Abstract
Galectin-1 (Gal-1) regulates leukocyte turnover by inducing the cell surface exposure of phosphatidylserine (PS), a ligand that targets cells for phagocytic removal, in the absence of apoptosis. Gal-1 monomer-dimer equilibrium appears to modulate Gal-1-induced PS exposure, although the mechanism underlying this regulation remains unclear. Here we show that monomer-dimer equilibrium regulates Gal-1 sensitivity to oxidation. A mutant form of Gal-1, containing C2S and V5D mutations (mGal-1), exhibits impaired dimerization and fails to induce cell surface PS exposure while retaining the ability to recognize carbohydrates and signal Ca2+ flux in leukocytes. mGal-1 also displayed enhanced sensitivity to oxidation, whereas ligand, which partially protected Gal-1 from oxidation, enhanced Gal-1 dimerization. Continual incubation of leukocytes with Gal-1 resulted in gradual oxidative inactivation with concomitant loss of cell surface PS, whereas rapid oxidation prevented mGal-1 from inducing PS exposure. Stabilization of Gal-1 or mGal-1 with iodoacetamide fully protected Gal-1 and mGal-1 from oxidation. Alkylation-induced stabilization allowed Gal-1 to signal sustained PS exposure in leukocytes and mGal-1 to signal both Ca2+ flux and PS exposure. Taken together, these results demonstrate that monomer-dimer equilibrium regulates Gal-1 sensitivity to oxidative inactivation and provides a mechanism whereby ligand partially protects Gal-1 from oxidation.
Immunological homeostasis relies on efficient contraction of activated leukocytes following an inflammatory episode. Several factors, including members of the galectin and tumor necrosis factor families (1, 2), regulate leukocyte turnover by inducing apoptotic cell death. In contrast, several galectin family members, in particular galectin-1 (Gal-1),2 uniquely regulate neutrophil turnover by inducing phosphatidylserine (PS) exposure, which normally sensitizes apoptotic cells to phagocytic removal (3, 4), independent of apoptosis, a process recently termed preaparesis (5).
Previous studies suggested that dimerization may be required for Gal-1-induced PS exposure, as a mutant form of Gal-1 (mGal-1) containing two point mutations within the dimer interface, C2S and V5D (C2S,V5D), displays impaired Gal-1 dimerization and fails to induce PS exposure (6). However, the manner in which monomer-dimer equilibrium regulates Gal-1 signaling remains unclear. Previous studies suggest that dimerization may be required for efficient cross-linking of functional receptors or the formation of signaling lattices (7–9). Consistent with this, monomeric mutants of several other galectins fail to induce PS exposure or signal leukocytes (4, 8). Gal-1 signaling of PS exposure requires initial signaling events, such as mobilization of intracellular Ca2+ followed by sustained receptor engagement (10). Although mGal-1 fails to induce PS exposure (6), whether mGal-1 can induce these initial signaling events remains unknown (10).
In addition to directly regulating signaling, monomer-dimer equilibrium may also regulate other aspects of Gal-1 function. Unlike many other proteins involved in the regulation of immunity, Gal-1 displays unique sensitivity to oxidative inactivation (11–15). Although engagement of ligand partially protects Gal-1 from oxidation (15), the impact of Gal-1 oxidation on signaling remains enigmatic. During oxidation, Gal-1 forms three distinct intramolecular disulfide bridges that facilitate profound conformational changes that preclude ligand binding and Gal-1 dimerization (12–14), suggesting that monomerdimer equilibrium may also regulate Gal-1 sensitivity to oxidative inactivation.
Previous studies utilized dithiothreitol (DTT) in treatment conditions to protect Gal-1 from oxidative inactivation (16, 17). Indeed, failure to include DTT precluded Gal-1-induced death in T cells (3, 18), suggesting that Gal-1 undergoes rapid oxidation in vivo in the absence of reducing conditions. However, DTT itself can induce apoptosis in leukocytes (19), leaving questions regarding the impact of Gal-1 oxidation on these signaling events. In contrast, recent studies utilizing iodoacetamide-alkylated Gal-1 (iGal-1), previously shown to protect Gal-1 from oxidative inactivation (20–29), demonstrated that DTT actually primes cells to become sensitive to Gal-1-induced apoptosis regardless of Gal-1 sensitivity to oxidation (5).
As the engagement of leukocyte ligands requires glycan recognition and oxidation precludes this binding (11, 15), understanding the impact of oxidation on Gal-1 signals will facilitate a greater appreciation of the factors that govern Gal-1 oxidation and therefore function. Our results demonstrate that Gal-1 monomer-dimer equilibrium provides a key regulatory point controlling both Gal-1 sensitivity to oxidation and its ability to signal PS exposure in leukocytes. These results provide novel insights into Gal-1 function and explain at a biochemical level the mechanisms regulating Gal-1 oxidative inactivation and signaling.
EXPERIMENTAL PROCEDURES
Preparation of Human Gal-1 and mGal-1—Gal-1 and the mutant Gal-1 with C2S,V5D mutations (mGal-1) were prepared as outlined previously (6, 30). Each recombinant galectin was purified by affinity chromatography on lactosyl-Sepharose and bound lectin was eluted with 100 mm lactose in PBS, 14 mm 2-ME. Prior to derivatization, 2-ME was removed from galectin samples by utilizing a PD10 gel filtration column (GE Healthcare), followed by the addition of lactose (100 mm final concentration) to help maintain the stability of each galectin and reduce the likelihood of adduct formation at or near the carbohydrate recognition domain. Alexa Fluor 488-labeled forms of galectins were prepared using either Alexa Fluor 488 C5-maleimide or Alexa Fluor 488 carboxylic acid, succinimidyl ester, dilithium salt reactive dyes (Molecular Probes) as described (30). Galectins were biotinylated by incubating 3–5 mg/ml of each galectin with 2 mm EZ-link™ Sulfo-NHS-LC-Biotin (sulfosuccinimidyl-6-(biotinamido) hexanoate) (Pierce) for 2 h at 4 °C. Unconjugated EZ-link Sulfo-NHS-LC-Biotin and free lactose were separated from galectin using a PD-10 gel filtration column. Galectins were re-chromatographed over lactosyl-Sepharose to remove any inactive material following labeling. Bound galectin was eluted with 100 mm lactose, then applied to a PD-10 gel filtration column to remove lactose, and stored at 4°C in 14 mm 2-ME in PBS until further use. Control lectins, Ricinus communis agglutinin and Lycopersicon esculentum agglutinin were purchased from Vector Labs.
Binding of Galectin to Aminoalkyl Glycosides Immobilized on Activated (N-Hydroxysuccinimidyl) Glass Surface—Glycan microarrays were prepared as described previously (31, 32) and obtained from the National Institutes of Health NIGMS-funded Consortium for Functional Glycomics (see www.functionalglycomics.org/static/index.shtml). For galectin recognition of glycans on the printed glycan microarray, a solution 7 μm Gal-1 or mGal-1 in PBS containing 0.005% Tween 20 and 14 mm 2-ME was incubated for 1 h at 25 °C. The slide was then immersed in PBS containing 0.005% Tween 20, drained, and then overlaid with fluorescein isothiocyanate-streptavidin. After 1 h at room temperature in a dark humid chamber, the slide was washed by successive immersion in PBS, 0.01% Tween 20 (three times) and PBS, 0.1% Tween 20 (three times). The slide was briefly rinsed with distilled water and dried under microfiltered air. An image of bound fluorescence was obtained using a microarray scanner (Scan Array Express, PerkinElmer Lifer Sciences). The integrated spot intensities were determined using Imagene software (BioDiscovery, El Segundo, CA).
