Abstract
Prolyl 4-hydroxylase (P4H) is a non-heme iron dioxygenase that catalyzes the post-translational hydroxylation of (2S)-proline (Pro) residues in protocollagen strands. The resulting (2S,4R)-4-hydroxyproline (Hyp) residues are essential for the folding, secretion, and stability of the collagen triple helix. P4H uses α-ketoglutarate and O2 as co-substrates, and forms succinate and CO2 as well as Hyp. Described herein is the first assay for P4H that continuously and directly detects turnover of the proline-containing substrate. This assay is based on (2S,4S)-4-fluoroproline (flp), a proline analogue that is transformed into (2S)-4-ketoproline (Kep) and inorganic fluoride by P4H. The fluoride ion, and thus turnover by P4H, is detected by a fluoride ion-selective electrode. Using this assay, steady-state kinetic parameters for the human P4H-catalyzed turnover of a flp-containing peptide were determined and found to be comparable to those obtained with a discontinuous HPLC-based assay. In addition, this assay can be used to characterize P4H variants, as demonstrated by a comparison of catalysis by D414A P4H and the wild-type enzyme. Finally, the use of the assay to identify small-molecule inhibitors of P4H was verified by an analysis of catalysis in the presence of 2,4-pyridine dicarboxylate, an analogue of α-ketoglutarate. Thus, the assay described herein could facilitate biochemical analyses of this essential enzyme.
Collagens are the major structural proteins of the extracellular matrix. Collagens undergo a number of post-translational modifications during biosynthesis. One major modification is the hydroxylation of certain prolyl residues. The repeating amino-acid sequence of collagen, wherein every third residue is a glycine (Gly): Xaa–Yaa–Gly, is rich in (2S)-proline (Pro). Indeed, Pro is the most common amino acid found in the Xaa position [1]. In the Yaa position, the most common amino is (2S, 4R)-4-hydroxyproline (Hyp). Hyp is formed by a post-translational modification of Pro catalyzed by prolyl 4-hydroxylase (P4H; EC 1.14.11.2).
Hyp is necessary for the stable formation of the triple-helical structure of collagen. Collagen with decreased levels of Hyp is defective in folding and secretion under physiological conditions [2–4]. P4H activity is required for the viability of the nematode Caenorhabditis elegans [5,6] and the mouse Mus musculus [7]. In vitro, P4H has been studied by availing enzyme via heterologous expression in insect cells, yeast, and (only recently [8,9]) bacteria.
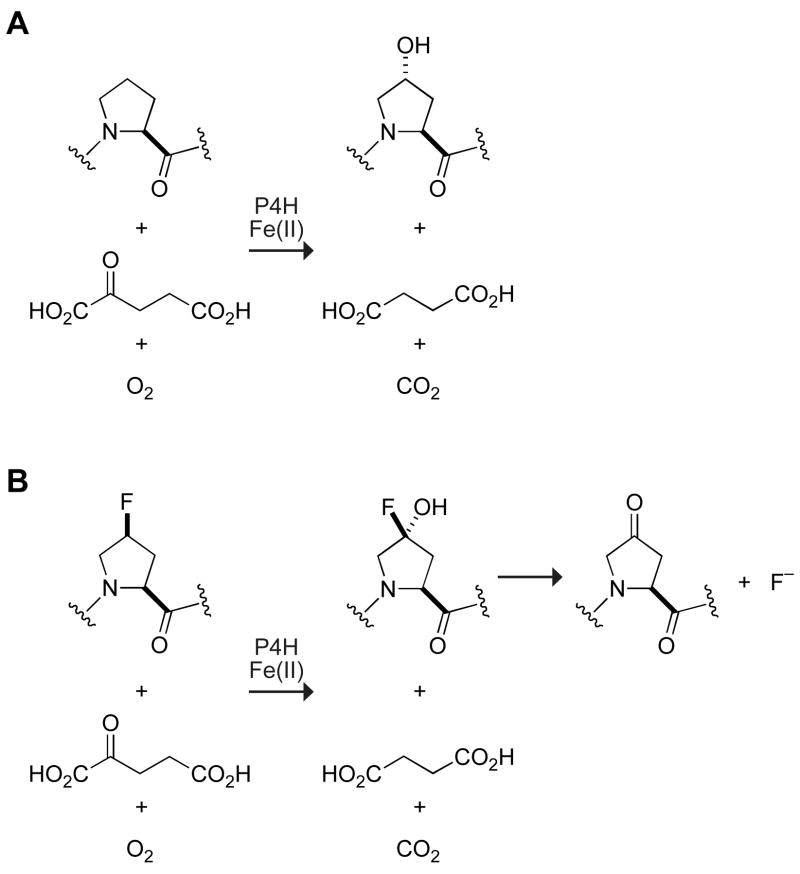
