Abstract
Cell cycle regulation and biochemical responses upon nutrients and growth factors are the major regulatory mechanisms for cell sizing in mammals. Recently, we identified that the death effector domain-containing DEDD impedes mitotic progression by inhibiting Cdk1 (cyclin-dependent kinase 1) and thus maintains an increase of cell size during the mitotic phase. Here we found that DEDD also associates with S6 kinase 1 (S6K1), downstream of phosphatidylinositol 3-kinase, and supports its activity by preventing inhibitory phosphorylation of S6K1 brought about by Cdk1 during the mitotic phase. DEDD-/- cells showed reduced S6K1 activity, consistently demonstrating decreased levels in activating phosphorylation at the Thr-389 site. In addition, levels of Cdk1-dependent inhibitory phosphorylation at the C terminus of S6K1 were enhanced in DEDD-/- cells and tissues. Consequently, as in S6K1-/- mice, the insulin mass within pancreatic islets was reduced in DEDD-/- mice, resulting in glucose intolerance. These findings suggest a novel cell sizing mechanism achieved by DEDD through the maintenance of S6K1 activity prior to cell division. Our results also suggest that DEDD may harbor important roles in glucose homeostasis and that its deficiency might be involved in the pathogenesis of type 2 diabetes mellitus.
Cell size is closely related to specialized cell function and to the specific patterning of tissues in the body. Cell sizing is regulated mainly by two mechanisms: cell cycle control and the biochemical response to nutrients and/or growth factors (1–5). During cell cycle progression, both the G1 (which is believed to be dominant) and the G2 periods are important for cells to increase their volume (6–9). In addition, we recently provided evidence that the mitotic period (M phase) also influences cell size, through analysis of DEDD-deficient mice (10, 11). The DEDD molecule was initially described as a member of the death effector domain (DED)2-containing protein family (12). Although the absence of DEDD did not apparently influence progression of apoptosis (10), we found that during mitosis, DEDD is associated with Cdk1-cyclin B1 and that it decreases the kinase activity of Cdk1. This response impedes the Cdk1-dependent mitotic program to shut off synthesis of ribosomal RNA (rRNA) and protein and is consequently useful in gaining sufficient cell growth prior to cell division. Depletion of DEDD consistently results in a shortened mitotic duration and an overall reduction in the amount of cellular rRNA and protein and, furthermore, in cell and body size (10, 11).
Of the biochemical responses responsible for cell sizing, the signaling cascade involving phosphatydilinositol 3-kinase (PI3K) and its downstream target of rapamycin (TOR) is most crucial (13–15). In mammals, upon stimulation by growth factors, including insulin, the mammalian TOR (mTOR) cooperates with PI3K-dependent effectors to activate S6K1, thereby phosphorylating the 40 S ribosomal protein S6, and subsequently enhances translation of the 5′-terminal oligopyrimidine sequences that encode components of the translational machinery. This reaction increases the number of ribosomes and the efficacy of protein synthesis, thus critically promoting cell growth (16–18). Therefore, mice deficient for S6K1 (S6K1-/-) had reduced cell and body size (19–23). This effect also involves S6K1 in maintenance of glucose tolerance. S6K1 significantly supports the size of insulin-producing β cells within pancreatic Langerhans islets (24, 25). Thus, in S6K1-/- mice, the insulin mass was diminished, which resulted in ineffective secretion of insulin upon glucose administration (21, 23).
The activation of S6K1 proceeds through chronological phosphorylation at various residues, toward the crucial phosphorylation of Thr-389, present within the linker domain between the catalytic domain and the carboxyl tail, to obtain maximal enzymatic activity (26). Interestingly, phosphorylation at several Ser/Thr residues within the C-terminal autoinhibitory tail appears to either activate or inhibit S6K1, depending on the cell cycle phase. Shah et al. (27) demonstrated that phosphorylation of those residues (featured by the Thr-421/Ser-424 site) during mitosis pursued by Cdk1 inactivates S6K1 to terminate protein synthesis prior to cell division (28). A recent report by Schmidt et al. (29) demonstrating that phosphorylation of Thr-421/Ser-424 is specifically increased during the G2/M phase may also support the finding, whereas during the G1 phase, there is consensus that the phosphorylation at the autoinhibitory domain is requisite for S6K1 activation (26), as also recently demonstrated by Hou et al. (30), where the Cdk5 phosphorylates the Ser-411 site, leading to activation of S6K1. In contrast to such inhibitory regulation of S6K1 during mitosis, however, a recent report by Boyer et al. (31) sharply demonstrated that the activity of S6K1 peaks at mitosis, suggesting that S6K1 may also have some roles during the mitotic phase. If so, how its activity is supported against the inhibitory regulation caused by Cdk1 remains an open question.
