Abstract
The Escherichia coli envelope stress response is controlled by the alternative sigma factor, σE, and is induced when unfolded outer membrane proteins accumulate in the periplasm. The response is initiated by sequential cleavage of the membrane-spanning antisigma factor, RseA. RseB is an important negative regulator of envelope stress response that exerts its negative effects onσE activity through its binding to RseA. In this study, we analyze the interaction between RseA and RseB. We found that tight binding of RseB to RseA required intact RseB. Using programs that performed global and local sequence alignment of RseB and RseA, we found regions of high similarity and performed alanine substitution mutagenesis to test the hypothesis that these regions were functionally important. This protocol is based on the hypothesis that functionally dependent regions of two proteins co-evolve and therefore are likely to be sequentially conserved. This procedure allowed us to identify both an N-terminal and C-terminal region in RseB important for binding to RseA. We extensively analyzed the C-terminal region, which aligns with a region of RseA coincident with the major RseB binding determinant in RseA. Both allele-specific suppression analysis and cysteine-mediated disulfide bond formation indicated that this C-terminal region of similarity of RseA and RseB identifies a contact site between the two proteins. We suggest a similar protocol can be successfully applied to pairs of non-homologous but functionally linked proteins to find specific regions of the protein sequences that are important for establishing functional linkage.
The Escherichia coli σE-mediated envelope stress response is the major pathway to ensure homeostasis in the envelope compartment of the cell (1-3). σE regulon members encode periplasmic chaperones and proteases, the machinery for inserting β-barrel proteins into the outer membrane and components controlling the synthesis and assembly of LPS (4-6). This pathway is highly conserved among γ-proteobacteria (6).
The σE response is initiated when periplasmic protein folding and assembly is compromised (7-9). During steady state growth, σE is inhibited by its antisigma factor, RseA, a membrane-spanning protein whose cytoplasmic domain binds to σE with picomolar affinity (10-13). Accumulation of unassembled porin monomers serves as a signal to activate the DegS protease to cleave RseA in its periplasmic domain (14, 15). This initiates a proteolytic cascade in which RseP cleaves periplasmically truncated RseA near or within the cytoplasmic membrane to release the RseAcytoplasmic-σE complex, and cytoplasmic ATP-dependent proteases complete the degradation of RseA thereby releasing active σE (16-19).
RseB, a second negative regulator of the envelope stress response (11, 20, 21), binds to the periplasmic domain of RseA with nanomolar affinity. RseB is an important regulator of the response (2, 22, 23). It prevents RseP from degrading intact RseA, thereby ensuring that proteolysis is initiated only when the DegS protease is activated by a stress signal (21). Additionally, RseB prevents activated DegS from cleaving RseA, suggesting that interaction of RseB with RseA must be altered before the signal transduction cascade is activated (23).
The goal of the present studies was to explore how RseB binds to RseA. The interaction partner of RseB is the unstructured periplasmic domain of RseA (RseA-peri). Within RseA-peri, amino acids ∼169-186 constitute a major binding determinant to RseB (23, 24). This peptide alone binds RseB with 6 μm affinity, and deleting this region abrogates binding to RseB (23). Additional regions of RseA-peri also contribute to RseB binding, as intact RseA-peri binds with 20 nm affinity to RseB (23). Much less is known about the regions of RseB required for interaction with RseA. RseB is homodimeric two-domain protein, whose large N-terminal domain shares structural homology with LolA, a protein that transports lipoproteins to outer membrane (24, 25). The smaller C-terminal domain is connected to the N-terminal domain by a linker, and the two domains share a large interface, which may facilitate interdomain signaling. Glutaraldehyde cross-linking studies indicate that the C-terminal domain interacts with RseA, but the regions of interaction were not identified (25).
In the present report, we study the interaction of RseB and RseA. We establish that both domains of RseB interact with RseA-peri. Using a global sequence alignment, we discovered several regions in RseA and RseB that had high sequence similarity, despite the low overall sequence similarity between these two proteins, a finding that was independently confirmed by a local sequence similarity algorithm. This suggested that these regions were functionally dependent, and we performed a set of mutagenesis experiments designed to test this idea. Our studies of the binding properties of these mutants revealed that regions in both the N terminus and C terminus of RseB modulate interaction with RseA. Moreover, genetic suppression analysis and cysteine-mediated disulfide bond formation suggest that the region of RseA/B with highest similarity (RseA residues 165-191 (major binding determinant in RseA) and RseB residues 233-258) are interacting partners.
EXPERIMENTAL PROCEDURES
Media and Antibiotics—Luria-Bertani (LB) was prepared as described (26). When required, the medium was supplemented with 30 μg/ml kanamycin (Kan), 20 μg/ml chloramphenicol (Cm), and/or 100 μg/ml ampicillin (Amp). A final concentration of 0.2% l-(+)-arabinose was used to induce the expression of cytochrome fusion of the C-terminal 50 amino acids of OmpC from the arabinose-inducible promoter Para. Isopropyl-β-d-galactoside (IPTG)2 at a final concentration of 0.1 mm was added to induce the expression of RseB from pFLAG-MAC and of RseA from constructs with Ptac and T7 promoters.
Plasmids—Plasmids used in this study are described in supplemental Table S1.
Cloning, Expression, and Purification of RseB—The coding sequence of mature RseB (residues 24-318) was amplified by PCR and cloned between HindIII and EcoRI sites of the vector pFLAG-MAC (Sigma-Aldrich) or pFLAG-ATS (Sigma-Aldrich) to make the construct pRseB-MAC or pRseB-ATS, respectively. Both these constructs have coding sequence for FLAG tag at the 5′-end of the gene, and the expression of the cloned gene is under the control of an IPTG-inducible tac promoter. The vector pFLAG-ATS additionally has a 63-bp OmpA signal sequence upstream of the FLAG tag coding sequence to allow periplasmic secretion of RseB. To clone RseB C-terminal domain, the coding sequence for residues 218-318 was amplified and cloned between HindIII and EcoRI sites of the vector pFLAG-MAC. For expression, pRseB-MAC (WT/mutant) was transformed into E. coli Top10 competent cells (Invitrogen). Cells bearing this plasmid were grown at 30 °C to an A600 of 0.3 in LB containing 100 μg/ml Amp and then induced with 0.5 mm IPTG. Induction was allowed for 3 h at 30 °C. The cells were then harvested by centrifugation (5000 × g, 15 min, 4 °C). The cell pellet was resuspended in buffer A (50 mm Tris, pH 7.4 with 150 mm NaCl) containing protease inhibitor mixture (Roche Applied Science), and cells were disrupted by sonication at 4 °C. Cell debris was removed by centrifugation (20,000 × g, 40 min, 4 °C), and the supernatant was mixed with 200 μl of anti-FLAG M2 affinity gel previously equilibrated with buffer A. Immunoprecipitation of RseB was allowed at 4 °C for 2 h on a rotary shaker. The gel was recovered by centrifugation at 500 × g and washed extensively with buffer A. RseB-FLAG fusion protein was then eluted by competition with 200 μg/ml of FLAG peptide. The eluted protein was dialyzed against phosphate-buffered saline (PBS) and 10% glycerol.
