Abstract
The life cycle of protein kinase C (PKC) is tightly controlled by mechanisms that mature the enzyme, sustain the activation-competent enzyme, and degrade the enzyme. Here we show that a conserved PXXP motif (Kannan, N., Haste, N., Taylor, S. S., and Neuwald, A. F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 1272–1277), in the C-terminal tail of AGC (c-AMP-dependent protein kinase/protein kinase G/protein kinase C) kinases, controls the processing phosphorylation of conventional and novel PKC isozymes, a required step in the maturation of the enzyme into a signaling-competent species. Mutation of both Pro-616 and Pro-619 to Ala in the conventional PKC βII abolishes the phosphorylation and activity of the kinase. Co-immunoprecipitation studies reveal that conventional and novel, but not atypical, PKC isozymes bind the chaperones Hsp90 and Cdc37 through a PXXP-dependent mechanism. Inhibitors of Hsp90 and Cdc37 significantly reduce the rate of processing phosphorylation of PKC. Of the two C-terminal sites processed by phosphorylation, the hydrophobic motif, but not the turn motif, is regulated by Hsp90. Overlay of purified Hsp90 onto a peptide array containing peptides covering the catalytic domain of PKC βII identified regions surrounding the PXXP segment, but not the PXXP motif itself, as major binding determinants for Hsp90. These Hsp90-binding regions, however, are tethered to the C-terminal tail via a “molecular clamp” formed between the PXXP motif and a conserved Tyr (Tyr-446) in the αE-helix. Disruption of the clamp by mutation of the Tyr to Ala recapitulates the phosphorylation defect of mutating the PXXP motif. These data are consistent with a model in which a molecular clamp created by the PXXP motif in the C-terminal tail and determinants in the αE-helix of the catalytic domain allows the chaperones Hsp90 and Cdc37 to bind newly synthesized PKC, a required event in the processing of PKC by phosphorylation.
Protein kinases, which comprise ∼2% of the human genome, are key signal transducers that regulate a wide variety of cellular processes, such as growth, proliferation, and metabolism, through catalysis of specific phosphorylation events (1). By integrating signals from extracellular stimuli and transmitting them to targeted downstream substrates, protein kinases serve as a pivotal point of regulation within the cell. Deregulation and mutation of protein kinases play a causal role in human pathology, notably cancer, poising kinases as important targets for the design of therapeutics (2–5). Therefore, understanding the mechanisms that regulate protein kinases, such as those important for maturation and processing, would be critical for designing therapeutics that would maintain the correct functioning of signal transduction pathways.
Heat shock proteins (Hsp),3 such as Hsp90, are ubiquitously expressed molecular chaperones that facilitate protein folding, regulate quality control, and guide protein turnover in an effort to maintain cellular homeostasis (6–8). Unlike other chaperones such as Hsp70, which non-specifically assists in folding of nascent polypeptide chains (7), Hsp90 works with a specific and discrete set of client proteins, particularly protein kinases (9). Many of the known client kinases of Hsp90, Src (10), Akt (11, 12), phosphoinositide dependent kinase-1 (PDK-1) (13), and ErbB2-/HER2 (14, 15), require the activity of Hsp90 to reach an activation-competent and mature state. Hsp90 is recruited to its kinase clients through interactions with cochaperones, such as Cdc37, which bridge the interaction between Hsp90 and the kinase client (16, 17); this mechanism is revealed in a structural analysis of the Cdc37-Cdk4-Hsp90 complex (18). Cdc37, originally identified in yeast (19), is a cochaperone specific to the kinome that not only assists Hsp90 function but can also recognize and stabilize clients independently of Hsp90 (20). By binding specific regions of the catalytic domains of these kinases, the Hsp90-Cdc37 complex utilizes ATP to promote and stabilize functional conformations of its clients (8, 16, 17, 21). Pharmacological inhibition of Hsp90 by ansamycin antibiotics such as 17-(allylamino)-17-demethoxygeldanamycin (17-AAG) leads to the destabilization and subsequent proteasomal degradation of its clients (6, 22). Recent studies have identified Hsp90 as a promising therapeutic target in cancer as levels of chaperones and activity of client kinases are frequently up-regulated (23–25).
