Abstract
The synthesis and storage of neutral lipids in lipid droplets is a fundamental property of eukaryotic cells, but the spatial organization of this process is poorly understood. Here we examined the intracellular localization of acyl-CoA:diacylglycerol acyltransferase 2 (DGAT2), an enzyme that catalyzes the final step of triacylglycerol (TG) synthesis in eukaryotes. We found that DGAT2 expressed in cultured cells localizes to the endoplasmic reticulum (ER) under basal conditions. After providing oleate to drive TG synthesis, DGAT2 also localized to near the surface of lipid droplets, where it co-localized with mitochondria. Biochemical fractionation revealed that DGAT2 is present in mitochondria-associated membranes, specialized domains of the ER that are highly enriched in lipid synthetic enzymes and interact tightly with mitochondria. The interaction of DGAT2 with mitochondria depended on 67 N-terminal amino acids of DGAT2, which are not conserved in family members that have different catalytic functions. This targeting signal was sufficient to localize a red fluorescent protein to mitochondria. A highly conserved, positively charged, putative mitochondrial targeting signal was identified in murine DGAT2 between amino acids 61 and 66. Thus, DGAT2, an ER-resident transmembrane domain-containing enzyme, is also found in mitochondria-associated membranes, where its N terminus may promote its association with mitochondria.
Most eukaryotic cells can synthesize neutral lipids, such as triacylglycerols (TGs)2 and sterol esters, and store them in cytosolic lipid droplets. Yet, a molecular understanding of this process and how it is spatially organized is lacking. For example, lipid substrates for TG synthesis (fatty acids and glycerolipid precursors) are found in the cytoplasm and membranes, energy for activating fatty acids (by converting to fatty acyl-CoA) comes from mitochondria, and the enzymes that catalyze TG formation are primarily found in the mitochondria and endoplasmic reticulum (ER). How the cell orchestrates this complex anabolic process to maximize lipid synthesis and storage during times of substrate excess is poorly understood.
In most cells, TG synthesis occurs via the glycerol 3-phosphate (Kennedy) pathway and involves multiple enzymatic reactions in different subcellular compartments (1). The fatty acids for TG synthesis must first be “activated” by acyl-CoA synthases, a family of enzymes that localize to membranes of different compartments, including the ER, mitochondria, and plasma membrane (2), and utilize ATP to ligate CoA to the fatty acyl chain. Next, these fatty acids enter the Kennedy pathway of glycerolipid synthesis, in which the first two reactions occur in both the ER and mitochondria. In the first reaction, glycerol 3-phosphate and a fatty acyl-CoA are combined to yield lysophosphatidic acid through the actions of glycerol-3-phosphate acyltransferase enzymes (1, 3). In the second reaction, 1-acylglycerol-3-phosphate O-acyltransferase enzymes catalyze the esterification of lysophosphatidic acid with fatty acyl-CoA to form phosphatidic acid (1, 4). Next, phosphatidic acid is dephosphorylated at membrane surfaces by phosphatidate phosphatase to yield diacylglycerol (1, 5, 6). All these steps are highly organized spatially, which is likely to be important for the efficiency of the pathway.
The final reaction of TG synthesis is catalyzed by acyl-CoA: diacylglycerol acyltransferase (DGAT) enzymes (7-9). The two mammalian DGATs, DGAT1 and DGAT2 (10, 11), which are encoded by genes of different families, have distinct roles in TG synthesis (12). DGAT2 is the major TG biosynthetic enzyme in eukaryotes. Dgat2-deficient mice die shortly after birth and are almost completely devoid of TG (13), indicating an essential requirement for DGAT2. Catalysis of TG synthesis is conserved in the DGAT2 gene family, with functional orthologs in many species, including Dga1p in Saccharomyces cerevisiae, which contributes to a major portion of TG synthesis (14-16).
Little is known about the intracellular localization of DGAT enzymes. DGAT activity is present in microsomes (7, 17, 18), but in vitro assays do not distinguish between DGAT1 and DGAT2. A DGAT2-green fluorescent fusion protein expressed in HeLa cells localized to the ER (19), and Dga1p activity in S. cerevisiae localizes to the ER and lipid droplets (16). DGAT1 and DGAT2 expressed in COS-7 cells localized primarily to the ER (20). A recent study of the subcellular localizations of tung tree DGAT1 and DGAT2 in tobacco BY-2 cells revealed that the enzymes are located in distinct, non-overlapping regions of the ER (21). Most recently, DGAT2 was reported to co-localize with lipid droplets in cultured adipocytes (22). As a step toward a better understanding of the cellular organization of processes that contribute to TG synthesis and storage, we determined the subcellular localization of murine DGAT2 in mammalian cells.
EXPERIMENTAL PROCEDURES
Construction of Plasmids—Murine DGAT2, DGAT1, and human acyl-CoA:monoacylglycerol acyltransferase-2 (MGAT2), with N-terminal FLAG epitopes (MGDYKDDDDG, epitope underlined) were cloned into pCDNA3.1 (Invitrogen). For co-expression of DGAT1 and DGAT2 in COS-7 cells, DGAT1 and DGAT2 were cloned in-frame into the pBudCE4.1 dual expression vector (Invitrogen). The resulting plasmid contained DGAT1 with a C-terminal myc tag (EQKLISEEDL) and DGAT2 with an N-terminal FLAG tag. Δ55, Δ30-67, and W3RKK3A DGAT2 mutants were generated, using DGAT2 as a template, in mutagenesis reactions with the QuikChange II site-directed mutagenesis kit (Stratagene). The plasmid, N67RFP, was constructed by cloning a PCR-amplified fragment of DGAT2 (amino acid residues 1-67), in-frame, to the N terminus of monomeric RFP (a gift from Dr. Roger Tsien) (23, 24). Mutants 1-30RFP, 30-67RFP, and 67RKKA3-RFP were generated by using N67RFP as a template in mutagenesis reactions and the QuikChange II kit. All plasmids were sequenced to confirm the presence of the desired mutations.
Cell Culture and Transfection—COS-7 cells (American Type Tissue Culture Collection) were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum in a 37 °C incubator with 5% CO2. For transfections, 3 μg of plasmid DNA was incubated with 9 μl of FuGENE 6 transfection reagent (Roche Diagnostics) in 200 μl of Dulbecco's modified Eagle's medium for 30 min at room temperature. The transfection mixture was added to a 100-mm culture dish containing 9 ml of medium and cells at ∼75% confluence. After 24 h, transfected cells were harvested and used for DGAT activity assays or subcellular fractionation experiments. McArdle rat hepatoma RH7777 cells stably expressing DGAT1 and DGAT2 were generated as described before (13).
DGAT Activity Assays—Transfected COS-7 cells were washed twice with ice-cold PBS, harvested by scraping, and collected by centrifugation (1000 × g). To lyse cells, the pellet was resuspended in 200 μl of 50 mm Tris-HCl (pH 7.4) and 250 mm sucrose and passed through a 27-gauge needle 15 times. Cell debris and nuclei were pelleted by centrifugation at 600 × g for 5 min. The supernatant or mouse liver membrane fractions (10-50 μg of protein) were used for in vitro DGAT activity assays, performed as described (13). In some experiments, cell lysates were preincubated with 0.5 mm DGAT1 inhibitor (2-((1s,4s)-4-(4-(4-amino-7,7-dimethyl-7H-pyrimido[4,5-b][1,4]oxazin-6-yl)phenyl)cyclohexyl)acetic acid) to reduce endogenous DGAT activity. The inhibitor was synthesized by Syngene International Ltd. (Bangalore, India).
