11 and 13, and exons 12 and 13.
Tissuses: 1 - adrenal cortex, 2 -adrenal medulla, 3-bladder, 4 -bone marrow, 5 – whole brain, 6 - brain (amygdala), 7 -brain (fetal), 8 - brain (caudate nucleus), 9 - brain (cerebellum), 10 -brain (cerebral cortex), 11 - brain (corpus callosum), 12 - brain (hippocampus), 13 - brain (postcentral gyrus), 14 - brain (thalamus), 15 - colon (descending), 16 - colon (transverse), 17 –duodenum, 18 –epididymis, 19 – heart, 20 – ileum, 21 – kidney, 22 - kidney (fetal), 23 – liver, 24 -liver (fetal), 25 – lung, 26 - lung (fetal), 27 – lymph node, 28 – pancreas, 29 – placenta, 30 –prostate, 31 – retina, 32 - salivary gland, 33 -skeletal muscle, 34 - spinal cord, 35 – spleen, 36 –stomach, 37 - testis, 38 – thymus, 39 – thyroid, 40 - tonsil, 41 – trachea, and 42 – uterus.
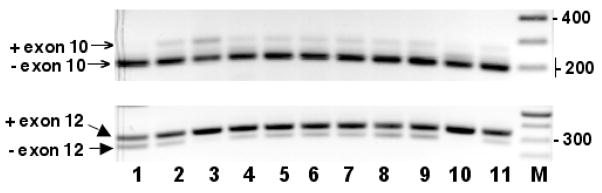
The presence and relative abundance of α, β, Δ, γ, and ε isoforms were further examined in mouse tissues by using PCR with primers designed to detect independently deletions of exons 10 and 9+10 (202bp and 124 bp with primers from exons 7 and 11) and deletion of exon 12 (281 bp with primers from exons 11 and 15). The gel profiles in Fig. 5 show that β is the major Gtf2i isoform, whereas ε isoform is not detectable in all tissues tested. α and Δ isoforms are absent in brain and testicle, and brain differs from other tissues by higher expression of γ isoform (similarly to human).
Fig. 5. Specific alternative splice isoforms of Gtf2i in mouse tissues.
RT was performed with gene-specific primer MGTF2IRT2 and cDNAs were then amplified with primers MTF2IE07 and MTF2E11R (upper gel) and with primers MTF2E11F and MTF2E15R (lower gel). Tissues analysed are as in Fig. 3A. DNA size marker is the 100 bp ladder (New England Biolabs).
To verify expression of other predicted isoforms (because a certain fraction of EST-derived sequences could represent splicing noise) and to characterize their expression profiles in different tissues, we performed more detailed RT-PCR analysis. For higher sensitivity and specificity, we used gene-specific primers in RT reactions. These primers were selected from three regions of each gene: from the beginning of ORF, from the middle of coding sequence, and from 3′ UTR (Table 1 and 2). However, other splice variants of Gtf2i appear to be minor and cannot be easily amplified in the presence of high-abundance transcript. Only one and undeleted variant of 3′ end is found to be used in Gtf2i mRNAs in all tissues examined (data not shown). Despite the possibility of diverse carboxyl termini and 3′ UTRs generated by alternative splicing (Fig. 1), the full-length sequence is found to be used as a principal GTF2I isoform (primers HTF2IE30 and HTF2IE35). 3′ UTRs with deleted optional intron I35 are seen as faint bands close to the major bands in lung, small intestine, skeletal muscle, spleen, thymus, and thyroid (data not shown).
3.5. Tissue distribution of GTF2IRD1 isoforms
To distinguish α and β variants of Gtf2ird1 transcripts, we performed reverse transcription with primers specific for each of two 3′ UTRs (Table 2). Although this RT-PCR analysis was designed to detect different isoforms rather than to compare their expression levels and, therefore, cannot provide accurate quantitative information, we observed some relative tissue-specific differences in alternative splicing patterns. Identity of obtained PCR products to the expected TFII-I sequences were confirmed by direct sequencing after their excising from the gels and PCR re-amplification.