Expression and Purification of RseA—To express His-tagged RseA-peri, the construct pLC234 was transformed in E. coli BL21(DE3) cells (11). Cells bearing this plasmid were grown at 30 °C to an A600 of 0.3 in LB containing 30 μg/ml Kan. IPTG was then added to a final concentration of 0.5 mm, and induction was allowed for 3 h at 30 °C. The cells were then harvested by centrifugation (5000 × g, 15 min, 4 °C). The cell pellet was resuspended in PBS containing Protease inhibitor mixture (Roche Applied Science), and cells were disrupted by sonication at 4 °C. Cell debris was removed by centrifugation (20,000 × g, 40 min, 4 °C), and the supernatant was loaded on a Ni-NTA column that had been previously equilibrated with PBS. After washing the column with 20 mm imidazole, RseA was eluted with 250 mm imidazole. The eluted protein was dialyzed against PBS and 10% glycerol. The expression and purification of His-Trx-tagged RseA 169-196 peptide was done as described previously by Kim et al. (24).
Mutagenesis of RseA and RseB—QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce the desired mutations into RseB using pRseB-MAC or pRseB-ATS as template, into RseA using pLC234 (RseA-peri; alanine substitutions), or pHis-Trx-RseA 169-196 (RseA-peptide; alanine substitutions) as template. Overlap extension PCR (27) was used to introduce cysteine substitution mutations in the His-tagged RseA periplasmic domain sequence, which were then cloned in pTrc99A vector.
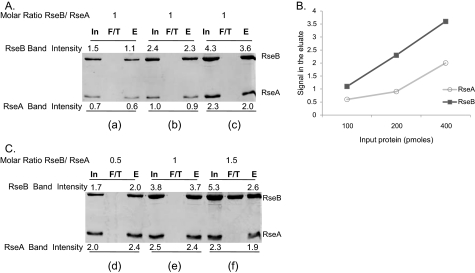
RseA-RseB Binding Assays—For the binding assays, purified His-tagged RseA-peri was immobilized on Ni-NTA resin that had been pre-equilibrated with PBS. After washing the column to remove unbound protein, equimolar amount of RseB (WT/mutant) was added to the column. RseB was allowed to interact with RseA for 15 min at room temperature, and then unbound protein was removed by washing the column with PBS. Bound protein was eluted with 250 mm imidazole and analyzed by SDS-PAGE. Fig. 1A shows SDS-PAGE analysis of in vitro binding of His-tagged RseA-peri to Flag-tagged RseB as the amounts of each are increased while maintaining a 1:1 ratio of the proteins, demonstrating that under these conditions RseB binds quantitatively to RseA-peri. Quantification of these results (Fig. 1B) clearly demonstrates that the data is within the linear range. Fig. 1C demonstrates that the binding is saturable, as RseB appears in the flow-through when the molar ratio of RseB/RseA exceeds 1:1. These experiments were repeated several times and comparable results were obtained. Relative binding of each RseB mutant was calculated by first quantifying the amount of RseA and RseB in the eluate with ImageJ software (NIH) and then expressing it as a percentage of RseBWTbound to RseA: (RseBmut/RseA)/(RseBWT/RseA) × 100.
FIGURE 1.
SDS-PAGE analysis and quantification of in vitro binding of His-tagged RseA-peri to Flag-tagged RseB. RseA was immobilized on Ni-NTA resin and was allowed to interact with RseB (input, In). Unbound protein was collected as flow through (F/T) and bound proteins were eluted with imidazole (eluate, E) after washing. Input, flow through, and eluate samples were run on SDS-PAGE and quantified with ImageJ software. Band intensities measurements were made by integrating pixel densities in the band of interest and subtracting the contribution of the background. A, determining the linear range of the assay. For panel a, 100 pmol of RseA and RseB were loaded onto the column and for panels b and c, quantities of RseA and RseB were successively doubled keeping their molar ratio constant. B, standard curve for the binding assay shown in Fig. 1A. The plot shows the relationship between the protein loaded on the column as input and the band intensity for protein eluted from the column. C, determining the saturation of the assay. For panels d, e, and f, RseB was titrated against a fixed quantity of RseA (400 pmol). Band intensities of the proteins are indicated.
In Vivo Analysis of RseA and RseB Binding by β-Galactosidase Assays—σE activity was assayed by monitoring β-galactosidase activity from a chromosomal σE-dependent lacZ reporter gene in ΦλrpoHP3::lacZ, carried in the ΔrseB strain CAG51050 (21) transformed with both plasmid pRseB-ATS (WT/mutant) and plasmid pBA174 (encodes the cytochrome fusion of C-terminal 50 residues of OmpC under control of the arabinose inducible promoter pBAD) (14). CAG51050 was grown at 30 °C in LB medium to an A600 of 0.1, when it was induced with 0.2% arabinose. Samples were collected for β-galactosidase analysis between OD600 of 0.2-0.4. β-galactosidase activity/0.5 ml cells was plotted against A600. The slope of the data is the differential rate of β-galactosidase synthesis and is used as the measure of σE activity, as described previously (28).
Disulfide Cross-linking between RseA-peri and RseB—Cultures overexpressing various cysteine-substituted RseB mutants were mixed with cultures overexpressing either wild-type RseA or cysteine-substituted RseA mutants and then sonicated at 4 °C. Cell lysates were clarified by centrifugation at 20,000 × g for 30 min at 4 °C and then loaded onto Ni-NTA columns that had been pre-equilibrated with PBS. The columns were washed with 20 mm imidazole and then eluted with 250 mm imidazole. The samples were then mixed either with SDS sample buffer containing no reducing agent or with sample buffers containing 5% β-mercaptoethanol. These samples were analyzed by SDS-PAGE and anti-RseB immunoblotting. We also determined whether cysteine-substituted RseB/A mutants when purified separately exhibited cross-linking. RseB mutants never exhibited cross-linking to themselves. RseA mutants formed dimers; however this is likely to be a very slow reaction because it was suppressed when the cultures were mixed with RseB prior to sonication.