P4H is a member of the α-ketoglutarate-dependent, non-heme iron(II) dioxygenase family of enzymes [10–12]. These enzymes require iron(II), α-ketoglutarate, and O2 for catalysis (Figure 1). To accomplish difficult oxidizing reactions, these enzymes employ a highly reactive iron(IV)-oxo species [13,14]. In P4H, this species abstracts the 4-proR hydrogen atom from a Pro substrate [15], and then transfers a hydroxyl radical to form the Hyp product. The key iron(IV)-oxo species is formed by the oxidative decarboxylation of α-ketoglutarate, which also results in the formation of succinate and CO2 [16]. During the reaction, one atom of molecular oxygen is incorporated into Hyp, and the other into succinate [17]. The turnover of α-ketoglutarate can occur without the formation of Hyp. This uncoupling of α-ketoglutarate decarboxylation from substrate hydroxylation leads to inactivated P4H. Ascorbate can reactivate the enzyme [18–20].
Fig. 1.
Reaction catalyzed by prolyl 4-hydroxylase when (A) a Pro-containing peptide or (B) a flp-containing peptide is the substrate.
Many non-heme iron dioxygenases catalyze subtle changes to large substrates, making their enzymatic activity difficult to assay. P4H is not an exception. Historically, its activity has been monitored by assays that employ radioactivity. Hydroxylation of the collagen substrate can be monitored directly by the detection of [14C]Hyp formed in collagen containing [14C]Pro after acid hydrolysis of the product [21]. A more rapid assay involves radiolabeling collagen with [3,4-3H]proline and detecting [3H]H2O after hydroxylation [22]. These assays are, however, not only discontinuous, but also require the time-consuming production of radiolabeled collagen.
P4H activity has been measured indirectly by monitoring the turnover of α-ketoglutarate. Assays have been developed to detect residual α-ketoglutarate substrate or the incipient succinate or CO2 product. Perhaps the most often used P4H assay quantifies the [14C]CO2 product of [1-14C]α-ketoglutarate decarboxylation [16,23]. Trapping of the [14C]CO2 can, however, be inefficient. The turnover of [5-14C] α-ketoglutarate produces [1-14C]succinate, which must be separated from unreacted α-ketoglutarate prior to analysis. Methods of separation by column chromatography [24] and chemical precipitation of α-ketoglutarate [25] have been reported previously.
Assays for P4H activity have also been developed that do not require radioactivity. The consumption of the oxygen co-substrate has been monitored using an O2-sensing electrode [26]. An assay developed more recently quantifies unreacted α-ketoglutarate by a post-reaction derivitization that forms a fluorescent product [27]. Another assay couples the formation of succinate to that of NAD+ formation via succinyl-coenzyme A synthetase, pyruvate kinase, and lactate dehydrogenase [28]. Although these assays are of general use for all α-ketoglutarate-dependent dioxygenases, they have the marked disadvantage of being indirect—they do not report on the hydroxylation of a substrate. Direct, but discontinuous, assays that monitor proline hydroxylation without using radioactivity have been developed with peptide substrates. In these assays, the product Hyp-containing peptide is separated from the substrate by thin-layer chromatography [29] or by HPLC [8,30].
