Abstract
Hedgehog signaling is often activated in tumors, yet it remains unclear how GLI2, a transcription factor activated by this pathway, acts as an oncogene. We show that GLI2 is a pleiotropic oncogene. Overexpression induces genomic instability and blocks differentiation, likely mediated in part by enhanced expression of the stem cell gene SOX2. GLI2 also induces TGFβ dependent transdifferentiation of foreskin and tongue, but not gingival fibroblasts into myofibroblasts, creating an environment permissive for invasion by keratinocytes, which are in various stages of differentiation having down regulated GLI2. Thus, up-regulated GLI2 expression is sufficient to induce a number of the acquired characteristics of tumor cells; however the stroma, in a tissue specific manner, determines whether certain GLI2 oncogenic traits are expressed.
Keywords: GLI2, organotypic culture, transdifferentiation, invasion
INTRODUCTION
Five-year survival for patients with oral SCC, at 40%, is among the worst of all sites in the body. While tobacco and alcohol are the major etiological agents, oral cancer commonly occurs in patients without a history of exposure to these agents (Schmidt et al., 2004). Thus, there is a need for improved understanding of the alterations in genes or pathways that lead to this disease in order to develop better therapeutic approaches and methods for early detection or chemoprevention.
Genome-wide DNA copy number profiling of oral SCC revealed that they characteristically amplify narrow regions of the genome, often < 3 Mb (Snijders et al., 2005). Although these amplicons are rare, (occurring in < 5% of tumors), they are highly informative for several reasons; (1) the narrow regions they span often contain only a small number of genes, (2) the genes highlighted by the amplicons may play a significant role in a large number of tumors, because their expression may be altered by mechanisms other than amplification (Albertson et al., 2003), and (3) up-regulation of the driver oncogenes in amplicons is expected to be under positive selection at the time the tumor was resected, since amplified DNA is unstable (Miele et al., 1989; Murnane & Sabatier, 2004; Roth & Andersson, 2004; Shimizu et al., 2005) and would otherwise disappear. Amplicons in oral SCC implicated alterations in integrin signaling, apoptosis, adhesion and migration, as well as deregulation of the hedgehog and notch pathways (Snijders et al., 2005).
Whereas genome-wide profiling of tumors highlights candidate disease genes, identifying the critical ones and how they promote tumor development remains difficult. To evaluate the role of GLI2, a gene mapping to one of the narrow amplicons in oral SCC, we co-cultured keratinocytes overexpressing GLI2 with fibroblasts in three dimensional organotypic cultures to allow communication between cell types and differentiation of the epithelium (Bell et al., 1981). GLI2 is a member of the GLI family of transcription factors, activated by hedgehog signaling. In the canonical pathway, a hedgehog ligand (e.g. sonic hedgehog, SHH) binds the transmembrane receptor Patched (PTCH1), which in turn relieves inhibition of a second transmembrane protein Smoothened (SMO) resulting in activation of the GLI transcription factors. Abnormal activation of the hedgehog pathway has been implicated in a variety of cancers, occurring either via the canonical pathway or by activation downstream of SMO (Lauth & Toftgard, 2007). In oral SCC, GLI2 is up-regulated both in tumors with amplification and also in a subset of oral SCC (~25%) without amplification. When overexpressed, GLI2 is biologically active in these tumors as evidenced by up-regulation of GLI1 and PTCH1, known targets of GLI2 (Snijders et al., 2005). Here, we report that overexpression of GLI2 promotes a number of the acquired characteristics of tumor cells that constitute the cancer phenotype.
RESULTS
Tumors with GLI2 amplification show basal-like cellular histology
Oral SCC with high level amplification of GLI2 (Figures 1A and B) display basal-like histology in routine hematoxylin and eosin (H&E) stained sections (Figures 1C and D), suggesting that GLI2 overexpression inhibits differentiation. In other aspects of histology, however, the tumors differed markedly. In addition, the genomic copy number profiles of the GLI2 amplifying tumors differed (Figure 1E), suggesting that amplification of GLI2 does not dictate the aberration spectrum in these tumors.
Figure 1. Tumors with GLI2 amplification display basal-like cellular histology, while also displaying tumor-specific histologic and genomic aberrations.
(A and B) Normalized genomic DNA copy number profiles for chromosome 2 from oral SCC showing high level amplification of the GLI2 locus. The tongue oral SCC 6929 was obtained from the UCSF Oral Cancer Tissue Bank, which collects and archives oral tissue samples after obtaining patient consent according to UCSF Institutional Review Board guidelines. The anterior floor of mouth oral SCC 1300C2 was provided by Drs. Gillian Hall and Richard Shaw, School of Cancer Studies, University of Liverpool, UK and was collected under approved procedures of that institution. Tumor DNA was extracted from formalin fixed paraffin embedded tissue sections following micro-dissection of tumor islands and subjected to BAC array CGH analysis according to our standard protocols (Snijders et al., 2005). The array CGH analysis of oral SCC 6929 was reported previously (Snijders et al., 2005).
(C and D) H&E stained sections of the GLI2 amplifying tumors. The tumor cells are small with hyperchromatic nuclei and scant cytoplasm. Keratinization is absent. In SCC 1300C2, mitotic figures are abundant (6.3 ± 2.5 mitotic figures per high power field) and comedo necrosis is present at the centers of the tumor islands, features which are absent in SCC 6929.
(E) Heatmap representation of copy number changes detected by array CGH in GLI2 amplifying tumors. Each column represents one tumor. Individual BAC clones are shown as rows and ordered according to their genome position (May 2004 freeze, UCSC Genome Browser). Copy number aberrations were assigned as described previously (Fridlyand et al., 2006). Losses are indicated in red, gains in green and amplifications as yellow dots.