Fluorescence Anisotropy—Fluorescein-labeling of RseA-peri was performed as described (23). Briefly, Ser-154 in RseA-peri was mutated to cysteine, the protein was expressed in E. coli, and purified by Ni-NTA chromatography. Purified RseA-peri S154C mutant was reduced with Tris(2-carboxyethyl)phosphine hydrochloride, mixed with a 10-fold molar excess of fluorescein-5-maleimide, and incubated overnight at 4 °C. Fluorescein-labeled RseA-peri was purified on reversephase high performance liquid chromatography. Eluted protein was resuspended in assay buffer (50 mm sodium phosphate pH 7.4, 200 mm potassium chloride, 10% glycerol). DegS cleavage assays and RseB binding assays were done to confirm that fluorescein-labeled RseA-peri was similar to the unlabeled RseA-peri.3 Dissociation kinetics measurements were performed in assay buffer at 25 °C using a PTI QM-2000-4SE spectrofluorometer. Fluorescein-labeled RseA-peri (50 nm) and wild-type or RLYSDL-mutant RseB (250 nm) were mixed, and the fluorescence anisotropy (467 nm excitation, 520 nm emission) was measured. Excess unlabeled RseA-peri was then added and mixed thoroughly, and fluorescence anisotropy was measured as a function of time.
RESULTS
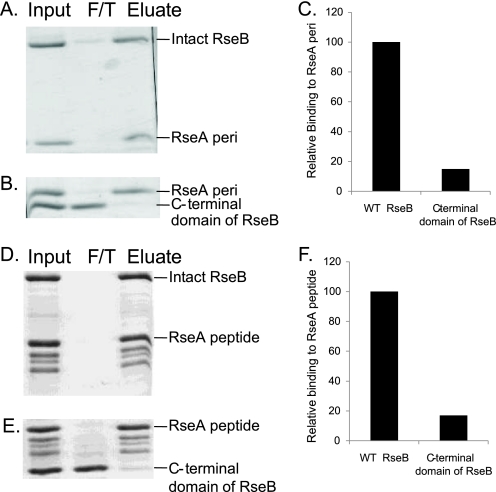
Intact RseB Is Necessary for Tight Binding to RseA (RseA-peri)—To examine RseB binding to RseA, N-terminally His-tagged RseA-peri (residues 121-216) was immobilized on Ni-NTA and then allowed to interact with an equimolar amount of either N-terminally Flag-tagged RseB (residues 24-318; N-terminal signal sequence removed) or the N-terminally Flag-tagged C-terminal domain of RseB (residues 218-318). Unbound RseB was removed by washing, and the bound proteins were co-eluted with imidazole. Under these conditions, RseB was essentially quantitatively bound to RseA-peri (Fig. 2, A and C), and about 15% of the C-terminal domain of RseB (Fig. 2, B and C) was bound to RseA-peri. Control experiments established that the band intensities analyzed were within the linear range for quantification (see “Experimental Procedures”). Note that we were unable to perform similar experiments with the N-terminal domain of RseB because that domain alone forms high molecular weight (MW) aggregates (25). We repeated these same experiments using an RseA fragment consisting of the major RseA binding determinant (RseA 169-196; RseA-peptide), carried in a His-Trx-RseA construct (24). Note that this construct is prone to C-terminal degradation (24), accounting for the three bands observable below His-Trx-RseA in the input sample (Fig. 2, D and E). Intact RseB bound more tightly than the C-terminal domain to RseA-peptide (Fig. 2, D-F). Taken together, these experiments indicate that both the periplasmic domain of RseA and its major binding determinant interact with the C-terminal domain of RseB. We argue in the discussion that some additional residues of RseB, outside of this domain are required for tight interaction with RseA.
FIGURE 2.
Intact RseB is required for complete binding to RseA. SDS-PAGE analysis of in vitro binding of His-tagged RseA-peri to Flag-tagged RseB (A) or Flag-tagged RseB C-terminal domain (B); and of His-Trx-tagged RseA 169-196 peptide to Flag-tagged RseB (D) or Flag-tagged RseB C-terminal domain (E). RseA was immobilized on Ni-NTA resin and was allowed to interact with an equimolar amount of RseB (shown as input, In). Unbound protein was collected as flow through (F/T), bound proteins were eluted with imidazole (eluate, E) after washing. C and F presents relative binding of RseB to RseA [(wt RseB or RseB C-terminal domain/RseA)/(wt RseB/RseA) × 100] after quantification of data with ImageJ software as described in Fig. 1 and “Experimental Procedures.” These experiments were repeated three times, and similar results were obtained.
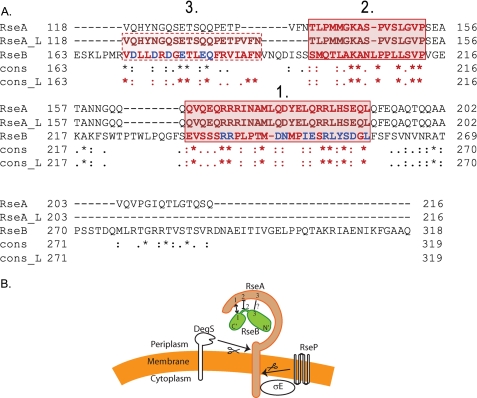
Alignment of RseB with RseA—Identifying regions of proteins that interact using experimental methods is a time consuming endeavor. Often, bioinformatics methods can be useful to formulate a hypothesis based on the results of computations as well as to suggest a set of experiments designed to validate it. We first aligned protein sequences of RseA and RseB to determine whether or not they are homologous. We used a global alignment software, T-Coffee, which, in turn, employs ClustalW algorithm for a pairwise alignment (29). ClustalW is perhaps the most widely accepted algorithm for sequence alignment (30). The overall sequence identity was low (14%) suggesting that the two proteins are not homologs. Surprisingly, we discovered an unusual sequence conservation between some residues of the RseA periplasmic domain and RseB. We then obtained a sequence alignment of the periplasmic domain of RseA and RseB using the same global alignment algorithm, T-Coffee. This alignment confirmed that the sequence of RseA-peri cannot be fully aligned with the sequence of RseB. Instead, the alignment determined two large locally aligned regions: RseA region 1 (residues 165-191) aligned to RseB region 1 (residues 233-258) (Fig. 3, lines 4 and 6); and RseA region 2 (residues 138-153) aligned to RseB region 2 (residues 197-213) (Fig. 3, lines 1 and 3). To independently confirm this result, we applied a specialized local alignment program JAligner that implements the Smith-Waterman algorithm for the local alignment of two sequences (31). In contrast to a global alignment, designed to align the entire input sequences, a local alignment algorithm focuses on optimizing the similarity measure and compares local segments of all possible lengths at all possible positions. The obtained local alignment fully confirmed our initial hypothesis, providing exactly the same alignment for the two previously identified regions 1 and 2 (Fig. 3, lines 5, 6 and 2, 3). In addition, aligning sequences locally, slightly improved the alignment of RseA region 3 (residues 118-137) to RseB region 3 (residues 170-189) suggesting that this region might be another functionally important sequence motif (Fig. 3, lines 2, 3). Notably, RseA region 1 is virtually coincident with the region of RseA identified as the major RseB binding determinant within RseA (∼residues 169-186) (23, 24). This finding encouraged us to pursue the idea that regions of high similarity between RseA and RseB might identify sites at which the two proteins interact. Based on the alignment consensus, we designed and constructed a set of alanine substitution mutations. By examining whether the mutant protein impaired interaction with its partner, we were able to test the importance of each of these three regions in mediating binding.
