Abstract
A cyclic imine conjugated to a fluorescent dansyl group was synthesized and used for covalent labeling of proteins. The covalent attachment to proteins was confirmed by gel electrophoresis and mass analysis.
Covalent labeling reactions of proteins are required for preparation of protein conjugates that are useful in research and medicine. For example, antibody-drug conjugates are more efficient therapeutics than antibody alone or drug alone.1 When conjugated to protein, small molecule drugs can be selectively delivered to targets, reducing side effects and extending the half-life of the drugs.1,2 Fluorescently labeled proteins can be traced by detection of the fluorescence.3 Currently available labeling methods include labeling at lysine with succinimidyl ester derivatives, labeling at cysteine with maleimide derivatives, and labeling of an unnatural amino acid residue incorporated in protein sequence during its expression.1c,d,4,5 Small peptide tag-based strategies for labeling of proteins have also been developed.6 Most of the methods have advantages and disadvantages.
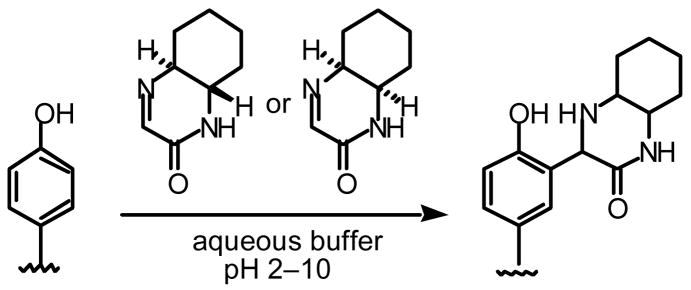
In order to provide labeling reactions complementary to the existing reactions, we have recently developed cyclic imines that covalently react with phenols, including tyrosine residues, in aqueous solutions7 (Scheme 1). The reactions of the imines with phenols proceeded over a wide pH range without the need of additional catalysts.7 The imines were relatively stable and were resistant to hydrolysis in aqueous solutions but efficiently reacted with phenols.7
Scheme 1.
Tyrosine residues are observed less often on the surface of folded proteins than lysines or carboxylic acid-containing residues,8 which are commonly used as labeling sites. Thus, accessible tyrosine is an attractive covalent labeling site.7,8,9 For proteins and peptides that do not have an accessible tyrosine, a tyrosine residue can be added by usual site-directed mutagenesis or by usual peptide synthesis. Although imines prepared from formaldehyde and aniline derivatives in situ have been used for labeling of proteins at tyrosine,8 the optimal pH range for this reaction is 5.5–6.5 and the reaction does not proceed above pH 8.8a In addition, formaldehyde cannot be used in many cases; for example, use of formaldehyde is not suitable for labeling of proteins at tyrosine inside living cells or in the presence of living cells. Reactions at tyrosine using reagents other than imines typically require additional catalysts or multistep conversions after the first reaction step on the phenol moiety of tyrosine.9a–d Use of the imine shown in Scheme 1 may bypass the limitations and disadvantages of the existing labeling methods.
To demonstrate the use of the cyclic imines shown in Scheme 1 for covalent attachment of non-native molecules to proteins and peptides, here, we have synthesized a conjugate of the imine with a fluorescent dansyl group and examined the reactions of the dansyl-conjugated imine with proteins and peptides.