Overexpression of GLI2 in keratinocytes in monolayer cultures does not provide a growth advantage
GLI2 acts both as a transcriptional activator and repressor. Therefore, to study the functional consequences of GLI2 up-regulation on cell growth and differentiation, we obtained tetracycline-inducible HaCaT cells expressing a constitutively active form of GLI2 (6xHis-GLI2ΔN), which lacks the N-terminal repressor domain (hereafter referred to as HaCaT GLI2) and control tetracycline responsive HaCaT cells lacking the GLI2ΔN expression construct (hereafter referred to as HaCaT Tet) from Dr. F. Aberger (Regl et al., 2004). The parental HaCaT cells have mutations in both TP53 alleles and cytogenetically abnormal, but stable karyotypes (Boukamp et al., 1988). Nevertheless, they retain the capability to differentiate in organotypic cultures and have been used extensively as a substitute for normal human keratinocytes (Schoop et al., 1999). Previous genome-wide expression profiling demonstrated that HaCaT cells and primary keratinocytes respond in a similar fashion to forced GLI expression (Regl et al., 2004). We confirmed that doxycycline induced high levels of expression of GLI2 and downstream targets, GLI1 and PTCH1, in HaCaT GLI2 cells in monolayer culture (Supplementary Figure 1A). We observed a reduction in proliferation in these cells (Supplementary Figure 1B), that was not due to an increased rate of apoptosis, as measured by the percentage Annexin V positive cells (data not shown). Similar results were obtained when we infected independent cultures of HaCaT cells with a 6xHis-GLI2ΔN retrovirus (Supplementary Figure 1C). We also found a dramatic reduction in the colony-forming efficiency of GLI2 expressing HaCaT GLI2 cells plated at low density (Supplementary Table 1). Allowing the HaCaT GLI2 cells to attach for 48 hours prior to inducing GLI2 expression did not significantly increase the number of colonies (Supplementary Table 1). Thus, overexpression of GLI2 in HaCaT cells in monolayer culture confers no growth advantage as measured by enhanced proliferation rate or capacity for autonomous growth. On the other hand, senescence and/or poor attachment of the GLI2 expressing cells to the substrate may be contributing to the reduced rate of increase in cell number (see below).
Overexpression of GLI2 induces genomic instability
Enhanced expression of oncogenes can lead to replication stress, up-regulation of the DNA damage response and genomic instability (Bartkova et al., 2005; Gorgoulis et al., 2005). To determine whether overexpression of GLI2 induces genomic instability, we asked whether GLI2 expression enhanced formation of methotrexate resistant colonies in HT1080, a cell line that reportedly does not give rise to methotrexate resistant colonies without prior exposure to a DNA damaging agent (Paulson et al., 1998). We generated HT1080 variants (see Supplementary Methods) expressing 6xHis-GLI2ΔN, enhanced green fluorescent protein (eGFP) and CCND1, an oncogene known to induce genomic instability in vitro (Nelsen et al., 2005; Zhou et al., 1996) and to be up-regulated prior to amplification in vivo in head and neck cancer (Izzo et al., 1998; Roh et al., 2000). Upon challenge with 25 nM methotrexate (2–3 × LD50), the GLI2 and CCND1 expressing HT1080 cells gave rise to equal numbers of drug resistant colonies and these numbers were significantly greater than the number recovered from either eGFP expressing or parental HT1080 cells (Supplementary Figures 2A and B), indicating that GLI2 overexpression like CCND1 overexpression induces genomic instability. We have shown previously that increased numbers or specific types of genomic alterations may be acquired by methotrexate resistant cells depending on genetic background (Snijders et al., 2003; Snijders et al., 2008). No specific types of chromosomal level instability were evident by array CGH in the methotrexate resistant cells overexpressing GLI2 (Supplementary Figure 2C). Similarly, we did not observe enhanced DNA breakage in GLI2 expressing cells as measured by the comet assay (data not shown). Thus, extensive DNA damage does not appear to occur in GLI2 overexpressing cells. At this time the mechanism of GLI2 induced genomic instability remains unknown.
Overexpression of GLI2 in keratinocytes in organotypic cultures recapitulates tumor histology
To assess the effects of GLI2 overexpession on differentiation, we cultured GLI2 expressing and control cells (HaCaT Tet and uninduced HaCaT GLI2 cells) with dermal fibroblasts in three dimensional organotypic cultures. In cultures with GLI2 expressing HaCaT GLI2 cells, we observed gross differences compared to controls. The epithelial layer of the GLI2 expressing tissue reconstructs adhered poorly to the collagen/fibroblast layer during routine tissue processing and there was a decrease in fibroblast surface area compared to controls (Supplementary Figure 3). These observations suggest that GLI2 expression reduced adhesive interactions between the epithelium and extracellular matrix and induced increased contraction of the collagen gel by the fibroblasts.