Hence, the two observations above that both DEDD-/- and S6K1-/- situations decrease the efficacy of ribosome and protein synthesis, resulting in smaller cell and body size, and that mitotic Cdk1 has a functional interaction with both S6K1 and DEDD led us here to assess a possible role of DEDD in the context of the functional regulation of S6K1.
EXPERIMENTAL PROCEDURES
Mice—DEDD-/- mice (10) had been backcrossed to C57BL/6 (B6) for 17 generations before they were used in experiments. Mice were maintained under a specific pathogen-free condition.
Antibodies—Antibodies used for experiments are as follows: anti-S6K1 phosphorylated at Thr-421/Ser-424, anti-S6K1 phosphorylated at Thr-389, anti-total rpS6 (clone 54D2), anti-rpS6 phosphorylated at Ser-240/244, anti-total Akt (clone 11E7), anti-Akt phosphorylated at Thr-308 (clone 244F9) (all are from Cell Signaling Technology, Beverly, MA); anti-S6K1 phosphorylated at Ser-411 (clone SC-7983R), anti-α-tubulin and anti-insulin (clone H-86) (from Santa Cruz Biotechnology, Inc., Santa Cruz, CA); anti-cyclin B1 (clone GNS-11) and anti-total S6K1 (clone 16) (from BD Biosciences); anti-Hsp90 (clone SPA-830) and anti-Cdk1 (clone A17) (from Stressgen (Victoria, Canada) and Zymed Laboratories Inc. (South San Francisco, CA)).
S6K1 Kinase Assay—DEDD+/+ or DEDD-/- mouse embryonic fibroblast (MEF) cells were lysed in lysis buffer (20 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm Na3VO4, 50 mm NaF, 5 mm EDTA, 0.1% Nonidet P-40, supplemented with a mixture of protease inhibitors). The cell lysates were clarified by centrifugation for 30 min at 14,000 rpm and immunoprecipitated with anti-S6K1 antibody preabsorbed to protein G-agarose beads at 4 °C for 4 h. The immune complexes were washed twice with lysis buffer and once with kinase assay buffer (20 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 0.1 mg/ml bovine serum albumin, and 0.4 mm dithiothreitol). The kinase reaction was performed at 30 °C for 15 min in the presence of 100 μm ATP, 200 μCi/ml [γ-32P]ATP, and 125 μm S6 peptide substrate (RRRLSSLRA; Upstate Biotechnology Inc., Lake Placid, NY) and was terminated by the addition of 20 μl of 250 mm EDTA and boiling for 5 min. Stopped reactions were loaded onto P-81 phosphocellulose membrane (Whatman, Maidstone, UK) and were washed with 75 mm phosphoric acid. The labeled probe was measured by liquid scintillation counting.
siRNA Transfection—Double-stranded siRNA targeting DEDD or Cdk1 were purchased from Applied Biosystems or Sigma, respectively. Wild-type MEF cells at a 50% confluent state were transfected with 10 μm siRNA using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, the cells were harvested and analyzed by Western blotting or reverse transcription-PCR. Sequences of the oligonucleotides were as follows: DEDD siRNA 1, 5-GCCCTGATCTTGTAGACAATT-3; DEDD siRNA 2, 5-AAATGGACGTGACTTCTTATT-3; Cdk1 siRNA 1, 5-CTATGATCCTGCCAAACGATT-3; Cdk1 siRNA 2, 5-GTTGTTACCGTTGGCTCTTT-3; Cdk1 siRNA 3, 5-CAATCAAACTGGCTGATTTTT-3. As a control, an oligonucleotide targeting the GFP sequence (Sigma) was used.
In Vitro Binding Assay—Glutathione S-transferase (GST)-fusion proteins containing mouse DEDD, Cdk1, cyclin B1, or S6K1 were produced in Escherichia coli M15 carrying pGEX-5X1-DEDD, Cdk1, cyclin B1, or S6K1. These proteins were purified according to protocols described in the GST·Bind kit instructions (Novagen, Madison, WI). V5-DEDD, Myc-cyclin B1, or HA-Cdk1 was efficiently expressed in the baculovirus/High Five cell system (Invitrogen). In each binding assay, 200 ng of GST fusion proteins or 100 μl of High Five cell lysates were used. GST fusion proteins or High Five cell extracts were incubated with the anti-S6K1 or anti-cyclin B1 antibodies in lysis buffer (20 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 1 mm Na3VO4, 25 mm NaF, 1 mm phenylmethylsulfonyl fluoride, 0.1% Nonidet P-40, supplemented with a mixture of protease inhibitors) at 4 °C for 4 h and precipitated with protein G-Sepharose beads (Sigma). The pellets were washed three times with lysis buffer. The immunoprecipitates and total cell lysates were analyzed by Western blotting using anti-DEDD antibody. Note that the anti-DEDD antibody was newly generated by us and that this antibody is available for recognition of recombinant DEDD, but not of endogenous DEDD by immunoblotting.