FIGURE 3.
Sequence analysis of RseA peri and RseB and a representation of regions of interactions. A, global and local sequence alignments of RseA-peri and RseB. The global and local sequence alignments were performed with the T-COFFEE and JAlign software packages, respectively. The periplasmic domain aligned to a specific region (170-289) of RseB. The global alignment is represented by sequences RseA and RseB, while the local alignment is represented by RseA_L and RseB. The degree of conservation for each aligned pair of residues between RseA and RseB and between RseA_L and RseB is indicated below the aligned pair: asterisk, denotes exact match; colon, conserved substitutions; dot, semi-conserved substitutions, and blank indicates that there is no match between the residues. While the overall sequence identity is only 14%, the alignment has revealed two sequence motif regions that are common for both proteins and are exactly the same in both alignments (red solid box, regions 1 and 2). Moreover, using the local alignment, we were able to identify another region with six conserved residues, that was recovered only partially when using global alignment (red dotted box, region 3). The alanine substitutions made in RseB during the course of this study are marked in blue. B, schematic representation summarizing our results of the interactions between RseA and RseB. N′ and C′ refer to N-terminal and C-terminal domains of RseB. We provide evidence that similarity region 1 of RseA and RseB interact and that similarity region 2 of RseA and RseB neither affect binding of the two proteins to each other nor interact. Altering similarity region 3 of RseB decreases binding to RseA but we do not know whether these two similarity regions interact.
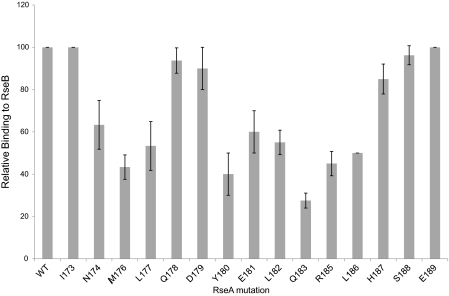
RseA Region 1 (Residues 165-191)—As RseA region 1 is coincident with the major binding determinant in RseA-peri, some mutagenesis of this region had already been performed. Changing arginine residues (Arg-170, Arg-171, Arg-172, Arg-184, Arg-185) to aspartate resulted in severe defects in binding of RseA-peri to RseB (24). However, these effects could also result from steric hindrance induced by charge reversal. When these arginine residues were changed to alanine, no single mutants exhibited an effect, but the double mutant R172A+R185A had a severe binding defect and R171A+R172A had a mild reduction in binding. To identify additional binding determinants, we mutated all of the remaining non-alanine residues to alanine starting with Ile-173 both in RseA-peri and in RseA-peptide (residues 169-196) and tested their binding to RseB. Using RseA-peptide sensitizes binding to small changes because it binds to RseB ∼300-fold more weakly than RseA-peri (23). When assayed in the context of RseA-peri, none of the single alanine substitutions showed decreased binding to RseB (data not shown). However, when assayed in the sensitized context of RseA-peptide, alanine substitution of residues located in two clusters (174-177 and 180-186) reduced binding to RseB (Fig. 4), significantly expanding the three previously identified residues (171, 172, 185) known to alter binding.
FIGURE 4.
Systematic identification of binding determinants in the RseA 169-196 peptide. The His-Trx-RseA 169-196 peptide containing single alanine substitution mutations were purified and tested in vitro for binding to RseB as described in the legend to Fig. 1 and “Experimental Procedures.” Relative binding of various mutant peptides to wild-type RseB is shown [(RseBwt/RseAmut)/(RseBwt/RseAwt) × 100].
RseA Region 2 (Residues 138-153)—The RseA region 2 includes the DegS cleavage site in RseA (between residues 148-149). Previous work strongly suggested that RseB binding does not directly occlude the DegS cleavage site, but did not directly test this proposition (23). We therefore tested whether deleting RseA residues 144-153 affects binding to RseB, either in the context of RseA-peri or in two sensitized mutant derivatives of RseA-peri (R171A+R172A and R172A+R185A), which already exhibited binding defects to RseB. Even in the sensitized backgrounds, the RseA deletion derivative did not decrease RseB binding further, confirming that the DegS cleavage site in RseA and immediately surrounding residues are not involved in binding to RseB (data not shown).
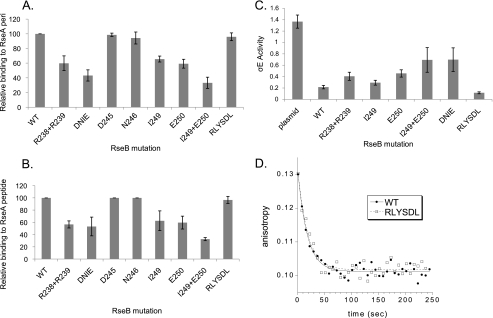
RseB Region 1 (Residues 233-258)—The C-terminal domain of RseB has a RseA binding site (25) (Fig. 2). As RseB region 1 is located in the C-terminal domain, it might contain the binding determinants located in this domain. To test this, we used the similarity scores generated by T-Coffee (high and moderate similarity are indicated by asterisk and colon under the residues in Fig. 3) to guide our substitutions. Initially, we examined the binding of 3 multiple alanine-substituted mutants of RseB to both RseA-peri and to RseA-peptide. Two substitutions, Arg-238, Arg-239 (RR) to Ala and Asp-245, Asn-246, Ile-249, Glu-250 (DNIE) to Ala exhibited binding defects both to RseA-peri and RseA-peptide (Fig. 5, A and B, lanes 2 and 3) whereas substitution of Arg-252, Leu-253, Tyr-254, Ser-255, Asp-256, Leu-258 (RLYSDL) to Ala does not (Fig. 5, A and B, lane 9). Further investigation revealed that both the I249A and E250A single mutants show binding defects (Fig. 5, A and B; lanes 6 and 7) and that the I249A+E250A double mutant has a larger binding defect than either alone (Fig. 5, A and B; lane 8) suggesting that both residues influence binding.
FIGURE 5.