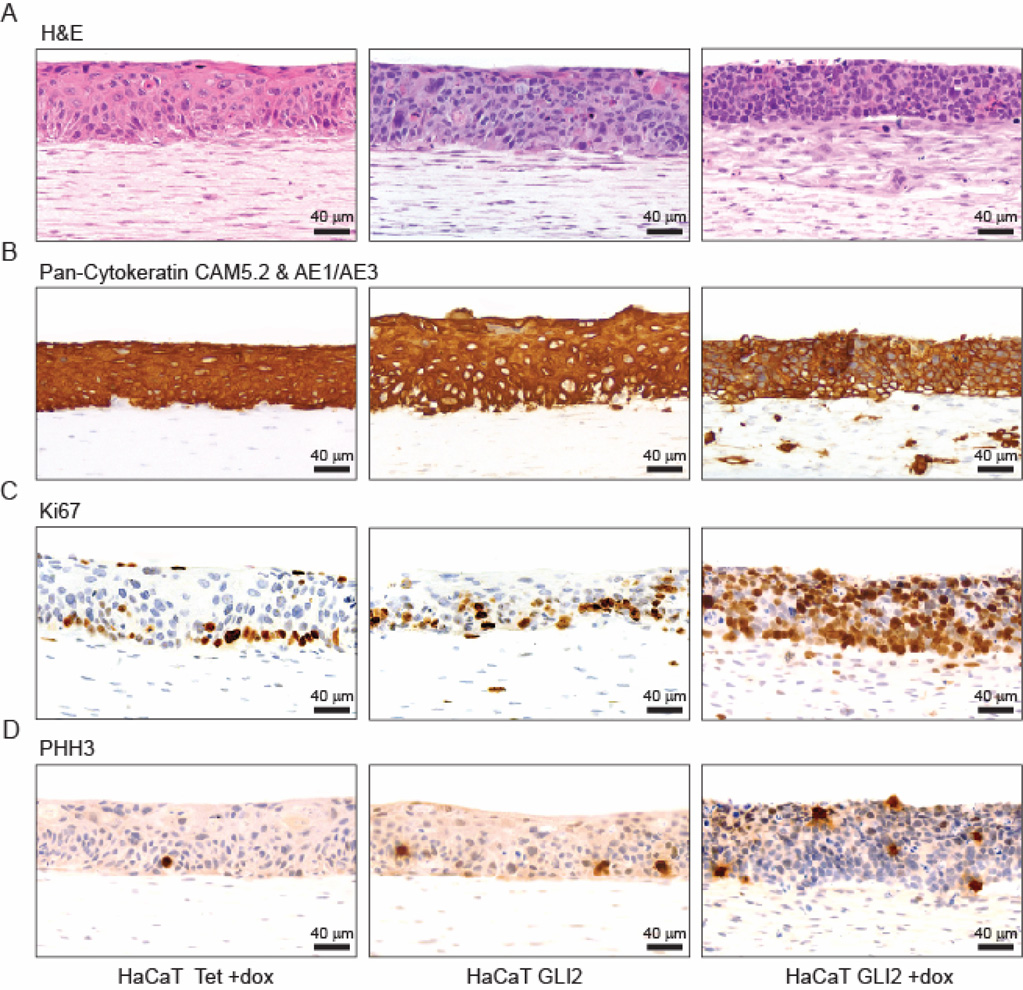
In routine H&E stained sections, GLI2 expressing HaCaT cells appeared basal-like (Figure 2A), recapitulating the histopathology of oral SCC with GLI2 amplification. We found both a decrease in keratinocyte nuclear size upon induction of GLI2 and an increase in nuclear density compared to controls (Supplementary Table 2). In addition, fibroblasts in the upper region of the dermal layer appeared less spindle shaped. Antibodies for pancytokeratin uniformly stained the epithelial layer in all reconstructs, and revealed the presence of individual or small groups of positively stained cells invading into the upper region of the collagen/fibroblast layer of GLI2 expressing HaCaT GLI2 reconstructs (Figure 2B).
Figure 2. Characteristics of control and HaCaT GLI2 organotypic cultures.
Sections of organotypic cultures of control HaCaT Tet (left), uninduced HaCaT GLI2 (middle) and induced HaCaT GLI2 cells (right) stained with H&E (A) or stained using horse radish peroxidase (HRP) and diaminobenzidine (DAB) detection and antibodies to epithelial cell specific pan-cytokeratin (Cam5.2 & AE1/AE3 (B), Ki67 (C) and PHH3 (D).
GLI2 overexpression does not accelerate proliferation in organotypic cultures
We investigated whether expression of GLI2 promoted proliferation by staining tissue reconstructs for proliferation (Ki67) and mitotic (phosphohistone H3, PHH3) markers. In GLI2 expressing HaCaT GLI2 reconstructs, staining was present throughout the entire epidermis, as well as in fibroblasts in the upper portion of the collagen/fibroblast layer, in stark contrast to the limited expression in only a few cells in the basal layer of the epidermis in controls (Figure 2C and Supplementary Figure 4). Although the proportion of Ki67 positive keratinocytes in induced HaCaT GLI2 organotypic cultufres was greater than in controls, no differences in the ratio of mitotic cells (PHH3 positive) to cycling Ki67 positive cells (Supplementary Table 3) were found for any pairwise comparison of the three cell types (Wilcoxon rank sum test). Thus, while cells overexpressing GLI2 are inappropriately in cycle in all layers of the epithelium, GLI2 overexpression does not promote more rapid cycling.
Overexpression of GLI2 opposes differentiation and impairs formation of a basement membrane zone
The basal-like phenotype of GLI2 expressing cells in the organotypic cultures and in the GLI2 amplifying tumors suggests that GLI2 opposes differentiation. Therefore, we stained sections from tissue reconstructs comprised of dermal fibroblasts and normal keratinocytes, control cells (HaCaT Tet and uninduced HaCaT GLI2 cells) and GLI2 expressing HaCaT GLI2 cells for differentiation markers cytokeratin 10/13 (CK10/13), involucrin and loricrin (Figure 3 and Supplementary Figure 4). Whereas staining for these markers was observed in the upper, more differentiated layers of the epidermis formed in reconstructs of normal dermal keratinocytes and controls, in GLI2 expressing reconstructs only sparse individual cells stained positively for CK10/13 or involucrin, but not for loricrin. We also observed that the invasive cells in the GLI2 expressing reconstructs stained positively for CK10/13 and in some cases for involucrin, but not loricrin (Figure 3).
Figure 3. GLI2 blocks epithelial differentiation and impairs formation of the basement membrane zone.
Sections of organotypic cultures of HaCaT Tet control (left), uninduced HaCaT GLI2 (middle) and induced HaCaT GLI2 cells (right) stained using HRP and DAB detection and antibodies to keratinocyte differentiation markers, cytokeratin 10/13 (A), involucrin (B), loricrin (C), and basement membrane components laminin 5 gamma-2 subunit (D), integrin β4 (E) and collagen type IV (F).
Expression of components of the basement membrane zone in GLI2 expressing HaCaT GLI2 cells was also abnormal (Figure 3D–F and Supplementary Figure 4). Integrin β4 (ITGB4), which participates in formation of hemi-desmosomes anchoring the epithelium to the stroma, was observed at the basal side of the basal keratinocyte layer in a linear pattern in controls, but was absent in GLI2 expressing HaCaT GLI2 reconstructs. Similarly, laminin 5 gamma-2 subunit (LAMC2) was only present in a few cells and not limited to the basal layer, whereas staining was intense in the cytoplasm of keratinocytes in the basal layer and at the dermal epidermal junction (DEJ) in controls. We also observed that ITGB4 or LAMC2 staining was present in some of the invading keratinocytes in the GLI2 expressing reconstructs. These observations and the poor adhesion of the GLI2 expressing HaCaT GLI2 cells to the collagen/fibroblast layer of the tissue reconstructs suggest that GLI2 overexpression causes defects in the structural link between the GLI2 expressing keratinocytes and the extracellular matrix. Indeed, in tissue reconstructs prepared from dermal fibroblasts and 1:1 mixtures of GLI2 expressing HaCaT GLI2 cells and control HaCaT Tet cells, we observed that the GLI2 expressing cells were exclusively located in the upper half of the epidermis, while control cells occupied the lower half (Figure 4).