The protein kinase C (PKC) family of Ser/Thr kinases serves as a paradigm of how conformation and processing by phosphorylation regulate activity, localization, and inter- and intramolecular interactions (26). The mammalian PKC family consists of 10 isozymes divided into three subclasses (conventional, novel, and atypical) based on their primary structure and second messenger mode of regulation. In the case of conventional PKC isozymes, newly synthesized enzyme is loosely engaged on the membrane in a conformation that exposes the activation loop for phosphorylation by the upstream kinase, PDK-1 (28). This phosphorylation triggers two sequential phosphorylations on the C terminus, one on the turn motif and one on the hydrophobic motif. Phosphorylation of the turn motif is required for phosphorylation of the hydrophobic motif and has recently been shown to depend on the mammalian target of rapamycin complex 2 (mTORC2), a complex consisting of the mammalian target of rapamycin (mTOR), Rictor, Sin1, and mLST8 (29, 30). Turn motif phosphorylation is rapidly followed by phosphorylation at the hydrophobic motif, a reaction that occurs by an intramolecular mechanism in vitro (31). Fully stey phosphorylated PKC is released into the cytosol in a closed conformation in which an autoinhibitory pseudosubstrate sequence occupies the substrate-binding cavity. Upon generation of the lipid second messenger diacylglycerol and elevation of intracellular Ca2+, conventional PKC isozymes (α, βI, βII, γ) translocate to membranes via their membrane-targeting C1 and C2 domains, where they adopt an open conformation in which the pseudosubstrate is expelled from the substrate-binding cavity, permitting phosphorylation of downstream substrates (32). Novel PKC isozymes (δ, ε, θ, and η) only respond to diacylglycerol, and their processing phosphorylations can occur through additional mechanisms (33, 34). Atypical PKC isozymes (ζ and ι/λ) do not respond to either Ca2+ or diacylglycerol but can also undergo regulation of their processing phosphorylations by external stimuli (35). In fact, atypical PKC isozymes contain a Glu at their hydrophobic motif site. Thus, although these three sites are conserved among PKC family members, additional layers of regulation generate specificity in how these kinases become signaling-competent enzymes.
The mechanisms of regulation of the activity and signaling properties of PKC by lipid second messengers and phosphorylation have been well characterized; however, mounting evidence suggests that there are many other regulatory inputs for PKC function. Recent analysis of the evolutionary constraints acting on AGC kinase sequences have underscored the importance of the C-terminal tail as a critical regulatory module (36). Deletion mutants of the C-terminal tail have been shown to abrogate PKC activity (37). In addition to containing the key regulatory phosphorylation sites and docking the upstream kinase PDK-1, the C-terminal tail contains key conserved motifs, found within all AGC kinases, that facilitate ATP binding, promote substrate binding, and structure the catalytic core (36). One such motif comprises the segment PXXP; this motif makes key contacts with the catalytic core, where it is important for modulating movement of the catalytic domain (36). Although this PXXP motif is conserved in all the PKC isozymes, its functional role is unknown.
In this study, we address the role of the conserved Pro residues in the PXXP motif of the C-terminal tail of PKC βII. We show that mutation of these two Pro to Ala (P616A and P619A) results in a kinase that is not processed by phosphorylation in cells and is thus inactive. Further analysis reveals that this mutant is not able to bind the chaperones Hsp90 and Cdc37, an event that is required for the processing of PKC by phosphorylation. Our peptide array data indicate that Hsp90 binds to regions of the catalytic core such as the αC-β4 loop and the αD-helix that serve as hinge points for C-helix movement (36). Structural analysis delineates that these hinge points are tethered to the C-terminal tail through a molecular clamp formed between the PXXP segment and AGC conserved residues in the αE-helix. Mutation of one of the “clamping” residues, a conserved Tyr (Tyr-446), recapitulates the defect resulting from mutation of the PXXP motif. Our data support a model in which the PXXP motif participates in an intramolecular clamp with determinants in the αE helix of the kinase core, by providing a recognition surface for Hsp90 to bind and facilitate the maturation of PKC, a required step in the processing of the enzyme.