Islet Isolation and Insulin Content—Isolation of islets from mice and assessment of insulin content within islet cells were carried out as described previously (32). Briefly, after clamping the common bile duct at a point close to its opening into the duodenum, liberase RI (Roche Applied Science) was injected into the pancreatic duct. The swollen pancreas was removed and incubated at 37 °C for 24 min. The pancreas was dispersed by pipetting, and dispersed cells were washed twice. Islets were manually collected through a stereoscopic microscope. These islets were used for the experiments immediately after isolation. Insulin content was measured as described previously (32). Isolated islets were extracted in acid ethanol at -20 °C, and after preincubation with the basal glucose concentration for 20 min, static incubation of 10 islets/tube was performed at 37 °C for 1 h. Insulin levels were determined with an insulin enzyme-linked immunosorbent assay kit (Morinaga).
Primers for Reverse Transcription-PCR—Primers used are as follows: Hsp90α, 5-GCGGCAAAGACAAGAAAAAG-3 (forward) and 5-CAAGTGGTCCTCCCAGTCAT-3 (reverse); Hsp90β, 5-CTGGGTCAAGCAGAAAGGAG-3 (forward) and 5-TCTCTGTTGCTTCCCGACTT-3 (reverse); Akt1, 5-CCACGCTACTTCCTCCTC-3 (forward) and 5-TGCCCTTGCCAACAGTCTGAAGCA-3 (reverse); Akt2, 5-GTCGCCAACAGTCTGAAGCA-3 (forward) and 5-GAGAGAGGTGGAAAAACAGC-3 (reverse); G3PDH, 5-ACCACAGTCCATGCCATCAC-3 (forward) and 5-TCCACCACCCTGTTGCTGTA-3 (reverse); β-actin, 5-GTGGCTACAGCTTCACCACCACAG-3 (forward) and 5-GCATCCTGTCAGCAATGCCTGGGT-3 (reverse); DEDD, 5-GCGGGATCCGCGGGCCTAAAGAGGC-3 (forward) and 5-GCGTCTAGAGTCTACAAGATCAGGGC-3 (reverse).
Quantification of Immunoblots—Quantification of the immunoblots was performed using NIH Image. Relative phosphorylation levels to those in control (shown as 1.0 ± S.E.) are presented. For all immunoblotting experiments, at least three independent blots were performed.
Statistical Analysis—A two-tailed Mann-Whitney test was used to calculate p values. **, p < 0.01; *, p < 0.05; error bars, S.E.
RESULTS AND DISCUSSION
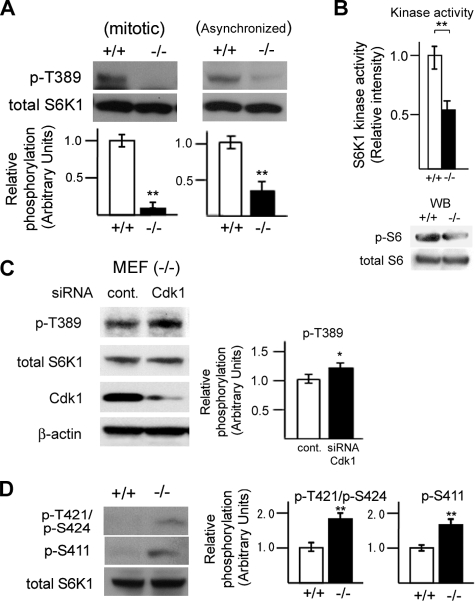
DEDD Prevents Inhibitory Phosphorylation of Mitotic S6K1 and Supports Its Activity—To test if DEDD is involved in regulation of S6K1 activity, we first investigated whether the lack of DEDD influences S6K1 activity. Significantly, levels of phosphorylation at Thr-389 of S6K1, a hallmark of active S6K1, was attenuated in DEDD-/- compared with DEDD+/+ MEF cells that had been enriched in the mitotic phase by a nocodazole block (Fig. 1A, left). Interestingly, reduction in Thr-389 phosphorylation was also observed in asynchronized DEDD-/- MEF cells (Fig. 1A, right). Thus, such a DEDD activating effect on S6K1 at the mitotic phase appears to influence the overall S6K1 activity in cells. Furthermore, a kinase activity assay based on the incorporation of 32P-labeled ATP demonstrated that S6K1 precipitated from DEDD-/- MEF cells had 50% less activity on a specific substrate of S6K1, rpS6, compared with that from DEDD+/+ cells (442.54 ± 30.79 (-/-) versus 850.67 ± 26.63 (+/+) cpm normalized by the amount of precipitated S6K1 protein; Fig. 1B (top) shows a summary of data). This result was also supported by a reduction in phosphorylation levels of rpS6 in DEDD-/- compared with DEDD+/+ MEF cells, when assessed by Western blotting (Fig. 1B, lower panels). The reduction of the rpS6 phosphorylation level in the absence of DEDD was less remarkable than expected by the result from the kinase activity assay. This might be due to a possible functional redundancy caused by S6K2 in the phosphorylation of rpS6 (33).