Measurement of Galectin Binding Affinity Using Surface Plasmon Resonance—All surface plasmon resonance experiments were preformed at 25 °C on a Biacore 3000 instrument (Biacore AB (part of GE Healthcare), Uppsala, Sweden) largely as outlined previously (31–34). Biotinylated glycosides were captured on research grade streptavidin-coated sensor chips (Sensor Chip SA, Biacore Inc.) that were pretreated according to the manufacturer's instructions. A solution of each biotinylated glycoside (10 fmol/ml) was injected at 2 ml/min in PBS, pH 7, containing 0.005% Tween 20 and containing 14 mm β-ME (running buffer) for varying lengths of time (3–7 min) until an optimal amount of glycan was captured on each independent surface. Three related glycosides were studied using one streptavidin sensor chip. A control (non-binding) glycan, arabinose, was also captured on the same sensor chip, and the specific binding of non-derivatized recombinant Gal-1 or mGal-1 for the test glycans was measured using the in-line reference subtraction feature of the Biacore 3000 instrument. Increasing concentrations of Gal-1 or mGal-1 (0.1–100 μm) were injected at a flow rate of 60 μl/min over all four surfaces of the sensor chip. Bound Gal-1 or mGal-1 was eluted with the running buffer after the injection was complete.
Cell Culture—Promyelocytic leukemia HL60 cells were obtained from ATCC and maintained at 37 °C and 5% CO2 in complete RPMI (RPMI 1640 supplemented with 10% fetal bovine serum, 2 mm glutamine, 100 milliunits/ml penicillin, 100 μg/ml streptomycin).
Enzymatic Deglycosylation—Prior to enzymatic deglycosylation, HL60 cells were fixed by washing three times in PBS at 4 °C, followed by resuspension in 2% paraformaldehyde buffered in PBS (pH 7.4) at 4 °C. Cells were allowed to fix overnight on a shaker at 4 °C. Following fixation, cells were washed three times in PBS and then two times in the appropriate buffer as recommended by the manufacturer. Cells were washed in 50 mm sodium acetate (pH 5.8) and incubated with 200 milliunits of Escherichia freundii endo-β-galactosidase (Seikagaku Kogyo) at 107 cells/ml for 24 h at 37 °C or washed in PBS (pH 7.0) and incubated with 100 milliunits of Arthrobacter ureafaciens neuraminidase for 2 h at 37 °C. Buffer control treatments lacking enzymes were used for each individual condition.
Galectin Binding to Cells—For lectin binding, cells were washed twice in PBS at 4 °C and incubated with biotinylated Gal-1, mGal-1, or the indicated plant lectins (L. esculentum agglutinin or R. communis agglutinin I, Vector Labs) at a concentration between 5 and 10 μg/ml at 4 °C for 1 h. As controls, cells were incubated with 50 mm lactose along with the galectins. Cells were washed 3 times and then incubated with Alexa Fluor 488-streptavidin or Alexa Fluor 633-streptavidin (Molecular Probes) at 4 °C for 1 h. Cells were then washed twice, followed by resuspension in 400 μl of PBS for analysis by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences). The bars in each graph represent the % change in binding when compared with the binding of control buffertreated cells from each enzymatic pair. Error bars in each graph represent standard deviation of duplicate analysis.
Measuring Galectin-induced PS Exposure—For annexin-V staining, 225 μl of cells at 1 × 106 cell/ml (unless otherwise indicated) resuspended in complete RPMI were treated with 25 μl of PBS, or the indicated concentrations of Gal-1, mGal-1, or camptothecin for the time and concentrations indicated at 37 °C and 5% CO2 followed by disengagement with 50 mm lactose and staining with annexin-V (CalTag) as outlined previously (3).
Ca2+ Flux Measurements—HL60 cells were loaded with 3 mm Fluor AM at 37 °C for 30 min in the presence of 4 mm probenecid, and inhibitor of anion transport, to minimize dye leakage. The cells were washed with Hanks' balanced salt solution, incubated for 30 min at room temperature to allow the Fluo-4 AM dye to completely de-esterify, washed twice more, and resuspended at 107 cells/ml Hanks' balanced salt solution with 0.5 mg/ml human serum albumin. Fluorescence readings were obtained in a stirring cell fluorometer (PerkinElmer Life Sciences LS-50) equipped with a water-jacketed cuvette holder. After obtaining the basal signal, fluorescence intensities were acquired at 0.1-s intervals for 10–15 min with continuous stirring of the cell suspension. The cells were lysed with 0.1% Triton X-100 to determine the maximum fluorescence. The minimum fluorescence was determined by adding EGTA to the lysed cells. These fluorescence measurements were converted to molar concentrations as previously described (35).
Agglutination Assay—HL60 cells were cultured as outlined above. Cells were plated in 96-well plates, mixed with serial dilutions of each galectin and allowed to agglutinate for 30 min. Cells were then graded on degree of agglutination from 4+ (high agglutination) to 0+ (no agglutination) as outlined previously (36, 37).
HPLC Analysis—For size exclusion HPLC, galectins were stored at 4 °C in described concentrations and subjected to size exclusion chromatography. Separation of monomeric and dimeric forms of Gal-1 or mGal-1 was accomplished by size exclusion HPLC as described (15), using a TSK-GEL SW 2000 column (Beckman) (7.5 mm × 30 cm) on a Beckman System Gold HPLC. For direct comparison of mGal-1 and imGal-1 a TSK-GEL G2000SW column was employed using a Shimadzu HPLC. The TSK-GEL SW 2000 column was calibrated using bovine γ-globulin, 158 kDa; bovine serum albumin, 67 kDa; chicken ovalbumin, 44 kDa; equine myoglobin, 25 kDa; chymotrypsinogen, 17 kDa; and ribonuclease A, 13.7 kDa, whereas the TSK-GEL G2000SW column was calibrated using bovine γ-globin, 158 kDa; chicken ovalbumin 44 kDa; equine myoglobin, 17 kDa; and vitamin B12, 1.3 kDa.
Determination of Gal-1 Activity by Affinity Chromatography and Soluble Protein Fraction Detection—To determine the amount of active protein at a given time point, each galectin was incubated at 37 °C for the indicated time followed by subjecting the soluble fraction to affinity chromatography utilizing lactosyl-Sepharose. Bound fraction was calculated by dividing the material eluted with lactose over the total material passed over the column. Alternatively, 40 μm Gal-1 or mGal-1 was incubated for the indicated times in a humidified incubator at 37 °C followed by centrifugation at 16,000 × g for 30 min to pellet the principle precipitated matter. The fraction of soluble protein was detected by dividing the absorbance of the soluble material at 280 nm by the absorbance of the starting material at the same wavelength.
Chemical Cross-linking—Both 2-ME and lactose were removed from Alexa Fluor-labeled Gal-1 or mGal-1 by gel filtration. In the case of Alexa Fluor-labeled galectin-1, prior to cross-linking, free cysteines were quenched with iodoacetamide to protect from activity loss as in the non-alkylated. Samples were incubated overnight at room temperature in PBS to allow each concentration to achieve equilibrium prior to cross-linking. Each sample was then incubated with 50-fold excess BS3 (Pierce) for 30 min at room temperature. Excess cross-linker was quenched using 50 mm Tris-HCl for 15 min, followed by analysis of the extent of covalently cross-linked dimer by SDS-PAGE in reducing conditions. Protein detection was accomplished using a Fluorochem imaging system.
Iodoacetamide Treatment of Galectin—Both 2-ME and lactose were removed from Gal-1 or mGal-1 samples by gel filtration. Gal-1 or mGal-1, at ∼2–5 mg/ml, was then re-suspended in 100 mm lactose in PBS containing 100 mm iodoacetamide and allowed to incubate at 4 °C overnight. Free iodoacetamide was removed following treatment by gel filtration using a PD-10 column as previously described (30). Activity of iodoacetamide-treated Gal-1 (iGal-1) was assessed by incubating iGal-1 for 24 h at 37 °C followed by addition to HL60 cells at 20 μm for 4 h followed by detection for PS exposure using flow cytometric analysis. Both iGal-1 and imGal-1 were incubated for 24 h at 37 °C followed by subjection to affinity chromatography over lactosyl-Sepharose to more accurately determine potential activity loss.