Herein, we present the first assay for P4H that is both direct and continuous. The assay utilizes an alternative substrate, (2S,4S)-4-fluoroproline (flp), that upon turnover by P4H forms (2S)-4-ketoproline (Kep) and releases inorganic fluoride [30]. The rate of substrate turnover is monitored continuously by using a fluoride ion-selective electrode, which has been employed previously in assays for alkaline phosphatase and chondroitin AC lyase [31,32]. Our assay for P4H yields kinetic parameters comparable to those from a discontinuous HPLC-based assay using the same peptide substrate. The assay is also able to reveal the effect of altering an active-site residue and that of a small-molecule inhibitor. Accordingly, the assay is likely to have a substantial impact on biochemical analyses of this essential enzyme.
Materials and methods
Materials
Boc-FlpOH and Boc-flpOH were from OmegaChem (Lévis, Québec). All other reagents were of reagent grade or better and were used without further purification.
Production and purification of P4H
P4H was produced and purified by using procedures reported previously [8]. cDNA encoding the D414A variant of P4H was created by oligonucleotide-mediated site-directed mutagenesis using the pBK1.PDI1.P4H7 plasmid described previously. P4H D414A was produced and purified by the method used for wild-type P4H.
Synthesis of PEGylated peptides
PEG-Gly–Tyr–Yaa–GlyOEt peptides, with Yaa = Flp or Pro, were synthesized as described previously [30]. The peptide with Yaa = flp was also synthesized as described, except for the use of Wang resin (Novabiochem, Gibbstown, NJ).
Fluoride ion-detection assay of enzymatic activity
Assays were performed at room temperature, which was (23 ± 2) °C, in glass vials with stirring. Assay solutions were 0.30 mL of 50 mM Tris–HCl buffer, pH 7.8, containing bovine serum albumin (1.0 mg/mL), catalase (0.10 mg/mL), dithiothreitol (0.10 mM), ascorbate (2.0 mM), FeSO4 (0.050 mM), P4H (90 nM), α-ketoglutarate (0.50 mM), and sodium fluoride (0.040 mM). The reaction mixture was allowed to equilibrate, and the tetrapeptide substrate was added from a stock solution in ethanol to initiate the reaction. The change in the concentration of fluoride ion was monitored with an Orion fluoride ion-selective electrode (Thermo Scientific, Waltham, MA) interfaced with a computer via an electrode amplifier (Vernier, Beaverton, OR). The data were fitted by linear-regression analysis to obtain initial rates. Fluoride ion concentrations were calculated by comparison to a standard curve with sodium fluoride in 50 mM Tris–HCl buffer, pH 7.8, which was found to generate the same signal as sodium fluoride in the assay solution described above.
HPLC-based assay of enzymatic activity
An HPLC-based assay described previously [8] was used to confirm product formation by P4H. Assays were performed for 5 min at room temperature. Assay solutions were 100 μL of 50 mM Tris–HCl buffer, pH 7.8, containing bovine serum albumin (1.0 mg/mL), catalase (0.10 mg/mL), dithiothreitol (0.10 mM), ascorbate (2.0 mM), FeSO4 (0.050 mM), P4H (90 nM), and α-ketoglutarate (0.50 μM).
Results
The P4H-catalyzed turnover of a flp-containing peptide produces a Kep-containing peptide and a fluoride ion (Figure 1). Hydroxylation of flp by P4H produces an α-fluorohydrin, which is known to fragment in an exothermic reaction [33]. The fluoride ion released, and thus P4H activity, is monitored by utilization of a fluoride ion-selective electrode. Sodium fluoride is added to the reaction mixture to put the initial fluoride concentration within the linear range of the fluoride ion-selective electrode. As measured by the HPLC-based assay, the enzymatic activity of P4H was found to be unaffected by the presence of salts, including 10 mM sodium fluoride (data not shown).
To assess the substrate specificity of the assay, we compared peptide substrates containing Pro, flp, or (2S,4R)-4-fluoroproline (Flp). Pro, the natural substrate is turned over, but does not produce fluoride and therefore is not detected by the fluoride ion-selective electrode (data not shown). Flp is a fluoride-containing proline analog that is not a substrate of P4H [30]. Assays in which 0.50 mM PEG-Gly–Tyr–Flp–GlyOEt is added produce no change in signal, as in assays that lack peptide (Figure 2). In contrast, addition of 0.50 mM of the analogous flp-containing peptide to the assay causes a large increase in signal.