Immunoblot Analyses—Protein samples were separated by SDS-PAGE on 10% polyacrylamide gels, transferred to polyvinylidene difluoride membranes (Bio-Rad), and incubated with the following antibodies: mouse anti-FLAG-M2 (Sigma, 1:2,000), rabbit anti-FLAG (Sigma, 1:2,000), rabbit anti-DGAT2 (1:2,000), anti-calnexin (Stressgen, 1:1,000), anti-c-Myc (9E10, Calbiochem, 1:1,000), anti-cytochrome c (Upstate Biotechnologies, 1:10,000), anti-heat-shock protein 70 (HSP70, Affinity Bioreagents, 1:2,000), anti-GAPDH (Covance, 1:2,000), anti-RFP (Chemicon, 1:1,000), anti-cytochrome c oxidase subunit IV (clone 20E8, Molecular Probes, 1:1,000), anti-mouse IgG (Amersham Biosciences, 1:2,000), and anti-rabbit IgG (Bio-Rad, 1:2,000). Protein-antibody complexes were visualized with the SuperSignal West Pico kit (Pierce). Membranes were exposed to Fuji RX film. Bands were quantified by scanning densitometry with ImageJ image analysis software (National Institutes of Health, rsb.info.nih.gov/ij/).
Immunofluorescence Microscopy—COS-7 cells in 6-well dishes were transfected with 1 μg of plasmid DNA and 3 μl of FuGENE 6. 24 h after transfection, cells were trypsinized and re-plated into 12-well plates containing glass coverslips and processed for immunofluorescence microscopy. In some experiments, cells were incubated with 0.5 mm oleate complexed to 0.5% fatty acid-free bovine serum albumin for 12 h to promote lipid droplet formation. Cells were fixed with 4% paraformaldehyde in PBS for 10 min and permeabilized with 0.2% Triton X-100 in PBS for 5 min. Cells were then incubated with 3% bovine serum albumin in PBS for 5 min to block nonspecific antibody binding and then incubated at room temperature for 1 h with the indicated primary antibodies: mouse anti-FLAG-M2 (1:500) and rabbit anti-calnexin (1:200), rabbit anti-Mito(1491) (a rabbit polyclonal antibody that recognizes an unknown mitochondrial protein; a generous gift from Dr. Peter Walter, University of California, San Francisco) (1:200), or rabbit anti-giantin. For some experiments, fixed cells were incubated with mouse anti-c-Myc (1:200) and rabbit anti-FLAG (1:200). After incubation with primary antibodies, cells were exposed to goat anti-rabbit Alexa Fluor 488 (1:200) and donkey anti-mouse 594 (1:200) secondary antibodies (Molecular Probes) for 30 min. All manipulations were performed at room temperature. To visualize lipid droplets, fixed cells were stained with the neutral lipid stain, BODIPY 493/503 (Molecular Probes), during incubation with donkey anti-mouse 594 secondary antibody. In some experiments, mitochondria were visualized by incubating cells expressing FLAG-tagged DGAT2 with 0.5 μm MitoTracker Red for 20 min. Cells were fixed as described above and incubated first with mouse anti-FLAG, and then with goat anti-mouse fluorescein isothiocyanate (Calbiochem). To visualize the lipid droplet protein, adipose differentiation-related protein (ADRP), COS-7 cells were co-transfected with plasmids containing an ADRP-GFP fusion protein (a gift from C. Sztalryd, University of Maryland) and FLAG-tagged DGAT2. Cells were fixed as described above, and DGAT2 was detected by staining cells with mouse anti-FLAG and donkey anti-mouse 594. ADRP was visualized detecting GFP fluorescence. Coverslips were mounted on glass slides with a drop of Immuno-Fluore mounting medium (ICN) and allowed to set overnight. Confocal images were acquired with a LSM510 confocal microscope (Zeiss). Images were processed with LSM Image Browser (Zeiss), ImageJ, and Adobe Photoshop. Lipid droplet diameters from 2 to 4 cells (9-17 lipid droplets) were measured with the LSM Image Browser. The number of lipid droplets per cell (3-5 cells) was determined with ImageJ (25). Images shown represent the majority of transfected cells in each experiment.
Subcellular Fractionation—Microsomal, mitochondria-associated membranes (MAM), and mitochondrial fractions were isolated from cultured cells as described (26). Cells were homogenized in isolation medium (250 mm mannitol, 5 mm Tris-Cl, pH 7.4, and 0.5 mm EGTA) with 15-20 up-and-down strokes in a Potter-Elvehjem motor-driven homogenizer. The homogenate was centrifuged at 600 × g for 5 min to remove cellular debris and nuclei, and the supernatant was centrifuged at 10,000 × g for 10 min to pellet the fraction containing mitochondria plus MAM. The 10,000 × g supernatant was centrifuged at 100,000 × g for 30 min in a Beckman Ti-70.1 rotor at 4 °C to pellet microsomes, which were resuspended in isolation medium. The mitochondria plus MAM pellet was resuspended in 800 μl of isolation medium and layered on top of 8 ml of Percoll medium (225 mm mannitol, 25 mm Tris-Cl, pH 7.4, 1 mm EGTA, and 30% Percoll (v/v)) and centrifuged for 30 min at 95,000 × g. A dense band containing purified mitochondria was recovered from ∼3/4 down the tube, diluted with isolation medium, and washed twice by centrifugation at 6300 × g for 10 min. The mitochondrial pellet was resuspended in isolation medium. Purified MAMs were recovered from the Percoll gradient as a diffuse white band located above the mitochondria. MAMs were diluted with isolation medium and centrifuged at 6300 × g for 10 min. The supernatant, containing MAMs, was then centrifuged at 100,000 × g for 30 min in a Beckman Ti-70.1 rotor. MAMs were resuspended in isolation medium.
Statistical Analyses—Data are presented as mean ± S.D. Differences between groups were compared by the Kruskal-Wallis test, followed by the Tukey test.
RESULTS
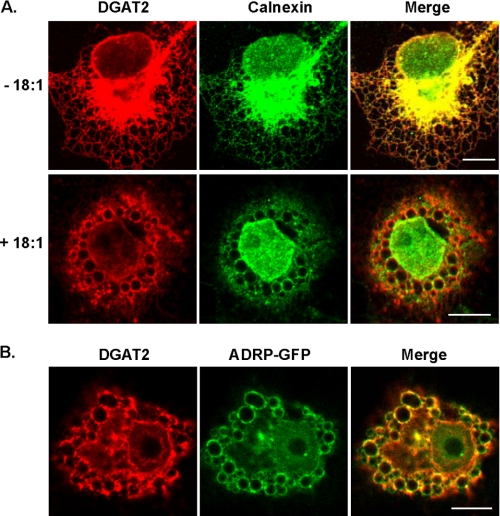
DGAT2 Is Localized to the ER and Associates with Lipid Droplets—To determine the subcellular localization of murine DGAT2, we expressed N-terminal FLAG-tagged DGAT2 in COS-7 cells and compared its distribution with that of calnexin, a resident ER membrane protein. Under basal culture conditions, DGAT2 exhibited a typical ER (reticular) staining pattern and co-localized extensively with calnexin (Fig. 1A, upper panel). DGAT2 and calnexin were also concentrated in the perinuclear region. When COS-7 cells transiently expressing DGAT2 were treated with 0.5 mm oleate for 12 h, both DGAT2 and calnexin were concentrated at the surface of large spherical structures that appeared to be lipid droplets (Fig. 1A, lower panel). Calnexin, but not DGAT2, has been identified as a lipid droplet-associated protein (27-30). Total cellular DGAT activity in McArdle RH7777 cells expressing DGAT2 and treated with 0.25 mm oleate for 12 h was not increased relative to untreated cells (data not shown), indicating that the enzymatic activity did not change upon the change in localization.
