Abstract
The matrix metalloproteinase stromelysin-3 (ST3) has long been implicated to play an important role in cell fate determination during normal and pathological processes. Using the thyroid hormone-dependent Xenopus laevis metamorphosis as a model, we have previously shown that ST3 is required for apoptosis during intestinal remodeling and that laminin receptor (LR) is an in vivo substrate of ST3 during this process. ST3 cleaves LR at two distinct sites that are conserved in mammalian LR. Human ST3 and LR are both associated with tumor development and cancer progression and human LR can also be cleaved by ST3, implicating a role of LR cleavage by ST3 in human cancers. Here, we carried out a series of mutational analyses on the two cleavage sites in LR. Our findings revealed that in addition to primary sequence at the cleavage site (positions P3-P3′, with the cleavage occurring between P1-P1′), flanking sequences/conformation also influenced the cleavage of LR by ST3. Furthermore, alanine substitution studies led to a surprising finding that surrounding sequence and/or conformation dictated the site of cleavage in LR by ST3. These results thus have important implications in our understanding of substrate recognition and cleavage by ST3 and argue for the importance of studying ST3 cleavage in the context of full-length substrates. Furthermore, the LR cleavage mutants generated here will also be valuable tools for future studies on the role of LR cleavage by ST3 in vertebrate development and cancer progression.
Keywords: stromelysin-3, laminin receptor, matrix metalloproteinase, Xenopus laevis, tumor invasion, extracellular matrix
Introduction
Matrix metalloproteinases (MMPs) form a group of structurally related zinc-dependent endopeptidases that are collectively capable of degrading all proteinaceous components of the extracellular matrix (ECM) (1-7). Most MMPs are synthesized as preenzymes and are secreted as the proenzymes into the extracellular matrix (ECM), accompanied by the removal of the pre-peptide. The proenzymes are activated upon proteolytic removal of the propeptide (1, 3, 6, 8-10). Other MMPs, such as stromelysin-3 (ST3) and membrane type MMPs (MT-MMPs) are activated intracellularly through a furin-dependent process due to the presence of the conserved furin-recognition motif RXKR in the propeptide and are thus secreted or inserted into the plasma membrane directly as active (mature) MMPs (4, 11, 12). The mature or activated MMPs have different but often overlapping substrate specificities (1, 4, 13, 14). While MMPs are known for their ability to degrade the proteinaceous components of the ECM, they are also capable of degrading non-ECM extracellular or membrane-bound proteins (1, 4, 13, 14). Many MMPs substrates have been identified in vitro. However, with a few exceptions, it is unknown whether MMPs cleave these substrates during normal or pathological processes in vivo.
We have been studying the role of MMPs in postembryonic tissue remodeling by using intestinal development during frog metamorphosis as a model (15). Tadpole intestine is a simple tubular organ with a single layer of larval epithelium surrounded by thin layers of connective tissue and muscles (16). During metamorphosis, massive apoptosis of the larval epithelium and concurrent proliferation and differentiation of the adult epithelium is accompanied by the remodeling of the basement membrane or basal lamina, the ECM that separates the epithelium from the connective tissue. These changes lead to the formation of a multiply folded frog epithelium surrounded by elaborate connective tissue and thick muscle layers. We and others have previously shown that a number of MMPs are upregulated in the intestine during Xenopus laevis metamorphosis (17, 18). In particular, we have shown that ST3 expression precedes the remodeling of the basal lamina, apoptosis in the larval epithelium, and morphogenesis of the adult intestinal epithelium (19-21), implicating a role of ST3 during intestinal remodeling. Indeed, organ culture and transgenic studies have shown that ST3 is necessary and sufficient for apoptosis and ECM remodeling during intestinal metamorphosis (22, 23).
Toward understanding how ST3 affects cell fate during intestinal metamorphosis, we have previous identified the 37 kd precursor of the 67 kd laminin receptor (LR) as a physiological substrate of Xenopus ST3 during development (the 67 kd laminin receptor is reported to consist of a homodimer of the 37 kd precursor or a heterodimer of this precursor with an, as yet, unknown partner (24-26). For simplicity, we will refer to both as LR in this paper). We have mapped the two ST3 cleavage sites to the extracellular domain of the protein, located in between the transmembrane domain and laminin binding sequence (27). Furthermore, LR is highly conserved and human LR can also be cleaved by ST3 at the same sites (27). More importantly, both mammalian ST3 and LR are associated with tumor development and cancer progression (25, 28-39). These findings suggest that LR is a conserved substrate of ST3 during developmental and pathological processes and that understanding LR cleavage by ST3 may offer not only insights into substrate recognition by ST3 but also valuable information for developing cleavage resistant LR mutants as tools for studying ST3 function in development and cancer progression. Toward this goal, we have carried out a detailed mutational analysis of the two cleavage sites in LR and discovered that both primary sequence at the cleavage site from P3 to P3′ and long-range interactions affect ST3 cleavage at the two sites in LR.
Materials and Methods
Construction of LR mutants and in vitro translation
The wild type Xenopus LR construct (W-W) for in vitro translation was the construct xLRpET30 as reported previously (27). The LR mutants were generated via two-step PCR. The sense (forward) primer A (5′ CGCACCGGTAGCATGTCCGGAGGTCTTGATGTC 3′) containing an AgeI restriction site (in italics) and initiation codon ATG (underlined), and an antisense (reverse) primer complimentary to the desired sequences were employed for the amplification of 5′-fragment for each mutant (Table 1). The 3′-fragment fused with the sequence for one copy of the FLAG tag at the 3′-end was amplified using indicated mutagenic forward primer (Table 1) and the primer B (5′GGGAGATCTACTTATCGTCGTCATCCTTGTCATCAGACCATTCAGTAGTGGTTCC 3′), which contained a BglII restriction site (in italics). The two PCR products were gel-purified and an aliquot of each was mixed together as the templates for the second round of PCR with primers A and B. The resulting product was inserted into pCR-BluntII-Topo vector (Invitrogen) and verified by sequencing. The middle portion of the LR encompassing the mutated region but without the FLAG-tag was excised from the pCR-BluntII-Topo vector by AleI and BmtI digestion and subcloned into xLRpET30 (27) predigested with AleI and BmtI. This resulted in a construct containing the coding region of the desired LR mutant fused to a N-terminal His-tag and S-tag under the control of T7 RNA polymerase promoter. The constructs for wild type and mutant LR were used for in vitro coupled transcription-translation (Promega). The resulting proteins were subjected to affinity chromatography purification with His-Select spin columns (Sigma). Briefly, the column was equilibrated with 600μl of Equilibration buffer (50mM Tris-HCl, 0.3M NaCl, pH 8.0), spun in a microcentrifuge at 1000 rpm for 1 min and then loaded with the in vitro translation sample. The column was spun again and the eluate was re-loaded onto the column, and the spin and reloading were repeated for a total of 3 times in order to ensure the maximal binding of the LR protein to the column. The column was then washed twice with 600μl washing buffer (50mn Tris-HCl, 0.3M NaCl, 5mM Imidaziole, pH 8.0). Finally, the protein was eluted with 100μl of the same buffer with 100 mM Imidazole.