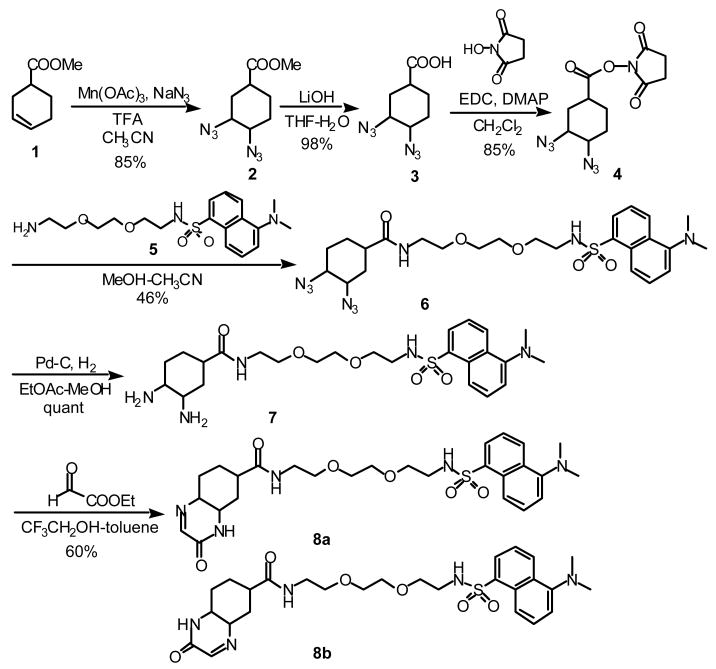
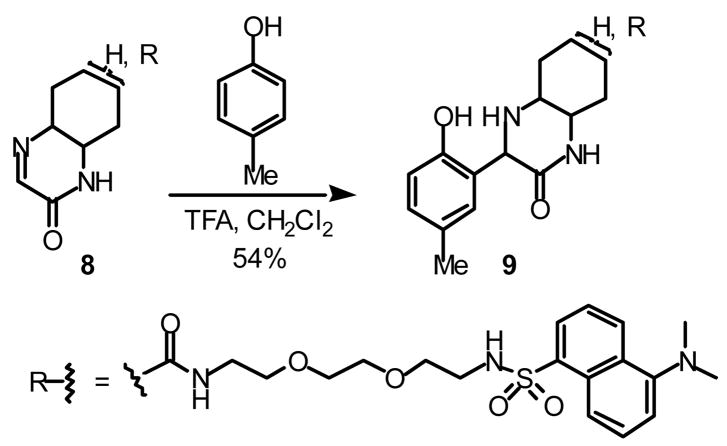
The imine derivative linked to a fluorescent dansyl group was synthesized as shown in Scheme 2.10 Diazidation of 1 afforded a mixture of stereoisomers of 2; the major isomer was expected to have trans-diazide groups due to the procedures used.11 Hydrolysis of the ester with LiOH afforded 3, which was converted to succinimidyl ester 4. Reaction of 4 with amine 5, prepared from the corresponding diamine and dansyl chloride, afforded compound 6. Diamine 7 was obtained by reduction of the azide groups by hydrogenation with Pd-C under H2 in EtOAc-MeOH (3:1). The solvents for this reduction were critical; reduction of 6 with Pd-C under H2 but in MeOH mainly afforded monoamine, which might have been generated via elimination of HN3. Diamine 7 was then transformed to imine 8 as a mixture of regioisomers (8a + 8b). To confirm the imine functionality of 8, reaction of 8 with p-cresol was performed in the presence of TFA (Scheme 3).12 Formation of addition product 9 confirmed the presence of the imine functionality in compound 8.
Scheme 2.
Scheme 3.