FIGURE 1.
Oleate loading alters the subcellular distribution of DGAT2. Confocal immunofluorescence microscopy of DGAT2 transiently expressed in COS-7 cells. A, COS-7 cells transfected with DGAT2 were treated with 0.5 mm oleate (+18:1) for 12 h or left untreated (-18:1) and then stained with anti-FLAG and anti-calnexin antibodies. (The nuclear calnexin staining (lower panel) likely reflects nuclear membrane staining in an image from a plane that did not bisect the nucleus.) B, DGAT2 co-localizes with ADRP-GFP. COS-7 cells were co-transfected with DGAT2 and ADRP-GFP. Transfected cells were then incubated with 0.5 mm oleate for 12 h. Cells were fixed and DGAT2 was visualized with anti-FLAG. ADRP was visualized by detecting GFP fluorescence. Scale bars, 10 μm.
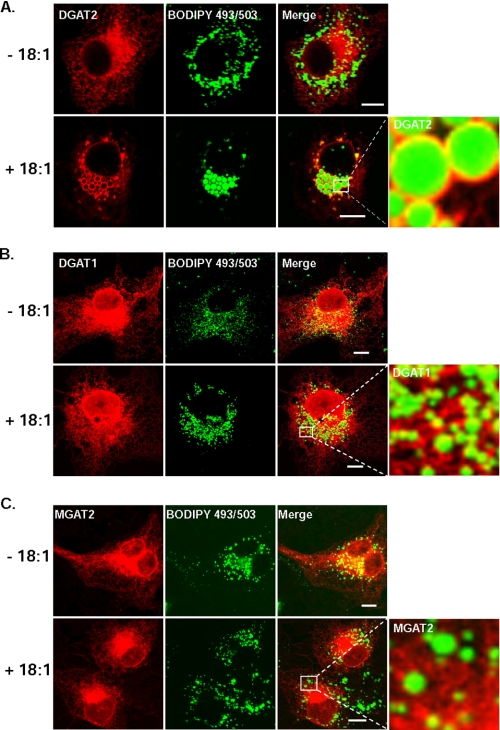
To confirm that the spherical structures were lipid droplets, we compared the localization of DGAT2 with that of a lipid droplet protein, ADRP. COS-7 cells were co-transfected with DGAT2 and a plasmid that expresses an ADRP-GFP fusion protein. In transfected cells that were treated with oleate, DGAT2 and ADRP co-localized at the surfaces of lipid droplets (Fig. 1B). We also co-stained DGAT2-transfected cells with the neutral lipid dye, BODIPY 493/503. In oleate-treated cells, DGAT2 had a striking ring-like localization around lipid droplets, which were concentrated in the perinuclear region (Fig. 2A). These findings agree with those reported by Kuerschner et al. (22), who also showed that DGAT2 co-localizes with lipid droplets in oleate-loaded COS-7 cells. In that study, a hemagglutinin epitope tag at the N or C termini of DGAT2 restricted its localization to the ER, whereas untagged DGAT2 localized predominantly to lipid droplets after lipid loading, suggesting that an epitope tag interferes with DGAT2 localization. This was apparently not the case for DGAT2 with the N-terminal FLAG-epitope that was used in our experiments. Both FLAG-epitope-tagged DGAT2 and untagged DGAT2 exhibited typical ER staining patterns and were present around lipid droplets in oleate-loaded cells (data not shown). We therefore performed all subsequent studies with FLAG-tagged DGAT2.
FIGURE 2.
DGAT2, but not DGAT1 or MGAT2, is associated with lipid droplets after oleate loading. Confocal immunofluorescence microscopy of DGAT2, DGAT1, and MGAT2 transiently expressed in COS-7 cells. Transfected COS-7 cells treated with 0.5 mm oleate (+18:1) for 12 h or left untreated (-18:1) and then stained with anti-FLAG and BODIPY 493/503 to visualize lipid droplets. Scale bars:10 μm.
Unlike DGAT2, the lipid acyltransferases DGAT1 and MGAT-2 showed predominantly an ER-staining pattern and did not redistribute to a similar degree around lipid droplets in oleate-treated cells (Fig. 2, B and C). We also examined the size and number of lipid droplets produced by cells expressing DGAT1, DGAT2, and MGAT2 that had been treated with 0.5 mm oleate for 12 h (Table 1). (The lipid droplet staining pattern of untransfected COS-7 cells treated with or without 0.5 mm oleate is shown in supplemental Fig. S1.) Lipid droplets in cells expressing DGAT1 or MGAT2 were similar in size to control cells but were ∼3-fold and ∼1.6-fold more numerous, respectively (supplemental Fig. S2). In contrast, lipid droplets in COS-7 cells expressing DGAT2 exhibited a 2-fold greater diameter than control cells and a marked reduction (∼60%) in the number of lipid droplets, which could result from coalescence of smaller droplets to produce fewer large lipid droplets. This is similar to the findings in McArdle RH7777 rat hepatoma cells stably expressing DGAT1 and DGAT2 (13), where DGAT1-transfected cells had numerous small lipid droplets, while DGAT2-transfected cells had only a few large cytosolic lipid droplets.
TABLE 1.
Analysis of lipid droplet size from cells expressing DGAT1, DGAT2, and MGAT2 that were incubated with 0.5 mm oleate for 12 h
|
Lipid droplet
|
Diameter (μm)
|
||||
|---|---|---|---|---|---|
| Control | DGAT2 | DGAT1 | MGAT2 | ||
| With oleate | 1.99 ± 0.33 | 4.11 ± 1.02a | 2.06 ± 0.20 | 1.89 ± 0.39 | |
|
Number/cell
|
|||||
| Without oleate | 131.2 ± 46.5 | 214.7 ± 60.7 | 147.0 ± 47.0 | 135.3 ± 9.5 | |
| With oleate | 88.0 ± 8.3 | 37.8 ± 11.7b | 269.7 ± 54.9c,d | 148.0 ± 43.7 | |
p < 0.001 versus control-transfected cells.
p < 0.001 versus DGAT2-expressing cells.
p < 0.001 versus oleate-loaded, control-transfected cells.
p < 0.05 versus DGAT1-expressing cells.