Figure 4. GLI2 expressing cells grown in the presence of control HaCaT Tet cells fail to adhere to the collagen/fibroblast layer.
Frozen sections of organotypic cultures prepared with 1:1 mixtures of control HaCaT Tet cells and HaCaT GLI2 cells infected with lentiviruses expressing eGFP (green) or RFP (red), respectively and grown in the absence or presence of doxycycline. Nuclei counterstained by DAPI are shown in blue.
Abnormal expression of collagen IV (COL4) was also present in GLI2 expressing HaCaT GLI2 reconstructs. In contrast to the continuous band of staining at the DEJ in normal keratinocytes and control HaCaT reconstructs (Figure 3F and Supplementary Figure 4), intense COL4 staining extended throughout the upper quarter to one-third of the collagen/fibroblast layer, encompassing several cell layers and appeared co-extant with the layer of more spindle shaped fibroblasts seen in the H&E stained sections (Figure 2A). Thus, overexpression of GLI2 both blocks differentiation and disrupts the normal expression pattern of keratinocyte derived proteins linking the epithelium to the stroma.
GLI2 induces expression of stem cell genes in keratinocytes
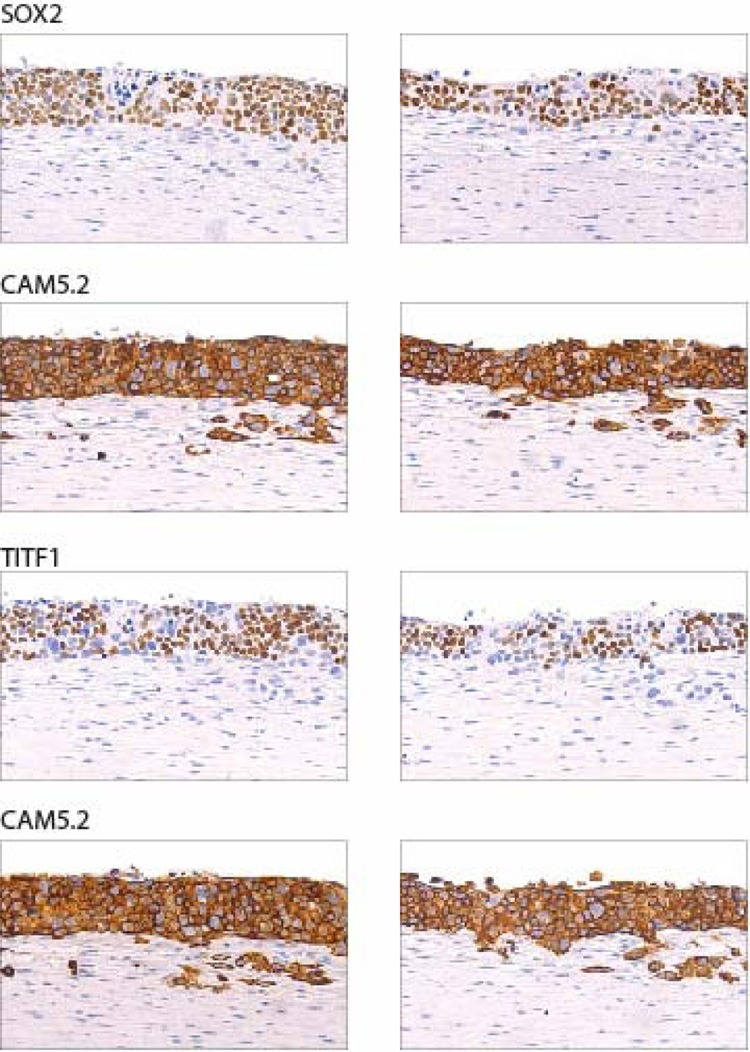
Stem cell gene expression signatures have been reported in poorly differentiated breast and other tumors (Ben-Porath et al., 2008; Chen et al., 2008), as well as in gliomas, which are associated with activated hedgehog signaling and GLI overexpression (Clement et al., 2007), suggesting that GLI2 might block keratinocyte differentiation by up-regulating stem cell genes. To investigate this possibility, expression profiling was carried on RNA extracted from isolated keratinocyte layers from the tissue reconstructs. This analysis revealed elevated expression of SOX2 in the GLI2 expressing keratinocytes (Supplementary Figure 5), which was also confirmed by staining reconstructs with antibodies to SOX2 (Figure 5). These observations suggest that SOX2 is a GLI2 responsive gene. Indeed, infection of primary keratinocytes with a GLI2 retrovirus resulted in induction of SOX2 RNA (Supplementary Figure 6A), and in HaCaT GLI2 cells, addition of doxcycline induced SOX2 expression with the same kinetics as the two known GLI2 downstream targets, GLI1 and PTCH1 (Supplementary Figure 6B). Moreover, a number of potential GLI2 consensus binding sites are present in SOX2 (Supplementary Table 4). In a separate analysis, we found that neither NANOG nor OCT4 (POU5F1) was up-regulated in GLI2 expressing cells (data not shown), whereas elevated RNA and protein expression of TITF1 (Nkx2.1) was observed (Figure 5). TITF1 is reciprocally regulated with SOX2 during early dorsal/ventral foregut patterning (Que et al., 2007). It is frequently amplified in lung cancer (Kendall et al., 2007; Kwei et al., 2008; Weir et al., 2007) and was amplified in one of 89 oral SCC (Snijders et al., 2005). Thus, induction of stem cell genes is likely to underlie the block in differentiation resulting from GLI2 overexpression, and is consistent with disruption of differentiation when SOX2 is expressed at high levels in basal epithelial cells of transgenic mice (Okubo et al., 2006).
Figure 5. GLI2 induces expression of stem cell genes.
Sections of organotypic cultures of GLI2 expressing HaCaT GLI2 cells stained using HRP and DAB detection and antibodies to SOX2 and TITF1. The locations of keratinocytes in the sections are indicated by staining adjacent sections with antibodies to CAM5.2. Two photomicrographs are shown from each section.