Table 1.
Forward (F) and reverse (R) primers used in PCR.
|
RT-W* F 5′– TGCTGGTGGTGACAGATCCCCGGGCAGATCATCAACGCACTACTGAAGCTTC –3′ R 5′– GGATCTGTCACCACCAGCAGCCGAGGCTCCCTAGTACGAGCCTGGATC –3′ |
|
RTF-W* F 5′–TGCTGGTGGTGACAGATCCCCGGGCAGATCATCAACCCATTACTGAAGCTTC–3′ R 5′–GGATCTGTCACCACCAGCAGCCGAGGAAACCTAGTACGAGCCTGGATC–3′ |
|
W-RT* F 5′– TGCTGGTGGTGACAGATCCCCGGGCAGATCATCAACGCACTACTGAAGCTTC –3′ R 5′– GGATCTGTCACCACCAGCAGCCGAGGCTCCCTAAAAGCAGCCTGGATC –3′ |
|
W-RTF* F 5′– TGCTGGTGGTGACAGATCCCCGGGCAGATCATCAACGCACTACTGAAGCTTC –3′ R 5′– GGATCTGTCACCACCAGCAGCCGAGGCTCCCTAAAAGCAGCCTGGATC –3′ |
|
RTF-RT* F 5′– TGCTGGTGGTGACAGATCCCCGGGCAGATCATCAACGCACTACTGAAGCTTC –3′ R 5′– GGATCTGTCACCACCAGCAGCCGAGGAAACCTAGTACGAGCCTGGATC –3′ |
|
RT-RTF* F 5′– TGCTGGTGGTGACAGATCCCCGGGCAGATCATCAACGCACTACTGAAGCTTC –3′ R 5′– GGATCTGTCACCACCAGCAGCCGAGGCTCCCTAGTACGAGCCTGGATC –3′ |
|
RTF-RTF* F 5′– TGCTGGTGGTGACAGATCCCCGGGCAGATCATCAACGCACTACTGAAGCTTC –3′ R 5′– GGATCTGTCACCACCAGCAGCCGAGGAAACCTAGTACGAGCCTGGATC –3′ |
|
RT–RT* F 5′– TGCTGGTGGTGACAGATCCCCGGGCAGATCATCAACGCACTACTGAAGCTTC –3′ R 5′– GGATCTGTCACCACCAGCAGCCGAGGAAACCTAGTACGAGCCTGGATC –3′ |
|
W-6A* F 5′– ATGCTGCTGCTGCTGCTGCTGCTTCATATGTCAACATTCCC –3′ R 5′– ACAGATCCCCGGGCAGATGCTGCTGCTGCTGCTGCTGCTTCA –3′ |
|
6A-W* F 5′– ATCGCTGCTGCTGCTGCTGCTCCTCGGCTGCTGGTGGTGA –3′ R 5′– GGTACCTTCACCAACCAGATCGCTGCTGCTGCTGCTGCTC –3′ |
|
W-aW* F 5′– ATCAGGCTGCTTTTAGGGAGGCTTCATATGTCAACATTCCC –3′ R 5′– ACAGATCCCCGGGCAGATCAGGCTGCTTTTAGGGAGGCTTCA –3′ |
|
W-aRT* F 5′– TCAGGCTCGTACTAGGGAGGCTTCATATGTCAACATTCC –3′ R 5′– GTGACAGATCCCCGGGCAGATCAGGCTCGTACTAGGGAGGC –3′ |
|
RT-aRT** F 5′– TCAGGCTCGTACTAGGGAGGCTTCATATGTCAACATTCC –3′ R 5′– GTGACAGATCCCCGGGCAGATCAGGCTCGTACTAGGGAGGC –3′ |
denotes that the template used for the reaction was W–W
denotes that the template used for the reaction was RT–W
In vitro cleavage and Western blot analysis
Purified His-tagged wild type and mutant LRs were incubated with ST3 catalytic domain (40) in the ST3 reaction buffer (400 mM NaCl, 20 mM CaCl2, 0.2 mM ZnCl2, 0.2% Brij-35, 100 mM Tris-HCl pH 7.5) at room temperature overnight (27). The reaction was stopped by adding SDS-PAGE sample buffer. The samples were separated by SDS-PAGE and blotted onto a PVDF membrane. The membrane was incubated with 1/10000 dilution of a polyclonal anti-Xenopus LR antibody (27). HRP-conjugated secondary antibody was used for detection by chemiluminescence.
Densitometry Analysis
Western blot films were scanned at 300 dpi using a flatbed scanner. The intensity of the full-length LR (migrating at a position of about 53 kDa) and its products from cleavage at the site a or b was quantified using the NIH ImageJ Software. A region without any bands in each lane was used as the background. After subtracting the background, the ratio of each cleavage product to the uncleaved 53 kd LR was used as a measure of the relative cleavage efficiency. For each experiment, we exposed the western blot membrane to an X-ray film for different lengths of time and for quantification, we chose two exposures that did not show saturation of the bands. We then calculated the average from these two different time points to represent this particular experiment. The results presented here were the average of two independent experiments.
Results
Amino acid substitutions have distinct effects on the two cleavage sites in LR
There has been no established consensus recognition sequence for ST3 cleavage. Thus, to investigate the sequence requirement of LR cleavage by ST3, we made use of our earlier analysis of the in vitro ST3 substrate α1-proteinase inhibitor (α1-PI, or α1-antitrypsin). We have shown that Xenopus ST3 is able to cleave human α1-PI, just like mammalian ST3, but not Xenopus α1-PI (41). The frog α1-PI contains an R and a T at the P1 and P1′ positions of the cleavage site, respectively, instead of an A and an M at the corresponding positions in human α1-PI. Mutagenic analyses have shown that substituting an R and T into the P1 and P1′ positions of human α1-PI inhibits its cleavage by ST3, and conversely substituting the R and T at the P1 and P1′ position in Xenopus α1-PI with an A and a M, respectively, leads to cleavage by ST3 (41). This suggests that an R at P1 and a T at P1′ inhibit ST3 cleavage. In addition, an additional change at P3′ position (L to F), also reduces the cleavage of human α1-PI by ST3 (41). Based on these findings, we generated a number of site-specific mutants of LR with amino acid substitutions at either one or both of the two ST3 cleavage sites located between the transmembrane and the laminin binding sequence (Fig. 1). To facilitate purification and detection, we fused a His-tag to the N-terminus. All proteins were made from in vitro coupled transcription-translation and purified by using the His-tag. The purified proteins were analyzed for their cleavage by ST3 to determine the effects of the mutations.