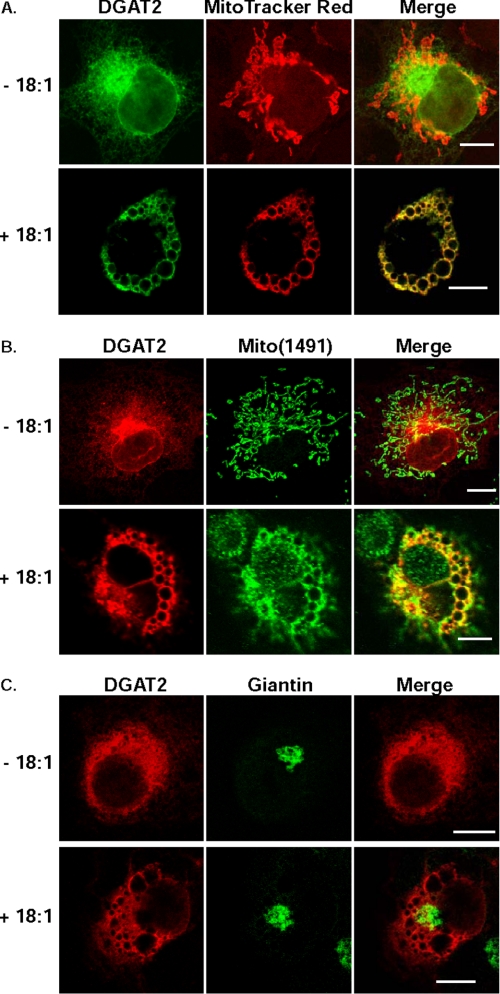
Co-localization of DGAT2 with Mitochondria after Lipid Loading—We next compared the localization of DGAT2 to that of other organelles, such as mitochondria and the Golgi apparatus. For mitochondria, we used the markers Mito-Tracker Red and Mito(1491). Mito(1491) is a protein with an undefined function that resides exclusively in the mitochondria and completely co-localizes with MitoTracker Red (supplemental Fig. S3). In COS-7 cells transfected with DGAT2 not treated with oleate, mitochondria displayed a typical worm-like staining pattern for both MitoTracker Red and Mito(1491) and partially co-localized with DGAT2 (Fig. 3, A and B, upper panels). Strikingly, when COS-7 cells expressing FLAG-DGAT2 were incubated with 0.5 mm oleate to promote lipid droplet formation, mitochondria underwent a dramatic re-organization around lipid droplets and co-localized almost completely with DGAT2 (Fig. 3, A and B, lower panels). To determine if the rearrangement around the lipid droplet surface was specific for mitochondria, we examined the subcellular distribution of the Golgi marker, giantin, under identical conditions. Unlike mitochondria, giantin did not undergo re-organization around the lipid droplet surface of oleate-loaded COS-7 cells expressing DGAT2 and displayed a typical Golgi staining pattern in both the absence and presence of oleate (Fig. 3C). Mitochondrial re-organization was not observed in COS-7 cells expressing MGAT2 or DGAT1 (data not shown).
FIGURE 3.
DGAT2 co-localizes with mitochondria after oleate loading. COS-7 cells expressing DGAT2 were incubated in the presence (+18:1) or absence (-18:1) of 0.5 mm oleate for 12 h. DGAT2 was visualized with anti-FLAG. Mitochondria were visualized with MitoTracker Red (A) or with anti-Mito(1491) (B). As a negative control, DGAT2-expressing COS-7 cells were also stained for the Golgi marker, giantin (C). Scale bars:10 μm.
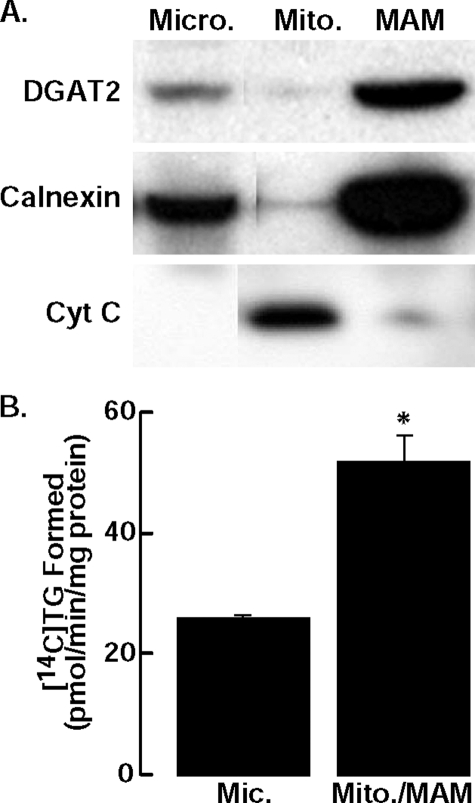
DGAT2 Is Present in MAM—DGAT activity is enriched in MAM (31), a subdomain of the ER that co-isolates with mitochondria and is enriched with lipid synthetic enzymes (26, 31, 32). The co-localization of DGAT2 with mitochondria by immunofluorescence suggested that DGAT2 is present in MAM. We therefore examined biochemical fractions of McArdle rat hepatoma cells, a cell line in which MAM has been well characterized (26), that stably expressed DGAT2 (13). Immunoblotting showed that DGAT2 and calnexin were present in the microsomal fraction and were also enriched several-fold in MAM (Fig. 4A). Our finding that calnexin is enriched in MAM is consistent with a report that the majority of calnexin in HeLa cells is present in MAM (33).
FIGURE 4.
DGAT2 is present in microsomes (Micro.) and enriched in MAM. A, subcellular fractions were isolated from McArdle cells stably expressing DGAT2. An equal amount of protein (30 μg) from each fraction was analyzed by immunoblotting with anti-DGAT2, anti-calnexin, and anti-cytochrome C (Cyt C) antibodies (Mito., mitochondria). B, in vitro DGAT activity of microsomes and mitochondria plus MAM (mito./MAM) from livers of DGAT knockout mice fed a high fat diet for 1 week. Data are from one experiment, performed in triplicate, which was repeated once with similar results. *, p = 0.038 versus mean for microsomes.
To confirm that DGAT2 is present in MAM, we also measured DGAT activity in microsomal and mitochondria plus MAM fractions that were isolated from livers from DGAT1 knockout mice that were fed a high fat diet. DGAT activity, which should reflect only that of DGAT2, was ∼2-fold higher in the mitochondria plus MAM fraction than in the microsomal fraction (Fig. 4B). These data are consistent with our immunolocalization studies. Thus, biochemical analyses of both cultured cells and murine livers show that DGAT2 is present in MAM.
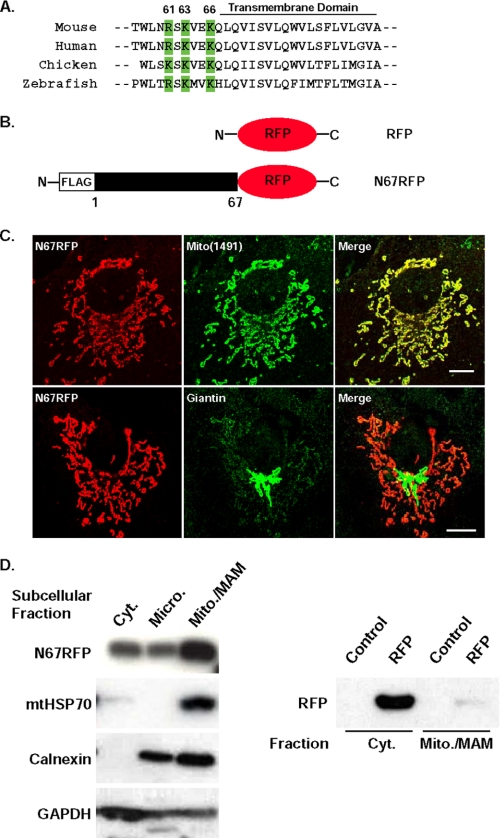
The N-terminal 67 Amino Acids of DGAT2 Targets RFP to Mitochondria—Because DGAT2 co-localized with mitochondria by immunofluorescence studies and was present in MAM, we hypothesized that it might contain a mitochondrial targeting sequence. Such sequences typically consist of a region of positively charged amino acids flanking a hydrophobic region (34). Unlike other members of the DGAT2 family, DGAT2 contains an N-terminal extension of 67 amino acids that is exposed to the cytosol (35). Within this stretch of 67 amino acids, we identified a candidate mitochondrial targeting sequence consisting of three positively charged amino acids (Arg-61, Lys-63, and Lys-66 of murine DGAT2) that immediately precede the membrane-embedded domain of DGAT2 and are highly conserved across species (Fig. 5A).