EXPERIMENTAL PROCEDURES
Plasmids—The cloning of rat PKC βII into the mammalian expression vector pCDNA3 and subsequent generation of the phosphorylation site mutants (T641E and S660E) has been previously described (38, 39). The construction of the N-terminal Myc-tagged PDK-1 in pCDNA3 has been previously described (40). Expression constructs for wild-type bovine PKC α and rat PKC ζ were generous gifts from Dr. Alex Toker (Harvard Medical School). Mouse PKC δ was a generous gift from Dr. Peter Blumberg (National Institutes of Health). PKC βII-YFP was cloned as described previously (41). PKC βII-RFP was constructed by PCR amplification and subsequent cloning into a pCDNA3 vector with monomeric RFP as a C-terminal tag (42). The other PKC mutants, PKC βII-K371R, PKC βII-P616A/P619A, PKC βII-P616A/P619A-YFP, PKC βII-Y446A, PKC α-P613A/P616A, PKC δ-P617A/P620A, and PKC ζ-P534A/P537A, were all generated using the QuikChange site-directed mutagenesis kit (Stratagene).
Materials—Human Hsp90α recombinant protein was purchased from Genway. Purified His-PDK-1 was a generous gift from Charles C. King. PKC substrate peptide (Ac-FKKSFKLNH2) was obtained from Anaspec. Oligonucleotides were purchased from Integrated DNA Technologies. Easy Tag [35S]Met/Cys (1175 Ci mmol–1) and [γ-32P]ATP (3000 Ci mmol–1) were purchased from PerkinElmer Life Sciences. Met/Cys-deficient DMEM was purchased from Invitrogen. 17-AAG was purchased from A.G. Scientific, Inc. Carbobenzoxyl-l-leucyl-l-leucyl-leucinal (MG-132), phorbol 12-myristate 13-acetate (PMA), and phorbol-12,13-dibutyrate (PDBu) were purchased from Calbiochem. Celastrol was purchased from Cayman Chemical. Polyclonal antibodies to PKC α, PKC βII, PKC δ, PKC ζ, and Cdc37 were obtained from Santa Cruz Biotechnology. Monoclonal antibodies to PKC α, PKC β, and Hsp90 were obtained from BD Transduction Laboratories. A phospho-specific antibody (pThr-500) that specifically recognizes the phosphorylated activation loop of PKC isozymes was characterized previously (43). A phospho-specific PKC antibody to the hydrophobic motif phosphorylation site (PKC βII Ser-660) and an antibody to the turn motif phosphorylation site (PKC α/βII Thr-638/641) were purchased from Cell Signaling Technology. An anti-Myc monoclonal (9E10) antibody was purchased from Covance. A monoclonal anti-β-actin antibody was purchased from Sigma-Aldrich. Ultra-Link protein A/G beads were obtained from Thermo Scientific. Electrophoresis reagents were obtained from Bio-Rad. All other materials and chemicals were reagent-grade.
Cell Culture and Transfection—tsA201, COS7, HeLa, and MCF7 cells were maintained in DMEM (Cellgro) containing 10% fetal bovine serum (Hyclone) and 1% penicillin/streptomycin at 37 °C in 5% CO2. MCF-10A cells were maintained in mammary epithelium growth medium (Clonetics) at 37 °C and 5% CO2. Transient transfection of tsA201 cells was carried out using Effectene transfection reagents (Qiagen), and transient transfection of COS7 cells was carried out using the FuGENE transfection reagent (Roche Applied Science).
Immunofluorescence—COS7 cells were transiently co-transfected with PKC βII-RFP and PKC βII-P616A/P619A-YFP for 24 h. Cells were washed once and imaged in Hanks' balanced salt solution with 1 mm Ca2+. Epifluorescent images were acquired on a Zeiss Axiovert microscope (Carl Zeiss Micro-Imaging, Inc.) using a Micro-Max digital camera (Roper-Princeton Instruments) controlled by Metafluor Software (Universal Imaging, Corp). Optical filters were obtained from Chromas Technologies and Semrock, Inc. Cells were treated with 200 nm PDBu, and the same cells were used to acquire YFP and RFP images through a 10% neutral density filter. YFP images were obtained using a 495/10-nm excitation filter, a 505-nm dichroic mirror, and a 535/25-nm emission filter. RFP images were obtained using a 560/25-nm excitation filter, a 593-nm dichroic mirror, and a 629/53-nm emission filter.