Fig. 2.
Fluoride ion-detection assay for P4H (90 nM). Assays were performed in the presence or absence of PEG-Gly–Tyr–Yaa–GlyOEt, where Yaa = flp (1.0 mM) or Flp (0.50 mM). The dashed line shows the fit of the Yaa = flp data by linear-regression analysis. Assay conditions are as described in the Materials and methods section.
To confirm that the defluorination of flp is due to P4H, the reaction rates were determined at varying P4H concentrations. Duplicate reactions were performed under standard reaction conditions containing 1.0 mM PEG-Gly–Tyr–flp–GlyOEt. With no P4H added to the reaction, there was no increase in fluoride ion concentration. In reactions including 10, 20, 45, or 90 nM P4H, the initial velocity of the reactions correlated linearly with the P4H concentration (Figure 3).
Fig. 3.
Dependence of the rate of fluoride-ion release from PEG-Gly–Tyr–flp–GlyOEt (1.0 mM) on the concentration of P4H. Assay conditions are as described in the Materials and methods section. Reactions were performed in duplicate. Data were fitted by linear-regression analysis. v = ∂[F−]/∂t.
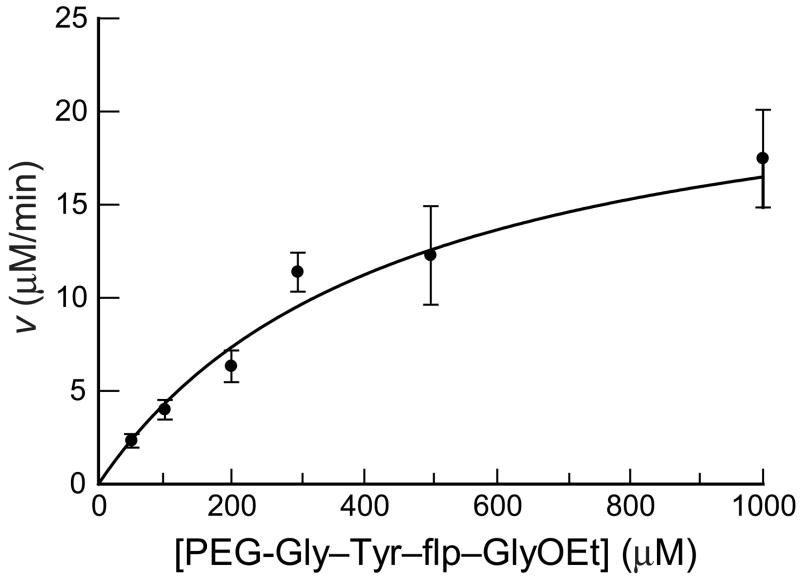
The fluoride ion-selective electrode assay was compared to the previously developed HPLC-based assay by determining the steady-state kinetic parameters for the P4H-catalyzed turnover of flp. The rate of fluoride ion formation was determined at varying concentrations of the flp-containing peptide (Figure 4, Table 1). Assays were performed in triplicate under standard conditions, and data were fitted to the Michaelis–Menten equation to obtain kinetic parameters. The kcat/KM value was determined to be (9.8 ± 2.8) × 103 M−1s−1 (Table 1, Figure 4). This value is similar to that from the previously developed HPLC-based assay.
Fig. 4.
Catalysis of fluoride ion-release from PEG-Gly–Tyr–flp–GlyOEt by P4H (90 nM). Assay conditions are as described in the Materials and methods section. Individual points are the average (±SE) of three reactions. Data were fitted to the Michaelis–Menten equation: v = ∂[F−]/∂t = kcat[P4H][peptide]/(KM + [peptide]).
Table 1.
Comparison of the turnover of PEG-Gly–Tyr–flp–GlyOEt by P4H (90 nM) determined by using the fluoride ion-detection assay or the HPLC-based assaya
| Assay | Vmax (μM·min−1) | kcat (min−1) | KM (mM) | kcat/KM (103 M−1s−1) |
|---|---|---|---|---|
| fluoride | 24 ± 3 | 270 ± 33 | 0.5 ± 0.1 | 9.8 ± 2.8 |
| HPLC | 32 ± 5 | 360 ± 53 | 1.2 ± 0.3 | 5.1 ± 1.4 |
Reaction components and conditions were as described in the Materials and methods section. Values represent the mean (±SE) of three replicates.