FIGURE 5.
The N-terminal 67 amino acids of DGAT2 target RFP to mitochondria. A, DGAT2 has three positively charged basic amino acids adjacent to the first transmembrane domain (green) that are highly conserved across species (Arg-61, Lys-63, and Lys-66 of mouse DGAT2). B, COS-7 cells were transfected with an expression plasmid containing amino acids 1-67 of DGAT2 fused to the N terminus of RFP (N67RFP). C, N67RFP was visualized with anti-FLAG. Mitochondria (upper panel) and Golgi (lower panel) were detected with anti-Mito(1491) and anti-giantin antibodies, respectively. D, cytosolic (Cyt.), microsomal (Micro.), and mitochondria plus MAM (Mito./MAM) fractions were isolated from cells expressing N67RFP. An equal amount of protein (15 μg) from each fraction was separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and immunoblotted with anti-FLAG, anti-HSP70, anti-calnexin, and anti-GAPDH antibodies (left panel). In cells expressing RFP alone, RFP was detected only in the cytosolic fraction with an anti-RFP antibody (right panel). Scale bars:10 μm.
To determine if the N terminus of DGAT2 is involved in mitochondrial targeting, we fused amino acids 1-67 to the N terminus of RFP (N67RFP) (Fig. 5B), expressed the fusion protein in COS-7 cells, and examined its subcellular localization. N67RFP had a worm-like staining pattern and completely co-localized with Mito(1491) (Fig. 5C, upper panel), whereas RFP alone exhibited a diffuse cytosolic expression pattern (not shown). N67RFP did not co-localize with the Golgi marker giantin (Fig. 5C, lower panel).
We also examined the subcellular distribution of N67RFP biochemically. As in the immunofluorescence studies, N67RFP protein was more abundant in the fraction containing mitochondria plus MAM than in the cytosolic and microsomal fractions (Fig. 5D, left panel). In contrast, RFP alone was found only in the cytosolic fraction and was not localized to mitochondria (Fig. 5D, right panel). The purity of each fraction was assessed by immunoblotting with known organelle markers (mitochondrial HSP70 (mtHSP70, mitochondria), calnexin (microsomes), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (cytosol)) (Fig. 5D, left panel). mtHSP70 was found exclusively in the mitochondria plus MAM fraction. Calnexin was detected in microsomes, but was most abundant in the mitochondria plus MAM fraction, likely because it contains MAM. GAPDH was most abundant in the cytosolic fraction. Thus, data from both immunolocalization and biochemical fractionation studies suggested that a mitochondrial targeting sequence resides within the first 67 amino acids of DGAT2.
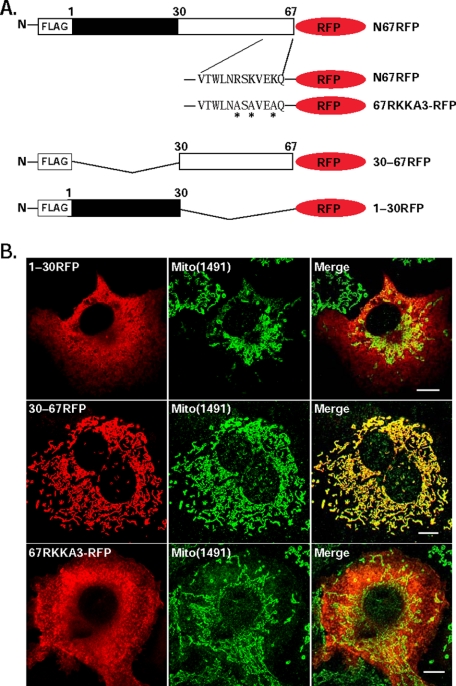
N67RFP Contains a Mitochondrial Targeting Signal—We next sought to identify the sequences within the N-terminal 67 amino acids of DGAT2 that mediate mitochondrial targeting. We initially examined the localization of constructs containing amino acids 1-30 (1-30RFP) or 30-67 (30-67RFP) of the N terminus (Fig. 6A). 1-30RFP displayed a diffuse cytoplasmic staining pattern and did not co-localize with Mito(1491) (Fig. 6B). In contrast, amino acids 30-67 were sufficient to localize RFP to mitochondria. To determine if amino acids Arg-61, Lys-63, and Lys-66 are required for targeting RFP to mitochondria, we generated a triple mutant in which these three amino acids were mutated to alanine (67RKKA3-RFP) (Fig. 6A). This mutant did not co-localize extensively with mitochondria (Fig. 6B), indicating that these three amino acids are required for mitochondrial targeting.
FIGURE 6.
Arg-61, Lys-63, and Lys-66 are required for targeting N67RFP to mitochondria. A, amino acids 1-30 (1-30RFP) and 30-67 (30-67RFP) of DGAT2 were fused to the N terminus of RFP. Additionally, amino acids Arg-61, Lys-63, and Lys-66 (a potential mitochondrial targeting sequence) were mutated (asterisks) to alanines (67RKKA3-RFP). B, RFP fusion proteins were expressed in COS-7 cells and visualized with anti-FLAG (red). Mitochondria were visualized with anti-Mito(1491) (green). Scale bars:10 μm.
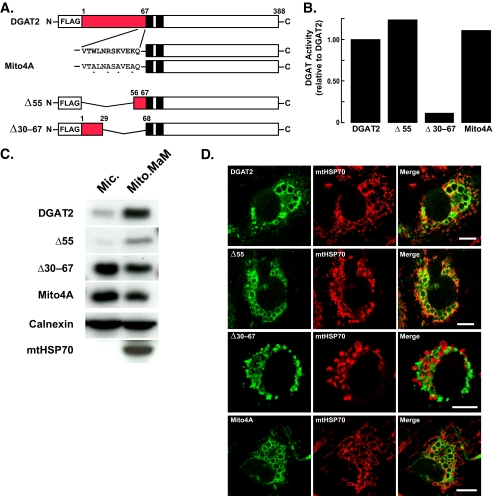
Mutagenesis of the Mitochondrial Targeting Sequence in DGAT2—To determine if the findings in studies of the RFP fusion proteins were also seen with full-length DGAT2, we expressed DGAT2 mutants in COS-7 cells. Two mutants included those in which either amino acids 1-55 (Δ55) or 30-67 (Δ30-67) were deleted and one in which key residues in the putative mitochondrial targeting signal were mutated to alanines (Arg-61, Lys-63, Lys-66, and a highly conserved tryptophan (Trp-58), termed Mito4A) (Fig. 7A). We first examined DGAT activity in cells expressing these mutants. Both the Δ55 and Mito4A mutants had levels of in vitro DGAT activity comparable to that of wild-type DGAT2 (Fig. 7B). However, the Δ30-67 deletion mutant was mostly inactive, with DGAT activity ∼90% lower than that of wild-type DGAT2 (Fig. 7B), suggesting that amino acids 56-67 are necessary for the full catalytic activity of DGAT2. The ability of DGAT2 expression to promote the formation of large lipid droplets correlated with activity levels (Fig. 7D, left panels, and supplemental Fig. S4).
