Abstract
A balance between ubiquitination and deubiquitination regulates numerous cellular processes and pathways, and specific deubiquitinating enzymes often play the decisive role of controlling this balance. We recently reported that the USP1 deubiquitinating enzyme, which regulates the Fanconi anemia pathway by deubiquitinating the central player of the pathway, FANCD2, is activated by the WD40-repeat containing UAF1 protein through formation of a stable USP1/UAF1 protein complex. Here we present the isolation of two novel multisubunit deubiquitinating enzyme complexes containing USP12 and USP46, respectively. Both complexes contain the UAF1 protein as a bona fide subunit. Interestingly, UAF1 regulates the enzymatic activity of both enzyme complexes, suggesting that this activator protein may regulate a subclass of human deubiquitinating enzymes. We postulate that additional WD40-containing proteins may also form complexes with other human deubiquitinating enzymes and thereby regulate their activity and substrate specificity.
Ubiquitination and deubiquitination regulate a number of essential biological processes such as gene transcription, DNA replication, and DNA repair (1). Ubiquitin modifications can be divided into three principally different types. First, monoubiquitination may alter the activity of the substrate, as described for the FANCD2 protein of the Fanconi anemia (FA)2 pathway (2) and the PCNA protein involved in Trans Lesion Synthesis (2, 3), or may alter the cellular localization of the protein (4). Second, polyubiquitination through K48-linkage typically targets the protein substrate for degradation by the proteasome (5). Third, polyubiquitination through the K63-linkage can alter the activity of the protein by modifying its protein-protein interaction properties. A recent example of K63 polyubiquitination is that of the histone variant H2AX, which is polyubiquitinated in response to DNA damage, and as such, is believed to orchestrate the recruitment of DNA repair factors to sites of DNA damage on the chromatin (6).
Processes regulated by ubiquitination are often controlled by the opposing enzymatic reaction, namely deubiquitination. First, accurate deubiquitination of the FANCD2 protein by the USP1/UAF1 complex is essential for an intact Fanconi anemia pathway. Loss of USP1 activity leads to accumulation of monoubiquitinated FANCD2, dysregulation of the FA pathway, and cellular hypersensitivity to DNA cross-linking agents (7-9). Second, failure to deubiquitinate Cdc20 as part of the APC-inhibitory Mad2-Cdc20 complex by USP44, leads to an anaphase entry defect (10). Third, the USP22 deubiquitinating enzyme, as a subunit of the SAGA complex, is critical for appropriate progression through the cell cycle due to its function in transcriptional regulation by deubiquitinating monoubiquitinated histone H2B (11).
There are ∼95 deubiquitinating enzymes in human (12). The family of deubiquitinating enzymes is divided into five subfamilies, including the USP subfamily (58 members), the UCH subfamily (4 members), the MJD subfamily (5 members), OTU subfamily (14 members), and the JAMM subfamily (14 members). The exact biological function of the majority of these enzymes is currently unknown. However, for those enzymes whose function has been uncovered, it has become apparent that regulation of their activities is essential for integrity of the pathways they regulate.
A critical question has remained unanswered, namely, how are deubiquitinating enzymes regulated. We previously reported that one deubiquitinating enzyme, USP1, which controls the Fanconi anemia pathway, is activated by a novel protein called UAF1. UAF1 forms a complex with USP1 in vivo, and this protein complex has high deubiquitinating enzyme activity, as opposed to free USP1, which by itself is nearly inactive. Thus, UAF1 regulates the activity of the USP1 enzyme. Here we describe the identification of two novel deubiquitinating enzyme complexes containing the USP12 and the USP46 enzymes, respectively. Interestingly, the UAF1 factor tightly regulates the activity of both complexes. Thus we describe a more general mechanism for regulation of human deubiquitinating enzymes.
EXPERIMENTAL PROCEDURES
Cell Lines, Antibodies, and Plasmids—HeLa cells were grown in Dulbecco's or Joklik's MEM (Invitrogen or Sigma, respectively) supplemented with 10% fetal bovine serum. Stable HeLa cells expressing USP1 or UAF1 knockdown plasmids were generated as previously described (9). shRNA target sequences against USP12 and USP46, were 5′-GAAGAGAGAAAGCAGGAAA-3′ and 5′-CCATGAAACTTACGCAGTA-3′, respectively. USP12 and USP46 cDNAs were cloned from a HeLa cDNA library, using standard cloning methods (13). Antibodies used were as follows: Rabbit anti-USP1 antibody (7); mouse anti-α-tubulin (CP06, Calbiochem or T5168, Sigma); mouse anti-HA (clone 12CA5); rabbit anti-UAF1 (9); rabbit anti-USP12 polyclonal antibodies were raised by immunizing a rabbit with an N-terminal His-tagged fusion protein of USP12 according to standard immunization methods (14).
Mass Spectrometric Analysis—Proteins were reduced with dithiothreitol, cysteine residues were derivatized with iodoacetamide, and the proteins were separated by SDS-PAGE. Proteins from Silver-stained gel bands were in-gel digested with trypsin (15). The generated peptide mixtures were subjected to LC-MS/MS using a hybrid linear ion trap/FT-ICR mass spectrometer (LTQ FT, Thermo Electron) essentially as described previously (16). MS/MS spectra were assigned by searching them with the SEQUEST algorithm (17) against the Human International Protein Index sequence data base.