FIGURE 1.
Absence of DEDD decreases S6K1 activity. A, phosphorylation (p-) at Thr-389 of S6K1 in DEDD+/+ or DEDD-/- MEF cells, tested by Western blotting. B (top), kinase activity of S6K1 precipitated from DEDD+/+ or DEDD-/- MEF cells. Results were normalized by the average of those for DEDD+/+ cells. Error bar, S.E. Bottom, phosphorylation at rpS6 in DEDD+/+ or DEDD-/- MEF cells, tested by Western blotting (WB). C, knockdown of Cdk1 increased phosphorylation at Thr-389 of S6K1 in mitotic DEDD-/- MEF cells. D, phosphorylation at Thr-421/Ser-424 and Ser-411 of S6K1 in mitotic DEDD+/+ or DEDD-/- MEF cells.
Since DEDD suppresses mitotic Cdk1 (10), increased Cdk1 activity in the absence of DEDD might enhance the inhibitory regulation of S6K1, leading to less S6K1 activity in DEDD-/- cells. Supporting this hypothesis, suppression of Cdk1 via siRNA expression increased Thr-389 phosphorylation in DEDD-/- cells (Fig. 1C). We then assessed the phosphorylation status of the mitosis-specific inhibitory residue of S6K1, Thr-421/Ser-424, which is targeted by mitotic Cdk1. As presented in Fig. 1D, phosphorylation at Thr-421/Ser-424 was enhanced in mitotic DEDD-/- MEF cells compared with DEDD+/+ MEF cells. In brief, in the absence of DEDD, the activity of S6K1 was substantially diminished due to hyperphosphorylation at the inhibitory Ser/Thr residues. The phosphorylation level at Ser-411 (also within the autoinhibitory tail) was also increased in DEDD-/- cells (Fig. 1D). Although this result is consistent with the observation by Shah et al. (28) suggesting the presence of multiple Cdk1-dependent inhibitory phosphorylation sites, whether the mitotic phosphorylation at Ser-411 certainly decreases S6K1 activity remains arguable.
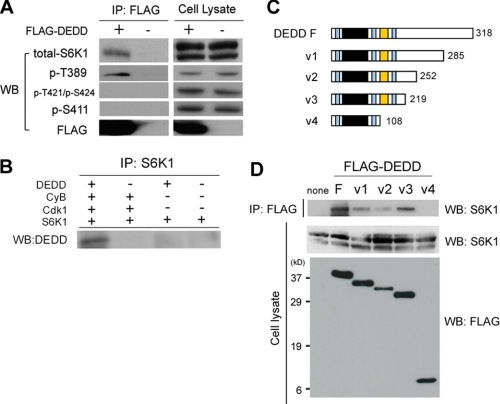
DEDD Associates with S6K1 through Its Proline-rich Region—We then tested whether DEDD associates with S6K1. We expressed FLAG-tagged DEDD in 293T cells and assessed whether S6K1 is co-precipitated with DEDD. Remarkably, S6K1 was bound to DEDD (Fig. 2A). Moreover, S6K1 co-precipitated with DEDD was deficient in phosphorylation at both Thr-421/Ser-424 and Ser-411, whereas the Thr-389 site was phosphorylated, suggesting that the association of DEDD with S6K1 prevents inhibitory phosphorylation of S6K1 that is caused by mitotic Cdk1 (Fig. 2A). An in vitro assay using tagged recombinant proteins supported the notion that DEDD associates with S6K1 through Cdk1-cyclin B1, since DEDD was co-precipitated with S6K1 only in the presence of both Cdk1 and cyclin B1 (Fig. 2B).
FIGURE 2.