Identification of binding determinants in the C-terminal domain of RseB. Relative binding of Flag-tagged RseB alanine-substituted mutants to His-tagged RseA-peri [(RseBmut/RseAperi)/(RseBwt/RseAperi) × 100] (A) or to the His-tagged RseA169-196 peptide [(RseBmut/RseApeptide)/(RseBwt/RseApeptide) × 100] (B). Multiple alanine substitutions in RseB were named as follows: R238A +R239A = RR; D245A+N246A+I249A+E250A = DNIE; and R252A+ L253A+Y254A+S255A+D256A+L258A = RLYSDL. C, σE activity in ΔRseB cells carrying vector only (no RseB; lane 1) or plasmid expressing wt RseB (lane 2) or various mutant RseB derivatives (lanes 3-8). Western blotting established that plasmid expressed RseB was present at the endogenous level.4 Samples were assayed for σE activity after inducing cells with 0.2% arabinose by monitoring the differential rate of β-galactosidase synthesis produced from a single copy ΦλrpoHP3::lacZ fusion as described under “Experimental Procedures.”σE activity of cells with wt RseB was 0.22 ± .03 and that of the RseB mutant RLYSDL was 0.12 ± .02. D, dissociation kinetics of wild-type and RseB mutant RLYSDL from fluorescein-labeled RseA-peri. Fluorescence anisotropy decreases over time as RseB dissociates from the fluorescein-labeled RseA-peri and binds to excess unlabeled RseA-peri. Data were fit to a simple exponential decay model with half-lives of 12.1 s for wild-type RseB and 12.5 s for RLYSDL RseB.
An important question is whether these RseB mutants exhibit binding defects in vivo (see schematic of signaling system in Fig. 3B). To answer this question, we developed an in vivo assay system that is sensitive to the presence of RseB. We observed that when σE is induced by overexpressing the C-terminal 50 amino acids of OmpC from the pBAD promoter, σE activity is 6-fold higher in a strain lacking RseB (Fig. 5C, lane 1) than when the ΔRseB phenotype of the parental strain is complemented by expression of RseB from a plasmid (Fig. 5C, lane 2). We reasoned that if the plasmid encoded RseB alleles were defective in binding to RseA, this defect should be manifested as higher σE activity. We tested this proposition. Importantly, quantitative Westerns established that in this system, both wt and mutant RseB alleles were expressed at the level of endogenous RseB.4 We observed that indeed, both the RR and DNIE mutants that were defective in binding to RseA in vitro, exhibited higher σE activity in vivo, as was expected from binding defective RseB mutants. Importantly, the in vivo analysis also mirrored in vitro studies when DNIE was deconvoluted to its important components; that is, both I249A and E250A showed binding defects and the combination of the two, I249A+E250A gave higher σE activity than the either single mutant. Moreover, the σE activity of I249A+E250A was approximately equivalent to that of DNIE (Fig. 5C) just as was true in our in vitro binding studies.
One multiple alanine-substituted mutant of RseB, RLYSDL had no binding defect in vitro (Fig. 5, A and B; lane 9). Interestingly, RLYSDL is about 2-fold more poorly induced than wt in vivo (Fig. 5C, lane 8). This could be explained if the RLYSDL mutant bound more tightly to RseA than the wt protein. Our binding assay cannot detect tighter binding as it already gives ∼100% binding of the two proteins. Therefore, we used a fluorescence based kinetic assay to compare dissociation of wt RseB or the RLYSDL mutant to RseA-peri (23). Results from this assay indicate that the two proteins bind indistinguishably to RseA-peri (Fig. 5D). We consider the implications of this phenotype in the “Discussion.”
RseB Region 2 (Residues 197-213)—RseB region 2 is in the linker region between the two RseB domains and has high similarity to RseA region 2, which we determined not to contribute to binding. A multiple alanine substitution in the linker of RseB (Leu-209, Leu-210, Ser-211, Val-212 to Ala) did not result in binding defect in vitro (data not shown), and this similarity region was not pursued further.
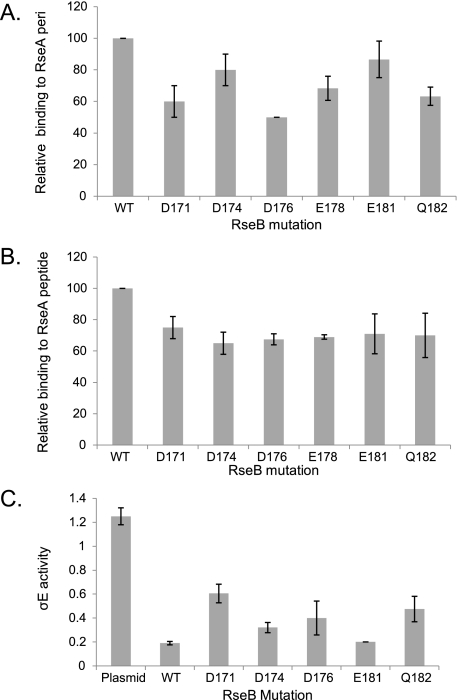
RseB Region 3 (Residues 170-189)—As the N-terminal domain of RseB contributes to RseA binding (Fig. 2) and RseB region 3 is located in the N-terminal domain of RseB, we tested whether RseB region 3 contributes to binding. A hexa-alanine-substituted mutant in this region (Asp-171, Asp-174, Asp-176, Glu-178, Glu-181, Gln-182 to Ala) had folding defects and copurified with GroEL (data not shown). We therefore made single alanine substitution mutations at each of these positions and examined their binding. Four mutants had binding defects (D171A, D176A, E178A, and Q182A); two others showed little to no binding defect (D174A and E181A) to RseA-peri (Fig. 6A). All of the mutants showed a relatively nonspecific reduction in binding to the RseA-peptide (Fig. 6B).
FIGURE 6.
Identification of binding determinants in the N-terminal domain of RseB. Relative binding of Flag-tagged alanine-substituted RseB mutants to His-tagged RseA-peri [(RseBmut/RseAperi)/(RseBwt/RseAperi) × 100] (A) or to the His-tagged RseA169-196 peptide [(RseBmut/RseApeptide)/(RseBwt/RseApeptide) × 100] (B), as described in the legend to Fig. 5, A and B. C, σE activity in ΔRseB cells carrying vector only (no RseB; lane 1) or plasmid expressing wt RseB (lane 2) or various mutant RseB derivatives (lanes 3-7). σE activity measurements were done as described previously in Fig. 5C. Western blotting confirmed that plasmid expressed RseB was present at the endogenous level.4
We used the in vivo assay system described above (Fig. 5C), to test whether the RseB mutants exhibit defects in vivo. The expression level of each of the tested mutant proteins was comparable to that of wt protein as judged by Western blotting (data not shown). There is good correlation between the in vivo effects of these RseB mutants and their in vitro binding to RseA-peri. Induction of σE is greater when the plasmid provides RseB proteins more defective in binding to RseA-peri (D171A, D176A, Q182A) than when it provides either the wt protein or relatively nondefective RseB mutants (D174A, E181A) (Fig. 6C, compare lanes 3, 5, 7 with lanes 2, 4, 6). The E178A mutant was not tested in vivo because the protein was unstable. As levels of all RseB mutants are comparable, these results suggest that induction of σE activity is reflective of a binding defect rather than of insufficient production of mutant protein.