FIGURE 7.
Mutagenesis of the mitochondrial targeting signal in DGAT2 disrupts mitochondrial targeting. A, map of mutant DGAT2 proteins. Red indicates the N-terminal 67 amino acids of DGAT2; black boxes indicate transmembrane domains in DGAT2; asterisks indicate point mutations. B, in vitro DGAT activities of lysates from COS-7 cells expressing DGAT2, Δ55, Δ30-67, and Mito4A. To reduce background DGAT activity, lysates from transfected cells were preincubated with 0.5 mm DGAT1 inhibitor for 20 min on ice before DGAT activity was determined. The activities of untransfected cells and cells transfected with wild-type DGAT2 were 5.9 and 317 pmol/mg protein/min, respectively. C, COS-7 cells were transfected with DGAT2, Δ55, Δ30-67, and Mito4A. Microsomal (Mic.) and mitochondrial plus MAM (Mito./MAM) fractions were isolated, and the amount of DGAT and various DGAT2 mutants in each fraction was determined by immunoblotting. D, COS-7 cells transfected with DGAT2, Δ55, Δ30-67, and Mito4A were treated with 0.5 mm oleate for 12 h and stained with anti-FLAG and BODIPY 493/503. Scale bars:10 μm.
Next, we looked for these DGAT2 mutants in biochemical fractions. Deletion of amino acids 1-55 did not appear to alter the mitochondrial association of DGAT2, because both Δ55 and DGAT2 were present in mitochondria plus MAM fraction in similar amounts (∼85-90%) (Fig. 7C). However, either deletion (Δ30-67) or point mutations (Mito4A) of the putative mitochondrial targeting sequence markedly reduced the proportions of DGAT2 in the mitochondria plus MAM fraction (Fig. 7C). Essentially all (∼98%) of the control protein HSP70 was present in the mitochondria plus MAM fraction.
We also utilized immunofluorescence to examine the co-localization of DGAT2 and the Δ55, Δ30-67, and Mito4A mutants with the mitochondrial marker HSP70. In agreement with the biochemical fractionation data, the Δ30-67 and Mito4A mutants, but not the Δ55 mutant, had reduced mitochondrial association (Fig. 7D and quantified for multiple cells in Table 2). Interestingly, the ability of DGAT2 to associate with lipid droplets was not affected by the Δ30-67, Δ55, or Mito4A mutations (Fig. 7D, left panels), nor was lipid droplet association dependent on the catalytic activity of these DGAT2 mutants.
TABLE 2.
Co-localization of mitochondria with DGAT2 and mutants in oleate-loaded COS-7 cells
| Mutant | Pearson's correlation coefficient |
|---|---|
| DGAT2 | 0.357 ± 0.184 |
| Δ55 | 0.291 ± 0.132 |
| Δ30-60 | −0.242 ± 0.190a |
| Mito4A | −0.001 ± 0.178a |
p < 0.001 versus control-transfected cells. Co-localization was determined in 3-7 cells/genotype. The degree of co-localization was calculated from 2-6 regions/cell, and each cell contained at least 3 droplets/region.
DISCUSSION
In this study, we examined the subcellular distribution of DGAT2, the major enzyme of TG biosynthesis in eukaryotes. By immunofluorescence microscopy and biochemical fractionation, we found that DGAT2 was present in the ER and enriched in MAM. DGAT2 also associated with the surface of lipid droplets and co-localized with mitochondria, particularly when cells were loaded with oleate. Targeting to the MAM and mitochondrial compartments appears to require in part a highly conserved, positively charged mitochondrial targeting signal that is present in the N terminus between amino acids 61 and 66 of murine DGAT2, where it likely promotes peripheral association of DGAT2 with mitochondria. Our findings suggest that DGAT2 is dynamically localized to different subcellular compartments, including the ER, lipid droplets, and MAM/mitochondria. This localization may promote the efficient synthesis and storage of TG in lipid droplets.
DGAT enzymes were reported decades ago to be present in the ER. Studies of microsomes from rat adipocytes or livers revealed DGAT activity (17, 18, 31, 36, 37). In this study, we focused on the localization of DGAT2, which was indeed present in the ER, as demonstrated by both immunofluorescence and biochemical fractionation studies. However, when cells were oleate-loaded to promote lipid droplet formation, DGAT2 became concentrated around lipid droplets. This localization was not found to a similar extent for either DGAT1 or MGAT2 and was not dependent on the size of the lipid droplets. Indeed, DGAT2 was present near the surfaces of both larger and smaller droplets, whereas DGAT1 and MGAT2 were not present around lipid droplets of similar size.
Our finding that DGAT2 localizes to lipid droplets agrees with a report of lipid-droplet localization for activity of Dga1p, the S. cerevisiae ortholog of DGAT2 (18). Furthermore, DGAT2 in the fungus Mortierella ramanniana was purified from the lipid body fraction (38). Recently, DGAT2 overexpressed in COS-7 cells and endogenous DGAT2 in 3T3-L1 adipocytes was reported to associate with lipid droplets (22).
Our studies show that DGAT2 co-localizes with ADRP, a lipid-droplet protein, when cells were oleate-loaded. However, because of the limited resolution of light microscopy, our studies do not distinguish between DGAT2 directly embedded on the surface of lipid droplets and DGAT2 in membrane bilayers of the ER in close proximity to lipid droplets. Murine DGAT2 has a long hydrophobic region spanning amino acids 66-115 that comprises either two transmembrane domains or a single extended domain that is embedded in the bilayer (35). The latter conformation could enable the protein to directly associate with the lipid droplet surface. Indeed, recent localization studies using immunoelectron microscopy showed partial localization of DGAT2 on purified lipid droplets (22). On the other hand, a conformation with two membrane-spanning domains would make it more likely that DGAT2 is present in a membrane bilayer in close proximity to the droplet. The latter possibility is suggested by the results of numerous analyses of the proteomes of lipid droplets isolated from cells and tissues (27-30, 39-44) that did not identify DGAT2 as a droplet component.
We also found that DGAT2 was present in MAM fractions isolated from McArdle RH7777 hepatoma cells, and DGAT activity in livers of DGAT1 knockout mice was enriched in mitochondria plus MAM fractions compared with microsomes. MAM is an ER-like membrane fraction that co-isolates with mitochondria and is enriched in activities of lipid biosynthetic enzymes, such as acyl-CoA:cholesterol acyltransferase, DGAT, and phosphatidylserine synthase-1 and -2 (26, 31, 32). MAM is hypothesized to serve as a membrane bridge between the ER and mitochondria and to facilitate the transport of lipids between these organelles (45-48). Our data show that DGAT2 accounts for at least a portion of DGAT activity in the MAM compartment.
An unexpected finding was the striking co-localization of DGAT2, but not DGAT1 or MGAT2, with mitochondria in cells that were oleate-loaded. These findings suggested that a portion of DGAT2 overexpressed in cells associates with mitochondria. Unfortunately, a polyclonal DGAT2 antibody did not recognize endogenous DGAT2 in cells such as 3T3-L1 adipocytes. Therefore, we were unable to determine whether endogenous DGAT2 undergoes the same mitochondrial association/rearrangement in oleate-loaded cells.