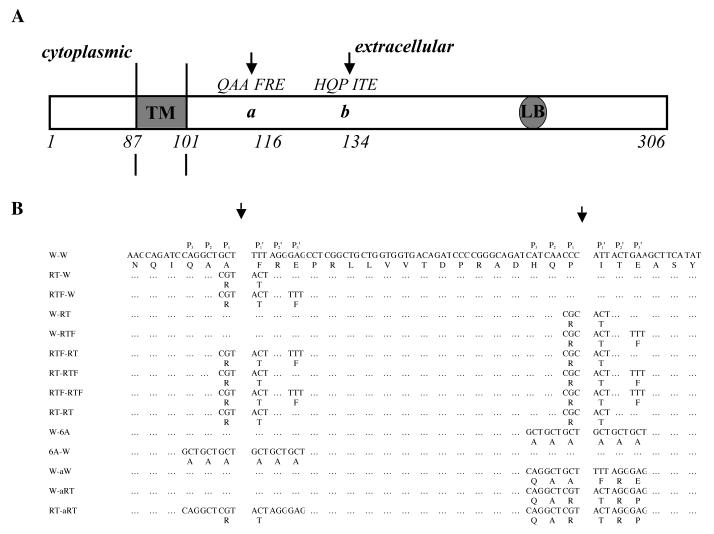
Fig. 1.
A. Schematic diagram showing ST3 cleavage sites between the transmembrane domain (TM) and the laminin binding sequence (LB) of LR (27). ST3 cleaves LR between A115 and F116 (site a), and P133 and I134 (site b) and these two cleavage sites by ST3 are indicated by two arrows (Based on (27)).
B. LR mutants used in this study.
First, we mutagenized individual cleavage sites by changing the 2 amino acids at the cleavage site, i.e., the P1 and P1′ amino acids, to R and T, respectively, to produce the mutants RT-W (mutated site a and wild type site b) and W-RT (wild type site a and mutated site b) (Fig. 1). The wild type (W-W) and mutant LRs were synthesized in vitro and subjected to cleavage by purified catalytic domain of Xenopus ST3. The cleavage products were analyzed by western blot with a polyclonal antibody against His-tagged Xenopus LR, which is specific for the C-terminal portion of the LR (27). As reported earlier, LR and the C-terminal fragments from ST3 cleavage have abnormal mobility on SDS-polyacrylamide gels (27). Thus, the wild type (W-W) LR migrated as a 53-kd band (Fig. 2A. Note that the higher molecular weight bands with variable amounts in different experiments, were likely due to homo- or hetero-dimerization/oligomerization during in vitro translation since the cleavage products were of homogenous sizes). Wild type LR was cleaved at both site a and site b by ST3 with site a as the preferred cleavage site (Fig. 2). RT mutations at either site severely inhibited the cleavage by ST3 (Fig. 2), consistent with the effect of RT substitutions on the cleavage of human α1-PI by ST3 (41). On the other hand, when an F residue was introduced into the P3′ position of either site in the RT mutants to produce RTF-W and W-RTF, we found that the additional change had distinct effects on LR cleavage by ST3. The RTF mutations at site a inhibited the cleavage of this site by ST3 (Fig. 2, RTF-W), just like in human α1-PI (41). In contrast, W-RTF was cleaved at site b by ST3 as efficiently as W-W (Fig. 2, W-RTF). Thus, the introduction of an F residue at the P3′ position had different effects on the two cleavage sites, i.e., having little effect on the RT mutant of site a but reversing the inhibitory effect of RT at site b. In addition, while RT or RTF mutations at site a had little effect on the cleavage of LR by ST3 at site b, RT or RTF mutations at site b resulted in a 2 fold inhibition of cleavage at the site a (Fig. 2), indicating that long range interaction and/or protein conformation has distinct effect on the cleavage of these two sites by ST3.
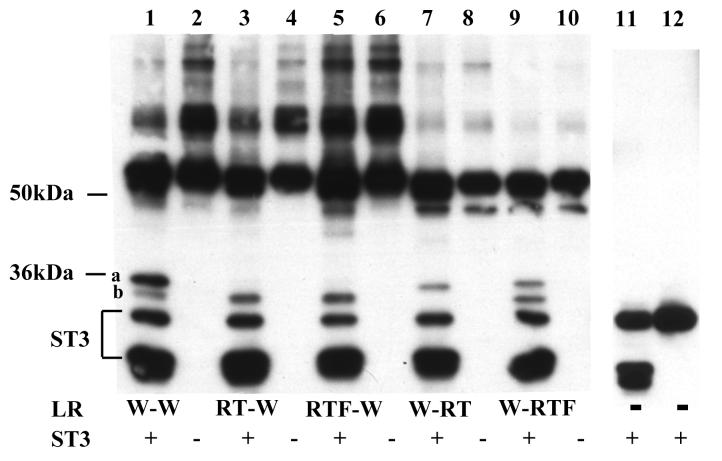
Fig. 2.
Differential effects of amino acid substitutions on the cleavage of the two sites in LR by ST3.
A. RT and RTF mutations at site a essentially abolished ST3 cleavage at this site (compare lanes 3 and 5, respectively, to lane 1). While RT mutation at site b also inhibited ST3 cleavage at this site (lane 7), an additional substitution of E to F at the P3′ position restored the ability of ST3 to cleave at this site (compare lane 1 to lane 9).
His-tagged wild type LR and indicated mutants were synthesized by in vitro translation, purified, and digested with purified Xenopus ST3 catalytic domain. The samples were mixed with SDS sample buffer and subjected to Western blotting with anti-Xenopus LR antibody. Note that the anti-LR antibody was made against His-tagged LR and also recognizes His-tagged ST3. During the incubation, ST3 auto degraded to produce the lower molecular weight bands that were present both in the presence (lanes 1, 3, 5, 7, 9) or absence of LR (11) but not in the ST3 sample without incubation (12) (lanes 11 and 12 were from a separate gel). The figure is a representative of 2 independent experiments with similar results.
B. Quantification of the data shown in Fig. A. Shown here is the average from 2 independent experiments.
Simultaneous mutations at both sites reveal interactions between the two cleavage sites in LR
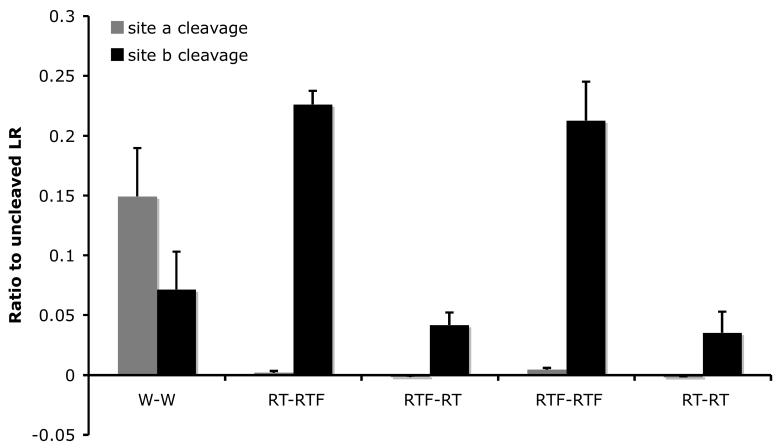
To further investigate the effects of the mutations on ST3 cleavage, we simultaneously mutagenized both cleavage sites and studied the cleavage of the mutant LR by ST3. As shown in Fig. 3, blocking the cleavage at site a with either RT or RTF mutations dramatically enhanced the cleavage of mutant site b with the RT or RTF substitutions (Fig. 3). Thus, while W-RT had little cleavage at site b (Fig. 2), RT-RT had about 50% of the wild type level of cleavage at site b (Fig. 3). Similarly, W-RTF had only about wild type levels of cleavage at site b (Fig. 2), but RT-RTF and RTF-RTF had nearly 3 times of the wild type levels of cleavage at site b (Fig. 3). These results indicate that when site a is inhibited, the cleavage at the mutant but not wild type site b by ST3 is enhanced. In contrast, when site b is inhibited by the RT mutations, the cleavage of the wild type site a is inhibited while the mutated site a remains non-cleavable.