The results show that both α and β mRNAs are present in all analyzed mouse tissues but their relative abundances vary: for example, α variant is more predominant in thymus, whereas β variant is more predominant in kidney (Fig. 6A). The PCR products demonstrate also that the deletion of cassette exons 26+27+28 is associated only with β isoform, whereas α isoform retains these exons. Pattern of other cassette deletions is the same for α and β variants suggesting that they are independent from other splice events in Gtf2ird1 mRNA processing. Amplification of region from exon 17 to exon 25 gives rise two major PCR products corresponding to the variant retaining all cassettes (678 bp) and to the variant skipping exon 23 (597 bp). These two variants are present in all tissues, however, the longer variant is more abundant in liver and testicle, the shorter variant is more frequent in total embryo, brain, and ovary, and both variant are represented approximately equally in mouse embryonic fibroblasts, heart, lung, thymus, spleen, liver, and kidney (Fig. 6A). Other splice variants are minor and appear as much weaker bands that hard to visualize.
Fig. 6. Expression profiles of specific alternative splice isoforms of Gtf2ird1 (A) and GTF2RD1 in mouse and human tissues.
RT was performed with α-specific primers MBENRT3 and HBENRT3 or with β-specific primers MBENRT2 and HBENRT2. Mouse cDNAs were amplified with primers pairs MBEN24A/MBENE31, MBEN24B/MBENE30, and MBENE17/MBENE25; human cDNAs were amplified with primer pairs HBENE24A/HBENE26 and HBENE24B/HBENE27. Tissues analyzed are as in Fig. 3A and 3B, respectively, and include two additional human tissues: breast normal (line 21) and breast tumor (line 22). DNA size marker is the 100 bp ladder (New England Biolabs). Numbers on the pictorial diagram correspond to the exon numbering of mouse Gtf2ird1.
Similarly to mouse, co-expression of α and β variants of GTF2IRD1 mRNAs are found in human tissues. The levels of α isoform are similar in all tissues tested, whereas levels of β variant vary significantly and are relatively higher in brain, kidney, cervix, esophagus, skeletal muscle, ovary, thyroid, and prostate (Fig. 6B).
3.6. Identification of alternative promoters in Gtf2i and Gtf2ird1 genes
Lower hybridization of TFII-I mRNAs with probes representing junctions between exons 1 and 2 in splicing-specific microarrays led us to the suggestion that such under-representation might reflect unknown alternative splicing at the 5′-end of transcripts. Within intron 1, we have found a few additional non-coding exons (Fig. 1). Human exon 1a (122 bp length) is optional and present in some transcripts extending their 5′ UTRs (for example, EST accession BG338017). Human exons 1b (242 bp length) or 1c (154 bp length) can serve as initial exons of transcripts (EST accessions AI346557 and DB454904, respectively). They splice correctly with exon 2 and provide alternative 5′ UTR in GTF2I mRNAs. Human exons 1a, 1b, and 1c have no homology in mouse Gtf2i gene, instead mouse intron 1 contains alternative initial exon 1a (> 873 bp length) and optional internal exon 1b (200 bp length). We also have obtained validations for existence of alternative transcriptional starts in the Cap-Analysis Gene Expression (CAGE) database: in addition to the main 5′ end of GTF2I mRNAs, significant clustering of CAGE tags was found within inton 1 at the positions corresponding alternative human exon 1b (tag cluster T07F0461FF860) and mouse exon 1a (tag cluster T05R07DEBE25).
Finally, we have identified two novel mouse exons (1a and 1b) encoding 5′ UTRs (Fig. 2): transcription of Gtf2ird1 can be started within intron 1 either from exon 1a (RefSeq accession NM_001081465) or from exon 1b (EST accession BY123668 and CF351704).