Protein Purification—The UAF1 complex was purified from nuclear extracts prepared from HeLa cells expressing N-terminal Flag- and HA epitope-tagged UAF1 as described (9). For density gradient sedimentation, 200 μl of the Flag peptide-eluted material was loaded onto a 4.8-ml glycerol gradient (10-40%) and spun at 55,000 rpm in a Beckman SW55Ti rotor for 2 h. 200-μl fractions were collected from the top of the gradient using a Brandel BR-184 density gradient fractionator.
Proteins purified from Sf9 cells were expressed using either the pFastBac-HTa vector (Invitrogen) containing an N-terminal His tag or the pFastBac vector (Invitrogen) with an engineered Flag tag and purified as described (9).
In Vitro Enzymatic Assays—In vitro enzymatic assays in the presence of 5 μm ubiquitin-VS (ubiquitin-vinyl sulfone; U-202; Boston Biochem) were performed 30 °C. In vitro enzymatic assays using ubiquitin-AMC (ubiquitin-7-amido-4-methylcoumarin; U-550; Boston Biochem) were performed in 50-100 μl of reaction buffer (20 mm HEPES-KOH pH 7.8, 20 mm NaCl, 0.1 mg/ml ovalbumin (A7641; Sigma), 0.5 mm EDTA, and 10 mm dithiothreitol) at 37 °C. Fluorescence was monitored in a Fluo-Star Galaxy Fluorometer (BMG Labtech Inc.).
RESULTS
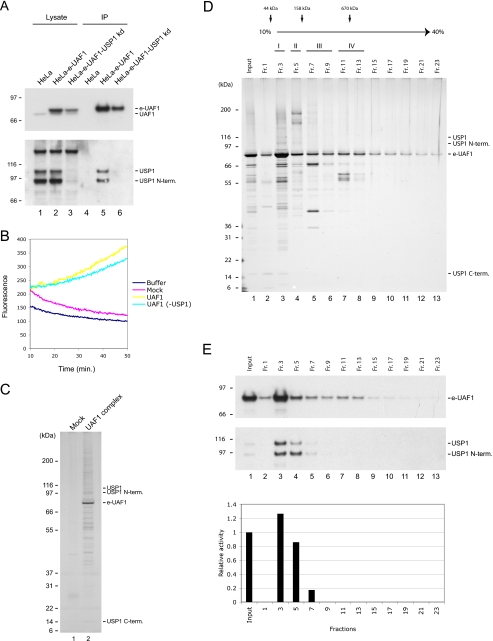
UAF1 Associates with a Deubiquitinating Enzyme Activity Different from USP1—We previously demonstrated that the WD40-repeat containing protein UAF1 forms a stable complex with the USP1 deubiquitinating enzyme in vivo and that the interaction both stabilizes and activates USP1 (9). To test the possibility that UAF1 regulates other deubiquitinating enzymes, we asked whether UAF1 co-purifies with deubiquitinating activity when purified from cells in which the USP1 protein in stably knocked down by shRNA (Fig. 1). As expected, purification of epitope-tagged e-UAF1 as a native complex from either wild-type HeLa cells or USP1 knockdown HeLa cells, demonstrated the presence or absence, respectively, of USP1 (Fig. 1A, lanes 5-6). However, when we assayed the deubiquitinating enzyme activity of the two UAF1 purifications, using ubiquitin-7-amido-4-methylcoumarin (ubiquitin-AMC) as a substrate, the activity was essentially identical, despite the absence of USP1 in the second purification (Fig. 1B). A mock purification from untransduced HeLa cells contained no detectable deubiquitinating enzyme activity. These data suggested that UAF1 interacts with other deubiquitinating enzymes in addition to USP1 in vivo.
FIGURE 1.
Purification of native UAF1 complexes containing USP1 and other deubiquitinating enzymes. A, Flag-HA-tagged UAF1 was immunoprecipitated from either HeLa cell extracts or cell extracts prepared from HeLa cells where the endogenous USP1 protein is stably knocked down by shRNA. The immunoprecipitate was analyzed by immunoblotting using the indicated antibodies. B, the immunoprecipitates from A were analyzed for associated deubiquitinating enzyme activity using ubiquitin-AMC as a substrate. C, silver stain of the UAF1 complexes. D, glycerol gradient sedimentation of the UAF1 complexes. Fractions were separated by SDS-PAGE, and proteins were visualized by silver stain. Subcomplexes as determined by sedimentation profile are indicated by roman numerals I-IV. Sedimentation profile of molecular weight markers is indicated. E, the fractions from C were analyzed by immunoblotting using the indicated antibodies (upper panels) and for deubiquitinating enzyme activity using ubiquitin-AMC (lower panel).