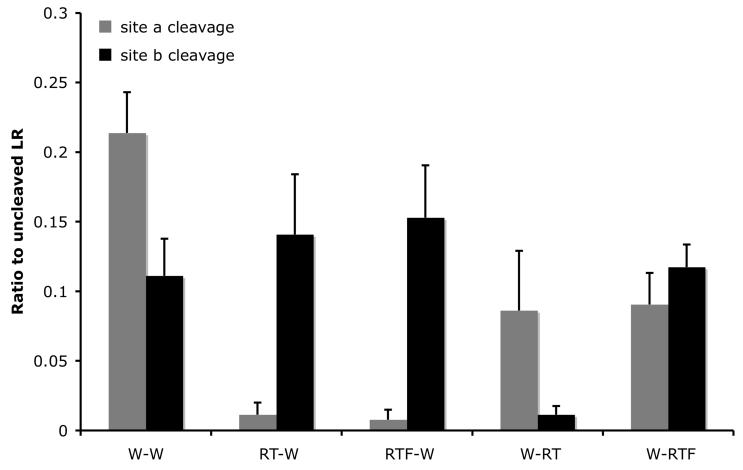
Fig. 3.
Mutations at site a enhance the cleavage of mutant site b by ST3. A. By inhibiting ST3 cleavage at site a with RT or RTF mutations, the cleavage at site b with RT or RTF mutations was enhanced compared to the wild-type LR (compare lane 1 to lanes 3 and 7) and to the single mutants (see Fig. 2).
His-tagged wild type LR and indicated mutants were synthesized by in vitro translation, purified, and digested with purified Xenopus ST3 catalytic domain. The samples were subjected to Western blotting with anti-Xenopus LR antibody. The figure is a representative of 2 independent experiments.
B. Quantification of the data shown in A. Shown here is the average from 2 independent experiments.
Surrounding sequence or conformation is important in the cleavage of the LR by ST3
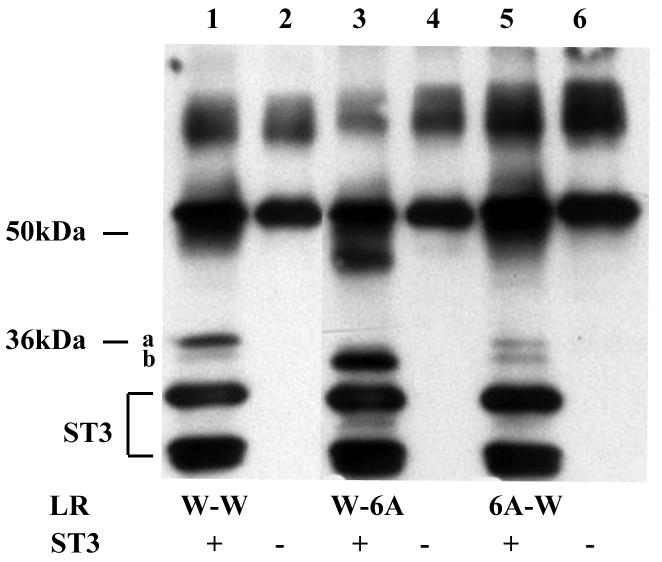
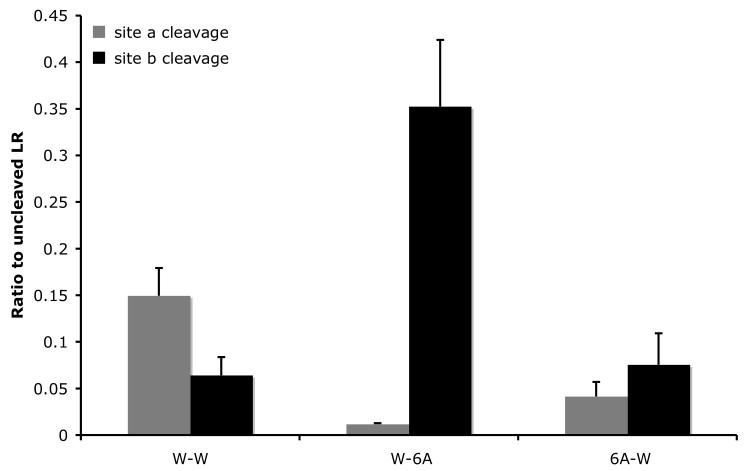
The differential effects of mutating one site on the cleavage of the wild type or mutant second site observed above suggest that the recognition of the cleavage site and/or efficiency of cleavage is influenced by surrounding sequence or conformation. To test this possibility, we substituted the six amino acids flanking the cleavage site at either site with alanines to produce 6A-W and W-6A, respectively. Analysis of the cleavage of the alanine substitution mutants by ST3 showed that surprisingly, 6 A residues in a row, when placed in the context of the LR cleavage sites, were cleaved right in the middle, judging by the identical sizes of the products from the cleavage of the mutants by ST3 as those observed with the wild type LR (Fig. 4, compare lane 3 or 5 to 1). This result suggests that the flanking sequence outside of P3-P3′ or protein conformation helps to determine the exact cleavage site by ST3 since the cleavage occurred between the 3rd and 4th A, the same location as in the wild type site a or b, but not in between any other two A residues. While the cleavage location was not affected by the alanine substitutions, the cleavage efficiency was altered in the mutants. The 6A substitution at site a inhibited the cleavage at this site by about 3 fold but did not affect the cleavage of the wild type site b in the same protein (Fig. 4) (similar to the lack of any effect on the cleavage of the wild type site b by RT mutations at site a, Fig. 2). In contrast, the 6A substitution at site b dramatically enhanced the cleavage at site b by ST3, such that no cleavage at site a was observed (Fig. 4, essentially only background signal was detected as shown in Fig. 4B) (again this effect of site b mutation on wild type site a cleavage by ST3 was similar to that observed with RT or RTF mutations at site b in Fig. 2).
Fig. 4.
Alanine substitution from P3-P3′ inhibits cleavage at site a but enhances that at site b. A. Six alanine (6A) were introduced to replace the 3 amino acids on either side of each cleavage site. His-tagged wild type LR and indicated mutants were synthesized by in vitro translation, purified, and digested with purified Xenopus ST3 catalytic domain. The samples were subjected to Western blotting with anti-Xenopus LR antibody. Note that all 6 lanes were run on the same gel but two lanes for a construct not relevant to this figure were removed. The figure is a representative of 2 independent experiments with similar results.