Analysis of Alkylated Galectin by Mass Spectrometry—Gal-1 or mGal-1 (1 μg) were analyzed for iodoacetamide incorporation by MALDI-TOF analysis as done previously (30) using a Voyager DE-RP BioSpectrometry Work station (PE Biosystems, Framingham, MA). Examination of tryptic fragments of Gal-1 following alkylation was also accomplished as outlined previously (20). Briefly, following trypsin digestion, all peptides were analyzed in the reflective, positive ion mode by delayed extraction using ESI mass spectrometry performed using a MSD Trap (Bruker Daltonics, Billerica, MA) in the positive ion mode. Reversed phase separated peptide fractions were reduced to a uniform volume of 100 μl and an equal volume of MeOH was added to all fractions. These peptide solutions were directly infused into the MSD system at 5 μl/min and the initial MS scan utilized a m/z range of 400 to 2,000, and the most abundant ions were selected for MS/MS analysis.
RESULTS
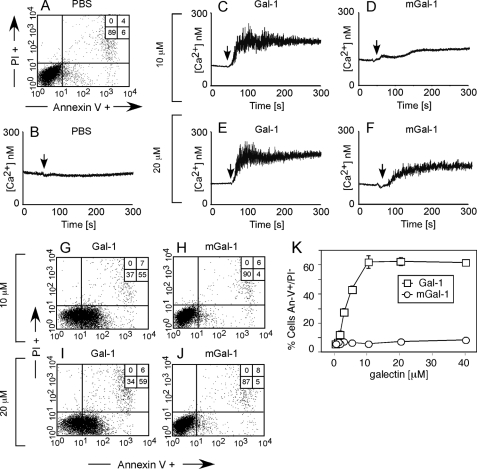
mGal-1 Induces Ca2+ Flux Yet Fails to Induce PS Exposure in HL60 Cells—Although Gal-1-induced PS exposure requires the induction of proximal signaling events, such as intracellular Ca2+ flux (6, 10), previous studies also demonstrated that Gal-1-induced PS exposure requires continuous engagement of functional cell surface receptors for PS to be realized (10). In contrast to Gal-1, a mutant form of Gal-1 (mGal-1), which contains two point mutations in the dimer interface that impair Gal-1 dimerization, C2S and V5D (C2S,V5D), fails to induce PS exposure regardless of the length of treatment (6). However, whether mGal-1 can engage early signaling events remains unknown (6). Thus, we determined whether mGal-1 induces Ca2+ mobilization in HL60 cells. Although mGal-1 failed to induce significant Ca2+ mobilization at 10 μm (Fig. 1D), mGal-1 produced robust Ca2+ mobilization at 20 μm (Fig. 1F), although at a reduced magnitude when compared with Ca2+ flux induced by Gal-1 (Fig. 1, C and E). However, consistent with previous results, mGal-1 failed to induce PS exposure over a wide range of concentrations (Fig. 1, H, J, and K) in contrast to Gal-1, which induces PS exposure at both 10 and 20 μm (Fig. 1, G and I). Taken together, these results demonstrate that although mGal-1 fails to induce PS exposure, it can induce significant Ca2+ flux in HL60 cells.
FIGURE 1.
mGal-1 induces proximal signaling events in leukocytes but fails to induce PS exposure. A, HL60 cells treated with PBS for 4 h were stained with annexin-V fluorescein isothiocyanate and propidium iodide (PI) to detect PS exposure followed by flow cytometric analysis. B, cells were loaded with Fluo-4 and analyzed for changes in intracellular Ca2+ using a fluorometer following addition of PBS at the indicated time (vertical arrow). C–F, HL60 cells were loaded with Fluo-4 as in B, followed by addition of: 10 μm Gal-1 (C), 10 μm mGal-1 (D), 20 μm Gal-1 (E), or 20 μm mGal-1 (F) as indicated by the arrows. G-J, HL60 cells treated with 10 μm Gal-1 (G), 10 μm mGal-1 (H), 20 μm Gal-1 (I), or 20 μm mGal-1 (J) for 4 h were used to label cells with annexin-V fluorescein isothiocyanate and PI to detect PS exposure followed by flow cytometric analysis. K, HL60 cells were treated with the indicated concentrations of Gal-1 or mGal-1 for 4 h followed by detection for PS exposure.
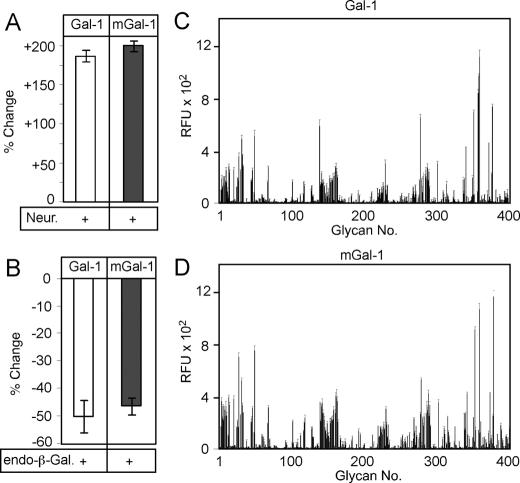
Gal-1 and mGal-1 Display Similar Glycan Recognition Properties—In an effort to elucidate the underlying mechanism responsible for the discordance between the ability of mGal-1 to induce Ca2+ flux while failing to induce PS externalization, we first examined the possibility that mutations in the dimer interface might significantly alter the carbohydrate binding specificity of Gal-1. To test this, we first examined Gal-1 and mGal-1 binding to cells following enzymatic removal of cell surface glycans as done previously to elucidate the binding specificity of different galectin family members (4, 29). Gal-1 and mGal-1 both bound leukocytes and inclusion of lactose inhibited binding, which demonstrated that both proteins required glycan recognition for cell surface binding (data not shown). Treatment of cells with neuraminidase, which enhances Gal-1 binding and cellular sensitivity to Gal-1-induced PS exposure (6, 29), resulted in a comparable increase in both Gal-1 and mGal-1 cell surface glycan recognition (Fig. 2A). Furthermore, treatment of cells with endo-β-galactosidase, which cleaves cell surface linear poly-N-acetyllactosamine sequences, a common galectin ligand (4, 29), resulted in comparable reduction in both Gal-1 and mGal-1 binding (Fig. 2B), suggesting that both proteins display a similar general preference for cell surface glycans. Furthermore, Gal-1 and mGal-1 displayed similar binding to carbohydrates on a chemically defined glycan microarray (Fig. 2, C and D, and supplemental Tables S1 and S2). These results show that Gal-1 and mGal-1 possess similar glycan recognition, and thus the observed differences in signaling between Gal-1 and mGal-1 do not likely reflect significant alterations in carbohydrate binding specificity of Gal-1 following introduction of the C2S,V5D mutations.
FIGURE 2.
Gal-1 and mGal-1 display similar glycan binding properties. A, quantification of mGal-1 and Gal-1 binding before and after treatment of cells with neuraminidase. B, quantification of mGal-1 and Gal-1 binding before and after treatment of cells with endo-β-galactosidase. C, Gal-1 binding toward distinct classes of glycan ligands on the glycan microarray. D, mGal-1 binding toward distinct classes of glycan ligands on the glycan microarray.
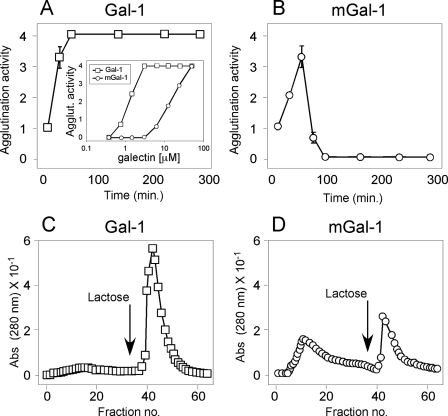
mGal-1 Displays Enhanced Sensitivity to Oxidative Inactivation—Although previous studies suggested that mGal-1 fails to dimerize (6, 15), we noticed that during incubation with mGal-1, leukocytes appeared to display varying levels of cellular agglutination, a process that requires functional bivalency of Gal-1. These results suggested that the ability of mGal-1 to signal Ca2+ flux might reflect residual dimerization not fully eliminated by the C2S,V5D mutation. To examine this in more detail, we incubated leukocytes with different concentrations of Gal-1 or mGal-1 followed by determination of agglutination. mGal-1 induced significant agglutination of cells at concentrations similar to those at which mGal-1 induced Ca2+ flux, although mGal-1 displayed significantly impaired agglutination when compared with Gal-1 (Fig. 3A, inset). Indeed, 10 μm mGal-1 produced very little agglutination (Fig. 3A, inset) and also failed to induce significant Ca2+ flux (Fig. 1D), which strongly suggested that proximal signaling requires Gal-1 dimerization.