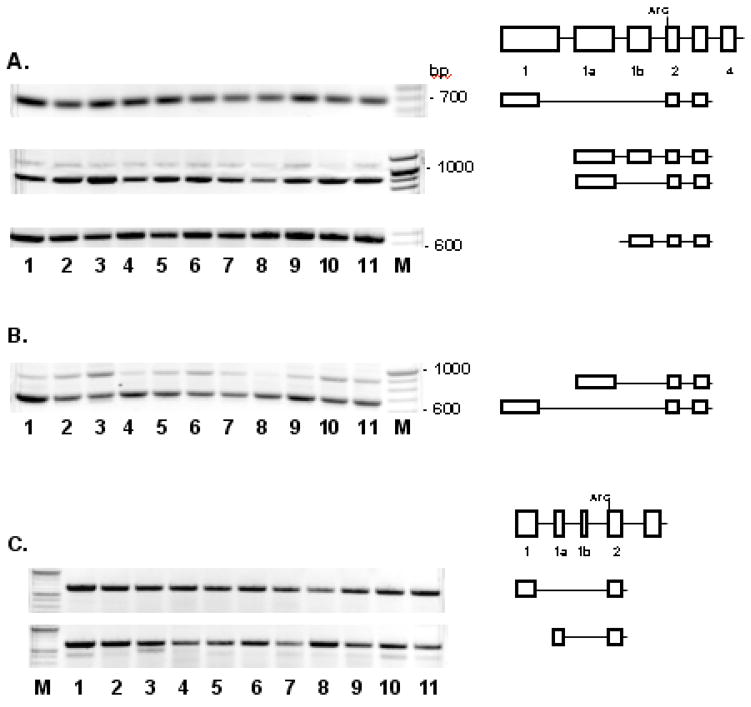
Our experimental data confirm that Gtf2i mRNAs can be started from both exon 1 and exon 1a in mouse tissues (Fig. 7A). Exon 1b serves as 5′ UTR extension relatively rare and only in transcripts starting from exon 1a. To compare promoter activity of the 5′-proximal regions of exons 1 and 1a, we performed three-primer PCR (MTF2IE01, MTF2IE1A, and MTF2IE5B) with cDNAs reverse transcribed using random decamer primers (Fig. 7B). The results demonstrate significantly higher expression level from promoter located upstream exon 1 in all tissues except brain, where activities of both alternative promoters were comparable. We failed to detect Gtf2i transcripts starting from exon 8a or exon 10a in tissue samples tested and in RNA prepared from mouse embryonic stem cells (data not shown).
Fig. 7. Alternative splicing at 5′-end of Gtf2i (A and B) and Gtf2ird1 (C) in mouse tissues.
Tissues analyzed are as in Fig. 3A. Numbers on the pictorial diagrams correspond to the exon numbering of mouse Gtf2i and Gtf2ird1 genes. DNA size marker is the 100 bp ladder (New England Biolabs).
A. RT was performed with gene-specific primer MGTF2IRT1; primers for PCR were MTF2IE01/MTF2I5A (upper gel), MTF2IE1A/MTF2IE5B (middle gel), and MTF2IE1B/MTF2I5A (lower gel).
B. RT was performed with random hexamers (Ambion); primers for PCR were MTF2IE01, MTF2IE1A, and MTF2IE5B (tree-primer PCR).
C. RT was performed with gene-specific primer MBENRT1; primers for PCR were MBENE01/MBENE5R (upper gel) and MBENE1A/MBENE5R (lower gel).
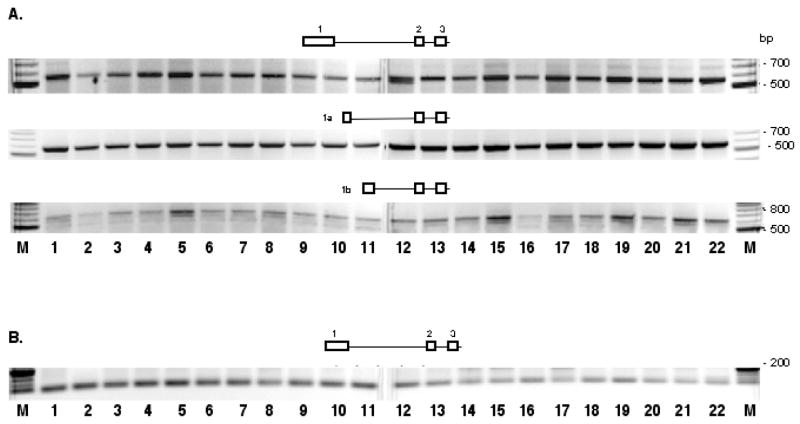
Human GTF2I mRNAs can be started from exon 1, exon1a, or exon 1b (Fig. 8A) but transcripts started from exon 1c were not detectable. Alternative promoter usage has been confirmed also for mouse Gtf2ird1 gene: similarly to Gtf2i, Gtf2ird1 mRNAs can be started from both exon 1 and exon 1a in mouse tissues (Fig. 7C). However, single transcription start has been detected for human GTF2IRD1 gene (Fig. 8B).