The presence of DGAT2 in MAM and the striking co-localization of DGAT2 with mitochondria in immunofluorescence studies prompted us to investigate whether DGAT2 had a mitochondrial targeting sequence. We focused on the N-terminal region of DGAT2, which contains a 67-amino acid region that is not found in other family members (e.g. acyl-MGATs or wax synthases). Amino acids 1-67 targeted RFP to the mitochondria, where it became peripherally associated with the outer mitochondrial membrane. Further, three positively charged amino acids, Arg-61, Lys-63, and Lys-66, were required for the mitochondrial association of RFP and promoted the mitochondrial association of DGAT2. These amino acids are located just proximal to the membrane-embedded regions of DGAT, conforming to the typical location of mitochondrial targeting signals (34).
Our findings raise the question of whether DGAT2 is a true mitochondrial enzyme or is present in a compartment, such MAM, which is associated with mitochondria. Based on our data, we favor a model in which DGAT2 is enriched in MAM, and DGAT2 in this compartment associates with mitochondria as a peripheral protein via its N terminus. In this model, DGAT2 would not be present directly in mitochondrial membranes. In support of this conjecture, ∼700 proteins have been identified in the mitochondrial proteomes from human and mouse tissues (49-51), but DGAT2 was not found.
Why DGAT2 in MAM associates with mitochondria is unclear. We speculate that this arrangement may serve as a bridge between the MAM and mitochondria that promotes the exchange of substrates and products for TG synthesis. Consistent with this notion, many enzymes of the glycerolipid (Kennedy) pathway of lipid synthesis, which is responsible for the synthesis of both TG and phospholipids, are located in both ER and mitochondria (1, 52). Furthermore, the close proximity of lipid droplets, mitochondria, and the ER has been reported by several other investigators (53-55). These findings provide further evidence that these three organelles are intimately associated, possibly to promote the efficient synthesis of lipids.
Our findings in this study provide insights into the spatial compartmentalization of lipid synthesis and storage. DGAT2, the major enzyme of TG synthesis, is not localized only to the ER; it is dynamically associated with lipid droplets and the MAM and mitochondrial compartments, which are important during active lipid synthesis. Our findings support the notion that robust lipid synthesis and storage are characterized by a reorganization of organelles to maximize this process. Through a bridging function, the N terminus of DGAT2 may promote this reorganization. Further studies of this enzymatic machinery and lipid droplet biogenesis will undoubtedly shed more light on this complex yet fascinating aspect of cell biology.
Supplementary Material
Acknowledgments
We thank R. Tsien for the RFP expression plasmid, P. Walter for use of a confocal microscope and the Mito(1491) antibody, C. Sztalryd for the ADRP-GFP expression plasmid, S. Ord-way and G. Howard for editorial assistance, D. Jones for manuscript preparation, and J. Vance for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01-DK065599 (to R. V. F.). This work was also supported by an American Heart Association scientist development grant (to S. S.), a Hill-blom postdoctoral fellowship (to M. L.), and the J. David Gladstone Institutes and National Institutes of Health/NCRR Grant CO6RR018928 to the J. David Gladstone Institutes. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S4.
Footnotes
The abbreviations used are: TG, triacylglycerol; ADRP, adipose differentiation-related protein; BODIPY 493/503, 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene; DGAT, acyl-CoA:diacylglycerol acyltransferase; ER, endoplasmic reticulum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; HSP70, heat-shock protein 70; MAM, mitochondria-associated membranes; MGAT, monoacylglycerol acyltransferase; 18:1, oleate; RFP, monomeric red fluorescent protein; PBS, phosphate-buffered saline.
References
- 1.Coleman, R. A., and Lee, D. P. (2004) Prog. Lipid Res. 43 134-176 [DOI] [PubMed] [Google Scholar]
- 2.Lewin, T. M., Kim, J.-H., Granger, D. A., Vance, J. E., and Coleman, R. A. (2001) J. Biol. Chem. 276 24674-24679 [DOI] [PubMed] [Google Scholar]
- 3.Gimeno, R. E., and Cao, J. (2008) J. Lipid Res. 49 2079-2088 [DOI] [PubMed] [Google Scholar]
- 4.Shindou, H., and Shimizu, T. (2009) J. Biol. Chem. 284 1-5 [DOI] [PubMed] [Google Scholar]
- 5.Han, G. S., Wu, W. I., and Carman, G. M. (2006) J. Biol. Chem. 281 9210-9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jamal, Z., Martin, A., Gomez-Muñoz, A., and Brindley, D. N. (1991) J. Biol. Chem. 266 2988-2996 [PubMed] [Google Scholar]
- 7.Bell, R. M., and Coleman, R. A. (1980) Annu. Rev. Biochem. 49 459-487 [DOI] [PubMed] [Google Scholar]
- 8.Brindley, D. N. (1991) in Biochemistry of Lipids, Lipoproteins and Membranes (Vance, D. E., and Vance, J. E., eds) pp. 171-203, Elsevier, Amsterdam
- 9.Gunstone, F. D., Harwood, J. L., and Padley, F. B. (eds) (1994) The Lipid Handbook, pp. 646-651, Chapman & Hall, London
- 10.Cases, S., Smith, S. J., Zheng, Y.-W., Myers, H. M., Lear, S. R., Sande, E., Novak, S., Collins, C., Welch, C. B., Lusis, A. J., Erickson, S. K., and Farese, R. V., Jr. (1998) Proc. Natl. Acad. Sci. U. S. A 95 13018-13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cases, S., Stone, S. J., Zhou, P., Yen, E., Tow, B., Lardizabal, K. D., Voelker, T., and Farese, R. V., Jr. (2001) J. Biol. Chem. 276 38870-38876 [DOI] [PubMed] [Google Scholar]
- 12.Yen, C. L., Stone, S. J., Koliwad, S., Harris, C., Farese, R. V., Jr. (2008) J. Lipid Res. 49 2283-2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stone, S. J., Myers, H. M., Watkins, S. M., Brown, B. E., Feingold, K. R., Elias, P. M., and Farese, R. V., Jr. (2004) J. Biol. Chem. 279 11767-11776 [DOI] [PubMed] [Google Scholar]