FIGURE 3.
mGal-1 displays enhanced sensitivity to oxidative inactivation. A, HL60 cells were incubated with 20 μm Gal-1 at 37 °C for the times indicated followed by assessment for the degree of agglutination. Inset, HL60 cells were incubated for 30 min at 37 °C with the concentrations of Gal-1 or mGal-1 indicated followed by assessment for agglutination. B, HL60 cells were incubated with 20 μm mGal-1 at 37 °C for the times indicated followed by assessment for the degree of agglutination. C, Gal-1 was incubated in PBS at 37 °C for 3 h followed by subjection to affinity chromatography over lactosyl-Sepharose. D, mGal-1 was incubated in PBS at 37 °C for 3 h followed by subjection to affinity chromatography over lactosyl-Sepharose.
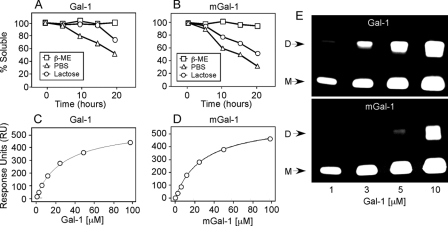
Although mGal-1-treated cells induced significant initial agglutination of cells, after 4 h, the time point at which evaluation for cell surface PS occurs, the cells incubated with mGal-1 were no longer agglutinated. Consistent with this, examination of mGal-1-treated cells over time demonstrated that although cells displayed significant agglutination when examined at earlier time points, cells displayed a gradual and spontaneous disengagement over time, with no detectable agglutination at 4 h (Fig. 3, A and B), suggesting that mGal-1 may display an enhanced sensitivity to oxidative inactivation, a process that precludes Gal-1 recognition of ligand (11, 39). Consistent with this, we also observed significant protein precipitation following prolonged incubation of cells with mGal-1, a hallmark of Gal-1 oxidation at higher concentrations. To examine this in more detail, we directly examined binding of Gal-1 or mGal-1 to lactosyl-Sepharose. Incubation of mGal-1 for 3 h at 37 °C resulted in a 50% reduction in activity, whereas Gal-1 lost less than 15% activity over the same time period (Fig. 3, C and D). Importantly, inclusion of 2-ME protected both proteins from activity loss (data not shown), which demonstrated that mGal-1 activity loss reflected oxidative inactivation.
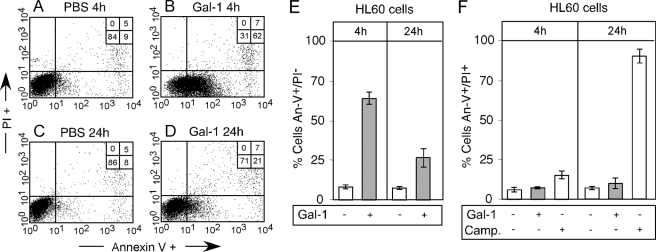
Prolonged Incubation of HL60 Cells with Gal-1 Results in PS Reversion—The enhanced sensitivity of mGal-1 to oxidative inactivation suggested that the inability of mGal-1 to signal PS exposure may reflect rapid oxidation and therefore inability to engage functional leukocyte receptors for the prolonged periods of time needed for full realization of PS exposure to occur (10) (Fig. 3B). However, previous studies also demonstrated that Gal-1 displays significant sensitivity to oxidation (11, 39). As a result, we next sought to examine the potential impact of oxidation on Gal-1-induced PS exposure. As expected, incubation of cells with Gal-1 for 4 h resulted in significant PS exposure (Fig. 4, A, B, and E). However, cells incubated in parallel for 24 h with Gal-1 displayed a significant reduction in cell surface PS (Fig. 4, C–E), without loss in cell viability (Fig. 4F). Similar to cells incubated with mGal-1 evaluated at 4 h, cells incubated with Gal-1 displayed a reversion of cellular agglutination at 24 h (data not shown), suggesting that reversion in Gal-1 signaling may also reflect Gal-1 oxidation. Taken together, these results demonstrate that whereas both Gal-1 and mGal-1 display sensitivity to oxidative inactivation, mGal-1 exhibits a significantly enhanced sensitivity to oxidation, although oxidative inactivation may impact signaling induced by both proteins.
FIGURE 4.
Oxidation of Gal-1 results in reversion of PS exposure. A–D, HL60 cells were treated with 20 μm Gal-1 for 4 or 24 h as indicated, followed by detection of PS exposure by flow cytometric analysis. E, quantification of the number of annexin-V+/PI– cells following Gal-1 treatment for the length of time indicated. F, quantification of the number of annexin-V+/PI+ cells following treatment of cells with either 20 μm Gal-1 or 20 μm camptothecin (Camp.) treatment for the length of time indicated. PI, propidium iodide.
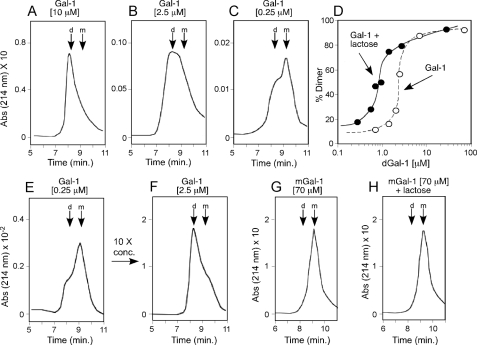
Ligand Shifts Gal-1 Monomer-Dimer Equilibrium in Favor of Dimerization—As mGal-1 displays an enhanced sensitivity to oxidative inactivation and ligand partially protects Gal-1 from oxidation (15), dimerization itself may be a mechanism through which ligand inhibits Gal-1 oxidation. Indeed, Gal-1 was found to exist in a reversible monomer-dimer equilibrium (Fig. 5, A–C, E, and F), whereas ligand shifted this equilibrium in favor of dimer formation (Fig. 5D). By contrast, mGal-1 behaved exclusively as a monomer in this assay and ligand failed to enhance dimerization (Fig. 5, G and H), suggesting that protection of Gal-1 from oxidation occurs through ligand-induced dimerization. Consistent with this, ligand not only failed to enhance mGal-1 dimerization, but also displayed a reduced capacity to protect mGal-1 from oxidative inactivation (Fig. 6, A and B), although mGal-1 and Gal-1 displayed similar binding to ligand (Fig. 6, C and D).
FIGURE 5.
Ligand modulates monomer-dimer equilibrium in favor of dimerization. A–C, gel filtration of Gal-1 at 10 (A), 2.5 (B), and 0.25 μm (C). D, quantification of the percent dimer formation following gel filtration for each indicated concentration in the presence or absence of ligand. E, gel filtration analysis of 0.25 μm Gal-1. Following analysis, monomeric fractions were concentrated to 2.5 μm Gal-1 and reanalyzed in F. G and H, mGal-1 was subjected to gel filtration analysis in either the absence (G) or presence (H) of ligand. D, dimer; M, monomer.
FIGURE 6.
mGal-1 displays impaired capacity to be stabilized by ligand. A, incubation of 40μm Gal-1 in PBS, with or without 2-ME or lactose as indicated followed by determination of the soluble fraction. B, incubation of 40 μm mGal-1 in PBS, with or without 2-ME or lactose as indicated followed by determination of the soluble fraction. C and D, surface plasmon resonance analysis of the binding of Gal-1 (C) or mGal-1 (D) to lactose. E, fluorescent image acquired following SDS-PAGE of BS3 chemically cross-linked Alexa Fluor 488-labeled Gal-1 or mGal-1. D, dimer; M, monomer.