To further explore the nature of the UAF1-associated deubiquitinating enzyme activity, we visualized the protein subunits of the native UAF1 complex by silver stain (Fig. 1C). As predicted, we observed full-length USP1 as well as both of its N- and C-terminal cleavage products (lane 2). The identities of these polypeptides were verified by immunoblotting (data not shown). We previously demonstrated the existence of an enzymatically competent ternary complex composed of UAF1 and these two cleavage products of USP1, in good agreement with the current data (9). In addition to the known USP1 derived polypeptides, we observed several additional polypeptides that were not present in a mock purification, suggesting that they are bona fide subunits of the UAF1 complex (Fig. 1C, lane 2). The relatively large number of polypeptides indicates that multiple protein subcomplexes exist rather than one large complex. To address this directly, we further fractionated the purified complexes by a 10-40% glycerol gradient sedimentation and analyzed the fractions by silver stain. The sedimentation profile suggested the existence of at least four distinct protein complexes containing UAF1 (complexes I-IV) (Fig. 1D). In good agreement with previous data showing that the USP1/UAF1 complex is a stoichiometric heterodimer, USP1 sedimented at a relatively low molecular weight, peaking in fractions 3-5 (Complex I, II) (Fig. 1D). Immunoblot blot analysis further confirmed that both UAF1 and USP1 peak in these fractions (Fig. 1E, fractions 3-5).
We next went on to assay the deubiquitinating enzyme activity in the various fractions using ubiquitin-AMC as a substrate. We detected activity in fractions 3-7 with a peak in fraction 3 (Fig. 1E), corresponding to the peak of USP1. These data suggested that the unidentified deubiquitinating enzyme complex has a molecular weight similar to the USP1/UAF1 complex and correspondingly co-sediments in the glycerol gradient sedimentation.
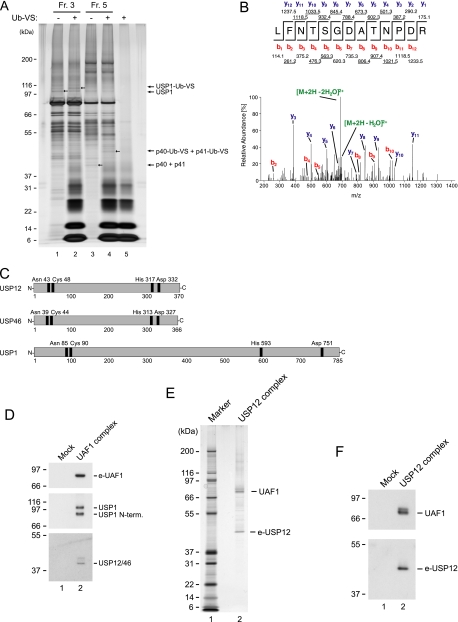
Identification of the UAF1-associated Deubiquitinating Enzyme Activity—To identify the UAF1-associated deubiquitinating enzyme(s), we used an affinity labeling approach. Many active deubiquitinating enzymes interact with and form a covalent complex with the ubiquitin derivative ubiquitin-vinyl sulfone (Ub-VS) upon incubation in vitro (18). The resulting deubiquitinating enzyme/ubiquitin complex will migrate ∼8 kDa slower in SDS-PAGE than the free deubiquitinating enzyme, corresponding to the molecular weight of the covalently attached ubiquitin protein. We subjected fractions 3 and 5 to an in vitro incubation with Ub-VS, and analyzed the resulting polypeptides by silver stain. As expected, we observed a shift of USP1 in fraction 3 where the protein is most abundant (Fig. 2A, lane 2). Importantly, we also observed a shift of two additional very closely migrating polypeptides with molecular weights of ∼40-41 kDa. The shift of these closely migrating polypeptides suggested that they may constitute the observed UAF1-associated deubiquitinating enzyme activity. Therefore, we excised and trypsinized the corresponding Silver stained band and analyzed the resulting peptides by mass spectrometry (LC-MS/MC). The analysis leads to the identification of the deubiquitinating enzyme USP12 (Fig. 2B).
FIGURE 2.
Identification of the USP12- and USP46-containing UAF1 complexes. A, fractions 3 and 5 fromtheglycerol-sedimented UAF1complex were incubated invitro with ubiquitin-VS and analyzed by silver stain. Arrows indicate USP1 and USP1-ubiquitin-VS complex (lanes 1-2). Likewise, arrows indicate putative deubiquitinating enzymes with molecular weights of 40 and 41 kDa, unbound or bound to ubiquitin-VS (lanes 3-4). In lane 5 only ubiquitin-VS is added. B, USP12 was identified by tandem mass spectrometry. The MS/MS spectrum of the peptide Leu-286-Arg-298 is shown. C, schematic of the USP12, UPS46 and USP1-deubiquitinating enzymes. Positions of the catalytic triad cysteine, histidine, and aspartic acid residues are indicated, as well as the position of the conserved asparagine. D, immunoblot analysis of the UAF1 complex using the indicated antibodies. E, silver stain of the USP12 complex. F, immunoblot analysis of the USP12 complex using the indicated antibodies.
USP12 is a 370-amino acid protein belonging to the USP family of cysteine ubiquitin proteases, whose exact biological function currently is unknown. USP12 is highly homologous to USP46, which is 366-amino acid long, and the overall homology between the two proteins is 88% (Fig. 2C). Like USP1, USP12, and USP46 contain the three conserved catalytical triad amino acid residues. The active site cysteine is located in the N-terminal end of all three proteins whereas the histidine and aspartic acid residues are located in the C-terminal ends of all three proteins (Fig. 2C).