Fig. 8. Alternative splicing at 5′-end of GTF2I (A) and GTF2IRD1 (B) in human tissues.
Tissues analyzed are as in Fig. 3B but include two additional human tissues: breast normal (line 21) and breast tumor (line 22). Numbers on the pictorial diagrams correspond to the exon numbering of human GTF2I and GTF2IRD1 genes. DNA size marker is the 100 bp ladder (New England Biolabs).
A. RT was performed with gene-specific primer HTF2IRT1; primers for PCR were HTF2IE01/HTF2IE5 (upper gel), HTF2IE1A/HTF2IE5 (middle gel), and HTF2IEB/HTF2IE5 (lower gel).
B. RT was performed with gene-specific primer HBENRT1; primers for PCR were HBENE01 and HBENE6R.
3.7. Promoter analysis of TFII-I genes
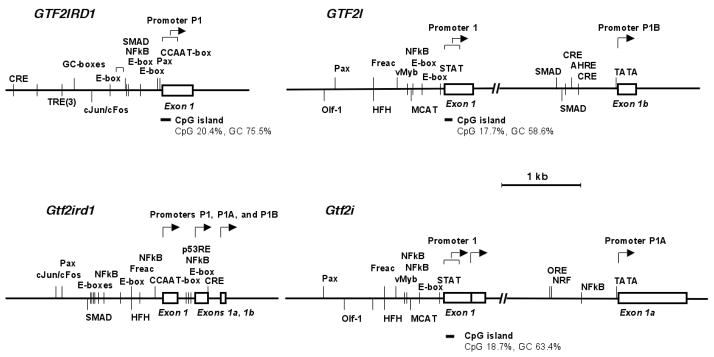
The RT-PCR detection of alternative 5′ non-translated exons 1 and 1a in the mouse Gtf2i gene and exons 1 and 1b in the human GTF2I gene indicates that each of them is preceded by a functional promoter. Therefore, 2 kb sequences upstream of these exons were examined for the presence of typical promoter elements and binding sites for transcription factors (TFBS).
Major (in terms of basal transcription activity) mouse and human P1 promoters are located upstream of exon 1 and immersed in CpG islands. They are both TATA-less and Initiator-less and show remarkable human-mouse conservation in assortment and relative positions of TFBS (Fig. 9). Shared cis-elements that are conserved in distance and orientation include E-boxes, muscle CAT element, fork head binding motifs (Pierrou et al., 1994), and TFBS for STAT, NFkB, vMyb, and Pax proteins.
Fig. 9. Comparative analysis of TFBS in proximal promoters of TFII-I genes.
Arrows indicate transcription start sites. Relative positions are shown for matches to the following consensus sequences: AHRE - aryl hydrocarbon receptor complex response element; CCAAT-box – RRCCAATYR consensus; GC-box – RGGCGKR consensus; cJun/cFos - cJun/cFos binding consensus (RSTGACTNMNW); CRE - cyclic AMP response element (TGASSTCA); E-box –palindromic sequence CANNTG; Freac – binding site for forkhead related activators (Pierrou 1994); HFH - fork head binding site (TRTTTRY); MCAT -muscle CAT element (CATNCYW); NFkB – binding site for activated NFkB complex; NRF - nuclear respiratory factor 1 binding site (YGCGCCAYGCGCR); Olf-1 – binding site for olfactory neuron-specific factor (Wang et al. 1993); ORF - osmotic response element (Ferraris and Garcia-Perez, 2001); p53RE - p53 response element (RRRCWWGYYY); TRE - TPA response element (TGASTCA); and vMyc - vMyb binding site (YAACGGH). TFBS on plus-strand are shown above each of the diagrams and TFBS on minus-strand are indicated below the diagrams.