B. Quantification of the data shown in A. Shown here is the average from 2 independent experiments.
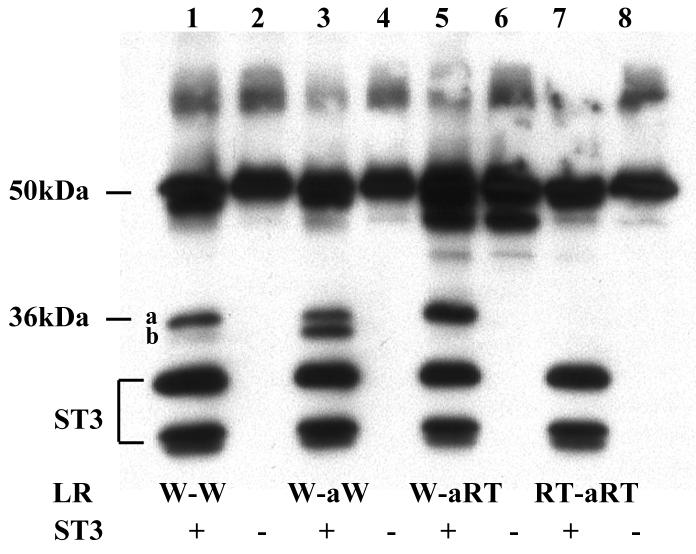
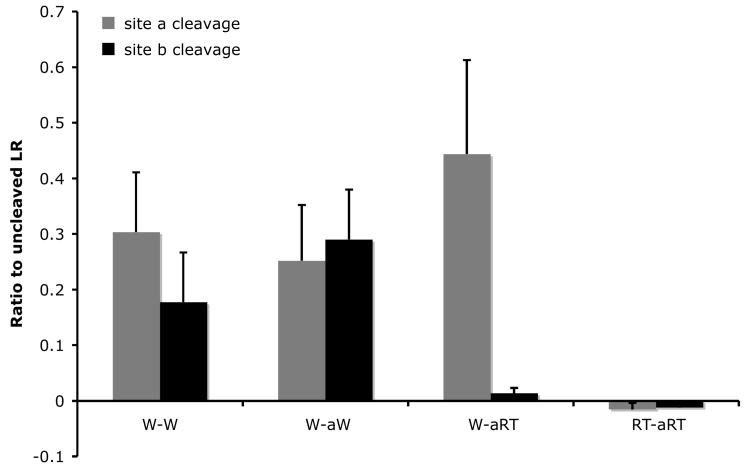
Our results above showed that site a was the preferred cleavage site in wild type LR and could be inhibited by both RT and RTF mutations regardless of the changes at site b. In contrast, site b cleavage could be inhibited by RT but not RTF mutations, and site a mutation enhanced the cleavage at mutant site b. In addition, one of our long-term goals is to generate a non-cleavable form of LR as a tool for studying the role of LR cleavage by ST3 in cell fate determination during development and pathogenesis. We, therefore, investigated the effect of substituting site b with site a. Thus, we replaced all 6 residues of site b from P3-P3′ in wild type LR with the corresponding sequence from wild type site a, or site a with RT mutations to produce the mutants W-aW or W-aRT, respectively (Fig. 1). In addition, we also introduced RT mutations into site a of W-aRT to produce RT-aRT, i.e., a mutant that had RT-mutated site a at both site a and site b locations (Fig. 1). The cleavage of these mutants by ST3 showed that when wild type site a was placed in the site b location, the resulting mutant was cleaved with similar efficiency as the site a in its original location in LR (Fig. 5, note that W-aW produced similar levels of the two products, generated from the cleavage from site a and site b location in LR, though both site now had site a sequence). When RT mutations were introduced into the site b location of W-aW to produce W-aRT (i.e., the RT mutation in the site a sequence now present at site b location), the cleavage of site b was inhibited (Fig. 5). Furthermore, when RT mutations were introduced into both sites of W-aW, the cleavage at both sites was blocked (Fig. 5, RT-aRT). This result is in contrast to that observed for RT-RT, where the RT mutations in site a led to an enhancement of the cleavage at site b where the site b sequence had the RT mutations (Fig. 3). Thus, in the context of site b, the sequence of site a played a dominant role in dictating its cleavage by ST3.
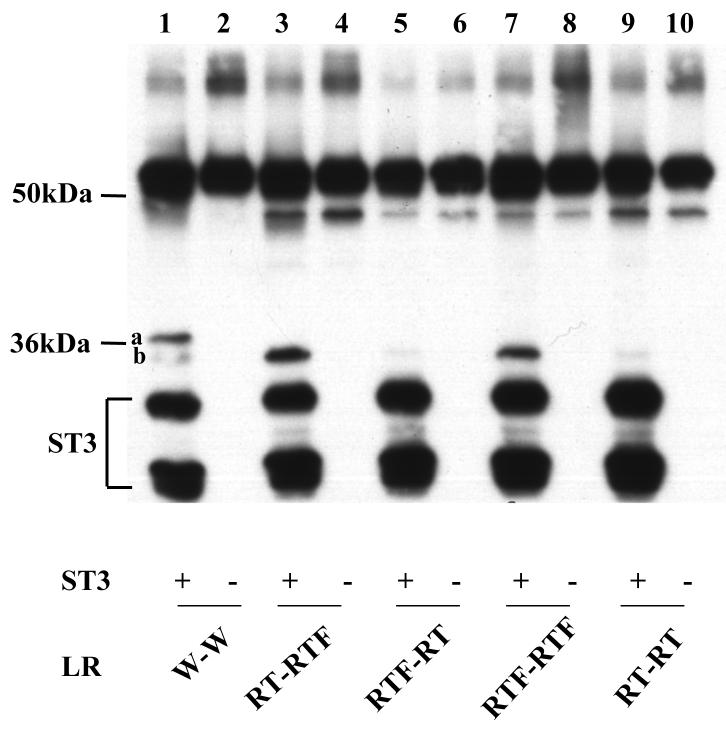
Fig. 5.
Substitution of the six amino acids at the cleavage site b with the corresponding ones at site a creates a cleavage site with site a characteristics. A. The 3 amino acids at either side of site b were replaced with the corresponding amino acids at site a to generate W-aW. RT mutations were then introduced into either one or both of the cleavage sites in W-aW to create the other mutants. His-tagged wild type LR and indicated mutants were synthesized by in vitro translation, purified, and digested with purified Xenopus ST3 catalytic domain. The samples were subjected to Western blotting with anti-Xenopus LR antibody. Note that RT mutation in the site b with the site a sequence completely blocked the cleavage by ST3 and RT mutation in site a failed to enhance the cleavage at this mutant site b (lane 7), in contrast to the corresponding mutants made from the wild type LR (Fig. 3, lane 9). The figure is a representative of 2 independent experiments with similar results.