DEDD forms a complex with S6K1 and Cdk1 via its proline-rich region. A, co-immunoprecipitation (IP) of FLAG-tagged DEDD (FLAG-DEDD) and S6K1, using anti-FLAG antibody. Precipitates were analyzed for the indicated items. Western blots (WB) of cell lysates for respective subjects are also presented. Note in 293T cell lysates, protein blot for total S6K1 using a mouse monoclonal antibody (clone 16; BD Biosciences) reveals double bands, as previously described (40, 41). B, in vitro binding assay using recombinant proteins. C, schematic diagram of the structure of DEDD variants. Light blue region, nuclear localizing signal; black region, DED domain; orange region, proline-rich region. The numbers indicate amino acids starting from the first methionine. D, co-immunoprecipitation of S6K1 and FLAG-tagged DEDD variants, using an anti-FLAG antibody.
Next, in order to determine the region involved in the association of DEDD with S6K1, we designed a number of variant DEDD molecules (Fig. 2C) and tested their association with S6K1. As depicted in Fig. 2D, a DEDD variant lacking the proline-rich region did not bind to S6K1, indicating a requirement of the proline-rich region of DEDD for the association with S6K1. Note that in multiple experiments, the expression level of the V2 DEDD variant was lower compared with that of other types when expressed in 293 cells (Fig. 2D, bottom). It is possible that the V2 DEDD variant may be structurally unstable.
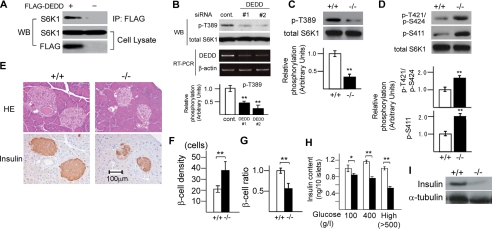
Reduction in β Cell Size and Insulin Mass Due to Decreased S6K1 Activity in DEDD-/- Pancreas—S6K1 is involved in control of glucose tolerance by supporting the size of insulin-producing β cells in the pancreas (21). As shown in Fig. 3A, the association of DEDD with S6K1 was certainly observed in β cells when tested using a mouse pancreatic β cell line, MIN6 (34). In line with this finding, activating phosphorylation of S6K1 at Thr-389 was significantly decreased in these cells when DEDD was knocked down by siRNA expression (Fig. 3B).
FIGURE 3.
Decreased insulin mass within pancreatic islets in DEDD-/- mice. A, co-immunoprecipitation (IP) of FLAG-DEDD and S6K1 in MIN6 cell lysates. B, DEDD knockdown decreased phosphorylation levels at the Thr-389 residue of S6K1 in MIN6 cells. Down-regulation of DEDD mRNA expression is also shown (reverse transcription-PCR; RT-PCR). C and D, phosphorylation at Thr-389 residue (C) or Thr-421/Ser-424 and Ser-411 (D) of S6K1 in DEDD+/+ or DEDD-/- pancreas. E, histological analysis of pancreatic Langerhans islets. Top, HE staining; bottom, immunostaining for insulin. F, β cell density assessed by the number of nuclei per 2.5 × 103 μm2 within the insulin-positive area. Ten independent islets for each genotype were analyzed. G, the ratio of insulin-stained area per whole islet. Twenty islets for each type of mice were analyzed. Results were normalized by the average of those for DEDD+/+ mice. H, insulin content. Islets were isolated from DEDD+/+ or DEDD-/- pancreas (four for each), and the insulin content within islets was analyzed. Data are averages of five groups of 10 islets. Error bar, S.E. The concentration of glucose used for incubation was 100 or 400 g/liter or higher (High). I, Western blotting (WB) for insulin using pancreas protein.
Also in vivo, diminishment of S6K1 phosphorylation at the Thr-389 site was apparent in the DEDD-/- pancreas compared with the DEDD+/+ pancreas (Fig. 3C). Furthermore, the DEDD-/- pancreas also exhibited an increase in phosphorylation levels at Thr-421/Ser-424 and Ser-411 residues, as seen in MEF cells (Fig. 3D). We then assessed whether pancreatic islets were smaller in DEDD-/- mice than in DEDD+/+ mice, as seen in S6K1-/- mice. According to histological analysis, the size of each β cell was reduced in DEDD-/- mice (Fig. 3E, top two panels), as also indicated by a higher density of β cells within the islet (Fig. 3F). Similarly, the insulin mass, determined by the ratio of β cells to whole islet mass after immunostaining of pancreatic sections for insulin, was reduced by 50% in DEDD-/- mice (Fig. 3, E (bottom two panels) and G). In agreement with this observation, insulin content within β cells from DEDD-/- mice was significantly decreased when assessed by in vitro measurement using isolated pancreatic islets (Fig. 3H). Furthermore, Western blot analysis demonstrated that the amount of insulin in the pancreas of DEDD-/- mice was decreased compared with that in the DEDD+/+ pancreas (Fig. 3I). Finally, Tdd-mediated dUTP-biotin nick-end labeling staining of the pancreas sections did not reveal any difference in DEDD-/- and DEDD+/+ islets, suggesting no influence of the lack of DEDD on apoptosis of islet cells (data not shown).