The inability of mGal-1 to appear as a dimer following HPLC gel filtration analyses likely reflects weak dimerization and rapid dissociation. In contrast, the ability of mGal-1 to agglutinate cells at higher concentrations suggested that mGal-1 might dimerize. To test this, we examined Gal-1 and mGal-1 following chemical cross-linking of the dimer using water-soluble BS3, a homobifunctional, water-soluble, non-cleavable cross-linker with a diameter of 11.4 Å, which allows trapping of weakly associated molecules. Cross-linking can alter protein detection when utilizing common protein staining procedures. To overcome this, we labeled Gal-1 and mGal-1 with Alexa-488 maleimide to ensure that dimer and monomer fractions retained detection sensitivity irrespective of cross-linking. Although reduced when compared with Gal-1, significant mGal-1 dimers could be trapped at higher concentrations (Fig. 6E), which indicated that although mGal-1 displays reduced dimer formation, mGal-1 can dimerize.
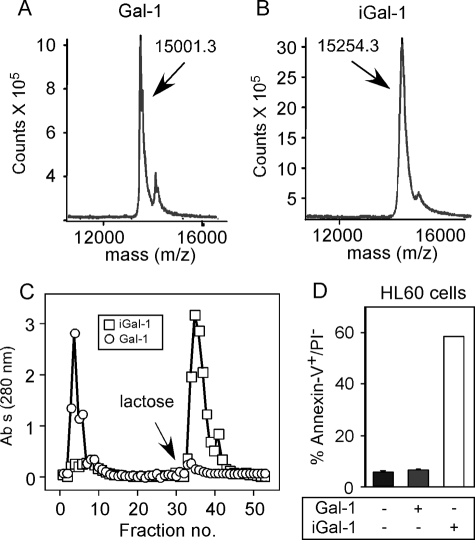
Carboxymethylation Protects Gal-1 from Oxidative Inactivation—These results suggest that the inability of mGal-1 to induce PS exposure and Gal-1 to sustain PS exposure likely reflects spontaneous oxidative inactivation during continual leukocyte incubation. Many previous studies utilized DTT in treatment conditions to prevent Gal-1 oxidative inactivation (16–18). Indeed, inclusion of 1 mm DTT prevented spontaneous disengagement of cells incubated with either Gal-1 or mGal-1 (data not shown), strongly suggesting that loss of agglutination reflected Gal-1 oxidation. However, DTT can induce significant alterations in cellular responses to Gal-1, making it difficult to separate the impact of DTT on cellular function from its ability to prevent Gal-1 oxidation (3, 19, 40). In contrast, previous studies alkylated Gal-1 with iodoacetamide followed by removal of free iodoacetamide, which protects Gal-1 from oxidation without introducing cells to a reducing environment (20–29). Importantly, these studies demonstrated that alkylation not only protects Gal-1 from oxidation, but also fails to alter biological activity or quaternary structure (20–29). Similar to previous results, alkylation with iodoacetamide failed to quantitatively alkylate all Cys residue, because we observed ∼5.4 mol of incorporation per subunit (Fig. 7, A and B), rather than 6.0 as expected (20). Examination of tryptic fragments of alkylated Gal-1 demonstrated a preference of iodoacetamide for Cys-2, -16, and -130 (data not shown), similar to previous findings (20). Importantly, alkylated Gal-1 (iGal-1) retained >90% activity and induced robust PS exposure in leukocytes (Fig. 7, C and D), consistent with previous findings (20–29). Taken together, these results demonstrate that alkylation can significantly protect Gal-1 from oxidative inactivation.
FIGURE 7.
Alkylation protects Gal-1 from oxidative inactivation. A and B, mass spectrometry analysis using MALDI-TOF of Gal-1 (A) without treatment or (B) following treatment with iodoacetamide. C, Gal-1 or iGal-1 were incubated for 24 h in PBS at 37 °C followed by subjection to affinity chromatography over lactosyl-Sepharose. D, Gal-1 or iGal-1 were incubated for 24 h in PBS at 37 °C followed by addition of 20 μm of each galectin to HL60 cells for 4 h. Cells were then stained with annexin-V fluorescein isothiocyanate and propidium iodide and analyzed for PS positivity by flow cytometric analysis.
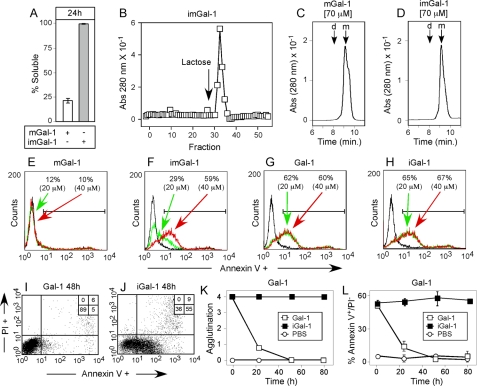
Carboxymethylation Enhances Gal-1 and mGal-1 Signaling of HL60 Cells—As alkylation protected Gal-1 from oxidative inactivation, we also alkylated mGal-1 (imGal-1) to directly examine whether alterations in cellular signaling reflected differential sensitivity of these proteins to oxidative inactivation. Similar to Gal-1, alkylation of mGal-1 with iodoacetamide resulted in incomplete Cys modification, with 1 mol less iodoacetamide incorporation than Gal-1, likely due to the C2S mutation in mGal-1 (data now shown). Similar to iGal-1, imGal-1 prevented oxidation-induced precipitation and when examined over lactosyl-Sepharose retained >90% activity following a 24-h incubation in the absence of 2-ME (Fig. 8, A and B). Furthermore, alkylation failed to alter mGal-1 dimerization as assessed following HPLC analysis (Fig. 8, C and D). To compare mGal-1 and imGal-1 directly, we examined PS exposure as done previously (2, 6, 41–43). Although Gal-1 and iGal-1 displayed similar signaling capacity following a 4-h incubation (Fig. 8, G and H), 20 and 40 μm imGal-1 now signaled significant PS exposure (Fig. 8F), although 20 μm signaled PS externalization at a reduced magnitude when compared with 20 μm Gal-1 (Fig. 8F), suggesting a preference for Gal-1 dimerization for full realization of PS exposure. In contrast, neither concentration of mGal-1 induced PS exposure (Fig. 8E). Furthermore, although cells incubated with mGal-1 spontaneously disengaged over time, imGal-1 sustained agglutination over the duration of the incubation (data not shown), further suggesting that mGal-1 fails to induce PS exposure due to rapid oxidation.
FIGURE 8.
Alkylation rescues Gal-1 and mGal-1 from oxidation. A, incubation of 40 μm mGal-1 or imGal-1 in PBS for 24 h followed by determination of the soluble fraction. B, imGal-1 was incubated in PBS at 37 °C for 24 h followed by subjection to affinity chromatography over lactosyl-Sepharose. C and D, gel filtration of mGal-1 (C) or imGal-1 (D) at 70 μm. D, dimer; M, monomer. E–H, HL60 cells were treated with either the indicated concentrations of mGal-1 (E), imGal-1 (F), Gal-1 (G), or iGal-1 (H) or PBS (black histogram in each data set) for 4 h followed by detection of PS exposure by flow cytometric analysis. I and J, HL60 cells were treated with PBS (I) or 20 μm iGal-1 (J) for 48 h followed by detection for PS exposure by flow cytometric analysis. K and L, HL60 cells were incubated with 20 μm iGal-1 or 20 μm Gal-1 for the indicated time at 37 °C followed by examination for (K) the agglutination state of cells and (L) detection for PS exposure by flow cytometric analysis.