Overexpression of GLI2 induces differentiation of fibroblasts into myofibroblasts
Alterations in cells in the upper region of the collagen/fibroblast layer of the tissue reconstructs expressing GLI2 are reminiscent of the desmoplastic response of the stroma adjacent to many tumors, wherein stromal cells display, among other characteristics, myofibroblastic features, including up-regulated synthesis of α smooth muscle actin (SMA), and increased proliferation, contractility and deposition of collagen. The myofibroblastic carcinoma associated fibroblasts (CAFs) may be derived from a variety of cell types in vivo (De Wever & Mareel, 2003; Kalluri & Zeisberg, 2006; Mueller & Fusenig, 2004; Zeisberg et al., 2007). Since expression of SMA is one of the hallmarks of CAFs, we assessed SMA expression and observed positive staining at the DEJ of tissue reconstructs prepared from dermal fibroblasts and normal dermal keratinocytes or control HaCaT Tet cells (Figure 6B and Supplementary Figure 4). By contrast in reconstructs prepared with HaCaT GLI2 cells expressing GLI2 and eGFP, we observed intense positive SMA staining in cells in the same upper region of the collagen/fibroblast layer that was also positive for collagen IV (Figure 6B). Moreover, no SMA positive cells also expressed eGFP (Figure 6B), indicating that the myofibroblasts originated from the co-cultured dermal fibroblasts and not from keratinocytes that had undergone epithelial to mesenchymal transition. Since our study of GLI2 was motivated by the observation of GLI2 amplification in oral SCC, we also prepared reconstructs with fibroblasts cultured from tongue and gingiva. Whereas, GLI2 expressing HaCaT GLI2 cells induced transdifferentiation of tongue fibroblasts in organotypic cultures, no transdifferentiation was observed with cultures of gingival fibroblasts (Figure 6C).
Figure 6. GLI2 induces TGFβ mediated differentiation of fibroblasts into myofibroblasts.
(A) Increased Ki67 staining in stromal fibroblasts in GLI2 expressing reconstructs. Immunofluorescent staining for Ki67 (red) in control HaCaT Tet reconstructs (left panel) and in GLI2 and eGFP expressing HaCaT GLI2 reconstructs (right panel). In GLI2 expressing reconstructs, most cells in the epithelial layer are positive for both Ki67 and eGFP, confirming their epithelial origin. In the stromal compartment of GLI2 expressing reconstructs, the majority of Kig positive cells do not co-express eGFP confirming their fibroblast origin.
(B) Staining for SMA is limited to the uppermost region in the dermal compartment, directly underneath the DEJe, which is demarcated by integrin β4 co-staining (green) in HaCaT Tet reconstructs (left panel). Intense SMA staining is present in the upper dermal compartment of reconstructs from GLI2 and eGFP expressing HaCaT GLI2 cells (right panel), which are also stained for GFP (green). No SMA positive cells co-expressed GFP.
(C) Organotypic cultures comprised of oral fibroblasts and HaCaT Tet (left) or GLI2 expressing HaCaT GLI2 cells (right) stained for SMA (brown, HRP and DAB detection) and counterstained with hematoxylin (blue). Cultures were prepared with tongue or gingival fibroblasts at passage 2 (top and bottom panels, respectively).
(D) GLI2-induced myofibroblast differentiation is dependent on active TGFβ signaling. Organotypic cultures from HaCaT GLI2 cells expressing GLI2 and eGFP were treated with either DMSO (left panels) or with the TGFβRI/II kinase inhibitor LY2109761 dissolved in DMSO at 2 µM (right panels). Sections were stained with H&E (upper panels) and for SMA and eGFP (bottom panels). The epithelial layer appears thinner in the drug treated sections due to less contraction of the collagen gel, which normally reduces the area covered by the GLI2 expressing HaCaT GLI2 cells.
These observations predict that SMA expression should be present in oral SCC with GLI2 amplification. This expectation was confirmed, as we observed high levels of expression of SMA adjacent to tumor cell islands in oral SCC with GLI2 amplification (Figure S7). We note, however, that SMA positive stroma is not restricted to tumors with GLI2 amplification, as we found 44% of oral SCC (11/25) from different oral subsites including gingiva to stain positively, while normal oral tissues, hyperkeratosis, and pre-malignant lesions (various grades of dysplasia and proliferative verrucous leukoplakia) were all negative, in agreement with other reports (Kellermann et al., 2007; Lewis et al., 2004).
GLI2 induced myofibroblast differentiation requires TGFβ signaling
Signaling via TGFβ has been implicated in promoting fibrosis and differentiation of fibroblasts into myofibroblasts in tumor associated desmoplasia (De Wever & Mareel, 2003; Tuxhorn et al., 2001). To determine whether TGFβ signaling underlies the phenotypes observed in the GLI2 expressing HaCaT organotypic cultures, we treated these cultures with the TGFβRI/II kinase inhibitor LY2109761 (Lacher et al., 2006) or DMSO. Both the LY2109761 and control, DMSO treated cultures displayed the basal-like epithelial cells characteristic of GLI2 expressing HaCaT GLI2 cultures, as well as similar proportions of Ki67 and PHH3 positive nuclei (0.85–0.88 and 0.24–0.29, respectively). In the dermal layer of the LY2109761 treated cultures, however, we observed noeither differentiation of fibroblasts into myofibroblasts nor keratinocyte invasion (Figure 6D). We obtained similar results when TGFβ signaling was abrogated in foreskin fibroblasts by introduction of a dominant negative TGFβ type II receptor construct lacking the kinase domain (Verona et al., 2007) (data not shown) or when the keratinocyte and fibroblast layers were separated by a 20–40 µm thick layer of acellular collagen (Supplementary Figure 8). Thus, TGFβ signaling in fibroblasts is required for their transdifferentiation and generation of a stromal environment permissive for invasion.