A variant of P4H and an inhibitor of P4H were used to probe the utility of this assay. Under standard assay conditions, the D414A variant of P4H showed no detectable catalytic activity at a concentration of 0.90 μM, which is 10-fold greater than normal (Figure 5A). The effect of the competitive inhibitor 2,4-pyridine dicarboxylate on the rate of reaction was investigated as well. In assays with 0.20 mM flp-containing peptide under standard conditions, 10, 33, and 100 μM concentrations of the inhibitor were shown to inhibit P4H increasingly (Figure 5B). Considering that the reported KM value of α-ketoglutarate is 22 μM [34] and its concentration in the assays herein is 0.50 mM, the inhibition determined by the decrease in the rate of fluoride ion production by P4H in the presence of the inhibitor is consistent with the reported Ki value of 2μM [35,36].
Fig. 5.
Catalysis of fluoride ion-release from PEG-Gly–Tyr–flp–GlyOEt (0.20 mM) by a P4H variant or in the presence of a small-molecule inhibitor. (A) Catalysis by wild-type P4H (90 nM) and its D414A variant (0.90 μM). (B) Inhibition of catalysis by 2,4-pyridine dicarboxylate. Assay conditions are as described in the Materials and methods section. v = ∂[F−]/∂t.
Discussion
The assay for P4H activity described herein monitors directly and continuously the turnover of a peptide substrate (Figures 2–4; Table 1). The steady-state kinetic parameters of a peptide substrate containing flp are comparable to those for the analogous peptide containing Pro, the natural substrate, making flp a suitable substrate [30]. The peptide substrate is readily accessible, as flp is available from commercial vendors or accessible by a facile synthetic route [37]. The assay has the advantages of being direct and continuous, in addition to using inexpensive instrumentation, avoiding the use of radioactivity, and not requiring additional enzymes.
Assays of enzymatic activity have identified a number of residues critical for P4H activity. Like most other non-heme iron(II) dioxygenases, the active site of P4H has a two histidine/one carboxylate motif that binds iron. Asp414 has been identified as the source of the carboxylate [38,39]. Recently, a subclass of non-heme iron(II) dioxygenases that perform halogenation reactions, instead of hydroxylations, has been identified [40]. These halogenases lack an active-site carboxylate, containing an alanine residue instead of the canonical aspartate or glutamate. The D414A P4H variant studied herein emulates the active site of a halogenase, though no halogenated product has been detected in a solution of high halide ion concentration (unpublished data). The D414A variant also lacks hydroxylase activity (Figure 5A), confirming the requirement of the aspartate for hydroxylase activity and validating the competence of the assay for reporting on P4H variants.
P4H plays a major role in the biosynthesis of collagen. Excessive collagen formation causes a number of fibrotic diseases, and P4H has been put forth as a target for beneficial intervention with chemotherapeutic agents [41–43]. Analogs of α-ketoglutarate are known to inhibit P4H competitively with respect to α-ketoglutarate [35,36,6]. The assay is able to reveal inhibition by one such analogue, 2,4-pyridine dicarboxylate (Figure 5B). We anticipate that the assay could be adapted to a high-throughput format with the use of appropriate fluorescent or colorimetric fluoride-sensing reagents. Such reagents are under development [44].
In addition to stabilizing collagen, the hydroxylation of proline residues also plays a critical role in the sensing of molecular oxygen [45,43]. For example, the formation of Hyp in the transcription factor hypoxia inducible factor (HIF) is catalyzed by the proline hydroxylase domain proteins (PHDs), which are distinct from the collagen P4H. Incorporating flp into a HIF-derived peptide substrate for PHD enzymes could enable the assay described herein to be used in the study of those enzymes as well.