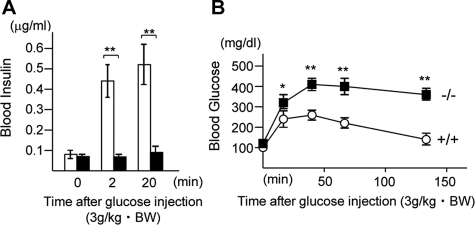
Attenuated Insulin Secretion upon Glucose Stimulation in DEDD-/- Mice—Last, we assessed insulin secretion and glucose tolerance in DEDD-/- mice in vivo. To this end, we first kinetically analyzed the blood insulin levels in response to glucose administration after mice were fasted for 2 h. As expected from the overt reduction in β cell size and insulin content (presented in Fig. 3), insulin levels were less elevated at 2 or 20 min after the glucose challenge in DEDD-/- mice compared with DEDD+/+ mice (Fig. 4A). Such inefficient insulin secretion in DEDD-/- mice resulted in glucose intolerance. As shown in Fig. 4B, at early times after the glucose challenge, blood glucose levels were more than 150% higher in DEDD-/- than in DEDD+/+ mice, although they were comparable in both types of mice before glucose administration (Fig. 4B). Two hours after the injection of glucose, when the blood glucose level decreased to the initial level in DEDD+/+ mice, it was still twice as high as that under the fasted condition in DEDD-/- mice (Fig. 4B). Thus, like in S6K1-/- mice, DEDD-/- mice suffered glucose intolerance due to inefficient insulin secretion upon glucose stimulation, which is accounted for by a reduction in insulin mass in pancreatic islets.
FIGURE 4.
Glucose intolerance in DEDD-/- mice. A, serum insulin levels after a glucose challenge. n = 5 each. Black bars, DEDD-/- mice; white bars, DEDD+/+ mice. B, blood glucose levels after glucose administration (3 g/kg body weight (BW)). Male DEDD-/- and their littermate DEDD+/+ mice at 6–8 weeks of age were used. n = 8 each. Black boxes, DEDD-/- mice; white circles, DEDD+/+ mice.
Conclusion—In this report, we demonstrated that DEDD is required for preservation of S6K1 activity during mitosis and that this reaction increases overall S6K1 activity in cells. This finding may suggest that the maintenance of mitotic S6K1 activity by the time of cytokinesis appears to be important to gain efficient cell and body size in mammals. This scenario is consistent with the observation by Boyer et al. (31), in which S6K1 activity achieves the maximal levels in the mitotic phase. As in our previous finding (10), DEDD appears to be involved in cell growth control prior to cell division via different mechanisms (i.e. impediment of mitosis progression and maintenance of S6K1 activity). Importantly, both effects are achieved through suppression of mitotic Cdk1, although further studies are required to fully understand the mechanism of how DEDD suppresses mitotic Cdk1 activity. Nonetheless, such new insights may further implicate the mitotic phase during the cell cycle as a crucial period involved in balancing mammalian cell size.
Although the DED-containing molecules were initially supposed to be involved in apoptosis regulation, evidence has accumulated that they have diverse functions (35, 36). For instance, Arechiga et al. (37) reported that FADD and caspase-8, also DED-containing family members, play essential roles in maintaining S6K1 activity during G1/S phases by modulating Cdk2 in T cells. Likewise, RIP1 (receptor-interacting protein 1), also a DED-containing kinase that mediates NFκB activation, regulates p27Kip1 levels through the PI3K-Akt-forkhead pathway, thereby promoting the G1/S transition (38). Together with our findings of the effects of DEDD in the mitotic phase, these observations may corroborate that the DED-containing elements appear to be widely involved in the control of the PI3K-signaling cascade as well as in the progression of different cell cycle phases. Because the PI3K pathway is crucial in the regulation of cell metabolisms, further studies might investigate the role of DED-containing proteins in various metabolic diseases. Since DEDD-/- mice revealed glucose intolerance, it might be worthwhile to assess whether any dysfunction of DEDD is present, either in the whole body or in specific tissues, in a subset of type 2 diabetes patients. Schumann et al. (39) reported that the Fas pathway, where many DED-containing elements are associated, is involved in β cell secretory function. Thus, DED-containing proteins might also influence metabolic homeostasis through apoptosis cascades, although DEDD-/- cells or mice showed no apparent defect in apoptosis (10, 11). Future analyses will clarify this issue more precisely.