Imine 8 was used for modification of proteins and peptides. Reaction of peptide was performed using GlyTyr or AlaTyrAla (90 mM) with 8 (20 mM) in 10% DMSO/100 mM sodium phosphate, pH 6.0 for 48 h at 37 °C. Imine 8 was completely consumed as judged by TLC analysis of the reaction mixture. Formation of the addition products were confirmed by mass analysis.13
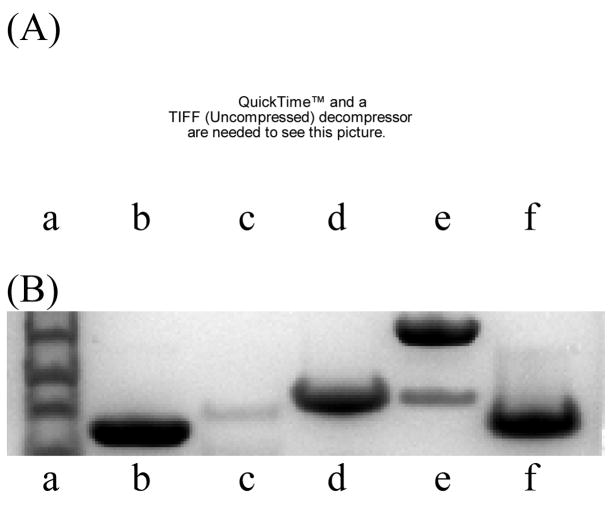
To test reaction of 8 with proteins, lysozyme from chicken egg white, α-chymotrypsinogen A type II from bovine pancreas, myoglobin from equine skeletal muscle, carbonic anhydrase isozyme II from bovine erythrocytes, and cytochrome C from horse heart were used. Reactions of proteins (475 μM) were performed in the presence of 8 (5 mM) in 5% DMSO/100 mM sodium phosphate, pH 6.0 for 4 days at 37 °C and were analyzed by gel electrophoresis (Figure 1). Gel bands corresponding to modified proteins were fluorescent under a UV lamp (312 nm). Unmodified proteins were not fluorescent on a gel under the UV lamp (data not shown). Gel analysis showed that lysozyme was most easily modified among proteins tested (lane b). Mass analysis of the reaction mixture of lysozyme with 8 showed the formation of the mono-addition product.14 We previously demonstrated that the imine derivative shown in Scheme 1, prepared from trans-diaminocyclohexane, selectively reacted at tyrosine and did not react with other natural amino acid residues except cysteine thiol.7 Thus, reaction of 8 with lysozyme probably occurred at tyrosine. Cytochrome C was not labeled with 8 under the conditions used (lane f). Myoglobin and carbonic anhydrase were slightly modified (lanes d and e).15 Chymotrypsinogen was hydrolyzed into small fragments by autolysis and by catalysis of generated chymotrypsin under the conditions used (lane c). Due to this hydrolysis, we were unable to evaluate labeling of chymotrypsinogen with 8. These results indicated that 8 did not inhibit the serine protease activity under the conditions used; this implies that the nucleophilic serine in the active site of chymotrypsin did not efficiently react with imine 8.
Figure 1.
Analysis of proteins modified with 8 after separation by gel electrophoresis. Lane a, protein standard marker; b, lysozyme; c, chymotrypsinogen (see text); d, myoglobin; e, carbonic anhydrase; f, cytochrome C. (A) Analysis of fluorescence of dansyl group attached to proteins under UV 312 nm. (B) Analysis of proteins by Coomassie staining.
Mass analysis of the reaction of 8 with lysozyme at 48 h suggested that the reaction of 8 was slower than that of the imine synthesized from trans-cyclohexanediamine shown in Scheme 1. We previously observed that reaction of the trans-isomer of the imine shown in Scheme 1 was faster than the corresponding cis-isomer.7 Relative stereochemistries of the two nitrogen functionalities and the amide carbonyl group on the cyclohexane ring of 8 may affect the reactivity of the imine moiety. In order to increase the reaction rate, stereoselective synthesis of 8 may be required. Alternatively, introduction of moieties that provide noncovalent interactions with the target protein may be used to enhance the reaction rate of the imine with the target.9e
In these reactions, excess of compound 8 was used compared to protein (5 mM versus 475 μM). After protein was separated by gel filtration or C18 column, compound 8 was recovered by extraction with organic solvent (such as CH2Cl2). This recoverable feature of the imine should be beneficial when a toxic drug or an expensive molecule is covalently attached to protein.