We next raised a rabbit antibody against USP12, which due to the high homology between USP12 and USP46, recognizes both proteins. Using this antibody, we analyzed the native UAF1 complex by immunoblotting. As expected, in addition to UAF1, we observed two very closely migrating bands of 40 and 41 kDa, corresponding to USP12 and USP46, respectively (Fig. 2D).
The data suggest that three UAF1 deubiquitinating enzyme complexes exist in vivo, namely the USP1/UAF1, USP12/UAF1, and USP46/UAF1 complexes. To further reinforce this notion, we stably expressed epitope-tagged USP12 and USP46 proteins in HeLa cells, and purified the two proteins together with associated proteins as native protein complexes. Silver stain analysis of the purified complexes revealed the presence of the epitope-tagged proteins but also an additional 80-kDa polypeptide, corresponding to the molecular weight of UAF1 (Fig. 2E and data not shown). Immunoblot analysis confirmed the identity of this polypeptide as UAF1 (Fig. 2F). The silver stain profile suggested that the proteins form a stoichiometric complex between UAF1 and the deubiquitinating enzyme, as we previously observed for the USP1/UAF1 complex (9). The predicted molecular weight of such heterodimeric complexes correlates well with the observed size sedimentation profile (Fig. 1D).
Taken together, we have identified three separate deubiquitinating enzyme complexes. Each complex contains a common subunit, namely the WD40-repeat protein UAF1 as well as a unique deubiquitinating enzyme subunit, either USP1, USP12, or USP46.
UAF1 Regulates the Enzymatic Activity of USP12 and USP46—We previously demonstrated that the USP1 deubiquitinating enzyme has weak activity by itself, and that its activity is stimulated 33-fold upon forming a heterodimeric complex with the UAF1 protein (9). We concluded that the enzymatic activity of USP1 is regulated by UAF1. The present data, demonstrating the identification of two additional deubiquitinating enzyme complexes, both containing UAF1 as a bona fide subunit, suggests that UAF1 may regulate the activity of these enzyme complexes as well. To address this question, we purified both USP12 and USP46, either alone or as heterodimeric protein complexes with UAF1, from Sf9 insect cells.
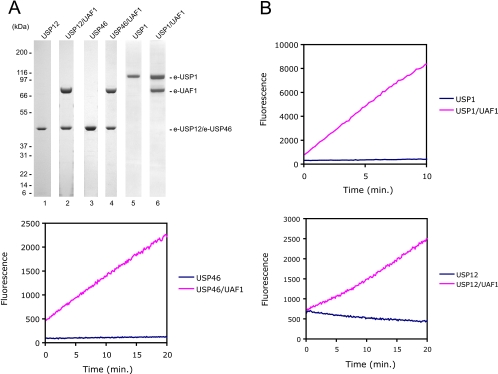
We were able to express and purify both USP12 and USP46 to homogeneity from Sf9 cells (Fig. 3A). As predicted, when the enzymes were co-expressed with UAF1, they were purified as stoichiometric heterodimeric protein complexes with UAF1 (Fig. 3A). These data confirm the existence of stable USP12/UAF1 and USP46/UAF1 complexes in vivo.
FIGURE 3.
UAF1 activates USP12 and USP46. A, Coomassie Blue stain of the proteins purified from Sf9 cells. B, in vitro enzymatic activity of USP12, USP12/UAF1, USP46, USP46/UAF1, USP1, and USP1/UAF1 was assayed using Ub-AMC as a substrate. The concentration of each protein was 100 nm except for USP1 and USP1/UAF1, which were 20 nm.
We next asked whether the enzymatic activity of USP12 and USP46 is regulated by UAF1. To this end we assessed the deubiquitinating enzyme activity of the enzymes, alone or as protein complexes with UAF1, by an in vitro deubiquitinating enzyme assay using ubiquitin-AMC as a substrate. USP12 and USP46 converted the substrate at a very low rate of turnover (Fig. 3B). The USP12/UAF1 and USP46/UAF1 complexes, on the other hand, possessed strong deubiquitinating enzyme activity. Therefore, UAF1 regulates the activity of both USP12 and USP46. Importantly, UAF1 did not stimulate the enzymatic activity of USP2, USP5, USP7, and USP14, underscoring the specificity of the activation (data not shown).
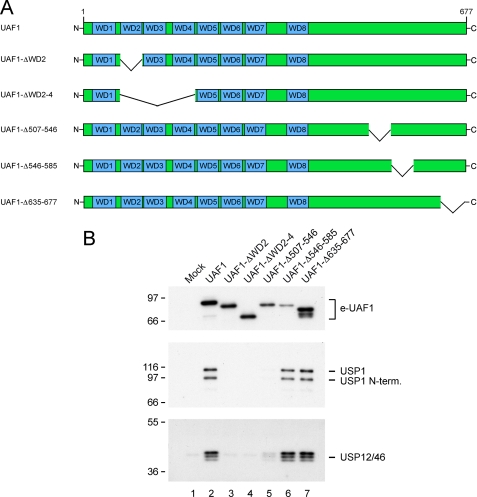
The WD40 Repeats of UAF1 Are Required for the Formation of the USP1/UAF1, USP12/UAF1, and USP46/UAF1 Complexes—Previous studies have shown that the large N-terminal region of UAF1, containing the eight predicted WD40 repeats, is required for binding and activation of USP1. We next determined the structural regions of UAF1 required for binding to USP12 and USP46. A series of Flag-tagged deletion mutant UAF1 polypeptides was expressed in 293T cells (Fig. 4A). Anti-Flag immunoprecipitates were blotted for bound endogenous USP1, USP12, and USP46 proteins (Fig. 4B). Deletion of only WD40 repeat 2 of UAF1 was sufficient to prevent detectable interaction with any of the three deubiquitinating enzymes. Only deletions in the proximal C-terminal part of UAF1 did not interfere with USP1 or USP12 and USP46 interaction. The data suggest that the interaction requires an intact propeller domain constituted by the 8 WD40 repeats. Taken together, these results indicate that UAF1 interacts with USP12 and USP46 in a manner similar to which it interacts with USP1.