B. Quantification of the data shown in A. Shown here is the average from 2 independent experiments.
Discussion
High levels of most MMPs are known to be associated with tumor invasion and metastasis (Tryggvason et al., 1987; Stetler-Stevenson et al., 1993; MacDaougall and Matrisian, 1995; Lochter and Bissell, 1999). In addition, during development, the expression of the mRNAs and proteins for various MMPs shows strong correlations with tissue remodeling and organogenesis in different animal species ranging from human to amphibians, although MMPs have little expression in most adult organs in mammals under normal physiological conditions (Matrisian and Hogan, 1990; Sang, 1998; Uria and Werb, 1998; Salamonsen and Woolley, 1999; Vu and Werb, 2000; Sarras et al., 2002). These findings suggest that MMPs play critical roles in the regulation of cell fate and behavior during developmental and pathological processes, although few in vivo functional studies are available.
ST3 was first isolated from a human breast tumor and has since been found to be expressed in a number of pathological processes, including most, if not all, human carcinomas (28-33). In addition, it is highly expressed in many developmental processes (13, 15, 19-22, 42-45). On the other hand, like other MMPs, the physiological and pathological roles of ST3 in vivo are largely unknown. By using thyroid hormone-dependent metamorphosis in Xenopus laevis as a model, we have previously shown in organ cultures and in developing tadpoles that ST3 is both necessary and sufficient at least for some aspects of intestinal metamorphosis, including the apoptosis of the larval epithelial cells (22, 23). We have further identified LR as a physiological substrate of ST3 in the intestine during metamorphosis (27, 46).
Compared to other MMPs, in vitro ST3 cleaves ECM proteins very weakly but cleaves a few non-ECM proteins such as α1-PI much more efficiently (47-49). In addition, a number of peptide substrates of ST3 have been isolated in vitro (Table 2) (50, 51). No consensus sequence for ST3 cleavage can be deduced from these substrates. Our results here suggest a possible underlying cause. This was made possible by the presence of two cleavage sites in LR. Mutations at the site a alone had little effect on the cleavage of wild type site b but enhanced the cleavage of site b with mutations. In contrast, mutations at site b reduced the cleavage at wild type site a and did not reverse the inhibitory effects of site a mutations on the cleavage at site a. These results indicate that mutations at one site can differentially affect the cleavage at the other site. Further structure and functional analyses are required to determine why the mutations at site a and site b have opposite effects on the cleavage at the other site. In addition to the interactions between the two sites, our analysis with the alanine substitution mutants demonstrates that flanking sequences and/or conformation help to dictate the exact cleavage site. In these mutants, there were 5 A-A bonds that may be cleaved by ST3 at site a or b. However, ST3 only cleaved the bond between the 3rd and 4th A based on the size of the cleavage products, i.e., the same location as in the wild type sites. In fact, for all mutations that could be cleaved by ST3, ST3 cleavage occurred at the same site as in the wild type site. That is, the mutations affected only the efficiency of the cleavage but the location, even in the case of 6A substitution, where only one of the 5 A-A bonds was cleaved. Thus, at least for the two cleavage sites in LR, the flanking sequence and/or conformation dictates where ST3 cleavage occurs. The combination of site-site interactions and the effects of flanking sequence and conformation may be the underlying cause for the inability to identify a simple consensus sequence for ST3 cleavage site. It argues, therefore, that it is important to identify and characterize substrates of ST3, and possibly other MMPs, in the contexts of full-length proteins.
Table 2.
Compilation of known ST3 cleavage site sequences.
| Good ST3 substrates | ||||
|---|---|---|---|---|
| P | P' | Reference (substrate) | ||
| 6 5 4 | 3 2 1 | 1 2 3 | 4 5 6 | |
| QAA | FRE | Amano et al 2005 (LR) | ||
| HQP | ITE | Amano et al 2005 (LR) | ||
| HQR | TTF | This study | ||
| RAD | AAA | AAA | ASY | This study |
| AGA | MFL | Pei et al 1994 (a1-PI) | ||
| VGF | YES | Pei et al 1994 (a2-macroglobulin) | ||
| ALH | VTN | Manes et al 1997 (IGFBP-1) | ||
| AE | LR | Pan et al. 2003 (peptide substrate) | ||
| GE | LR | Pan et al. 2003 (peptide substrate) | ||
| AAN | LVR | Pan et al. 2003 (peptide substrate) | ||
| AAN | LTR | Pan et al. 2003 (peptide substrate) | ||
| YAE | LRM | Pan et al. 2003 (peptide substrate) | ||
| GQA | YVK | Pan et al. 2003 (peptide substrate) | ||
| DnsPLA | L/M | Mucha et al. 1998 (peptide substrate) | ||
| DnsAAA | L/M | Mucha et al. 1998 (peptide substrate) | ||
| Poor/non-cleavable ST3 substrates | ||||
| P | P' | Reference (substrate) | ||
| 6 5 4 | 3 2 1 | 1 2 3 | 4 5 6 | |
| PLG | LYA | Pan et al. 2003 (peptide substrate) | ||
| PLA | LWA | Pan et al. 2003 (peptide substrate) | ||
| QPR | GVW | Pan et al. 2003 (peptide substrate) | ||
| TDA | WLS | Pan et al. 2003 (peptide substrate) | ||
| QAR | TRE | This study | ||
| QAR | TRF | This study | ||
| HQR | TTE | This study | ||
| NQI | AAA | AAA | PRL | This study |
MMPs have long been studied in a large part due to the high levels of MMP expression in cancerous but not normal tissues, suggesting the possibility of targeting MMP function for cancer treatment. Unfortunately, human clinical trials using various MMP inhibitors have yet to produce any positive results (52). One possible cause for this is that ECM remodeling/degradation is not the determining factor in tumor invasion and metastasis, since metastasis occurs after secondary genetic changes take place in the tumor cells. Inhibiting MMP activity may simply force tumor cells to find other means, e.g., employing other proteases, to get through the ECM barrier for metastasis. The fact that many MMPs can also cleave non-ECM substrates raises the possibility of targeting those non-ECM pathways for disease prevention and treatment. In this regard, the ST3 substrate LR is highly conserved from frog to human and both human and Xenopus LR can be cleaved by Xenopus ST3 at the same two conserved sites located between the transmembrane domain and laminin binding sequence in the extracellular half of the protein (27). In addition, LR, like ST3, is also highly expressed in cancers (25, 34-39), raising the possibility of targeting LR cleavage by ST3 to inhibit cancer development and metastasis. Thus, understanding how ST3 cleaves LR will help to determine if and how LR cleavage by ST3 contributes to human pathogenesis. In particular, the ST3-resistant LR mutants of should be powerful tools toward this end. It will be of interest in the future to determine if LR is cleaved by ST3 in human cancers and whether the LR mutants generated here can affect cancer development and tumor invasion.
Acknowledgement
We thank Dr. L. Notari and Dr. A. Antignani for suggestions on protein purification. This research was supported by the Intramural Research Program of NICHD, NIH.