In summary, we have synthesized a fluorescent conjugate of cyclic imines that we have recently developed for covalent reactions with phenols, including accessible tyrosine. We have demonstrated that the imine derivative can be used to fluorescently label proteins, although the reaction of the imine was relatively slow. The imine derivative was stable in aqueous solutions and recoverable after reactions. Reactions of the imine derivative proceeded without the need of additional catalysts. This system does not require formaldehyde or multistep conversions after the first reaction step on tyrosine and thus is an improvement over previously reported labeling systems. Further studies and improvements on the imine derivatives described here are under investigation.
Acknowledgments
This study was supported by NIH R21 GM078447. We thank Prof. Peter K. Vogt for helpful technical discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.(a) Schrama D, Reisfeld RA, Becker JC. Nature Rev Drug Discovery. 2006;5:147. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]; (b) Wu AM, Senter PD. Nat Biotechnol. 2005;23:1137. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]; (c) Hamann PR, Hinman LM, Hollander I, Beyer CF, Lindh D, Holcomb R, Hallett W, Tsou HR, Upeslacis J, Shochat D, Mountain A, Flowers DA, Bernstein I. Bioconjugate Chem. 2002;13:47. doi: 10.1021/bc010021y. [DOI] [PubMed] [Google Scholar]; (d) Lillo AM, Sun C, Gao C, Ditzel H, Parrish J, Gauss GM, Moss J, Felding-Habermann B, Wirsching P, Boger DL, Janda KD. Chem Biol. 2004;11:897. doi: 10.1016/j.chembiol.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 2.Vincent S, Thomas A, Brasher B, Benson JD. Nat Biotechnol. 2003;21:936. doi: 10.1038/nbt844. [DOI] [PubMed] [Google Scholar]
- 3.Keppler A, Pick H, Arivoli C, Vogel H, Johnsson K. Proc Natl Acad Sci USA. 2004;101:9955. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Prescher JA, Bertozzi CR. Nat Chem Biol. 2005;1:13. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]; (b) Antos JM, Francis MB. Curr Opin Chem Biol. 2006;10:253. doi: 10.1016/j.cbpa.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 5.(a) Link AJ, Tirrell DA. J Am Chem Soc. 2003;125:11164. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]; (b) Agard NJ, Prescher JA, Bertozzi CR. J Am Chem Soc. 2004;126:15046. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]; (c) Saxon E, Bertozzi CR. Science. 2000;287 doi: 10.1126/science.287.5460.2007. 2007. [DOI] [PubMed] [Google Scholar]; (d) Dirksen A, Hackeng TM, Dawson PE. Angew Chem, Int Ed. 2006;45:7581. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]; (e) Crich D, Banerjee A. J Am Chem Soc. 2007;129:10064. doi: 10.1021/ja072804l. [DOI] [PubMed] [Google Scholar]; (f) Tanaka K, Masuyama T, Hasegawa K, Tahara T, Mizuma H, Wada Y, Watanabe Y, Fukase K. Angew Chem, Int Ed. 2008;47:102. doi: 10.1002/anie.200702989. [DOI] [PubMed] [Google Scholar]