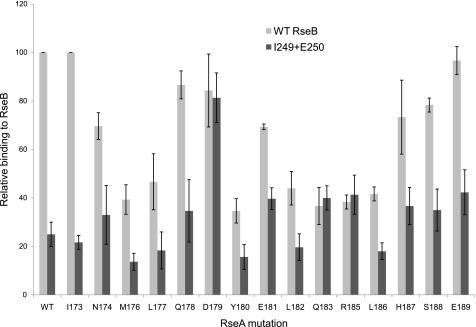
A Genetic Analysis of the Interaction between RseA Region 1 and RseB Region 1—Our alignment programs indicated very high similarity between RseA region 1 (previously identified as a major binding determinant in RseA) and RseB region 1. Mutational analysis indicated that altering these regions decreased the binding of RseA/B to each other. We now addressed whether these regions interact with each other. Allele-specific suppression analysis is often used to investigate the possibility that two proteins interact. The rationale is as follows: If two residues interact and mutating one of them to alanine eliminates the interaction then mutating the second residue of the interacting pair to alanine will not reduce binding further. We therefore tested whether any of the single alanine substitution mutations in the RseA peptide showed equivalent binding to wt RseB and to the RseB mutant I249A+E250A. Three RseA alanine substitution mutations, D179A, Q183A, and R185A showed this phenotype (Fig. 7). This result is consistent with the idea that these residues in the two proteins directly interact and therefore provides support for the possibility that the similarity region 1 identifies a point of contact between RseA and RseB. We note that the RseA R185A mutant binds significantly better to RseB I249A+E250A mutant than to wt RseB. Possibly, removal of RseA residue Arg-185 allows the mutant RseB protein to make alternative contacts not possible with wt RseB.
FIGURE 7.
Allele-specific suppression analysis of RseA and RseB. The His-Trx-RseA 169-196 peptide containing single alanine substitution mutations were purified and tested in vitro for binding to Flag-tagged WT RseB (light gray) [(RseBwt/RseAmut)/(RseBwt/RseAwt) × 100] or Flag-tagged I249A+E250A RseB mutant (dark gray) [(RseBmut/RseAmut)/(RseBwt/RseAwt) ×100].
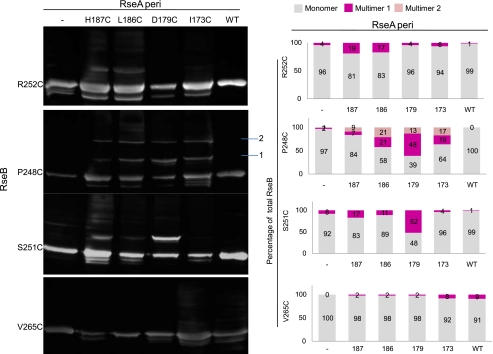
Disulfide Cross-linking Indicates That RseA Region 1 and RseB Region 1 Are in Close Proximity—Disulfide cross-linking mediated by cysteine residues has been used to identify protein regions in close proximity. This approach is particularly convenient for the RseA/B proteins since the periplasmic domain of RseA lacks cysteines and RseB has only a single cysteine (Cys-141) that is buried near a cleft in the protein (24, 25). Using the crystal structure of RseB as a guide, we chose residues facing both in the direction of Glu-250/Arg-238 (Pro-248, Arg-252, and Val-265) and Arg-239/Ile-249 (Ser-251) for cysteine substitution. As there is no structural information for RseA-peri, we used results from our scanning alanine mutagenesis of RseA region 1 to determine which residues to mutate to cysteine. We mutated four RseA residues (Ile-173, Asp-179, Leu-186, and His-187) to cysteine. Three of these, Ile-173, Asp-179, and His-187 showed little or no defect in binding when mutated to alanine, but were directly adjacent to residues implicated in binding to RseB (Fig. 4) and one of them, Leu-186, had a 2-fold binding defect. Two of the RseB cysteine substitution mutations (P248C and S251C) showed convincing evidence of disulfide bond formation with a RseA cysteine partner protein on non-reducing SDS-PAGE gels. When probed with RseB antibody, P248C and S251C alone or incubated with wt RseA-peri showed only a single band that corresponds in MW to an RseB monomer (Fig. 8). In sharp contrast, when incubated with RseA-peri cysteine-substituted partner proteins, these same RseB mutant proteins showed significant formation of higher MW bands (Fig. 8), which were eliminated when the samples were run on reducing SDS-PAGE gels (supplemental Fig. S1). Both the fact that P248C and S251C showed distinct patterns of reactivity and the fact that R252C and V265C showed weak or no reactivity suggest that RseA region 1 and RseB region 1 are in close proximity with a specific orientation.
FIGURE 8.
Disulfide cross-linking between RseA cysteine mutants and RseB cysteine mutants. Cultures overexpressing Flag-tagged cysteine-substituted RseB mutant proteins were mixed with cultures overexpressing either wt or cysteine-substituted His-tagged RseA, sonicated, cleared of cell debris by centrifugation, loaded onto Ni-NTA column, washed, eluted with imidazole, run on SDS-PAGE under non-reducing conditions, and immunoblotted with anti-RseB antibodies. The first lane shows the mobility of the cysteine-substituted RseB mutants treated identically except that they were purified on M2-agarose from cultures that were not premixed with cultures expressing RseA. High MW protein complexes obtained upon incubation of RseB mutant P248C with the various cysteine-substituted RseA mutants, migrated to two distinct positions (marked as 1 and 2) on SDS-PAGE/immunoblot analysis under non-denaturing condition. Multimer position 1 (∼50 kDa) corresponds to an approximate position where a complex consisting of single molecules of RseA and RseB is expected to migrate (left panel). Band intensities were calculated using ImageJ software, and the percentage of RseB cysteine mutant in monomeric/multimeric form is presented (right panel).
DISCUSSION
The RseB protein is an integrator of the σE response pathway, binding very tightly to the RseA antisigma factor to control its degradation. The principal contribution of the present work was to further define how RseB interacts with RseA. This work demonstrates the utility of employing bioinformatics methods to predict functionally important sequence conservation between two proteins as well as to guide the experimental determination of how the proteins are functionally linked.