In contrast to P1, mouse promoter P1A located upstream of exon 1a and human promoter P1B located upstream of exon 1b use canonic TATA-boxes and do not reside within CpG islands. Just as non-homologous exons 1a and 1b, these auxiliary promoters lack conservation in assortment of associated TFBS (Fig. 9). Consensus binding site for the activated NFkB complex, osmotic response element, and nuclear respiratory factor 1 binding site were found in proximal promoter P1A, whereas P1B contains TFBS for SMADs, two classical cyclic AMP response elements, and aryl hydrocarbon receptor complex response element.
Despite the absence of reliable evidences for transcriptional initiation within introns 8 and 10 in mouse Gtf2i gene, our search for core promoter elements upstream of mouse exons 8a and 10a has revealed the presence of the putative TATA-box, Initiator, and X Core Promoter Element 1 (XCPE1; Tokusumi et al., 2007) (data not shown).
We also performed comparative analysis for human and mouse GTF2IRD1 promoters (Fig. 9). They are both TATA-less and Initiator-less but contain the perfect CCAAT-box. Both mouse and human GTF2IRD1 promoters harbor multiple E-boxes, cJun/cFos binding consensus, and TFBS for STAT, NFkB, and Pax proteins. In addition, p53 response element and cyclic AMP response elements were found upstream mouse exons 1a and 1b, whereas human P1 promoter contains additional cyclic AMP response elements and tandem of replicated TPA response element (Fig. 9).
4. Discussion
Our comprehensive analysis of GTF2I and GTF2IRD1 mRNA structures has revealed a complex pattern of alternative splicing and alternative promoters corresponding to the modern appreciation of the complexity of the mammalian transcriptome. We have found that alternative splicing of TFII-I genes affect all categories of the structural changes: changes in the protein domain and 3D architecture, introduction of stop codons, and changes in the 5′ or 3′UTRs.
Among this diverse range of mature mRNAs, some isoforms are abundant in many tissues, whereas others are rare variants that are difficult to detect. In particular, mouse Gtf2i mRNAs starting from exons 8a and 10a are present in cDNA libraries prepared from mouse blastocysts and from 17 days pregnant adult female amnion, respectively. We show that these transcripts are absent in the limited array of tested tissues, but our data do not rule out a possibility that such isoforms are expressed only in particular cells or within a narrow time window during development and, therefore, serve for highly specific functions.
The absence of mouse-human conservation of some minor isoforms casts doubt upon their functionality. Also, the frameshifting isoforms encoding severely truncated GTF2I and GTF2IRD1 proteins are likely to represent transcripts where exons are erroneously skipped, and such mRNAs are candidates for elimination by nonsense-mediated decay. However, large-scale computational study demonstrated that as much as one-third of reliable mRNA isoforms (supported by multiple ESTs) may be subject to nonsense-mediated decay and suggest that RUST (regulated unproductive splicing and translation) is one of the major regulatory mechanisms for establishing the right level of gene expression (Lewis at al., 2003). In addition, many minor splice variants are hypothesized to function as dominant negative isoforms that regulate the pathways in which the main functional form is involved (Arinobu et al., 1999; Stojic et al., 2007).