Acknowledgments
We thank Drs. I. Takamoto, T. Kubota, M. Tsunoda, M. Kaneko, and Genostaff Inc. (in particular, K. Ohkubo) and Tosoh Corp. for technical assistance; Dr. J.-I. Miyazaki for MIN6 cells; and M. Egami for preparing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 5RO1AI50948-05. This work was also supported by grants-in-aid from the Ministry of Education, Sports, Culture, Science, and Technology, Japan, the Takeda Science Foundation (to T. M. and S. A.), the Mitsubishi Pharma Research Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Danone Institute of Nutrition for Health, the Uehara Memorial Foundation (to T. M.), the Kanae Foundation for the Promotion of Medical Science, and the Kanzawa Medical Research Foundation (to S. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DED, death effector domain; rRNA, ribosomal RNA; S6K1, S6 kinase 1; PI3K, phosphatidylinositol 3-kinase; TOR, target of rapamycin; mTOR, mammalian TOR; MEF, mouse embryonic fibroblast; siRNA, small interfering RNA; GST, glutathione S-transferase; rpS6, S6 ribosomal protein.
References
- 1.Conlon, I., and Raff, M. (1999) Cell 96 235-244 [DOI] [PubMed] [Google Scholar]
- 2.Montagne, J. (2000) Mol. Cell. Biol. Res. Commun. 4 195-202 [DOI] [PubMed] [Google Scholar]
- 3.Cooper, S. (2004) BMC Cell Biol. 5 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorgensen, P., and Tyers, M. (2004) Curr. Biol. 14 1014-1027 [DOI] [PubMed] [Google Scholar]
- 5.De Virgilio, C., and Loewith, R. (2006) Oncogene 25 6392-6415 [DOI] [PubMed] [Google Scholar]
- 6.Jorgensen, P., Nishikawa, J. L., Breitkreutz, B. J., and Tyers, M. (2002) Science 297 395-400 [DOI] [PubMed] [Google Scholar]
- 7.Rupes, I. (2002) Trends Genet. 18 479-485 [DOI] [PubMed] [Google Scholar]
- 8.Kellogg, D. R. (2003) J. Cell Sci. 116 4883-4890 [DOI] [PubMed] [Google Scholar]
- 9.Jorgensen, P., Rupes, I., Sharom, J. R., Schneper, L., Broach, J. R., and Tyers, M. (2004) Genes Dev. 18 2491-2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arai, S., Miyake, K., Voit, R., Nemoto, S., Wakeland, E. K., Grummt, I., and Miyazaki, T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2289-2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyazaki, T., and Arai, S. (2007) Cell Cycle 6 1419-1425 [PubMed] [Google Scholar]
- 12.Stegh, A. H., Schickling, O., Ehret, A., Scaffidi, C., Peterhänsel, C., Hofmann, T. G., Grummt, I., Krammer, P. H., and Peter, M. E. (1998) EMBO J. 17 5974-5986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oldham, S., and Hafen, E. (2003) Trends Cell Biol. 13 79-85 [DOI] [PubMed] [Google Scholar]
- 14.Valentinis, B., and Baserga, R. (2001) Mol. Pathol. 54 133-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogt, P. K. (2001) Trends Mol. Med. 7 482-484 [DOI] [PubMed] [Google Scholar]
- 16.Huang, S., and Houghton, P. J. (2003) Curr. Opin. Pharmacol. 3 371-377 [DOI] [PubMed] [Google Scholar]
- 17.Shamji, A. F., Nghiem, P., and Schreiber, S. L. (2003) Mol. Cell 12 271-280 [DOI] [PubMed] [Google Scholar]
- 18.Fingar, D. C., Salama, S., Tsou, C., Harlow, E., and Blenis, J. (2002) Genes Dev. 16 1472-1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shima, H., Pende, M., Chen, Y., Fumagalli, S., Thomas, G., and Kozma, S. C. (1998) EMBO J. 17 6649-6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montagne, J., Stewart, M. J., Stocker, H., Hafen, E., Kozma, S. C., and Thomas, G. (1999) Science 285 2126-2129 [DOI] [PubMed] [Google Scholar]
- 21.Pende, M., Kozma, S. C., Jaquet, M., Oorschot, V., Burcelin, R., Le Marchand-Brustel, Y., Klumperman, J., Thorens, B., and Thomas, G. (2000) Nature 408 994-997 [DOI] [PubMed] [Google Scholar]