- 14.Oelkers, P., Cromley, D., Padamsee, M., Billheimer, J. T., and Sturley, S. L. (2002) J. Biol. Chem. 277 8877-8881 [DOI] [PubMed] [Google Scholar]
- 15.Sandager, L., Gustavsson, M. H., Stahl, U., Dahlqvist, A., Wiberg, E., Banas, A., Lenman, M., Ronne, H., and Stymne, S. (2002) J. Biol. Chem. 277 6478-6482 [DOI] [PubMed] [Google Scholar]
- 16.Sorger, D., and Daum, G. (2002) J. Bacteriol. 184 519-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehner, R., and Kuksis, A. (1996) Prog. Lipid Res. 35 169-201 [DOI] [PubMed] [Google Scholar]
- 18.Wilgram, G. F., and Kennedy, E. P. (1963) J. Biol. Chem. 238 2615-2619 [PubMed] [Google Scholar]
- 19.Wakimoto, K., Chiba, H., Michibata, H., Seishima, M., Kawasaki, S., Okubo, K., Mitsui, H., Torii, H., and Imai, Y. (2003) Biochem. Biophys. Res. Commun. 310 296-302 [DOI] [PubMed] [Google Scholar]
- 20.Cao, J., Cheng, L., and Shi, Y. (2007) J. Lipid Res. 48 583-591 [DOI] [PubMed] [Google Scholar]
- 21.Shockey, J. M., Gidda, S. K., Chapital, D. C., Kuan, J. C., Dhanoa, P. K., Bland, J. M., Rothstein, S. J., Mullen, R. T., and Dyer, J. M. (2006) Plant Cell 18 2294-2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuerschner, L., Moessinger, C., and Thiele, C. (2008) Traffic 9 338-352 [DOI] [PubMed] [Google Scholar]
- 23.Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (2002) Proc. Natl. Acad. Sci. U. S. A 99 7877-7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N. G., Palmer, A. E., and Tsien, R. Y. (2004) Nat. Biotech. 22 1567-1572 [DOI] [PubMed] [Google Scholar]
- 25.Abramoff, M. D., Magelhaes, P. J., and Ram, S. J. (2004) Biophotonics Int. 11 36-42 [Google Scholar]
- 26.Stone, S. J., and Vance, J. E. (2000) J. Biol. Chem. 275 34534-34540 [DOI] [PubMed] [Google Scholar]
- 27.Brasaemle, D. L., Dolios, G., Shapiro, L., and Wang, R. (2004) J. Biol. Chem. 279 46835-46842 [DOI] [PubMed] [Google Scholar]
- 28.Sato, S., Fukasawa, M., Yamakawa, Y., Natsume, T., Suzuki, T., Shoji, I., Aizaki, H., Miyamura, T., and Nishijima, M. (2006) J. Biochem. (Tokyo) 139 921-930 [DOI] [PubMed] [Google Scholar]
- 29.Turro, S., Ingelmo-Torres, M., Estanyol, J. M., Tebar, F., Fernandez, M. A., Albor, C. V., Gaus, K., Grewal, T., Enrich, C., and Pol, A. (2006) Traffic 7 1254-1269 [DOI] [PubMed] [Google Scholar]
- 30.Wan, H.-C., Melo, R. C. N., Jin, Z., Dvorak, A. M., and Weller, P. F. (2007) FASEB J. 21 167-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusinol, A. E., Cui, Z., Chen, M. H., and Vance, J. E. (1994) J. Biol. Chem. 269 27494-27502 [PubMed] [Google Scholar]
- 32.Cui, Z., Vance, J. E., Chen, M. H., Voelker, D. R., and Vance, D. E. (1993) J. Biol. Chem. 268 16655-16663 [PubMed] [Google Scholar]
- 33.Myhill, N., Lynes, E. M., Nanji, J. A., Blagoveshchenskaya, A. D., Fei, H., Simmen, K. C., Cooper, T. J., Thomas, G., and Simmen, T. (2008) Mol. Biol. Cell 19 2777-2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rapaport, D. (2003) EMBO Rep. 4 948-952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stone, S. J., Levin, M. C., and Farese, R. V., Jr. (2006) J. Biol. Chem. 281 40273-40282 [DOI] [PubMed] [Google Scholar]
- 36.Andersson, M., Wettesten, M., Borén, J., Magnusson, A., Sjöberg, A., Rustaeus, S., and Olofsson, S.-O. (1994) J. Lipid Res. 35 535-545 [PubMed] [Google Scholar]
- 37.Coleman, R., and Bell, R. M. (1976) J. Biol. Chem. 251 4537-4543 [PubMed] [Google Scholar]
- 38.Lardizabal, K. D., Mai, J. T., Wagner, N. W., Wyrick, A., Voelker, T., and Hawkins, D. J. (2001) J. Biol. Chem. 276 38862-38869 [DOI] [PubMed] [Google Scholar]
- 39.Beller, M., Riedel, D., Jansch, L., Dieterich, G., Wehland, J., Jackle, H., and Kuhnlein, R. P. (2006) Mol. Cell Proteomics 5 1082-1094 [DOI] [PubMed] [Google Scholar]
- 40.Cermelli, S., Guo, Y., Gross, S. P., and Welte, M. A. (2006) Curr. Biol. 16 1783-1795 [DOI] [PubMed] [Google Scholar]
- 41.Fujimoto, Y., Itabe, H., Sakai, J., Makita, M., Noda, J., Mori, M., Higashi, Y., Kojima, S., and Takano, T. (2004) Biochim. Biophys. Acta 1644 47-59 [DOI] [PubMed] [Google Scholar]
- 42.Liu, P., Ying, Y., Zhao, Y., Mundy, D. I., Zhu, M., and Anderson, R. G. (2004) J. Biol. Chem. 279 3787-3792 [DOI] [PubMed] [Google Scholar]
- 43.Umlauf, E., Csaszar, E., Moertelmaier, M., Schuetz, G. J., Parton, R. G., and Prohaska, R. (2004) J. Biol. Chem. 279 23699-23709 [DOI] [PubMed] [Google Scholar]
- 44.Wu, C. C., Howell, K. E., Neville, M. C., Yates, J. R., III, and McManaman, J. L. (2000) Electrophoresis 21 3470-3482 [DOI] [PubMed] [Google Scholar]
- 45.Ardail, D., Gasnier, F., Lerme, F., Simonot, C., Louisot, P., and Gateau-Roesch, O. (1993) J. Biol. Chem. 268 25985-25992 [PubMed] [Google Scholar]
- 46.Ardail, D., Lerme, F., and Louisot, P. (1991) J. Biol. Chem. 266 7978-7981 [PubMed] [Google Scholar]
- 47.Shiao, Y. J., Lupo, G., and Vance, J. E. (1995) J. Biol. Chem. 270 11190-11198 [DOI] [PubMed] [Google Scholar]
- 48.Vance, J. E. (1990) J. Biol. Chem. 265 7248-7256 [PubMed] [Google Scholar]
- 49.Johnson, D. T., Harris, R. A., French, S., Blair, P. V., You, J., Bemis, K. G., Wang, M., and Balaban, R. S. (2007) Am. J. Physiol. 292 C689-C697 [DOI] [PubMed] [Google Scholar]
- 50.Mootha, V. K., Bunkenborg, J., Olsen, J. V., Hjerrild, M., Wisniewski, J. R., Stahl, E., Bolouri, M. S., Ray, H. N., Sihag, S., Kamal, M., Patterson, N., Lander, E. S., and Mann, M. (2003) Cell 115 629-640 [DOI] [PubMed] [Google Scholar]
- 51.Taylor, S. W., Fahy, E., Zhang, B., Glenn, G. M., Warnock, D. E., Wiley, S., Murphy, A. N., Gaucher, S. P., Capaldi, R. A., Gibson, B. W., and Ghosh, S. S. (2003) Nat. Biotech. 21 281-286 [DOI] [PubMed] [Google Scholar]
- 52.Coleman, R. A., Lewin, T. M., and Muoio, D. M. (2000) Annu. Rev. Nutr. 20 77-103 [DOI] [PubMed] [Google Scholar]
- 53.Cinti, S. (2001) Proc. Nutr. Soc. 60 319-328 [DOI] [PubMed] [Google Scholar]
- 54.Stemberger, B. H., Walsh, R. M., and Patton, S. (1984) Cell Tissue Res. 236 471-475 [DOI] [PubMed] [Google Scholar]
- 55.Sturmey, R. G., O'Toole, P. J., and Leese, H. J. (2006) Reproduction 132 829-837 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.