- 6.(a) Tanaka F, Fuller R, Asawapornmongkol L, Warsinke A, Gobuty S, Barbas CF., III Bioconjugate Chem. 2007;18:1318. doi: 10.1021/bc070080x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tanaka F, Fuller R. Bioorg Med Chem Lett. 2006;16:4059. doi: 10.1016/j.bmcl.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Minakawa M, Guo HM, Tanaka F. J Org Chem. 2008;73:8669. doi: 10.1021/jo8017389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Joshi NS, Whitaker LR, Francis MB. J Am Chem Soc. 2004;126:15942. doi: 10.1021/ja0439017. [DOI] [PubMed] [Google Scholar]; (b) McFarland JM, Joshi NS, Francis MB. J Am Chem Soc. 2008;130:7639. doi: 10.1021/ja710927q. [DOI] [PubMed] [Google Scholar]
- 9.(a) Hooker JM, Kovacs EW, Francis MB. J Am Chem Soc. 2004;126:3718. doi: 10.1021/ja031790q. [DOI] [PubMed] [Google Scholar]; (b) Tilley SD, Francis MB. J Am Chem Soc. 2006;128:1080. doi: 10.1021/ja057106k. [DOI] [PubMed] [Google Scholar]; (c) Hooker JM, Kovacs EW, Francis MB. J Am Chem Soc. 2004;126:3718. doi: 10.1021/ja031790q. [DOI] [PubMed] [Google Scholar]; (d) Hass JA, Frederick MA, Fox BG. Protein Expr Purif. 2000;20:274. doi: 10.1006/prep.2000.1293. [DOI] [PubMed] [Google Scholar]; (e) Koshi Y, Nakata E, Miyagawa M, Tsukiji S, Ogawa T, Hamachi I. J Am Chem Soc. 2008;130:245. doi: 10.1021/ja075684q. [DOI] [PubMed] [Google Scholar]
- 10.Compound 2. To a mixture of Mn(OAc)3.2H2O (16.1 g, 60 mmol) and NaN3 (6.50 g, 100 mmol) in CH3CN (180 mL), compound 1 (2.75 mL, 20 mmol) was added followed by trifluoroacetic acid (TFA, 20 mL) at −20 °C under Ar.11 After 5 min, the mixture was added to saturated NaHSO3 and extracted with CH2C12. Usual workup and purification by silica gel flash column chromatography afforded 27 (3.83 g, 85%). Compound 3. To a solution of 2 (3.38 g, 17 mmol) in THF (20 mL) was added a solution of LiOH (1.43 g, 34 mmol) in water (20 mL) at room temperature and the mixture was stirred for 15 h. After THF was evaporated in vacuo, 3 N HCl was added to adjust the pH to 3 and the mixture was extracted with CH2C12. Usual workup and purification by silica gel flash column chromatography (EtOAc/hexane) afforded 3 (3.27 g, 98%). 1H NMR (400 MHz, CDCl3):δ 11.1 (brs, 1H), 3.94–3.49 (m, 1H), 3.42–3.21 (m, 1H), 2.82–2.68 (m, 1H), 2.46–2.27 (m, 1H), 2.16–1.85 (m, 2H), 1.83–1.40 (m, 3H). Compound 4. A mixture of 3 (2.64 g, 12.6 mmol), N-hydroxysuccinimide (2.17 g, 18.9 mmol), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) (3.62 g, 18.9 mmol), and DMAP (5.0 mg, 0.04 mmol) in CH2Cl2 (40 mL) was stirred at room temperature for 24 h. The reaction mixture was added to H2O and extracted with CH2Cl2. Usual workup and purification by silica gel flash column chromatography (EtOAc/hexane) afforded 4 (3.28 g, 85%). 