- 22.Fingar, D. C., and Blenis, J. (2004) Oncogene 23 3151-3171 [DOI] [PubMed] [Google Scholar]
- 23.Ohanna, M., Sobering, A. K., Lapointe, T., Lorenzo, L., Praud, C., Petroulakis, E., Sonenberg, N., Kelly, P. A., Sotiropoulos, A., and Pende, M. (2005) Nat. Cell Biol. 7 286-294 [DOI] [PubMed] [Google Scholar]
- 24.Um, S. H., D'Alessio, D., and Thomas, G. (2006) Cell Metab. 3 393-402 [DOI] [PubMed] [Google Scholar]
- 25.Dann, S. G., Selvaraj, A., and Thomas, G. (2007) Trends Mol. Med. 13 252-259 [DOI] [PubMed] [Google Scholar]
- 26.Pullen, N., and Thomas, G. (1997) FEBS Lett. 410 78-82 [DOI] [PubMed] [Google Scholar]
- 27.Papst, P. J., Sugiyama, H., Nagasawa, M., Lucas, J. J., Maller, J. L., and Terada, N. (1998) J. Biol. Chem. 273 15077-15084 [DOI] [PubMed] [Google Scholar]
- 28.Shah, O. J., Ghosh, S., and Hunter, T. (2003) J. Biol. Chem. 278 16433-16442 [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, T., Wahl, P., Wüthrich, R. P., Vogetseder, A., Picard, N., Kaissling, B., and Le Hir, M. (2007) Histochem. Cell Biol. 127 123-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou, Z., He, L., and Qi, R. Z. (2007) J. Biol. Chem. 282 6922-6928 [DOI] [PubMed] [Google Scholar]
- 31.Boyer, D., Quintanilla, R., and Lee-Fruman, K. K. (2007) Mol. Cell. Biochem. 307 59-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubota, N., Tobe, K., Terauchi, Y., Eto, K., Yamauchi, T., Suzuki, R., Tsubamoto, Y., Komeda, K., Nakano, R., Miki, H., Satoh, S., Sekihara, H., Sciacchitano, S., Lesniak, M., Aizawa, S., Nagai, R., Kimura, S., Akanuma, Y., Taylor, S. I., and Kadowaki, T. (2000) Diabetes 49 1880-1889 [DOI] [PubMed] [Google Scholar]
- 33.McMullen, J. R., Shioi, T., Zhang, L., Tarnavski, O., Sherwood, M. C., Dorfman, A. L., Longnus, S., Pende, M., Martin, K. A., Blenis, J., Thomas, G., and Izumo, S. (2004) Mol. Cell. Biol. 24 6231-6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazaki, J., Araki, K., Yamato, E., Ikegami, H., Asano, T., Shibasaki, Y., Oka, Y., and Yamamura, K. (1990) Endocrinology 127 126-132 [DOI] [PubMed] [Google Scholar]
- 35.Tibbetts, M. D., Zheng, L., and Lenardo, M. J. (2003) Nat. Immunol. 4 404-409 [DOI] [PubMed] [Google Scholar]
- 36.Gudur Valmiki, M., and Ramos, J. W. Cell. Mol. Life Sci., in press [DOI] [PMC free article] [PubMed]
- 37.Arechiga, A. F., Bell, B. D., Leverrier, S., Weist, B. M., Porter, M., Wu, Z., Kanno, Y., Ramos, S. J., Ong, S. T., Siegel, R., and Walsh, C. M. (2007) J. Immunol. 179 5291-5300 [DOI] [PubMed] [Google Scholar]
- 38.Park, S., Ramnarain, D. B., Hatanpaa, K. J., Mickey, B. E., Saha, D., Paulmurugan, R., Madden, C. J., Wright, P. S., Bhai, S., Ali, M. A., Puttaparthi, K., Hu, W., Elliott, J. L., Stuve, O., and Habib, A. A. (2008) EMBO Rep. 9 766-773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schumann, D. M., Maedler, K., Franklin, I., Konrad, D., Størling, J., Böni-Schnetzler, M., Gjinovci, A., Kurrer, M. O., Gauthier, B. R., Bosco, D., Andres, A., Berney, T., Greter, M., Becher, B., Chervonsky, A. V., Halban, P. A., Mandrup-Poulsen, T., Wollheim, C. B., and Donath, M. Y. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2861-2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson, R. B., Dennis, P. B., Han, J. W., Williamson, N. A., Kozma, S. C., Wettenhall, R. E., and Thomas, G. (1995) EMBO J. 14 5279-5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, Q., Inoki, K., Kim, E., and Guan, K. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 6811-6816 [DOI] [PMC free article] [PubMed] [Google Scholar]