Although the biological relevance of non-conserved and frameshifting isoforms is not clear, the functional importance of major GTF2I and GTF2IRD1 splice variants is obvious. In TFII-I, the segment between I-repeats 1 and 2 contains nuclear localization signal, two tyrosine phosphorylation sites, D-box, and polyproline II domain (Roy, 2007), and alternative splicing within this region should ultimately affect the functional properties of TFII-I. The opposing functions of such internally deleted proteins in growth factor-induced gene expression has been already shown (Hakre et al. 2006). Under basal conditions, TFII-I isoform Δ is predominantly located in the cytoplasm, and, upon growth factor signaling, it interacts with Erk1/2, becomes tyrosine phosphorylated, and translocates to the nucleus to activate c-Fos. In contrast, isoform β is constitutively recruited to the c-Fos promoter interacting with HDAC3, resulting in repression of the c-Fos transcriptional activity, but upon growth factor signaling, TFII-Iβ moves out of the nucleus. It was also demonstrated that some of GTF2IRD1 isoforms differentially regulate muscle fibre-type-specific promoter (Tay et al., 2003). However, the functional significance of the majority of TFII-I isoforms remains to be elucidated.
Human exons 1a, 1b, and 1c have no homology in mouse Gtf2i, and analysis of their sequences reveals their relation to AluSg/x, AluJo, and AluY repeats, respectively. Exonization of Alu elements is not unusual in the human genome (Nekrutenko and Li, 2001; reviewed in Sorek,, 2007). Screening the human transcriptome for the presence of Alu-derived sequences transcribed as a part of human mRNAs has allowed us to identify 291 Alu RNAs embedded in 5′ UTRs of 244 different transcripts, and 2142 Alu RNAs embedded in 3′UTRs of 1548 different mRNAs (www.unige.ch/sciences/biologie/bicel/Strub/research/Alu.html). Therefore, our findings of novel Alu-derived exons of GTF2I serve as yet another example of Alu repeat exonization.
To trace the evolutionary history of this event, we have found sequences similar to human exons 1a, 1b, and 1c within intron 1 of GTF2I in other primates. In Pan troglodytes all these sequences have acquired splice sites, whereas in Macaque mulatta only sequence corresponding to exon 1b and belonging to the oldest AluJ subfamily of Alu repeats (spread in the genome 80–100 mya) has features of a functional exon. Consistently, the RT-PCR data show that exons 1a and 1b are used frequently in all human tissues, whereas exon 1c does not show appreciate expression: it belongs to the youngest AluY subfamily of Alu repeats (spread in the genome 25 mya), and its function-gaining appears not to be finished yet.
In mice, B1 mobile elements are similar to human Alu repeats, and comparison of Alu in human and B1 in mice shows a significant correlation between their distributions with respect to orthologs (Polak and Domany, 2006). However, our analysis has revealed that only a part (27–34%) of rodent-specific exon 1a and 1b of Gtf2i corresponds to B1F and ID_B1 sequences.
Alu elements contain many binding sites for transcription factors and their incorporation upstream of coding sequences of human genes may play a role in developmental regulation. This hypothesis has been already supported by bioinformatics analysis (Polak and Domany, 2006). In particular, it has been shown that Alu-mediated expansion of consensus retinoic acid response elements DR2 contributes to the evolution of gene regulation by RARs and other nuclear receptors in primates and humans (Laperriere et al., 2007).
Because we can detect an activity of TFII-I auxiliary promoters P1A and P1B in all tissues examined, they may potentially serve as means for producing specific isoforms that are responsive to distinct upstream regulatory signals. Consistent with this idea, the promoter region we have found upstream of exons 1a and 1b possesses consensus sequences for a variety of signals (Fig. 9). The identification of these features would provide a direct link to regulatory pathways. It is interesting that among pairs of TFBS conserved in P1 of TFII-I orthologous genes we have also found consensus sequence for olfactory neuron-specific factor Olf-1, which mediates specific gene expression in the olfactory epithelium (Wang et al., 1993). This can explain an observation of higher expression of Gtf2i in the olfactory bulbs (data not shown). In contrast to GTF2I, GTF2IRD1 promoters demonstrates both constitutive and inducible features (Fig. 9).
With the repertoire of alternative transcripts described here, we can now address their biological relevance to further elucidate the functional heterogeneity of TFII-I proteins. In addition, because TFII-I proteins are the prime candidates responsible for craniofacial and cognitive abnormalities in the Williams-Beuren syndrome, our findings of non-coding variations in these genes lead us to examine the effect of different haplotypes on their promoter functions.