Conclusions
In summary, we have developed a new assay for the activity of P4H that utilizes flp as the substrate. Upon turnover, the release of fluoride ion is monitored by a fluoride ion-selective electrode. This assay for P4H is the first that is both direct and continuous. We have demonstrated the utility of the assay in characterizing P4H variants and identifying inhibitors. We anticipate the modification of this assay into a high-throughput format suitable for the discovery of novel inhibitors of this important enzyme.
Acknowledgments
This work was supported by grant AR044276 (NIH). K.L.G. was supported by Chemistry–Biology Interface training grant T32 BM008505 (NIH).
Footnotes
Abbreviations used: Flp, (2S,4R)-4-fluoroproline; flp, (2S,4S)-4-fluoroproline; Gly, glycine; HPLC, high-performance liquid chromatography; Hyp, (2S,4R)-4-hydroxyproline; Kep, (2S)-4-ketoproline; PEG, poly(ethylene glycol); P4H, prolyl 4-hydroxylase; Pro, (2S)-proline.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prockop DJ, Kivirikko KI. Collagens: Molecular biology, diseases, and potentials for therapy. Annu Rev Biochem. 1995;64:403–434. doi: 10.1146/annurev.bi.64.070195.002155. [DOI] [PubMed] [Google Scholar]
- 2.Berg RA, Prockop DJ. The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple helix of collagen. Biochem Biophys Res Comm. 1973;52:115–120. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- 3.Chopra RK, Ananthanarayanan VS. Conformational implications of enzymatic proline hydroxylation in collagen. Proc Natl Acad Sci USA. 1982;79:7180–7184. doi: 10.1073/pnas.79.23.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulleid NJ, Wilson R, Lees JF. Type-III procollagen assembly in semi-intact cells: Chain association, nucleation and triple-helix folding do not require formation of inter-chain disulphide bonds but triple-helix nucleation does require hydroxylation. Biochem J. 1996;317:195–202. doi: 10.1042/bj3170195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter AD, Page AP. Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans. Mol Cell Biol. 2000;20:4084–4093. doi: 10.1128/mcb.20.11.4084-4093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman L, Higgin JJ, Moulder G, Barstead R, Raines RT, Kimble J. Prolyl 4-hydroxylase is required for viability and morphogenesis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2000;97:4736–4741. doi: 10.1073/pnas.97.9.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holster T, Pakkanen O, Soininen R, Sormunen R, Nokelainen M, Kivirikko KI, Myllyharju J. Loss of assembly of the main basement membrane collagen, Type IV, but not fibril-forming collagens and embryonic death in collagen prolyl 4-hydroxylase I null mice. J Biol Chem. 2007;282:2512–2519. doi: 10.1074/jbc.M606608200. [DOI] [PubMed] [Google Scholar]
- 8.Kersteen EA, Higgin JJ, Raines RT. Production of human prolyl 4-hydroxylase in Escherichia coli. Protein Exp Purif. 2004;38:279–291. doi: 10.1016/j.pep.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Neubauer A, Neubauer P, Myllyharju J. High-level production of human collagen prolyl 4-hydroxylase in Escherichia coli. Matrix Biol. 2005;24:59–68. doi: 10.1016/j.matbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Guzman NA, editor. Prolyl Hydroxylase, Protein Disulfide Isomerase, and Other Structurally Related Proteins. Marcel Dekker; New York: 1998. [Google Scholar]
- 11.Fox BG. Catalysis by non-heme iron. In: Sinnott M, editor. Comprehensive Biological Catalysis. Academic Press; New York: 1998. pp. 261–348. [Google Scholar]
- 12.Kivirikko KI, Pihlajaniemi T. Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv Enzymol Relat Areas Mol Biol. 1998;72:325–98. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