Invading keratinocytes down regulate GLI2 responsive genes and are only locally invasive within the myofibroblast modified collagen matrix
Transdifferentiation of stromal myofibroblasts via TGFβ1 is also associated with up-regulated secretion of HGF, which acts in a paracrine manner to activate its receptor, c-Met on keratinocytes by inducing its autophosphorylation. Activation of MET results in enhanced proliferation, motility and invasiveness of keratinocytes. We confirmed that HGF transcription was up-regulated in GLI2 expressing HaCaT GLI2 reconstructs (data not shown). In addition, the GLI2 expressing HaCaT GLI2 cells in the tissue reconstructs stained positively for both c-Met and phosphorylated c-Met, whereas control HaCaT Tet cells were only positive for c-Met (Figures 7A and B). Thus, c-Met signaling is activated in the GLI2 expressing cells with the potential to induce keratinocyte migration and invasion.
Figure 7. Invading epithelial cells in GLI2 expressing tissue reconstructs express keratinocyte differentiation markers and down regulate GLI2 responsive genes.
Sections of organotypic cultures of control HaCaT Tet (left) and induced HaCaT GLI2 cells (right) were stained using immunohistochemistry or immunofluorescence with antibodies for (A) c-Met, (B) phosphorylated c-Met, (C) E-cadherin, (D) β-catenin, (E) BCL2 and (F) the apoptosis marker caspase 3 (green) and epithelial marker pan-cytokeratin (CAM5.2 and AE1/AE3, red). Nuclei were counterstained with hematoxylin (immunohistochemistry) or DAPI (immunofluorescence) and are shown in blue.
All keratinocytes invading into the collagen/fibroblast layer of GLI2 expressing HaCaT GLI2 tissue reconstructs appeared to be positive for pancytokeratin (Figure 2C) and were uniformly negative for E-cadherin (Figure 7C), while cells in the epithelial layer showed membranous staining for both E-cadherin and β-catenin (Figure 7D), suggesting that invasion is dependent on the loss of E-cadherin as is commonly observed in other systems. Enhanced expression of β-catenin was also noted in the myofibroblasts in GLI2 expressing HaCaT GLI2 reconstructs. It was notable that the invading cells down regulated expression of the GLI2 responsive gene BCL2 (Figures 7E and Supplementary Figure 9), as well as SOX2 and TITF1 (Figure 4), and some cells stained positively for differentiation markers (Figure 3) or markers of apoptosis (caspase 3 or TUNEL, Figure 7F and data not shown). We also observed a lack of sprouting from spheroids formed from GLI2 expressing HaCaT GLI2 cells embedded in collagen I matrices (Figure 8). These observations suggest that GLI2 expressing keratinocytes are non-invasive and that the capabilities to both invade and differentiate are acquired upon down regulation of GLI2. Nevertheless, invading cells appear to remain confined to the myofibroblast region. These observations together with the lack of invasion in tissue reconstructs lacking myofibroblasts (e.g. due to inhibition of TGFβ1 signaling) suggest that remodeling of the collagen I matrix by myofibroblasts is required for local invasion of HaCaT GLI2 cells that have down regulated GLI2 responsive genes.
Figure 8. GLI2 expressing HaCaT cells do not migrate into collagen I gels.
Spheroid cultures of eGFP expressing control HaCaT Tet cells (green), RFP expressing HaCaT GLI2 cells (red) and a 1:1 mixture of eGFP expressing HaCaT Tet cells (green) and RFP expressing HaCaT GLI2 cells (red) were embedded in a collagen I matrix and grown for 13 days with or without doxycycline to induce GLI2 expression.
DISCUSSION
We have shown that GLI2 is a pleiotropic oncogene, and thus, up-regulated GLI2 expression alone is sufficient to induce a number of the hallmarks of cancer (Hanahan & Weinberg, 2000). Nevertheless, the differential responses of fibroblasts to GLI2 overexpressing keratinocytes indicate that the stroma, in a tissue specific manner, determines whether certain GLI2 oncogenic traits are expressed.
The roles of other oncogenes have been evaluated in HaCaT organotypic cultures, including HRAS, BCL2 (Delehedde et al., 2001), MYC, TERT (Cerezo et al., 2003), CCND1 (Burnworth et al., 2006), and SHH (Bigelow et al., 2005). While expression of the GLI2 target, BCL2 had little effect on differentiation, expression of other genes resulted in phenotypes shared with GLI2, including extension of proliferating Ki67 positive cells from the basal layer to upper epithelial layers (CCND1, HRAS, SHH) and absence or abnormal expression of differentiation markers (TERT, CCND1). The absence of the basement membrane zone, however, may be unique to the GLI2 phenotype as its formation was reported to be normal in HaCaT reconstructs overexpressing TERT, MYC or CCND1.
Modification of fibroblasts and keratinocyte invasion have been reported in organotypic cultures of SHH expressing HaCaT cells (Bigelow et al., 2005) and GLI2 overexpressing hTERT immortalized keratinocytes (Marsh et al., 2008). In contrast to our studies, however, invasion occurred as fingers of cells rather than as individual cells or small groups of cells. On the one hand, although SHH and GLI2 are the proximal and distal ends of the hedgehog pathway, the different phenotypes may be attributed to the fact that additional signaling pathways are induced by SHH expression (Riobo et al., 2006) or by differences in the level of GLI2 expression, considered a critical determinant of the outcome of hedgehog signaling (Grachtchouk et al., 2003; Hooper & Scott, 2005; Riobo et al., 2006). On the other hand, the differences in invasive phenotype may be attributed to the source of fibroblasts used in the organotypic cultures. Embryonic (NIH3T3) or human fetal foreskin fibroblasts were used to prepare the organotypic cultures with SHH expressing HaCaT cells and GLI2 overexpressing hTERT immortalized keratinocytes, respectively. It is well recognized that fetal wounds heal without scarring, suggesting that the different patterns of invasion observed in response to activation of the hedgehog pathway may reflect tissue specific behavior of fibroblasts. In our studies, the differential response of fibroblasts to TGFβ1 may be attributed to the embryological origins of the cells (e.g. neural crest and pharyngeal arches, gingival and tongue, respectively), repertoire of genes expressed (Chang et al., 2002), and signaling pathways involved in inducing SMA expression in response to TGFβ1 (Black et al., 2007). While further work is required to dissect the different signaling pathways and responsiveness of dermal, tongue and gingival fibroblasts to TGFβ1, these studies, in accord with Paget’s “seed and soil” hypothesis, highlight the importance of co-evolution of carcinoma cells and stroma, and the importance of the stroma in determining the invasive properties of the tumor.