1H NMR (400 MHz, CDCl3):δ 3.73–3.69 (m, 1H), 3.44–3.28 (m, 1H), 3.12–3.08 (m, 1H), 2.85 (s, 4H), 2.41–2.35 (m, 1H), 2.24–2.14 (m, 1H), 2.04–1.92 (m, 1H), 1.83–1.66 (m, 3H). Compound 5. To a solution of 1,2-bis(2-aminoethoxy)ethane (2.5 mL, 17.0 mmol) in CH2Cl2 (20 mL), a solution of dansyl chloride (467 mg, 1.73 mmol) in CH2Cl2 (20 mL) was added dropwise at 0 °C. After stirring at room temperature for 1 h, the mixture was acidified with 1N HCl and washed with CH2Cl2. The aqueous layer was basified (pH 9) with 3N NaOH and extracted with CH2Cl2. Organic layers were combined, washed with brine, dried over Na2SO4, filtered, and concentrated in vacuo to afford 5 (630 mg, 95%). 1H NMR (400 MHz, CDCl3):δ 8.52 (d, J = 8.8 Hz, 1H), 8.34 (d, J = 8.8 Hz, 1H), 8.23 (dd, J = 7.2 Hz, 1.2 Hz, 1H), 7.56–7.48 (m, 2H), 7.17 (d, J = 7.6 Hz, 1H), 3.50–3.46 (m, 4H), 3.41–3.39 (m, 4H), 3.11–3.09 (m, 4H), 2.87 (s, 6H). Compound 6. A mixture of 5 (612 mg, 1.60 mmol) and 4 (493 mg, 1.60 mmol) in CH3CN (0.5 mL)-MeOH (1.5 mL) was stirred for 15 h at room temperature. Solvents were evaporated in vacuo and the residue was purified by silica gel flash column chromatography (CH2Cl2/EtOAc = 4:1) to afford 6 (322 mg, 47%). 1H NMR (400 MHz, CDCl3):δ 8.55 (d, J = 8.8 Hz, 1H), 8.30 (d, J = 8.8 Hz, 1H), 8.24 (d, J = 7.2 Hz, 1H), 7.57–7.50 (m, 2H), 7.19 (d, J = 7.6 Hz, 1H), 6.58 (brs, 1H), 5.90 (t, J = 5.6 Hz, 1H), 3.85–3.80 (m, 1H), 3.59–3.57 (m, 2H), 3.54–3.45 (m, 8H), 3.37–3.32 (m, 1H), 3.12–3.08 (m, 2H), 2.89 (s, 6H), 2.56–2.54 (m, 1H), 2.22–2.16 (m, 1H), 1.90–1.81 (m, 2H), 1.76–1.71 (m, 1H), 1.61–1.53 (m, 2H). 13C NMR (100 MHz, CDCl3):δ 174.7, 152.1, 134.0, 130.8, 129.8, 129.6, 129.3, 128.6, 123.1, 118.2, 115.2, 115.2, 63.7, 63.6, 60.1, 45.3, 43.0, 39.1, 38.5, 29.8, 26.9, 25.3, 24.6. HRMS: calcd for C25H36N9O5S (MH+) 574.2555, found 574.2561. Compound 7. A mixture of 6 (322 mg, 0.56 mmol) and 10% Pd/C (32 mg) in MeOH (5 mL)-EtOAc (15 mL) was stirred under H2 for 24 h at room temperature. The reaction mixture was filtered through celite and concentrated in vacuo to afford 7 (290 mg, quant). 1H NMR (400 MHz, CD3OD):δ 8.52 (d, J = 8.8 Hz, 1H), 8.31 (d, J = 8.8 Hz, 1H), 8.17 (dd, J = 7.2 Hz, 1.2 Hz, 1H), 7.56–7.51 (m, 2H), 7.23 (d, J = 7.6 Hz, 1H), 3.78–3.74 (m, 1H), 3.46–3.36 (m, 2H), 3.36–3.33 (m, 2H), 3.30–3.25 (m, 7H), 3.02–2.98 (m, 2H), 2.83 (s, 6H), 2.55–2.52 (m, 1H), 2.09–2.03 (m, 1H), 1.83–1.76 (m, 2H), 1.64–1.51 (m, 3H). 