In contrast to mGal-1, Gal-1 induces PS exposure following a 4-h incubation, although this PS externalization gradually reverted, a process that paralleled gradual and spontaneous disengagement of cells (Fig. 4). To examine whether the inability of Gal-1 to sustain PS exposure over a prolonged incubation period also reflected oxidation, we evaluated cells after 48 h of treatment with Gal-1 or iGal-1. Although Gal-1-treated cells completely reverted PS exposure (Fig. 8I), iGal-1-treated cells displayed significant PS positivity (Fig. 8J). Indeed, iGal-1-treated cells displayed continuous PS exposure for 72 h of treatment, whereas Gal-1-treated cells displayed a significant loss of PS following 24 h accompanied by spontaneous disengagement (Fig. 8, K and L).
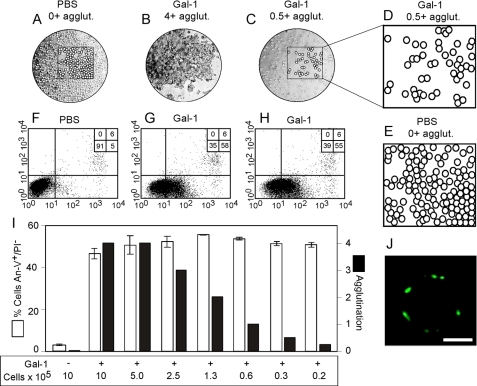
Gal-1 Signals PS Exposure Independent of Cellular Agglutination—Although alkylation allowed mGal-1 to signal PS, imGal-1-induced PS exposure occurred at a reduced magnitude when compared with cells incubated with Gal-1, which suggested that dimerization not only protects Gal-1 from oxidation but also facilitates Gal-1 signaling as suggested previously (6). Although the requirement for Gal-1 dimerization strongly suggests cross-linking of functional cell surface receptors, we observed that Gal-1-induced agglutination paralleled Gal-1-induced PS exposure. Agglutination may mediate the association of other receptors not directly bound by Gal-1, allowing Gal-1 to indirectly signal PS exposure through receptor approximation instead of directly inducing signaling. To test this, we serially diluted cells in the presence of uniform Gal-1 that resulted in reduced agglutination as a function of reduced cell number. If Gal-1-induced PS exposure required agglutination, a significant reduction in agglutination would be expected to reduce Gal-1-induced PS externalization. However, Gal-1 induced equivalent PS exposure regardless of the agglutination state of the cells (Fig. 9, A–I). Similarly, cytospin-induced adhesion of leukocytes followed by Gal-1 incubation resulted in PS positivity of single cells following Gal-1 treatment (Fig. 9J). These results demonstrate that Gal-1-induced PS exposure likely reflects a need for cross-linking of functional receptors and can occur independently of cell-cell agglutination.
FIGURE 9.
Gal-1-induced PS exposure occurs independent of Gal-1-induced agglutination. A–C, cells treated with PBS (106 cells) (A), 20 μm Gal-1 (106 cells) (B), or 20 μm Gal-1 (0.3 × 105 cells) (C) at ×10 magnification are shown. D, demarcation of cells in C. E, demarcation of cells in A. F–H, HL60 cells treated with PBS (106 cells) (F), 20 μm Gal-1 (106 cells) (G), or 20 μm Gal-1 (0.3 × 105 cells) (H) were incubated for 4 h followed by detection for PS exposure by flow cytometric analysis. I, the indicated number of HL60 cells was treated with 20 μm Gal-1 followed by detection for agglutination (black bars). Following determination of cellular agglutination, cells were analyzed for PS exposure by flow cytometric analysis (white bars). J, HL60 cells were fixed to slides using cytospin followed by incubation with 20 μm Gal-1 for 4 h and confocal analysis using annexin-V-Alexa Fluor-488. White bar, 5 μm.
DISCUSSION
These results demonstrate that Gal-1 undergoes oxidation when co-incubated with leukocytes and that this oxidation significantly impacts Gal-1 signaling. Furthermore, our results demonstrate that glycan ligand regulates Gal-1 sensitivity to oxidation by shifting the monomer-dimer equilibrium in favor of dimerization, providing an explanation whereby ligand may protect Gal-1 from oxidative inactivation.
Although the unique sensitivity of Gal-1 to oxidative inactivation has been known for many years (11, 15, 44–46), the underlying mechanism responsible for this sensitivity remained enigmatic. Previous studies demonstrated that following oxidation, each subunit of Gal-1 forms three discrete intramolecular disulfide bridges (13, 14). Disulfide bridge formation results in a significant conformational change (20, 47, 48), which prohibits ligand recognition and prevents dimerization (13). Crystallographic studies strongly suggest impaired conformational rotation of Cys residues during dimerization (49–51). As a result, dimerization likely limits the conformational freedom needed to successfully form intramolecular disulfide bonds. Taken together, these results suggest that the dimer interface itself likely protects Gal-1 from oxidation by locking Cys residues in positions that make intramolecular disulfide bond formation unfavorable.
The ability of ligand to enhance dimerization provides a mechanism whereby ligand protects Gal-1 from oxidative inactivation, an observation made shortly after the initial discovery of Gal-1. Although crystallographic and solution-based CD experiments failed to detect significant changes in Gal-1 conformation following ligand binding (49–52), previous studies utilizing chaotropic denaturation demonstrated that ligand can significantly impact the nature and pathway of folding intermediates (53), providing some insight into Gal-1 oxidation. Introduction of ligand enhances Gal-1 stability and results in denaturation of dimeric Gal-1 directly into unfolded monomers (53). By contrast, in the absence of ligand, Gal-1 undergoes an unfolding reaction that involves the formation of monomeric intermediates prior to full denaturation and required lower concentration of chaotropic denaturant (53), suggesting that monomer formation and alterations in monomer conformation are favored in the absence of ligand. Given the present results, Gal-1 oxidation likely proceeds through a similar pathway. Ligand enhanced dimerization likely precludes conformational changes needed to form critical intramolecular disulfide bridges. However, absence of ligand allows increased formation of monomeric Gal-1 and therefore monomeric metastable intermediates. These monomeric intermediates likely possess greater conformational freedom, increasing the likelihood of intramolecular disulfide bond formation. The monomeric nature of oxidized Gal-1 corroborates this finding and strongly suggests a monomeric intermediate in this pathway (13). Furthermore, the enhanced sensitivity of mGal-1 to oxidation strongly suggests that, just as ligand enhances dimerization and thereby reduces sensitivity to oxidative inactivation, mutations that impair dimerization and therefore increase monomer formation favor oxidation.
The ability of ligand to enhance dimerization not only suggests a pathway for protecting Gal-1 oxidation, but also demonstrates that Gal-1 exists in a monomer-dimer equilibrium, in contrast to previous studies that suggested that Gal-1 exists as an irreversible dimer (54). Gal-1 dilution resulted in monomer formation, whereas concentrating Gal-1 allowed monomeric Gal-1 to reform dimers. As the present results suggest that monomer-dimer equilibrium likely regulates both Gal-1 activity and the ability to signal, regulation of monomer-dimer equilibrium likely provides a key regulatory point governing Gal-1 function.
Although alkylation protected mGal-1 from oxidative inactivation, imGal-1 still exhibited impaired signaling compared with iGal-1, which suggests a requirement for dimerization to effect full signaling. Furthermore, the inability of mGal-1 to induce PS externalization corroborates previous results demonstrating a requirement for continual engagement of leukocyte ligands for PS to be realized (10). Once PS exposure occurs, continual ligand binding must also occur for sustained PS exposure, as cells treated with Gal-1 failed to maintain PS following Gal-1 oxidation. Reversion of PS during Gal-1 incubation does not likely reflect removal of cells following potential Gal-1-induced death, as cells undergoing apoptosis were readily detected in this assay and iGal-1 induced sustained PS exposure over prolonged periods. These results also demonstrate that loss of PS following prolonged Gal-1 incubation does not likely reflect cellular insensitivity to Gal-1 over time. Given the oxidative nature of the extracellular environment, oxidation of Gal-1 may be an irreversible event. As a result, cellular movement into different redox environments may actually facilitate Gal-1 oxidation and therefore allow cells initially targeted for phagocytic removal to become re-engaged in host defense following PS reversion. In contrast, leukocyte-mediated damage of viable tissue may facilitate the release of reduced Gal-1 from intracellular stores that then engage leukocytes and induce their turnover (6, 55).