Our initial experiments indicated that RseA-peri binds more tightly to intact RseB than to the RseB C-terminal domain alone. This finding is not likely to be explained by postulating that the C-terminal domain is somewhat misfolded. The crystal structure of RseB indicates that this region is an independent domain and previous biochemical studies showed that the domain is well behaved, fractionating as a single entity (25). Our conclusion that the N terminus of RseB is likely to contain RseA binding determinants is consistent with a previous study demonstrating that the presence of RseA disaggregated the higher MW aggregates of the N-terminal domain that exist in the absence of RseA-peri (25). Moreover, we have shown that overexpression of a periplasmically targeted C-terminal domain of RseB is insufficient to mediate repression of σE activity,4 providing in vivo validation for the requirement for intact RseB for tight binding to RseA. Interestingly, the small tight binding peptide (RseA-peptide; residues 169-196) within RseA-peri also binds more weakly to the C-terminal domain than to intact RseB, suggesting that this peptide may interact with both domains. To begin identification of the regions in RseB that might interact with RseA, we turned to a bioinformatic analysis of how these proteins align.
We used both the global alignment program, T-Coffee (29) and local alignment program JAligner (31) (http://jaligner.sourceforge.net) to examine the alignment of RseB to RseA, and found essentially congruent results with both. The two proteins did not align globally; instead, alignment was determined by a few locally aligned regions. That the alignment was independently confirmed by both global and local alignment algorithms, as well as the fact that no overall sequence similarity between RseB and the periplasmic domain of RseA could be detected, strongly suggested that these motifs were likely to have co-evolved due to their functional dependence. Therefore, we proceeded to test the functional dependence hypothesis by site directed mutagenesis of these regions in RseB. Indeed, our mutagenesis experiments indicated that 2 of the 3 aligned regions (RseB region 1 and 3) alter binding and provide evidence that aligned region 1 identifies a contact site between RseB and RseA. In addition a subregion of RseB region 1 has an additional role that may be involved in signal sensing.
RseA region 1 is virtually coincident with the region encompassed by the tight binding RseA-peptide, which is required for the specific interaction between RseA and RseB (23, 24). This makes it critical to define the RseB contacts of RseA region 1. Two small segments of the proximal RseB region 1, residues 238-9 and 249-50 (which are close to each other in the crystal structure; Fig. 9) are strongly implicated in binding to RseA region 1 by three independent lines of experimentation. First, alanine-substitution mutations at these positions showed reduced binding to RseA-peri and RseA-peptide in vitro and commensurate defects in our in vivo binding assay. Second, allele-specific suppression analysis indicates that three different alanine substitution mutations in RseA region 1 (D179A, Q183A, and R185A) bound equivalently to wt RseB and the RseB region 1 mutant I249A/E250A The finding of allele-specific suppression strongly suggests of interaction between region 1 of RseA and RseB. Finally, the most direct evidence for proximity comes from demonstration of disulfide bonds between cysteine residues introduced in both RseA and RseB. RseB residue Ser-251 when mutated to cysteine specifically forms a disulfide bond with a cysteine substitution mutation in Asp-179, providing strong evidence that the two are in close proximity with a specific orientation. Likewise, RseB residue Pro-248 when mutated to cysteine, also interacts most strongly with cysteine-substituted Asp-179, and somewhat less strongly with cysteine-substituted Ile-173, Leu-186, and His-187. Taken together, this evidence strongly favors the idea that this similarity region defines a contact site between RseB and RseA.
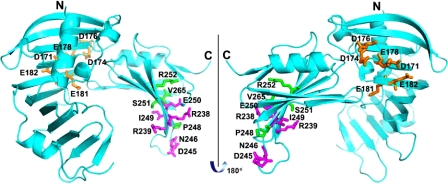
FIGURE 9.
Crystal structure of RseB with alanine and cysteine substitutions marked. The three groups of substitutions shown in color are depicted using a ball and stick representation: 1) C-terminal residues R238, R239, D245, N246, I249, and E250, which were mutated to alanine are colored magenta, 2) N-terminal residues D171, D174, D176, E178, E181, and Q182, which were mutated to alanine are colored orange, and 3) C-terminal residues P248, S251, R252, and V265, which were mutated to cysteine are colored green.
The crystal structure indicates that residues Arg-238 and Glu-250 of RseB are close together and on the same face, whereas residues Arg-239 and Ile-249 of RseB are close together and on a different face. Our data are consistent with the idea that the RseA region 1 interacts with both faces of RseB. First, alanine-scanning mutagenesis of RseA region 1 indicated that contiguous residues affect binding. However, one would actually expect residues binding to a single face would exhibit a spacing dependent on the structure of this region; i.e. every 3-4 residues if the region were to be α-helical. More directly, we observed specific disulfide bond formation both with P248C, which faces in the same direction as residues Arg-238 and Glu-250 and with S251C, which faces in the same direction as residues Arg-239 and Ile-249. Biochemical studies indicate that RseA-peri exists in a molten globule-like state in the absence of RseB (14). Some portions of RseA might become structured upon RseB binding, which could involve several different interactions between region 1 of RseA and RseB, possibly explaining why different faces of RseB specifically interact with this portion of RseA. We note that our binding studies suggest that RseA region 1 interacts with the N-terminal domain of RseB as well (Fig. 2, D-F), arguing that RseA region 1 might have multiple, sequential binding partners. The RseB region 3 is located in the N-terminal domain and mutations in that domain showed reduced binding with RseA-peptide, suggesting the possibility that RseB region 3 might be a partner. However, the pattern of binding defects of RseB region 3 mutants with RseA-peptide (Fig. 6B) was not consistent with the binding defects observed with RseA-peri (Fig. 6A) or with our in vivo assay (Fig. 6C), suggesting a nonspecific defect. We therefore did not pursue this possible interaction further.
The distal portion of the RseB region 1 (residues 252-258) comprises the region of highest similarity between the two proteins. Substituting 6 of these residues with alanine produced no change in the in vitro binding of RseB to RseA, as determined both by our assay and by a high-resolution fluorescence anisotropy assay (Fig. 5, A, B, and D). However, this mutant did have a provocative phenotype when tested in our in vivo assay. Whereas every other RseB mutant tested either induced σE activity to the same extent or better than the wt, this mutant showed significantly less induction (∼2-fold) than the wt (Fig. 5C). As this phenotype does not reflect either an altered amount of the mutant protein4 or altered binding to RseA, this mutant must be defective in some other aspect of RseB function required for induction. We are currently pursuing the hypothesis that this mutant is defective in sensing a signal required for release of RseB from RseA.
Traditionally, bioinformatics methods are used to detect evolutionarily conserved residues among homologous proteins that may identify binding site residues (32) or residues involved in particular biological functions (33). In our case, we discovered residues that are conserved between two non-homologous protein sequences. Like conservation among homologous proteins, conservation between non-homologous proteins could be caused by an evolutionary pressure to preserve these residues. For example, similar sequences in the two unrelated proteins p107 and E2F1 target these proteins to a common binding partner (34) The similarity between some host and pathogen proteins has been explained as molecular mimicry (35, 36). What could be the evolutionary pressure for similarity between RseA and RseB? We tested the hypothesis that residues in the three conserved regions directly contribute to establishing functional linkage between RseA and RseB. Indeed, we found that a portion of similarity region 1 defines a major binding interface between the two proteins, and similarity region 3 may also be involved in binding. It is possible that the conserved residues mimic an interface of a homodimer. Intriguingly, the remainder of similarity region 1 appears to contribute to signaling in an unknown way. We will continue to characterize the molecular mechanisms that explain the necessity for the mutual conservation of regions in RseA and RseB.