- 13.Costas M, Mehn MP, Jensen MP, Que L., Jr Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chem Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 14.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Jr, Krebs C. Direct spectroscopic detection of a C–H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc Natl Acad Sci USA. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita Y, Gottlieb A, Peterkofsky B, Udenfriend S, Witkop B. The preparation of cis-and trans-4-H3-L-prolines and their use in studying the mechanism of enzymatic hydroxylation in chick embryos. J Am Chem Soc. 1964;86:4709–4716. [Google Scholar]
- 16.Rhoads RE, Udenfriend S. Decarboxylation of α-ketoglutarate coupled to collagen proline hydroxylase. Proc Natl Acad Sci USA. 1968;60:1473–8. doi: 10.1073/pnas.60.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardinale GJ, Rhoads RE, Udenfriend S. Simultaneous incorporation of 18O into succinate and hydroxyproline catalyzed by collagen proline hydroxylase. Biochem Biophys Res Commun. 1971;43:537–543. doi: 10.1016/0006-291x(71)90647-4. [DOI] [PubMed] [Google Scholar]
- 18.Myllylä R, Kuutti-Savolainen ER, Kivirikko KI. The role of ascorbate in the prolyl hydroxylase reaction. Biochem Biophys Res Commun. 1978;83:441–448. doi: 10.1016/0006-291x(78)91010-0. [DOI] [PubMed] [Google Scholar]
- 19.de Jong L, Albracht SP, Kemp A. Prolyl 4-hydroxylase activity in relation to the oxidation state of enzyme-bound iron. The role of ascorbate in peptidyl proline hydroxylation. Biochim Biophys Acta. 1982;704:326–332. doi: 10.1016/0167-4838(82)90162-5. [DOI] [PubMed] [Google Scholar]
- 20.Myllylä R, Majamaa K, Gunzler V, Hanauske-Abel HM, Kivirikko KI. Ascorbate is consumed stoichiometrically in the uncoupled reactions catalyzed by prolyl 4-hydroxylase and lysyl hydroxylase. J Biol Chem. 1984;259:5403–5405. [PubMed] [Google Scholar]
- 21.Peterkofsky B, Udenfriend S. Enzymatic hydroxylation of proline in microsomal polypeptide leading to formation of collagen. Proc Natl Acad Sci USA. 1965;53:335–42. doi: 10.1073/pnas.53.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutton JJ, Jr, Trappel AL, Udenfriend S. A rapid assay for collagen proline hydroxylase. Anal Biochem. 1966;16:384–394. [Google Scholar]
- 23.Kivirikko KI, Myllylä R. Posttranslational enzymes in the biosynthesis of collagen: Intracellular enzymes. Methods Enzymol. 1982;82:245–304. doi: 10.1016/0076-6879(82)82067-3. [DOI] [PubMed] [Google Scholar]
- 24.Cunliffe CJ, Franklin TJ, Gaskell RM. Assay of prolyl 4-hydroxylase by the chromatographic determination of [14C]succinic acid on ion-exchange minicolumns. Biochem J. 1986;240:617–619. doi: 10.1042/bj2400617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaule G, Gunzler V. Assay for 2-oxoglutarate decarboxylating enzymes based on the determination of [1-14C]succinate: Application to prolyl 4-hydroxylase. Anal Biochem. 1990;184:291–297. doi: 10.1016/0003-2697(90)90683-z. [DOI] [PubMed] [Google Scholar]
- 26.Nietfeld JJ, Kemp A. Properties of prolyl 4-hydroxylase containing firmly-bound iron. Biochim Biophys Acta. 1980;613:349–358. doi: 10.1016/0005-2744(80)90089-3. [DOI] [PubMed] [Google Scholar]
- 27.McNeill LA, Bethge L, Hewitson KS, Schofield CJ. A fluorescence-based assay for 2-oxoglutarate-dependent oxygenases. Anal Biochem. 2005;336:125–131. doi: 10.1016/j.ab.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 28.Luo LS, Pappalardi MB, Tummino PJ, Copeland RA, Fraser ME, Grzyska PK, Hausinger RP. An assay for Fe(II)/2-oxoglutarate-dependent dioxygenases by enzyme-coupled detection of succinate formation. Anal Biochem. 2006;353:69–74. doi: 10.1016/j.ab.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 29.Tandon M, Wu M, Begley TP. Substrate specificity of human prolyl-4-hydroxylase. Bioorg Med Chem Lett. 1998;8:1139–1144. doi: 10.1016/s0960-894x(98)00183-8. [DOI] [PubMed] [Google Scholar]