The oncogenic properties of GLI2 revealed in organotypic cultures are directly relevant to the behavior of tumors in vivo. The cultures recapitulated the basal-like histology of the GLI2 amplifying tumors and correctly predicted that SMA positive cells should be present in the stroma of these tumors. These studies also revealed that invasive keratinocytes had down regulated GLI2 responsive genes, which may have been necessary to overcome GLI promoted cell-cell adhesion (Neill et al., 2008); however concomitant with reduction in GLI2 expression is the apparent release of the block in the differentiation program, allowing the cells to differentiate and die. These cells are likely to be inefficient at seeding metastases, consistent with the low frequency of metastasis associated with basal cell carcinomas (BCC), a GLI2 overexpressing skin tumor. Phenotypic differences in invasive cells of BCCs have been noted previously (Svensson et al., 2003). Thus, the organotypic culture model allows specific oncogene associated molecular alterations to be identified in both the carcinoma and the stroma. In the case of GLI2 amplifying or overexpressing tumors, phenotypes observed in the organotypic cultures suggest that benefit could be gained from therapeutic approaches targeting both the consequences of GLI2 overexpression in the carcinoma cells, as well as TGFβ signaling in the stroma, for which a number of agents have already been identified (De Wever & Mareel, 2003; Lauth et al., 2007; Mueller & Fusenig, 2004).
MATERIALS AND METHODS
Establishment of primary keratinocyte and fibroblast cultures
Human neonatal foreskin tissue was obtained from the UCSF Well Baby Nursery following UCSF guidelines for obtaining and handling human tissue. Human oral tissues were obtained from patients undergoing routine third molar extraction at the UCSF Dental Clinic or were obtained from clinically normal tissue at the time of surgical excision of dysplasia or SCC in the UCSF Department of Oral and Maxillofacial Surgery. Patients ranged in age from 17 to 43. Only discarded tissue was used and all identifiers were removed from the samples prior to collection. Epithelial keratinocyte and fibroblasts cultures were prepared and maintained as described in the Supplementary Methods.
Cell culture
All HT1080 and HaCaT cells were cultured in DME H-16 supplemented with 10% fetal calf serum at 37°C in 5% CO2 unless otherwise specified. Cells transfected with various constructs were cultured in the presence of appropriate antibiotics as described in the Supplementary Methods.
Organotypic cultures were prepared by adding oral or dermal fibroblasts (7.5 × 104) to collagen, plating them in 6-well culture plates and culturing for 48 hours to allow the gel to contract before adding 5 × 105 keratinocytes to the surface. After 48–72 hours, the cultures were raised to the air/liquid interface and the rafts cultured for a further 2–3 weeks, after which time they were fixed and processed for routine histology as described in detail in the Supplementary Methods.
Immunohistochemistry and Immunofluorescence
Sections were deparaffinized by three five minute washes in xylene, followed by two washes for five minutes each through a graded alcohol series to distilled water. Different antigen retrieval methods and processing for immunohistochemistry and immunofluorescence were used as appropriate for each antibody (Supplementary Tables 5 and 6).
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Gillian Hall and Richard Shaw, University of Liverpool for providing oral SCC 1300C2, Dr. Fritz Aberger, University of Salzburg for providing the HaCaT Tet cells, HaCaT GLI2 cells and the 6xHis-GLI2ΔN pcDNA4/TO vector construct, and Dr. Rosemary Akhurst, UCSF Helen Diller Family Comprehensive Cancer Center (HDFCCC) and Dr. J. Yingling (Eli Lilly and Company, Indianapolis, IN) for providing the TGFβ receptor kinase inhibitor LY2109761. The UCSF HDFCCC Microarray, Tissue, Immunohistochemistry/Molecular Pathology and Genome Analysis Shared Resources provided assistance. This work was supported by NIH grant CA118323. AMS was the recipient of a postdoctoral fellowship from the California Tobacco-Related Disease Research Program (14FT-0011) and a trainee of the NCI-Sponsored Tumor Microenvironment Training Program: Techniques in the Establishment and Manipulation of Organotypic Model Systems. BLS is an appointee of the Western Oral Research Consortium (NIH K12 DE14609).
REFERENCES
- Albertson DG, Collins C, McCormick F, Gray JW. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bell E, Ehrlich HP, Buttle DJ, Nakatsuji T. Science. 1981;211:1052–1054. doi: 10.1126/science.7008197. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow RL, Jen EY, Delehedde M, Chari NS, McDonnell TJ. J Invest Dermatol. 2005;124:457–465. doi: 10.1111/j.0022-202X.2004.23590.x. [DOI] [PubMed] [Google Scholar]
- Black SA, Jr, Palamakumbura AH, Stan M, Trackman PC. J Biol Chem. 2007;282:15416–15429. doi: 10.1074/jbc.M610432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnworth B, Popp S, Stark HJ, Steinkraus V, Brocker EB, Hartschuh W, Birek C, Boukamp P. Oncogene. 2006;25:4399–4412. doi: 10.1038/sj.onc.1209474. [DOI] [PubMed] [Google Scholar]
- Cerezo A, Stark HJ, Moshir S, Boukamp P. J Invest Dermatol. 2003;121:110–119. doi: 10.1046/j.1523-1747.2003.12304.x. [DOI] [PubMed] [Google Scholar]
- Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, Brown PO. Proc Natl Acad Sci U S A. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, Sun L, Yang X, Wang Y, Zhang Y, Shang Y. J Biol Chem. 2008 doi: 10.1074/jbc.M802917200. [DOI] [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wever O, Mareel M. J Pathol. 2003;200:429–447. doi: 10.1002/path.1398. [DOI] [PubMed] [Google Scholar]