In addition to regulating Gal-1 signaling, monomer-dimer equilibrium may also regulate Gal-1 secretion. Gal-1 exists in a monomer-dimer equilibrium both inside and outside the cell (15), although secreted Gal-1, which exits through an incompletely defined ER-independent pathway or pathways, occurs primarily as a monomer (15, 39, 56, 57). In the absence of extracellular ligand, Gal-1 readily undergoes inactivation into oxidized monomers following secretion (13, 39), suggesting that Gal-1 may be secreted as an inactive or partially folded monomer. Engagement of ligand, either on the secreting cell or target cell, likely facilitates dimerization, thereby protecting Gal-1 from oxidative inactivation while also facilitating signaling events. In this way, ligand may not only stabilize Gal-1, but also enhance Gal-1-induced signaling.
Although the present study focused on Gal-1 oxidation as an inactivating process, oxidized Gal-1 appears to have significant biological activity independent of glycan ligand recognition. Oxidized Gal-1 enhances peripheral nerve regeneration both in vitro and in vivo (58). As a result, oxidation not only regulates the immunomodulary lectin-dependent activities of Gal-1, but also determines when the bioactivities of oxidized Gal-1 become apparent. In this way, Gal-1 provides another example of a morpheein, a protein capable of adopting different conformations capable of regulating distinct biological processes (59).
Given the significant number of studies suggesting a role for Gal-1 in the regulation of immunity, the sensitivity of Gal-1 to oxidative inactivation likely evolved as an intrinsic regulatory mechanism responsible for governing Gal-1 activity once secreted from the cell (60). Such a sensor may be important in dictating the distribution and longevity of Gal-1 signaling. For example, whereas Gal-1 induces turnover of neutrophils (3, 6, 40), inhibits leukocyte chemotaxis and induces immunosuppressive cytokine secretion in both naive and activated T cells (40, 61), premature engagement of infiltrating leukocyte by Gal-1 could ameliorate an otherwise productive and necessary inflammatory response. The highly oxidative environment surrounding inflammation likely facilitates the oxidation of Gal-1 released during primary tissue injury prior to significant leukocyte recruitment (1), allowing leukocytes to successfully neutralize pathogen or remove necrotic tissue without being impeded by the immunosuppressive effects of Gal-1. However, as leukocytes encroach on viable tissue surrounding an area of tissue injury, leukocyte-mediated damage may release reduced and therefore active Gal-1, allowing Gal-1 to inhibit leukocyte chemotaxis and induce their turnover (38, 62). Future studies will explore these intriguing possibilities.
Supplementary Material
Acknowledgments
We thank Sandy Cummings for technical assistance. We also thank Drs. Tongzhong Ju, Baoyun Xia, and Jamie Heimburg-Molinaro for helpful discussions and manuscript review.
This work was supported, in whole or in part, by National Institutes of Health Grant HL085607 and NIGMS Grant GM62116. This work was also supported by the Consortium for Functional Glycomics (Core D and Core H). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
Footnotes
The abbreviations used are: Gal-1, human galectin-1; mGal-1, C2S,V5D mutant of human galectin-1; 2-ME, 2-mercaptoethanol; DTT, dithiothreitol; PS, phosphatidylserine; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; iGal-1, iodoacetamide-treated Gal-1; PBS, phosphate-buffered saline; HPLC, high pressure liquid chromatography; imGal-1, alkylated iodoacetamide-treated Gal-1.
References
- 1.Simon, H. U. (2003) Immunol. Rev. 193 101–110 [DOI] [PubMed] [Google Scholar]
- 2.Toscano, M. A., Bianco, G. A., Ilarregui, J. M., Croci, D. O., Correale, J., Hernandez, J. D., Zwirner, N. W., Poirier, F., Riley, E. M., Baum, L. G., and Rabinovich, G. A. (2007) Nat. Immunol. 8 825–834 [DOI] [PubMed] [Google Scholar]
- 3.Stowell, S. R., Karmakar, S., Stowell, C. J., Dias-Baruffi, M., McEver, R. P., and Cummings, R. D. (2007) Blood 109 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stowell, S. R., Arthur, C. M., Slanina, K. A., Horton, J. R., Smith, D. F., and Cummings, R. D. (2008) J. Biol. Chem. 283 20547–20559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stowell, S. R., Qian, Y., Karmakar, S., Koyama, N. S., Dias-Baruffi, M., Leffler, H., McEver, R. P., and Cummings, R. D. (2008) J. Immunol. 180 3091–3102 [DOI] [PubMed] [Google Scholar]
- 6.Dias-Baruffi, M., Zhu, H., Cho, M., Karmakar, S., McEver, R. P., and Cummings, R. D. (2003) J. Biol. Chem. 278 41282–41293 [DOI] [PubMed] [Google Scholar]
- 7.Demetriou, M., Granovsky, M., Quaggin, S., and Dennis, J. W. (2001) Nature 409 733–739 [DOI] [PubMed] [Google Scholar]
- 8.Nieminen, J., Kuno, A., Hirabayashi, J., and Sato, S. (2007) J. Biol. Chem. 282 1374–1383 [DOI] [PubMed] [Google Scholar]
- 9.Brewer, C. F., Miceli, M. C., and Baum, L. G. (2002) Curr. Opin. Struct. Biol. 12 616–623 [DOI] [PubMed] [Google Scholar]
- 10.Karmakar, S., Cummings, R. D., and McEver, R. P. (2005) J. Biol. Chem. 280 28623–28631 [DOI] [PubMed] [Google Scholar]
- 11.Hirabayashi, J., and Kasai, K. (1991) J. Biol. Chem. 266 23648–23653 [PubMed] [Google Scholar]
- 12.Miura, T., Takahashi, M., Horie, H., Kurushima, H., Tsuchimoto, D., Sakumi, K., and Nakabeppu, Y. (2004) Cell Death Differ. 11 1076–1083 [DOI] [PubMed] [Google Scholar]
- 13.Inagaki, Y., Sohma, Y., Horie, H., Nozawa, R., and Kadoya, T. (2000) Eur. J. Biochem. 267 2955–2964 [DOI] [PubMed] [Google Scholar]
- 14.Tracey, B. M., Feizi, T., Abbott, W. M., Carruthers, R. A., Green, B. N., and Lawson, A. M. (1992) J. Biol. Chem. 267 10342–10347 [PubMed] [Google Scholar]
- 15.Cho, M., and Cummings, R. D. (1995) J. Biol. Chem. 270 5198–5206 [DOI] [PubMed] [Google Scholar]
- 16.Pace, K. E., Hahn, H. P., and Baum, L. G. (2003) Methods Enzymol. 363 499–518 [DOI] [PubMed] [Google Scholar]
- 17.Perillo, N. L., Pace, K. E., Seilhamer, J. J., and Baum, L. G. (1995) Nature 378 736–739 [DOI] [PubMed] [Google Scholar]
- 18.Carlow, D. A., Williams, M. J., and Ziltener, H. J. (2003) J. Immunol. 171 5100–5106 [DOI] [PubMed] [Google Scholar]
- 19.Tartier, L., McCarey, Y. L., Biaglow, J. E., Kochevar, I. E., and Held, K. D. (2000) Cell Death Differ. 7 1002–1010 [DOI] [PubMed] [Google Scholar]
- 20.Clerch, L. B., Whitney, P., Hass, M., Brew, K., Miller, T., Werner, R., and Massaro, D. (1988) Biochemistry 27 692–699 [DOI] [PubMed] [Google Scholar]