FIGURE 4.
The WD40 repeats of UAF1 are required for the interaction with USP12 and USP46. A, scheme of wild-type UAF1 and deletion mutants thereof. B, Flag-tagged deletion mutants of UAF1 were transiently expressed in 293T cells. The Flag-tagged proteins were immunoprecipitated from cell lysates using anti-Flag antibodies. The immunoprecipitates were analyzed using the indicated antibodies.
UAF1 Forms Separate and Distinct Complexes with USP1, USP12, and USP46—Our data suggest that UAF1 forms separate and distinct complexes with USP1, USP12, and USP46, and that each complex is unique in its enzyme subunit composition. To further address this question, we analyzed the presence of any of the three deubiquitinating enzymes in the USP1, USP12, and USP46 complexes purified from HeLa cells. As expected, immunoblot analysis of the three complexes clearly demonstrated that the USP1 complex contained only USP1 and UAF1, but not USP12 and USP46 (Fig. 5A). Likewise, we found that the USP12 and USP46 complexes contained only USP12, USP46, and UAF1, but not USP1 (Fig. 5A). Importantly, we detected all three deubiquitinating enzymes in the UAF1 purification, which contains all three subcomplexes (Fig. 5A).
FIGURE 5.
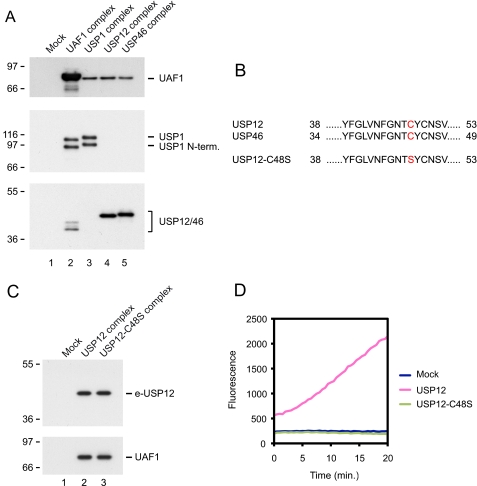
UAF1 forms three distinct deubiquitinating enzyme complexes with USP1, USP12, and USP46. A, native UAF1, USP1, USP12, and USP46 complexes were purified from HeLa cells and analyzed by immunoblot analysis using the indicated antibodies. B, alignment of the amino acids surrounding the catalytic cysteine residue in USP12 and USP46. C, immunoblot analysis of the USP12 and USP12-C48S complexes using the indicated antibodies. D, USP12 and USP12-C48S complexes were analyzed for associated deubiquitinating enzyme activity using ubiquitin-AMC as a substrate.
To further reinforce the notion that UAF1 forms three separate protein complexes in vivo, where each complex contains a distinct deubiquitinating enzyme, we asked whether a protein complex, formed by a catalytically inactive version of USP12, possesses any deubiquitinating enzyme activity. The amino acid consensus sequence surrounding the catalytical cysteine is highly conserved among members of the USP subfamily of deubiquitinating enzymes (12). Thus, the catalytic cysteine of USP12 is readily detectable as amino acid 48 (Fig. 5B). We expressed a mutant USP12 protein in HeLa cells, where the cysteine has been mutated to a serine, USP12-C48S (Fig. 5B). We purified USP12-C48S as a protein complex and confirmed that it forms a stable complex with UAF1, with similar stoichiometry as we observed in the USP12 complex (Fig. 5C). We subjected the USP12-C48S complex to an in vitro deubiquitinating enzyme assay, using ubiquitin-AMC as a substrate. Whereas the USP12 complex contained robust deubiquitinating enzyme activity, the USP12-C48S complex contained no detectable activity (Fig. 5D).
Taken together, these data confirm the notion that three separate UAF1 deubiquitinating enzyme complexes exist in vivo, and that each complex contains only one deubiquitinating enzyme species.
USP12/UAF1 and USP46/UAF1 Complexes Do Not Regulate FANCD2 Ubiquitination or the FA Pathway—The cellular functions and substrates of the USP12/UAF1 and USP46/UAF1 deubiquitinating complexes are currently unknown. Previous studies have indicated that the USP1/UAF1 complex regulates the deubiquitination of monoubiquitinated Fanconi anemia protein, Ub-FANCD2. Cellular knockdown of USP1 results in accumulation of monoubiquitinated FANCD2 (Ub-FANCD2) and disruption of the FA pathway (7, 9). Moreover, USP1 knockout results in cellular hypersensitivity to the cross-linking agent, Mitomycin C(8).