- Delehedde M, Cho SH, Hamm R, Brisbay S, Ananthaswamy HN, Kripke M, McDonnell TJ. J Invest Dermatol. 2001;116:366–373. doi: 10.1046/j.1523-1747.2001.01260.x. [DOI] [PubMed] [Google Scholar]
- Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, Dairkee S, Tokuyasu T, Ljung BM, Jain AN, McLennan J, Ziegler J, Chin K, Devries S, Feiler H, Gray JW, Waldman F, Pinkel D, Albertson DG. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Grachtchouk V, Grachtchouk M, Lowe L, Johnson T, Wei L, Wang A, de Sauvage F, Dlugosz AA. Embo J. 2003;22:2741–2751. doi: 10.1093/emboj/cdg271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hooper JE, Scott MP. Nat Rev Mol Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- Izzo JG, Papadimitrakopoulou VA, Li XQ, Ibarguen H, Lee JS, Ro JY, El-Naggar A, Hong WK, Hittelman WN. Oncogene. 1998;17:2313–2322. doi: 10.1038/sj.onc.1202153. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Zeisberg M. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- Kellermann MG, Sobral LM, da Silva SD, Zecchin KG, Graner E, Lopes MA, Kowalski LP, Coletta RD. Oral Oncol. 2007 doi: 10.1111/j.1365-2559.2007.02873.x. [DOI] [PubMed] [Google Scholar]
- Kendall J, Liuz Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Proc Natl Acad Sci U S A. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, Lee J, Choi YL, Sato M, Wang P, Hernandez-Boussard T, Gazdar AF, Petersen I, Minna JD, Pollack JR. Oncogene. 2008 doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacher MD, Tiirikainen MI, Saunier EF, Christian C, Anders M, Oft M, Balmain A, Akhurst RJ, Korn WM. Cancer Res. 2006;66:1648–1657. doi: 10.1158/0008-5472.CAN-05-2328. [DOI] [PubMed] [Google Scholar]
- Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Proc Natl Acad Sci U S A. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauth M, Toftgard R. Cell Cycle. 2007;6:2458–2463. doi: 10.4161/cc.6.20.4808. [DOI] [PubMed] [Google Scholar]
- Lewis MP, Lygoe KA, Nystrom ML, Anderson WP, Speight PM, Marshall JF, Thomas GJ. Br J Cancer. 2004;90:822–832. doi: 10.1038/sj.bjc.6601611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D, Dickinson S, Neill GW, Marshall JF, Hart IR. Cancer Res. 2008;68:3295–3303. doi: 10.1158/0008-5472.CAN-08-0174. [DOI] [PubMed] [Google Scholar]
- Miele M, Bonatti S, Menichini P, Ottaggio L, Abbondandolo A. Mutat Res. 1989;219:171–178. doi: 10.1016/0921-8734(89)90012-x. [DOI] [PubMed] [Google Scholar]
- Mueller MM, Fusenig NE. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- Murnane JP, Sabatier L. Bioessays. 2004;26:1164–1174. doi: 10.1002/bies.20125. [DOI] [PubMed] [Google Scholar]
- Neill GW, Harrison WJ, Ikram MS, Williams TD, Bianchi LS, Nadendla SK, Green JL, Ghali L, Frischauf AM, O'Toole EA, Aberger F, Philpott MP. Carcinogenesis. 2008;29:738–746. doi: 10.1093/carcin/bgn037. [DOI] [PubMed] [Google Scholar]
- Nelsen CJ, Kuriyama R, Hirsch B, Negron VC, Lingle WL, Goggin MM, Stanley MW, Albrecht JH. J Biol Chem. 2005;280:768–776. doi: 10.1074/jbc.M407105200. [DOI] [PubMed] [Google Scholar]
- Okubo T, Pevny LH, Hogan BL. Genes Dev. 2006;20:2654–2659. doi: 10.1101/gad.1457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson TG, Almasan A, Brody LL, Wahl GM. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regl G, Kasper M, Schnidar H, Eichberger T, Neill GW, Ikram MS, Quinn AG, Philpott MP, Frischauf AM, Aberger F. Oncogene. 2004;23:1263–1274. doi: 10.1038/sj.onc.1207240. [DOI] [PubMed] [Google Scholar]
- Riobo NA, Lu K, Emerson CP., Jr Cell Cycle. 2006;5:1612–1615. doi: 10.4161/cc.5.15.3130. [DOI] [PubMed] [Google Scholar]
- Roh HJ, Shin DM, Lee JS, Ro JY, Tainsky MA, Hong WK, Hittelman WN. Cancer Res. 2000;60:6496–6502. [PubMed] [Google Scholar]
- Roth JR, Andersson DI. Res Microbiol. 2004;155:342–351. doi: 10.1016/j.resmic.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Schmidt BL, Dierks EJ, Homer L, Potter B. J Oral Maxillofac Surg. 2004;62:1055–1058. doi: 10.1016/j.joms.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Schoop VM, Mirancea N, Fusenig NE. J Invest Dermatol. 1999;112:343–353. doi: 10.1046/j.1523-1747.1999.00524.x. [DOI] [PubMed] [Google Scholar]
- Shimizu N, Shingaki K, Kaneko-Sasaguri Y, Hashizume T, Kanda T. Exp Cell Res. 2005;302:233–243. doi: 10.1016/j.yexcr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Fridlyand J, Mans DA, Segraves R, Jain AN, Pinkel D, Albertson DG. Oncogene. 2003;22:4370–4379. doi: 10.1038/sj.onc.1206482. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Hermsen MA, Baughman J, Buffart TE, Huey B, Gajduskova P, Roydasgupta R, Tokuyasu T, Meijer GA, Fridlyand J, Albertson DG. Genes Chromosomes Cancer. 2008;47:71–83. doi: 10.1002/gcc.20509. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, Albertson DG. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- Svensson S, Nilsson K, Ringberg A, Landberg G. Cancer Res. 2003;63:1737–1742. [PubMed] [Google Scholar]
- Tuxhorn JA, Ayala GE, Rowley DR. J Urol. 2001;166:2472–2483. [PubMed] [Google Scholar]
- Verona EV, Elkahloun AG, Yang J, Bandyopadhyay A, Yeh IT, Sun LZ. Cancer Res. 2007;67:5737–5746. doi: 10.1158/0008-5472.CAN-07-0444. [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba Watson MA, II, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg EM, Potenta S, Xie L, Zeisberg M, Kalluri R. Cancer Res. 2007;67:10123–10128. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- Zhou P, Jiang W, Weghorst CM, Weinstein IB. Cancer Res. 1996;56:36–39. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.