References
- 1.Barrett JA, Rawloings ND, Woessner JF. Handbook of proteolytic enzymes. Academic Press; NY: 1998. [Google Scholar]
- 2.Alexander CM, Werb Z. Extracellular matrix degradation. In: Hay ED, editor. Cell Biology of Extracellular Matrix. 2nd ed. Plenum Press; New York: 1991. pp. 255–302. [Google Scholar]
- 3.Birkedal-Hansen H, Moore WGI, Bodden MK, et al. Matrix metalloproteinases: a review. Crit. Rev. in Oral Biol. and Med. 1993;4:197–250. doi: 10.1177/10454411930040020401. [DOI] [PubMed] [Google Scholar]
- 4.McCawley LJ, Matrisian LM. Matrix metalloproteinases: they're not just for matrix anymore! Current Opinion in Cell Biology. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 5.Parks WC, Mecham RP. Matrix metalloproteinases. Academic Press; New York: 1998. [Google Scholar]
- 6.Nagase H. Cell surface activation of progelatinase A (proMMP-2) and cell migration. Cell Res. 1998;8:179–86. doi: 10.1038/cr.1998.18. [DOI] [PubMed] [Google Scholar]
- 7.Pei D. Leukolysin/MMP25/MT6-MMP: a novel matrix metalloproteinase specifically expressed in the leukocyte lineage. Cell Res. 1999;9:291–303. doi: 10.1038/sj.cr.7290028. [DOI] [PubMed] [Google Scholar]
- 8.Murphy G, Stanton H, Cowell S, et al. Mechanisms for pro matrix metalloproteinase activation. Apmis. 1999;107:38–44. doi: 10.1111/j.1699-0463.1999.tb01524.x. [DOI] [PubMed] [Google Scholar]
- 9.Nagase J, Suzuki K, Morodomi T, Englhild JJ, Salvesen G. Activation Mechanisms of the Precursors of Matrix Metalloproteinases 1,2, and 3. Matrix Supple. 1992;1:237–244. [PubMed] [Google Scholar]
- 10.Kleiner DE, Jr., Stetler-Stevenson WG. Structural biochemistry and activation of matrix metalloproteases. Curr Opin Cell Biol. 1993;5:891–7. doi: 10.1016/0955-0674(93)90040-w. [DOI] [PubMed] [Google Scholar]
- 11.Pei D, Weiss SJ. Furin-dependent intracellular activation of the human stromelysin-3 zymogen. Nature. 1995;375:244–7. doi: 10.1038/375244a0. [DOI] [PubMed] [Google Scholar]
- 12.Sato H, Seiki M. Membrane-Type Matrix Metallproteinases (MT-MMPs) in Tumor Metastasis. J. Biochem. 1996;119:209–215. doi: 10.1093/oxfordjournals.jbchem.a021223. [DOI] [PubMed] [Google Scholar]
- 13.Uria JA, Werb Z. Matrix metalloproteinases and their expression in mammary gland. Cell Res. 1998;8:187–94. doi: 10.1038/cr.1998.19. [DOI] [PubMed] [Google Scholar]
- 14.Overall CM. Molecular determinants of metalloproteinase substrate specificity. Molecular Biotechnology. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y-B, Ishizuya-Oka A. Thyroid hormone regulation of apoptotic tissue remodeling: Implications from molecular analysis of amphibian metamorphosis. Progress in Nucleic Acid Research and Molecular Biology. 2001;65:53–100. doi: 10.1016/s0079-6603(00)65002-x. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y-B, Ishizuya-Oka A. Biphasic intestinal development in amphibians: Embryogensis and remodeling during metamorphosis. Current Topics in Develop. Biol. 1996;32:205–235. doi: 10.1016/s0070-2153(08)60429-9. [DOI] [PubMed] [Google Scholar]
- 17.Amano T, Fu L, Ishizuya-Oka A, Shi Y-B. Thyroid hormone induced apoptosis during amphibian metamorphosis. In: Shi Y, et al., editors. Molecular Mechanisms of Programmed Cell Death. Kluwer Academic/Plenum; New York: 2003. pp. 9–19. [Google Scholar]
- 18.Shi Y-B. Amphibian Metamorphosis: From morphology to molecular biology. John Wiley & Sons, Inc.; New York: 1999. [Google Scholar]
- 19.Damjanovski S, Ishizuya-Oka A, Shi YB. Spatial and temporal regulation of collagenases-3, -4, and stromelysin - 3 implicates distinct functions in apoptosis and tissue remodeling during frog metamorphosis. Cell Res. 1999;9:91–105. doi: 10.1038/sj.cr.7290009. [DOI] [PubMed] [Google Scholar]
- 20.Ishizuya-Oka A, Ueda S, Shi Y-B. Transient expression of stromelysin-3 mRNA in the amphibian small intestine during metamorphosis. Cell Tissue Res. 1996;283:325–9. doi: 10.1007/s004410050542. [DOI] [PubMed] [Google Scholar]
- 21.Patterton D, Hayes WP, Shi YB. Transcriptional activation of the matrix metalloproteinase gene stromelysin-3 coincides with thyroid hormone-induced cell death during frog metamorphosis. Dev Biol. 1995;167:252–62. doi: 10.1006/dbio.1995.1021. [DOI] [PubMed] [Google Scholar]
- 22.Ishizuya-Oka A, Li Q, Amano T, Damjanovski S, Ueda S, Shi Y-B. Requirement for matrix metalloproteinase stromelysin-3 in cell migration and apoptosis during tissue remodeling in Xenopus laevis. J Cell Biol. 2000;150:1177–88. doi: 10.1083/jcb.150.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu L, Ishizuya-Oka A, Buchholz DR, Amano T, Matsuda H, Shi YB. A causative role of stromelysin-3 in extracellular matrix remodeling and epithelial apoptosis during intestinal metamorphosis in Xenopus laevis. J Biol Chem. 2005;280:27856–65. doi: 10.1074/jbc.M413275200. [DOI] [PubMed] [Google Scholar]
- 24.Landowski TH, Dratz EA, Starkey JR. Studies of the Structure of the Metastasis-Associated 67 kDa Laminin Binding Protein: Fatty Acid Acylation and Evidence Supporting Dimerization of the 32 kDa Gene Product To Form the Mature Protein. Biochemistry. 1995;34:11276–11287. doi: 10.1021/bi00035a037. [DOI] [PubMed] [Google Scholar]
- 25.Menard S, Castronovo V, Tagliabue E, Sobel ME. New insights into the metastasis-associated 67 kD laminin receptor. Journal of Cellular Biochemistry. 1997;67:155–165. [PubMed] [Google Scholar]
- 26.Buto S, Tagliabue E, Ardini E, et al. Formation of the 67-kDa laminin receptor by acylation of the precursor. Journal of Cellular Biochemistry. 1998;69:244–251. doi: 10.1002/(sici)1097-4644(19980601)69:3<244::aid-jcb2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 27.Amano T, Kwak O, Fu L, Marshak A, Shi Y-B. The matrix metalloproteinase stromelysin-3 cleaves laminin receptor at two distinct sites between the transmembrane domain and laminin binding sequence within the extracellular domain. Cell Research. 2005;15:150–159. doi: 10.1038/sj.cr.7290280. [DOI] [PubMed] [Google Scholar]