Supplementary Material
Acknowledgments
We thank Dr. Tania Baker for valuable suggestions. We thank members of the Gross laboratory for critically reading the manuscript. We also thank Dr. Virgil Rhodius for helpful discussions and Sonia Diaz for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants GM-32678 (to C. A. G.) and AI-16892 (to R. T. S.). This work was also supported by FPR08B2-270 of the 21C Frontier Functional Proteomics Project (to K. K. K.) from the Korean Ministry of Education, Science, and Technology. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
Footnotes
The abbreviations used are: IPTG, isopropyl-1-thio-β-d-galactopyranoside; PBS, phosphate-buffered saline; wt, wild type; NTA, nitrilotriacetic acid.
B. Cezairliyan, unpublished data.
N. Ahuja, data not shown.
References
- 1.De Las Penas, A., Connolly, L., and Gross, C. A. (1997) J. Bacteriol. 179 6862-6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connolly, L., De Las Penas, A., Alba, B. M., and Gross, C. A. (1997) Genes Dev. 11 2012-2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raivio, T. L., and Silhavy, T. J. (2001) Annu. Rev. Microbiol. 55 591-624 [DOI] [PubMed] [Google Scholar]
- 4.Dartigalongue, C., Missiakas, D., and Raina, S. (2001) J. Biol. Chem. 276 20866-20875 [DOI] [PubMed] [Google Scholar]
- 5.Rezuchova, B., Miticka, H., Homerova, D., Roberts, M., and Kormanec, J. (2003) FEMS Microbiol. Lett. 225 1-7 [DOI] [PubMed] [Google Scholar]
- 6.Rhodius, V. A., Suh, W. C., Nonaka, G., West, J., and Gross, C. A. (2006) PLoS Biol. 4 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mecsas, J., Rouviere, P. E., Erickson, J. E., Donohue, T. J., and Gross, C. A. (1993) Genes Dev. 7 2618-2628 [DOI] [PubMed] [Google Scholar]
- 8.Raina, S., Missiakas, D., and Georgopoulos, C. (1995) EMBO J. 14 1043-1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouviere, P. E., De Las Penas, A., Mecsas, J., Lu, C. Z., Rudd, K. E., and Gross, C. A. (1995) EMBO J. 14 1032-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ades, S. E., Connolly, L. E., Alba, B. M., and Gross, C. A. (1999) Genes Dev. 13 2449-2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Las Penas, A., Connolly, L., and Gross, C. A. (1997) Mol. Microbiol. 24 373-385 [DOI] [PubMed] [Google Scholar]
- 12.Missiakas, D., Mayer, M. P., Lemaire, M., Georgopoulos, C., and Raina, S. (1997) Mol. Microbiol. 24 355-371 [DOI] [PubMed] [Google Scholar]
- 13.Campbell, E. A., Tupy, J. L., Gruber, T. M., Wang, S., Sharp, M. M., Gross, C. A., and Darst, S. A. (2003) Mol. Cell 11 1067-1078 [DOI] [PubMed] [Google Scholar]
- 14.Walsh, N. P., Alba, B. M., Bose, B., Gross, C. A., and Sauer, R. T. (2003) Cell 113 61-71 [DOI] [PubMed] [Google Scholar]
- 15.Wilken, C., Kitzing, K., Kurzbauer, R., Ehrmann, M., and Clausen, T. (2004) Cell 117 483-494 [DOI] [PubMed] [Google Scholar]
- 16.Alba, B. M., Leeds, J. A., Onufryk, C., Lu, C. Z., and Gross, C. A. (2002) Genes Dev. 16 2156-2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehara, K., Ito, K., and Akiyama, Y. (2002) Genes Dev. 16 2147-2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flynn, J. M., Neher, S. B., Kim, Y. I., Sauer, R. T., and Baker, T. A. (2003) Mol. Cell 11 671-683 [DOI] [PubMed] [Google Scholar]
- 19.Chaba, R., Grigorova, I. L., Flynn, J. M., Baker, T. A., and Gross, C. A. (2007) Genes Dev. 21 124-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alba, B. M., and Gross, C. A. (2004) Mol. Microbiol. 52 613-619 [DOI] [PubMed] [Google Scholar]
- 21.Grigorova, I. L., Chaba, R., Zhong, H. J., Alba, B. M., Rhodius, V., Herman, C., and Gross, C. A. (2004) Genes Dev. 18 2686-2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collinet, B., Yuzawa, H., Chen, T., Herrera, C., and Missiakas, D. (2000) J. Biol. Chem. 275 33898-33904 [DOI] [PubMed] [Google Scholar]
- 23.Cezairliyan, B. O., and Sauer, R. T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 3771-3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, D. Y., Jin, K. S., Kwon, E., Ree, M., and Kim, K. K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8779-8784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wollmann, P., and Zeth, K. (2007) J. Mol. Biol. 372 927-941 [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., and Russell, D. W. (2001) Molecular Cloning: a Laboratory Manual, p. A2.2, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 27.Higuchi, R., Krummel, B., and Saiki, R. K. (1988) Nucleic Acids Res. 16 7351-7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. (1972) Experiments in Molecular Genetics, pp. 352-355, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 29.Notredame, C., Higgins, D. G., and Heringa, J. (2000) J. Mol. Biol. 302 205-217 [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, T. F., and Waterman, M. S. (1981) J. Mol. Biol. 147 195-197 [DOI] [PubMed] [Google Scholar]
- 32.Lichtarge, O., Bourne, H. R., and Cohen, F. E. (1996) J. Mol. Biol. 257 342-358 [DOI] [PubMed] [Google Scholar]
- 33.Hannenhalli, S. S., and Russell, R. B. (2000) J. Mol. Biol. 303 61-76 [DOI] [PubMed] [Google Scholar]
- 34.Adams, P. D., Sellers, W. R., Sharma, S. K., Wu, A. D., Nalin, C. M., and Kaelin, W. G., Jr. (1996) Mol. Cell. Biol. 16 6623-6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham, M. W., McCormack, J. M., Fenderson, P. G., Ho, M. K., Beachey, E. H., and Dale, J. B. (1989) J. Immunol. 143 2677-2683 [PubMed] [Google Scholar]
- 36.Stebbins, C. E., and Galan, J. E. (2001) Nature 412 701-705 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.