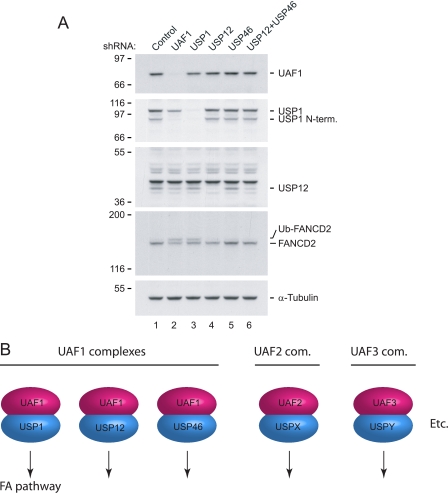
We next determined whether the USP12/UAF1 and USP46/UAF1 complexes also participate in the regulation of the FA pathway. We established HeLa cell lines where UAF1, USP1, USP12, USP46, or both USP12 and USP46 where stably knocked down by shRNA. The efficiency and specificity of the various protein knockdowns were assayed by immunoblot analysis (Fig. 6A). Knockdown of UAF1, USP12, or USP12 and USP46 resulted in a decrease in USP12/46 protein levels (Fig. 6A, lanes 2, 4, and 6). As expected, knockdown of USP1 or UAF1 both resulted in deregulation of the FA pathway, as visualized by an increase in the level of monoubiquitinated FANCD2 protein (Fig. 6A, lanes 2 and 3). On the contrary, knockdown of either USP12 or USP46 did not affect the levels of monoubiquitinated FANCD2 (Fig. 6A, lanes 4-6), and had no effect on the Mitomycin C sensitivity of these cells (data not shown). Our data suggest distinct and separate functions of the three deubiquitinating enzyme complexes.
FIGURE 6.
USP12/UAF1 and USP46/UAF1 complexes do not regulate FANCD2 ubiquitination or the FA pathway. A, immunoblot analysis of protein lysates prepared from stable HeLa cell lines expressing shRNA constructs targeting UAF1, USP1, USP12, or USP46. The lysates were analyzed using the indicated antibodies. B, model for various deubiquitinating complexes. The USP1/UAF1, USP12/UAF1, and USP46/UAF1 complexes are shown schematically as indicated. The speculated protein complexes containing other human deubiquitinating enzymes and their activating subunits are shown on the right.
DISCUSSION
Regulation of Deubiquitinating Enzyme Activity—Deubiquitinating enzymes regulate numerous fundamental biological processes such as protein degradation, gene transcription, cell cycle control, and genomic stability (12, 19, 20). However, the mechanism of regulation for the majority of these critical enzymes has remained elusive.
Several mechanisms of regulation for deubiquitinating enzymes have been described. First, regulation may be intrinsic to the enzyme itself. For instance, some deubiquitinating enzymes such as USP7 (HAUSP) have low activity catalytic sites, which are rearranged into an active conformation upon interaction with the substrate (21). Second, some proteasome-associated deubiquitinating enzymes such as yeast Ubp6 have relatively low activity by themselves, but are activated upon binding to the proteasome (22). Third, some deubiquitinating enzymes such as Uch37 have low activity, but are stimulated by binding partners that enhance the recruitment to their substrates (23, 24). Fourth, some deubiquitinating enzymes are regulated at the transcriptional level (9). Finally, some deubiquitinating enzymes have a relatively high baseline activity, but are recruited to their substrates by their own substrate binding domain, as the UIM domain of DUB-A (25) or by a chaperone protein (26).
We recently described a novel mechanism of regulation for one of these essential enzymes, namely the USP1 deubiquitinating enzyme (9, 27). We found that USP1 is activated upon forming a complex with the novel WD40-domain containing activator protein UAF1. The active USP1/UAF1 complex in turn controls the cellular levels of monoubiquitinated FANCD2 DNA repair protein, a regulation that is instrumental for integrity of the Fanconi anemia DNA repair pathway.
Interestingly, uncovering the molecular mechanism of the activation revealed that UAF1 enhances the catalytic turnover (kcat) of USP1 upon formation of a stable USP1/UAF1 protein complex, while only affecting enzyme affinity for the substrate (Km) slightly. This novel mechanism of activation coupled with the biological importance of human deubiquitinating enzymes in general, stimulated us to search for other deubiquitinating enzymes regulated by UAF1.
UAF1 Regulates the Enzymatic Activity of the USP12/UAF1 and USP46/UAF1 Complexes—We identified two new human UAF1 complexes. One, the USP12/UAF1 complex, contains the USP12 deubiquitinating enzyme as a bona fide subunit whereas the other, the USP46/UAF1 complex, contains the USP46 deubiquitinating enzyme. Detailed analysis of these complexes in vitro and in vivo, clearly demonstrate that UAF1 serves as an activator of both protein complexes. Recombinant USP12 and USP46 enzymes possess only very low deubiquitinating enzyme activity, whereas the USP12/UAF1 and USP46/UAF1 complexes have robust activity.