- 30.Gorres KL, Edupuganti R, Krow GR, Raines RT. Conformational preferences of substrates for human prolyl 4-hydroxylase. Biochemistry. 2008;47:9447–9455. doi: 10.1021/bi8009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venetz WP, Mangan C, Siddiqi IW. Kinetic determination of alkaline-phosphatase activity based on hydrolytic cleavage of the P–F bond in monofluorophosphate and fluoride ion-selective electrode. Anal Biochem. 1990;191:127–132. doi: 10.1016/0003-2697(90)90398-s. [DOI] [PubMed] [Google Scholar]
- 32.Rye CS, Withers SG. Development of an assay and determination of kinetic parameters for chondroitin AC lyase using defined synthetic substrates. Anal Biochem. 2002;308:77–82. doi: 10.1016/s0003-2697(02)00223-3. [DOI] [PubMed] [Google Scholar]
- 33.Seppelt K. Trifluoromethanol, CF3OH. Angew Chem Int Ed. 1977;16:322–323. [Google Scholar]
- 34.Myllylä R, Tuderman L, Kivirikko KI. Mechanism of the prolyl hydroxylase reaction. 2. Kinetic analysis of the reaction sequence. Eur J Biochem. 1977;80:349–357. doi: 10.1111/j.1432-1033.1977.tb11889.x. [DOI] [PubMed] [Google Scholar]
- 35.Majamaa K, Hanauske-Abel HM, Gunzler V, Kivirikko KI. The 2-oxoglutarate binding site of prolyl 4-hydroxylase. Identification of distinct subsites and evidence for 2-oxoglutarate decarboxylation in a ligand reaction at the enzyme-bound ferrous ion. Eur J Biochem. 1984;138:239–245. doi: 10.1111/j.1432-1033.1984.tb07907.x. [DOI] [PubMed] [Google Scholar]
- 36.Majamaa K, Gunzler V, Hanauske-Abel HM, Myllylä R, Kivirikko KI. Partial identity of the 2-oxoglutarate and ascorbate binding sites of prolyl 4-hydroxylase. J Biol Chem. 1986;261:7819–7823. [PubMed] [Google Scholar]
- 37.Chorghade MS, Mohapatra DK, Sahoo G, Gurjar MK, Mandlecha MV, Bhoite N, Moghe S, Raines RT. Practical syntheses of 4-fluoroprolines. J Fluor Chem. 2008;129:781–784. doi: 10.1016/j.jfluchem.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamberg A, Pihlajaniemi T, Kivirikko KI. Site-directed mutagenesis of the α subunit of human prolyl 4-hydroxylase. Identification of three histidine residues critical for catalytic activity. J Biol Chem. 1995;270:9926–9931. doi: 10.1074/jbc.270.17.9926. [DOI] [PubMed] [Google Scholar]
- 39.Myllyharju J, Kivirikko KI. Characterization of the iron- and 2-oxoglutarate-binding sites of human prolyl 4-hydroxylase. EMBO J. 1997;16:1173–1180. doi: 10.1093/emboj/16.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krebs C, Galonic Fujimori D, Walsh CT, Bollinger JM., Jr Non-heme Fe(IV)-oxo intermediates. Acc Chem Res. 2007;40:484–92. doi: 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franklin TJ. Therapeutic approaches to organ fibrosis. Int J Biochem Cell Biol. 1997;29:79–89. doi: 10.1016/s1357-2725(96)00121-5. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu I. Antifibrogenic therapies in chronic HCV infection. Curr Drug Targets Infect Disord. 2001;1:227–240. doi: 10.2174/1568005014606053. [DOI] [PubMed] [Google Scholar]
- 43.Myllyharju J. Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann Med. 2008;40:402–417. doi: 10.1080/07853890801986594. [DOI] [PubMed] [Google Scholar]
- 44.Gunnlaugsson T, Glynn M, Tocci GM, Kruger PE, Pfeffer FM. Anion recognition and sensing in organic and aqueous media using luminescent and colorimetric sensors. Coord Chem Rev. 2006;250:3094–3117. [Google Scholar]
- 45.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]