5. Conclusions
This study has revealed that the alternative splicing in human and mouse TFII-I genes is more complicated than considered before and reports identification of new Gtf2i and Gtf2ird1 isoforms. In particular, mouse Gtf2i mRNAs, human GTF2IRD1 mRNAs, and mouse Gtf2ird1 mRNAs have diverse sets of 5′UTRs and they are transcribed from alternative promoters that can be regulated differently.
Acknowledgments
This work was supported by the NIH grants R01 DE017205 and K02 DE18412, and KSEF-1122-RDE-009 and to DB.
Abbreviations
- cDNA
DNA complementary to RNA
- EST
expressed sequence tag
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- ORF
open reading frame
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arinobu Y, Atamas SP, Otsuka T, Niiro H, Yamaoka K, Mitsuyasu H, Niho Y, Hamasaki N, White B, Izuhara K. Antagonistic effects of an alternative splice variant of human IL-4, IL-4delta2, on IL-4 activities in human monocytes and B cells. Cell Immunol. 1999;191:161–167. doi: 10.1006/cimm.1998.1431. [DOI] [PubMed] [Google Scholar]
- Bayarsaihan D, Ruddle FH. Isolation and characterization of BEN, a member of the TFII-I family of DNA-binding proteins containing distinct helix-loop-helix domains. Proc Natl Acad Sci USA. 2000;97:7342–7347. doi: 10.1073/pnas.97.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayarsaihan D, Dunai J, Greally JM, Kawasaki K, Sumiyama K, Enkhmandakh B, Shimizu N, Ruddle FH. Genomic organization of the genes Gtf2ird1, Gtf2i, and Ncf1 at the mouse chromosome 5 region syntenic to the human chromosome 7q11.23 Williams syndrome critical region. Genomics. 2002;79:137–143. doi: 10.1006/geno.2001.6674. [DOI] [PubMed] [Google Scholar]
- Bayarsaihan D, Bitchevaia N, Enkhmandakh B, Tussie-Luna MI, Leckman JF, Roy A, Ruddle F. Expression of BEN, a member of TFII-I family of transcription factors, during mouse pre- and postimplantation development. Gene Expr Patterns. 2003;3:579–589. doi: 10.1016/s1567-133x(03)00118-2. [DOI] [PubMed] [Google Scholar]
- Cheriyath V, Roy AL. Alternatively spliced isoforms of TFII-I: complex formation, nuclear translocation, and differential gene regulation. J Biol Chem. 2000;275:26300–26308. doi: 10.1074/jbc.M002980200. [DOI] [PubMed] [Google Scholar]
- Danoff SK, Taylor HE, Blackshaw S, Desiderio S. TFII-I, a candidate gene for Williams syndrome cognitive profile: parallels between regional expression in mouse brain and human phenotype. Neuroscience. 2000;123:931–938. doi: 10.1016/j.neuroscience.2003.08.038. [DOI] [PubMed] [Google Scholar]
- Doi-Katayama Y, Hayashi F, Inoue M, Yabuki T, Aoki M, Seki E, Matsuda T, Kigawa T, Yoshida M, Shirouzu M, et al. Solution structure of the general transcription factor 2I domain in mouse TFII-I protein. Protein Sci. 2007;16:1788–1792. doi: 10.1110/ps.072792007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dralyuk I, Brudno M, Gelfand MS, Zorn M, Dubchak I. ASDB: database of alternatively spliced genes. Nucleic Acids Res. 2000;28:296–297. doi: 10.1093/nar/28.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkhmandakh B, Bitchevaia N, Ruddle F, Bayarsaihan D. The early embryonic expression of TFII-I during mouse preimplantation development. Gene Expr Patterns. 2004;4:25–28. doi: 10.1016/s1567-133x(03)00155-8. [DOI] [PubMed] [Google Scholar]
- Ferraris JD, Garcia-Perez A. Osmotically Responsive Genes: The Mammalian Osmotic Response Element (ORE) American Zoologist. 2001;41:734–742. [Google Scholar]