13C NMR (100 MHz, CDCl3):δ 174.6, 151.8, 135.1, 130.3, 130.2, 129.8, 129.6, 129.1, 128.18, 128.15, 128.12, 128.0, 123.1, 118.9, 115.16, 115.14, 70.23, 70.20, 70.2, 70.1, 70.0, 69.4, 56.1, 53.4, 45.3, 42.8, 39.5, 39.2, 26.1. HRMS: calcd for C25H40N5O5S (MH+) 522.2745, found 522.2745. Compound 8 (8a + 8b). To a solution of 7 (406 mg, 0.78 mmol) in CF3CH2OH (15 mL), a solution of ethyl glyoxylate polymer form (45–50% in toluene, 218 μL, 0.78 mmol) in CF3CH2OH (15 mL) was added dropwise over 24 h at room temperature. The mixture was further stirred for 6 days. After solvents were removed in vacuo, the residue was purified by silica gel flash column chromatography (CH2Cl2/EtOH = 20:1) to afford 8 (259 mg, 60%). 1H NMR (400 MHz, CDCl3):δ 8.56–8.53 (m, 1H), 8.31–8.29 (m, 1H), 8.25–8.22 (m, 1H), 7.72–7.68 (m, 1H), 7.56–7.50 (m, 2H), 7.19–7.17 (m, 1H), 6.80–6.39 (m, 1H), 6.30 (m, 1H), 6.28–6.25 (m, 1H), 3.72–3.40 (m, 12H), 3.13–3.07 (m, 2H), 2.89 (s, 6H), 2.75–1.30 (m, 7H). 13C NMR (100 MHz, CDCl3):δ 174.5, 173.8, 157.8, 157.5, 156.49, 156.47, 151.97, 151.93, 134.8, 134.4, 130.5, 130.4, 129.8, 129.58, 129.55, 129.4, 129.3, 128.16, 128.13, 123.1, 118.8, 118.7, 115.2, 115.1, 70.4, 70.3, 70.2, 70.1, 69.3, 69.2, 62.8, 59.2, 54.0, 50.4, 45.3, 42.9, 42.8, 39.4, 39.2, 39.1, 38.1, 33.0, 32.2, 28.0, 27.5, 27.4, 25.5. HRMS: calcd for C27H38N5O6S (MH+) 560.2537, found 560.2543.
- 11.Snider BB, Lin H. Synth Commun. 1998;28:1913. [Google Scholar]
- 12.Compound 9. A mixture of compound 8 (136 mg, 0.24 mmol) and 4-methylphenol (39.4 mg, 0.36 mmol) in CF3COOH (1 mL)-CH2Cl2 (1 mL) was stirred for 24 h at room temperature. After solvents were removed in vacuo, residue was dissolved in CH2Cl2 and was washed with saturated NaHCO3. The aqueous layer was extracted with CH2Cl2. Usual workup and purification by silica gel flash column chromatography (CH2Cl2/MeOH = 30:1) afforded 9 (88 mg, 54%). 1H NMR (400 MHz, CDCl3):δ 8.54 (d, J = 8.8 Hz, 1H), 8.30–8.27 (m, 1H), 8.24–8.20 (m, 1H), 7.55–7.49 (m, 2H), 7.17 (d, J = 7.2 Hz, 1H), 7.06 (s, 1H), 6.99–6.93 (m, 1H), 6.78–6.75 (m, 1H + 1H × 1/2), 6.65 (m, 1H × 1/2), 6.45 (s, 1H), 6.36 (brs, 1H × 1/2), 6.20 (brs, 1H × 1/2), 4.96 (s, 1H × 1/2), 4.91 (s, 1H × 1/2), 3.62–3.42 (m, 12H), 3.09–3.04 (m, 3H), 2.88 (s, 6H), 2.70–1.30 (m, 8H), 2.26 (s, 3H × 1/2), 2.23 (s, 3H × 1/2). HRMS: calcd for C34H46N5O7S (MH+) 668.3112, found 668.3115.
- 13.GlyTyr labeled with 8. HRMS: calcd for C38H51N7O10S (MH+) 798.3491, found 798.3495. AlaTyrAla labeled with 8. HRMS: calcd for C42H58N8O11S (MH+) 883.4018, found 883.4015.
- 14.Lysozyme labeled with 8. ESI-MS: 14865 (mono-addition product); unmodified 14306.
- 15.Myoglobin labeled with 8. ESI-MS: 17512 (mono-addition product); unmodified 16953. Modification of myoglobin, which lacks surface-accessible tyrosine,8,9b may have occurred when the protein was denatured during the 4-day reaction time. We previously showed that myoglobin was not modified with the cyclic imine derivative shown in Scheme 1 in aqueous buffer, pH 7.0 at 37°C for 48 h.7