- 28.Basset P, Bellocq JP, Lefebvre O, et al. Stromelysin-3: a paradigm for stroma-derived factors implicated in carcinoma progression. Crit Rev Oncol Hematol. 1997;26:43–53. doi: 10.1016/s1040-8428(97)00010-3. [DOI] [PubMed] [Google Scholar]
- 29.Lochter A, Bissell MJ. An odyssey from breast to bone: multi-step control of mammary metastases and osteolysis by matrix metalloproteinases. APMIS. 1999;107:128–36. doi: 10.1111/j.1699-0463.1999.tb01535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tetu B, Brisson J, Lapointe H, Bernard P. Prognostic significance of stromelysin 3, gelatinase A, and urokinase expression in breast cancer. Human Pathology. 1998;29:979–985. doi: 10.1016/s0046-8177(98)90204-0. [DOI] [PubMed] [Google Scholar]
- 31.Ahmad A, Hanby A, Dublin E, et al. Stromelysin 3: an independent prognostic factor for relapse-free survival in node-positive breast cancer and demonstration of novel breast carcinoma cell expression. Am. J. Pathol. 1998;152:721–8. [PMC free article] [PubMed] [Google Scholar]
- 32.Chenard MP, OSiorain L, Shering S, et al. High levels of stromelysin-3 correlate with poor prognosis in patients with breast carcinoma. International Journal of Cancer. 1996;69:448–451. doi: 10.1002/(SICI)1097-0215(19961220)69:6<448::AID-IJC5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 33.Anderson IC, Sugarbaker DJ, Ganju RK, et al. Stromelysin-3 is overexpressed by stromal elements in primary non-small-cell lung cancers and regulated by retinoic acid in pulmonary fibroblasts. Cancer Res. 1995;55:4120–4126. [PubMed] [Google Scholar]
- 34.Yow HK, Wong JM, Chen HS, et al. Increased mRNA expression of a laminin-binding protein in human colon carcinoma: Complete sequence of a full-length cDNA encoding the protein. Proc. Natl. Acad. Sci. USA. 1988;85:6394–6398. doi: 10.1073/pnas.85.17.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hand PH, Thor A, Schlom J, Rao CN, Liotta LA. Expression of laminin receptor in normal and carcinomatous human tissues as defined by a monoclonal antibody. Cancer Res. 1985;45:2713–2719. [PubMed] [Google Scholar]
- 36.Sanjuan X, Fernandez PL, Miquel R, et al. Overexpression of the 67-kD laminin receptor correlates with tumor progression in human colorectal carcinoma. J. Pathol. 1996;179:376–380. doi: 10.1002/(SICI)1096-9896(199608)179:4<376::AID-PATH591>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 37.Sobel ME. Differential expression of the 67 kDa laminin receptor in cancer. Semin. Cancer Biol. 1993;4:311–317. [PubMed] [Google Scholar]
- 38.Martignone S, Menard S, Bufalino R, et al. Prognostic significance of the 67-kilodalton laminin receptor expression in human breast carcinomas. J. Natl. Cancer Inst. 1993;85:398–402. doi: 10.1093/jnci/85.5.398. [DOI] [PubMed] [Google Scholar]
- 39.Castronovo V. Laminin receptors and laminin-binding proteins during tunor invasion and matastasis. Invasion Metastasis. 1993;13:1–30. [PubMed] [Google Scholar]
- 40.Damjanovski S, Amano T, Li Q, Pei D, Shi Y-B. Overexpression of Matrix Metalloproteinases Leads to Lethality in Transgenic Xenopus Laevis: Implications for Tissue-Dependent Functions of Matrix Metalloproteinases during Late Embryonic development. Dev. Dynamics. 2001;221:37–47. doi: 10.1002/dvdy.1123. [DOI] [PubMed] [Google Scholar]
- 41.Amano T, Fu L, Sahu S, Markey M, Shi Y-B. Substrate specificity of Xenopus matrix metalloproteinase stromelysin-3. International Journal of Molecular Medicine. 2004;14:233–239. [PubMed] [Google Scholar]
- 42.Lefebvre O, Regnier C, Chenard MP, et al. Developmental expression of mouse stromelysin-3 mRNA. Development. 1995;121:947–55. doi: 10.1242/dev.121.4.947. [DOI] [PubMed] [Google Scholar]
- 43.Chin JR, Werb Z. Matrix metalloproteinases regulate morphogenesis, migration and remodeling of epithelium, tongue skeletal muscle and cartilage in the mandibular arch. Development. 1997;124:1519–1530. doi: 10.1242/dev.124.8.1519. [DOI] [PubMed] [Google Scholar]
- 44.Basset P, Bellocq JP, Wolf C, et al. A novel metalloproteinase gene specifically expressed in stromal cells of breast carcinomas. Nature. 1990;348:699–704. doi: 10.1038/348699a0. [DOI] [PubMed] [Google Scholar]
- 45.Lefebvre O, Wolf C, Limacher JM, et al. The breast cancer-associated stromelysin-3 gene is expressed during mouse mammary gland apoptosis. J Cell Biol. 1992;119:997–1002. doi: 10.1083/jcb.119.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amano T, Fu L, Marshak A, Kwak O, Shi YB. Spatio-temporal regulation and cleavage by matrix metalloproteinase stromelysin-3 implicate a role for laminin receptor in intestinal remodeling during Xenopus laevis metamorphosis. Dev Dyn. 2005;234:190–200. doi: 10.1002/dvdy.20511. [DOI] [PubMed] [Google Scholar]
- 47.Murphy G, Segain J-P, O'Shea M, et al. The 28-kDa N-terminal domain of mouse stromelysin-3- has the general properties of a weak metalloproteinase. J. Biol. Chem. 1993;268:15435–15441. [PubMed] [Google Scholar]
- 48.Pei D, Majmudar G, Weiss SJ. Hydrolytic inactivation of a breast carcinoma cell-derived serpin by human stromelysin-3. J Biol Chem. 1994;269:25849–55. [PubMed] [Google Scholar]
- 49.Manes S, Mira E, Barbacid MD, et al. Identification of insulin-like growth factor-binding protein-1 as a potential physiological substrate for human stromelysin-3. J. of Biol. Chem. 1997;272:25706–25712. doi: 10.1074/jbc.272.41.25706. [DOI] [PubMed] [Google Scholar]
- 50.Pan W, Arnone M, Kendall M, et al. Identification of peptide substrates for human MMP-11 (stromelysin-3) using phage display. J Biol Chem. 2003;278:27820–7. doi: 10.1074/jbc.M304436200. [DOI] [PubMed] [Google Scholar]
- 51.Mucha A, Cuniasse P, Kannan R, et al. Membrane type-1 matrix metalloprotease and stromelysin-3 cleave more efficiently synthetic substrates containing unusual amino acids in their P1′ positions. J Biol Chem. 1998;273:2763–8. doi: 10.1074/jbc.273.5.2763. [DOI] [PubMed] [Google Scholar]
- 52.Coussens LM, Fingleton BM, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]