- Hakre S, Tussie-Luna MI, Ashworth T, Novina CD, Settleman J, Sharp PA, Roy AL. Opposing functions of TFII-I spliced isoforms in growth factor-induced gene expression. Mol Cell. 2006;24:301–308. doi: 10.1016/j.molcel.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Ji H, Zhou Q, Wen F, Xia H, Lu X, Li Y. AsMamDB: an alternative splice database of mammals. Nucleic Acids Res. 2001;29:260–263. doi: 10.1093/nar/29.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Laperriere D, Wang TT, White JH, Mader S. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007;8:23. doi: 10.1186/1471-2164-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Nekrutenko A, Li WH. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- Ohazama A, Sharpe PT. TFII-I gene family during tooth development: candidate genes for tooth anomalies in Williams syndrome. Dev Dyn. 2007;236:2884–2888. doi: 10.1002/dvdy.21311. [DOI] [PubMed] [Google Scholar]
- Palmer SJ, Tay ES, Santucci N, Cuc Bach TT, Hook J, Lemckert FA, Jamieson RV, Gunnning PW, Hardeman EC. Expression of Gtf2ird1, the Williams syndrome-associated gene, during mouse development. Gene Expr Patterns. 2007;7:396–404. doi: 10.1016/j.modgep.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Pierrou S, Hellqvist M, Samuelsson L, Enerbäck S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994;13:5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:13. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisil H, Herrmann A, Bortfeldt RH, Reich JG. EASED: extended alternatively spliced EST database. Nucleic Acids Res. 2004;32:D70–D74. doi: 10.1093/nar/gkh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AL. Signal-induced functions of the transcription factor TFII-I. Biochim Biophys Acta. 2007;1769:613–621. doi: 10.1016/j.bbaexp.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R. The birth of new exons: mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Riethoven JJM, Le Texier V, Gopalakrishnan C, Kumanduri V, Tang Y, Barbosa-Morais NL, Thanaraj TA. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojic J, Stoihr H, Weber BH. Three novel ABCC5 splice variants in human retina and their role as regulators of ABCC5 gene expression. BMC Mol Biol. 2007;8:42–50. doi: 10.1186/1471-2199-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, Carette M, Wilmot C, Donnai D, Read AP, Metcalfe K. A transcription factor involved in skeletal muscle gene expression is deleted in patients with Williams syndrome. Europ J Hum Genet. 1999;7:737–747. doi: 10.1038/sj.ejhg.5200396. [DOI] [PubMed] [Google Scholar]
- Tay ESE, Guven KL, Subramaniam N, Polly P, Issa LL, Gunning PW, Hardeman EC. Regulation of alternative splicing of Gtf2ird1 and its impact on slow muscle promoter activity. Biochem J. 2003;374:359–367. doi: 10.1042/BJ20030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokusumi Y, Ma Y, Song X, Jacobson RH, Takada S. The new core promoter element XCPE1 (X Core Promoter Element 1) directs activator-, mediator-, and TATA-binding protein-dependent but TFIID-independent RNA polymerase II transcription from TATA-less promoters. Mol Cell Biol. 2007;27:1844–1858. doi: 10.1128/MCB.01363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vullhorst D, Buonanno A. Multiple GTF2I-like repeats of general transcription factor 3 exhibit DNA binding properties. Evidence for a common origin as a sequence-specific DNA interaction module. J Biol Chem. 2005;280:31722–31731. doi: 10.1074/jbc.M500593200. [DOI] [PubMed] [Google Scholar]
- Wang MM, Tsai RY, Schrader KA, Reed RR. Genes encoding components of the olfactory signal transduction cascade contain a DNA binding site that may direct neuronal expression. Mol Cell Biol. 1993;13:5805–5813. doi: 10.1128/mcb.13.9.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hubbell E, Hu JS, Mei G, Cline M, Lu G, Clark T, Siani-Rose MA, Ares M, Kulp DC, Haussler D. Gene structure-based splice variant deconvolution using a microarray platform. Bioinformatics. 2003;19(suppl 1):1315–1322. doi: 10.1093/bioinformatics/btg1044. [DOI] [PubMed] [Google Scholar]