- 21.van den Brule, F., Califice, S., Garnier, F., Fernandez, P. L., Berchuck, A., and Castronovo, V. (2003) Lab. Investig. 83 377–386 [DOI] [PubMed] [Google Scholar]
- 22.He, L., Andre, S., Siebert, H. C., Helmholz, H., Niemeyer, B., and Gabius, H. J. (2003) Biophys. J. 85 511–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasano, K., and Hirakawa, Y. (1997) J. Histochem. Cytochem. 45 275–283 [DOI] [PubMed] [Google Scholar]
- 24.Whitney, P. L., Powell, J. T., and Sanford, G. L. (1986) Biochem. J. 238 683–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaltner, H., Seyrek, K., Heck, A., Sinowatz, F., and Gabius, H. J. (2002) Cell Tissue Res. 307 35–46 [DOI] [PubMed] [Google Scholar]
- 26.Andre, S., Kaltner, H., Lensch, M., Russwurm, R., Siebert, H. C., Fallsehr, C., Tajkhorshid, E., Heck, A. J., von Knebel Doeberitz, M., Gabius, H. J., and Kopitz, J. (2005) Int. J. Cancer 114 46–57 [DOI] [PubMed] [Google Scholar]
- 27.Powell, J. T., and Whitney, P. L. (1984) Biochem. J. 223 769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell, J. T. (1988) Biochem. J. 252 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stowell, S. R., Arthur, C. M., Mehta, P., Slanina, K. A., Blixt, O., Leffler, H., Smith, D. F., and Cummings, R. D. (2008) J. Biol. Chem. 283 10109–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stowell, S. R., Dias-Baruffi, M., Penttila, L., Renkonen, O., Nyame, A. K., and Cummings, R. D. (2004) Glycobiology 14 157–167 [DOI] [PubMed] [Google Scholar]
- 31.Blixt, O., Head, S., Mondala, T., Scanlan, C., Huflejt, M. E., Alvarez, R., Bryan, M. C., Fazio, F., Calarese, D., Stevens, J., Razi, N., Stevens, D. J., Skehel, J. J., van Die, I., Burton, D. R., Wilson, I. A., Cummings, R., Bovin, N., Wong, C. H., and Paulson, J. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bochner, B. S., Alvarez, R. A., Mehta, P., Bovin, N. V., Blixt, O., White, J. R., and Schnaar, R. L. (2005) J. Biol. Chem. 280 4307–4312 [DOI] [PubMed] [Google Scholar]
- 33.van Liempt, E., Bank, C. M., Mehta, P., Garcia-Vallejo, J. J., Kawar, Z. S., Geyer, R., Alvarez, R. A., Cummings, R. D., Kooyk, Y., and van Die, I. (2006) FEBS Lett. 580 6123–6131 [DOI] [PubMed] [Google Scholar]
- 34.Nam, H. J., Gurda-Whitaker, B., Yee Gan, W., Ilaria, S., McKenna, R., Mehta, P., Alvarez, R. A., and Agbandje-McKenna, M. (2006) J. Biol. Chem. 281 25670–25677 [DOI] [PubMed] [Google Scholar]
- 35.Grynkiewicz, G., Poenie, M., and Tsien, R. Y. (1985) J. Biol. Chem. 260 3440–3450 [PubMed] [Google Scholar]
- 36.Galton, M. M., Sulzer, C. R., Santarosa, C. A., and Fields, M. J. (1965) Appl. Microbiol. 13 81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbons, R. J., and Fitzgerald, R. J. (1969) J. Bacteriol. 98 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kakimoto, K., Matsukawa, A., Yoshinaga, M., and Nakamura, H. (1995) Cell Immunol. 165 26–32 [DOI] [PubMed] [Google Scholar]
- 39.Cho, M., and Cummings, R. D. (1995) J. Biol. Chem. 270 5207–5212 [DOI] [PubMed] [Google Scholar]
- 40.Stowell, S. R., Qian, Y., Karmakar, S., Koyama, N. S., Dias-Baruffi, M., Leffler, H., McEver, R. P., and Cummings, R. D. (2008) J. Immunol. 180 3091–3102 [DOI] [PubMed] [Google Scholar]
- 41.Kopcow, H. D., Rosetti, F., Leung, Y., Allan, D. S., Kutok, J. L., and Strominger, J. L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 18472–18477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smrz, D., Lebduska, P., Draberova, L., Korb, J., and Draber, P. (2008) J. Biol. Chem. 283 10904–10918 [DOI] [PubMed] [Google Scholar]
- 43.Smrz, D., Draberova, L., and Draber, P. (2007) J. Biol. Chem. 282 10487–10497 [DOI] [PubMed] [Google Scholar]
- 44.Barondes, S. H. (1984) Science 223 1259–1264 [DOI] [PubMed] [Google Scholar]
- 45.Teichberg, V. I. (1978) Methods Enzymol. 50 291–302 [DOI] [PubMed] [Google Scholar]
- 46.Teichberg, V. I., Silman, I., Beitsch, D. D., and Resheff, G. (1975) Proc. Natl. Acad. Sci. U. S. A. 72 1383–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pande, A. H., Gupta, R. K., Sumati, and Hajela, K. (2003) Protein Pept. Lett. 10 265–275 [DOI] [PubMed] [Google Scholar]
- 48.Levi, G., and Teichberg, V. I. (1981) J. Biol. Chem. 256 5735–5740 [PubMed] [Google Scholar]
- 49.Bourne, Y., Bolgiano, B., Liao, D. I., Strecker, G., Cantau, P., Herzberg, O., Feizi, T., and Cambillau, C. (1994) Nat. Struct. Biol. 1 863–870 [DOI] [PubMed] [Google Scholar]
- 50.Liao, D. I., Kapadia, G., Ahmed, H., Vasta, G. R., and Herzberg, O. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 1428–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez-Lucendo, M. F., Solis, D., Andre, S., Hirabayashi, J., Kasai, K., Kaltner, H., Gabius, H. J., and Romero, A. (2004) J. Mol. Biol. 343 957–970 [DOI] [PubMed] [Google Scholar]
- 52.Shahwan, M., Al-Qirim, M. T., Zaidi, S. M., and Banu, N. (2004) Biochemistry (Mosc.) 69 506–512 [DOI] [PubMed] [Google Scholar]
- 53.Iglesias, M. M., Elola, M. T., Martinez, V., Fink, N., and Wolfenstein-Todel, C. (2003) Biochim. Biophys. Acta 1648 164–173 [DOI] [PubMed] [Google Scholar]
- 54.Giudicelli, V., Lutomski, D., Levi-Strauss, M., Bladier, D., Joubert-Caron, R., and Caron, M. (1997) Glycobiology 7 viii–x [DOI] [PubMed] [Google Scholar]
- 55.Cerri, D. G., Rodrigues, L. C., Stowell, S. R., Araujo, D. D., Coelho, M. C., Oliveira, S. R., Bizario, J. C., Cummings, R. D., Dias-Baruffi, M., and Costa, M. C. (2008) Glycobiology 18 842–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cleves, A. E., Cooper, D. N., Barondes, S. H., and Kelly, R. B. (1996) J. Cell Biol. 133 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooper, D. N., and Barondes, S. H. (1990) J. Cell Biol. 110 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horie, H., Kadoya, T., Sango, K., and Hasegawa, M. (2005) Curr. Drug Targets 6 385–394 [DOI] [PubMed] [Google Scholar]
- 59.Jaffe, E. K. (2005) Trends Biochem. Sci 30 490–497 [DOI] [PubMed] [Google Scholar]
- 60.Than, N. G., Romero, R., Erez, O., Weckle, A., Tarca, A. L., Hotra, J., Abbas, A., Han, Y. M., Kim, S. S., Kusanovic, J. P., Gotsch, F., Hou, Z., Santolaya-Forgas, J., Benirschke, K., Papp, Z., Grossman, L. I., Goodman, M., and Wildman, D. E. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 15819–15824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.La, M., Cao, T. V., Cerchiaro, G., Chilton, K., Hirabayashi, J., Kasai, K., Oliani, S. M., Chernajovsky, Y., and Perretti, M. (2003) Am. J. Pathol. 163 1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carden, D. L., and Korthuis, R. J. (1996) Am. J. Physiol. 271 H1947–H1952 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.