We show that the three deubiquitinating enzyme complexes are different and distinct in their enzyme subunits, in that none of the complexes contain more than one of the deubiquitinating enzymes. The substrate for the USP1/UAF1 complex is well described, namely, the monoubiquitinated FANCD2 protein, whereas the substrates for the USP12/UAF1 and USP46/UAF1 complexes have not been defined at present.
The Cellular Function of the USP12/UAF1 and USP46/UAF1 Complexes Is Unknown—The USP1/UAF1 complex has a clear function in the regulation of the DNA damage response by controlling the cellular levels of monoubiquitinated FANCD2 protein. In contrast, the cellular function of the USP12/UAF1 and USP46/UAF1 complexes is unknown. ShRNA knockdown of USP12 or USP46 (or both) did not result in increased levels of monoubiquitinated FANCD2. Elucidation of the cellular function of these deubiquitinating enzymes may therefore depend on the successful identification of their ubiquitin substrates in the future.
Interestingly, USP12 and USP46 have extensive amino acid identity with each other and with two highly related Saccharomyces cereviciae deubiquitinating enzymes, namely, Ubp9 and Ubp13. The cellular function of these yeast enzymes also remains unknown. Previous studies indicate that Ubp9 and Ubp13 are not essential genes in yeast, and that single deletion of these genes has no effect on growth under standard conditions (28). Interestingly, Ubp9 and Ubp13, co-purify with a yeast WD40-containing protein, Yol087c, a protein containing structural similarity to human UAF1 (9, 29, 30). Indeed, in agreement with these genomic and proteomic data, deletion of Yol087c leads to a common growth phenotype as a double deletion of the yeast Ubp9 and Ubp13.3 We are therefore led to hypothesize that the USP12/UAF1 and USP46/UAF1 complexes described in this report may be functional orthologues of the Ubp9/Yol087c and Ubp13/Yol087c proteins in yeast.
General Mechanism of Activation for Human Deubiquitinating Enzymes—Our data describe a mechanism of regulation for three of the 95 human deubiquitinating enzymes. In addition to USP1, USP12, and UPS46, a number of other human deubiquitinating enzymes have also been shown to possess very low enzymatic activity as recombinant proteins, including USP2, USP7, USP14, and USP44 (10, 22) (data not shown). Therefore, several members of the USP subfamily of deubiquitinating enzymes may require an activating protein.
The low intrinsic activity of these and perhaps other deubiquitinating enzymes, particularly of the USP subfamily, suggest the presence of other regulatory subunits for these enzymes. Specifically, we postulate that other WD40-repeat-containing proteins (i.e. UAF2, UAF3, etc.) may exist in protein complexes with other deubiquitinating enzymes and thereby regulate their activity. The proposed combinatorial assortment of USP enzymes and UAF activators is described schematically in Fig. 6B.
Acknowledgments
We thank members of the D'Andrea laboratory for helpful discussions and Dr. Rosine Haguen auer-Tsapis for communicating unpublished data.
This work was supported, in whole or in part, by National Institutes of Health Grant 5P01CA092584. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FA, Fanconi anemia; AMC, 7-amido-4-methylcoumarin; Ub, ubiquitin; VS, vinyl sulfone.
D. Bernard, S. Kanga, D. Urban-Grimal, and R. Haguenauer-Tsapis, personal communication.
References
- 1.Pickart, C. M., and Eddins, M. J. (2004) Biochim. Biophys. Acta 1695 55-72 [DOI] [PubMed] [Google Scholar]
- 2.Kennedy, R. D., and D'Andrea, A. D. (2005) Genes Dev. 19 2925-2940 [DOI] [PubMed] [Google Scholar]
- 3.Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G., and Jentsch, S. (2002) Nature 419 135-141 [DOI] [PubMed] [Google Scholar]
- 4.Sigismund, S., Polo, S., and Di Fiore, P. P. (2004) Curr. Top. Microbiol. Immunol. 286 149-185 [DOI] [PubMed] [Google Scholar]
- 5.Hanna, J., and Finley, D. (2007) FEBS Lett. 581 2854-2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang, X. H., and Zou, L. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 20645-20646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nijman, S. M., Huang, T. T., Dirac, A. M., Brummelkamp, T. R., Kerkhoven, R. M., D'Andrea, A. D., and Bernards, R. (2005) Mol. Cell 17 331-339 [DOI] [PubMed] [Google Scholar]
- 8.Oestergaard, V. H., Langevin, F., Kuiken, H. J., Pace, P., Niedzwiedz, W., Simpson, L. J., Ohzeki, M., Takata, M., Sale, J. E., and Patel, K. J. (2007) Mol. Cell 28 798-809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohn, M. A., Kowal, P., Yang, K., Haas, W., Huang, T. T., Gygi, S. P., and D'Andrea, A. D. (2007) Mol. Cell 28 786-797 [DOI] [PubMed] [Google Scholar]
- 10.Stegmeier, F., Rape, M., Draviam, V. M., Nalepa, G., Sowa, M. E., Ang, X. L., McDonald, E. R., 3rd, Li, M. Z., Hannon, G. J., Sorger, P. K., Kirschner, M. W., Harper, J. W., and Elledge, S. J. (2007) Nature 446 876-881 [DOI] [PubMed] [Google Scholar]
- 11.Zhang, X. Y., Varthi, M., Sykes, S. M., Phillips, C., Warzecha, C., Zhu, W., Wyce, A., Thorne, A. W., Berger, S. L., and McMahon, S. B. (2008) Mol. Cell 29 102-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nijman, S. M., Luna-Vargas, M. P., Velds, A., Brummelkamp, T. R., Dirac, A. M., Sixma, T. K., and Bernards, R. (2005) Cell 123 773-786 [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: a Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 14.Harlow, E., and Lane, D. (1988) Antibodies: a Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 15.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996) Anal. Chem. 68 850-858 [DOI] [PubMed] [Google Scholar]
- 16.Haas, W., Faherty, B. K., Gerber, S. A., Elias, J. E., Beausoleil, S. A., Bakalarski, C. E., Li, X., Villen, J., and Gygi, S. P. (2006) Mol. Cell Proteomics 5 1326-1337 [DOI] [PubMed] [Google Scholar]
- 17.Eng, J. K., McCormack, A. L., and Yates, J. R., 3rd. (1994) J. Am. Soc. Mass Spectrom. 5 976-989 [DOI] [PubMed] [Google Scholar]
- 18.Borodovsky, A., Kessler, B. M., Casagrande, R., Overkleeft, H. S., Wilkinson, K. D., and Ploegh, H. L. (2001) EMBO J. 20 5187-5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Andrea, A., and Pellman, D. (1998) Crit. Rev. Biochem. Mol. Biol. 33 337-352 [DOI] [PubMed] [Google Scholar]
- 20.Weake, V. M., and Workman, J. L. (2008) Mol. Cell 29 653-663 [DOI] [PubMed] [Google Scholar]
- 21.Hu, M., Li, P., Li, M., Li, W., Yao, T., Wu, J. W., Gu, W., Cohen, R. E., and Shi, Y. (2002) Cell 111 1041-1054 [DOI] [PubMed] [Google Scholar]
- 22.Leggett, D. S., Hanna, J., Borodovsky, A., Crosas, B., Schmidt, M., Baker, R. T., Walz, T., Ploegh, H., and Finley, D. (2002) Mol. Cell 10 495-507 [DOI] [PubMed] [Google Scholar]
- 23.Qiu, X. B., Ouyang, S. Y., Li, C. J., Miao, S., Wang, L., and Goldberg, A. L. (2006) EMBO J. 25 5742-5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao, T., Song, L., Xu, W., DeMartino, G. N., Florens, L., Swanson, S. K., Washburn, M. P., Conaway, R. C., Conaway, J. W., and Cohen, R. E. (2006) Nat. Cell Biol. 8 994-1002 [DOI] [PubMed] [Google Scholar]
- 25.Kayagaki, N., Phung, Q., Chan, S., Chaudhari, R., Quan, C., O'Rourke, K. M., Eby, M., Pietras, E., Cheng, G., Bazan, J. F., Zhang, Z., Arnott, D., and Dixit, V. M. (2007) Science 318 1628-1632 [DOI] [PubMed] [Google Scholar]
- 26.Kee, Y., Lyon, N., and Huibregtse, J. M. (2005) EMBO J. 24 2414-2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ventii, K. H., and Wilkinson, K. D. (2008) Biochem. J. 414 161-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amerik, A. Y., Li, S. J., and Hochstrasser, M. (2000) Biol. Chem. 381 981-992 [DOI] [PubMed] [Google Scholar]
- 29.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., Yang, L., Wolting, C., Donaldson, I., Schandorff, S., Shewnarane, J., Vo, M., Taggart, J., Goudreault, M., Muskat, B., Alfarano, C., Dewar, D., Lin, Z., Michalickova, K., Willems, A. R., Sassi, H., Nielsen, P. A., Rasmussen, K. J., Andersen, J. R., Johansen, L. E., Hansen, L. H., Jespersen, H., Podtelejnikov, A., Nielsen, E., Crawford, J., Poulsen, V., Sorensen, B. D., Matthiesen, J., Hendrickson, R. C., Gleeson, F., Pawson, T., Moran, M. F., Durocher, D., Mann, M., Hogue, C. W., Figeys, D., and Tyers, M. (2002) Nature 415 180-183 [DOI] [PubMed] [Google Scholar]
- 30.Krogan, N. J., Cagney, G., Yu, H., Zhong, G., Guo, X., Ignatchenko, A., Li, J., Pu, S., Datta, N., Tikuisis, A. P., Punna, T., Peregrin-Alvarez, J. M., Shales, M., Zhang, X., Davey, M., Robinson, M. D., Paccanaro, A., Bray, J. E., Sheung, A., Beattie, B., Richards, D. P., Canadien, V., Lalev, A., Mena, F., Wong, P., Starostine, A., Canete, M. M., Vlasblom, J., Wu, S., Orsi, C., Collins, S. R., Chandran, S., Haw, R., Rilstone, J. J., Gandi, K., Thompson, N. J., Musso, G., St Onge, P., Ghanny, S., Lam, M. H., Butland, G., Altaf-Ul, A. M., Kanaya, S., Shilatifard, A., O'Shea, E., Weissman, J. S., Ingles, C. J., Hughes, T. R., Parkinson, J., Gerstein, M., Wodak, S. J., Emili, A., and Greenblatt, J. F. (2006) Nature 440 637-643 [DOI] [PubMed] [Google Scholar]