Kinase Assays—For the PKC activity assay, detergent-solubilized lysates (20 mm HEPES, pH 7.5, 0.1% Triton X-100, 2 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride) from untransfected tsA201 cells and tsA201 cells transfected with empty vector, WT PKC βII, or PKC βII-P616A/P619A were assayed for PKC activity for 8 min by monitoring 32P incorporation from [γ-32P]ATP into a synthetic PKC-selective peptide (Ac-FKKSFKL-NH2) using a paper assay as described previously (38). The standard reaction mix contained 20 mm HEPES, pH 7.5, 2 mm dithiothreitol, 5 mm MgCl2, 100 μm ATP, 50 μm peptide substrate, and 100 μCi of [γ-32P]ATP. Non-activating conditions included 2 mm EGTA. Activating conditions included 140 μm phosphatidylserine, 3.8 μm diacylglycerol membranes, and 500 μm CaCl2. The activity of the transfected kinase was determined by first subtracting 32P incorporation in control (vector-transfected) lysate samples and then normalizing the Ca2+/lipid-stimulated peptide phosphorylation to PKC expression (determined by Western blot analysis); activity is expressed in relative units. For the PDK-1 activity assay, COS7 cells expressing empty vector, WT PKC βII, or PKC βII-P616A/P619A were lysed in Buffer A (50 mm Tris, pH 7.4, 1% TritonX-100, 50 mm NaF, 10 mm Na4P2O7, 100 mm NaCl, 5 mm EDTA, 1 mm Na3VO4, and 1 mm phenylmethylsulfonyl fluoride), and detergent-solubilized lysates were incubated with a monoclonal anti-PKC α antibody (cross-reactive with PKC β) overnight at 4 °C to allow immune complex formation. The immune complexes were collected with Ultra-Link protein A/G beads. After washing the immune complexes with Buffer A, they were resuspended in the kinase buffer (20 mm HEPES, pH 7.4, 2 mm dithiothreitol). The standard reaction mix contained the immune complexes, 20 mm HEPES, 500 μm ATP, and 2 mm MgCl2. The reaction was performed in the absence or presence of purified His-PDK-1 (0.5 nm) and in non-activating or activating PKC conditions. Non-activating conditions included 2 mm EGTA. Activating conditions included 140 μm phosphatidylserine, 3.8 μm diacylglycerol membranes, and 500 μm CaCl2. The reaction was allowed to proceed for 30 min at 30 °C and stopped by the addition of Laemmli sample buffer. The samples were separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and analyzed for pThr-500 phosphorylation by Western blotting.
PMA Time Course—COS7 cells were transiently transfected with WT PKC βII or PKC βII-P616A/P619A. Approximately 24 h after transfection, cells were treated with 200 nm PMA for 0, 15, 30, and 90 min. The cells were lysed in Buffer A, and whole cell lysates were analyzed by SDS-PAGE and Western blotting.
Pulse-Chase Analysis—COS7 cells were transfected with WT PKC βII, PKC βII-P616A/P619A, PKC βII T641E, or PKC βII S660E. At 24–30 h after transfection, cells were incubated with Met/Cys-deficient DMEM for 30 min at 37 °C. The cells were then pulse-labeled with 0.5 mCi ml–1 [35S]Met/Cys in Met/Cys-deficient DMEM for 7 min at 37 °C, media were removed, and cells were chased with DMEM containing 5 mm unlabeled methionine and 5 mm unlabeled cysteine (44). At the indicated times, cells were lysed in Buffer A and centrifuged at 16,000 × g for 5 min at 22 °C, and PKC βII in the supernatant was immunoprecipitated with an anti-PKC α monoclonal antibody (cross-reactive with PKC βII) overnight at 4 °C. The immune complexes were collected with Ultra-Link protein A/G beads, washed with Buffer A, separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and analyzed by autoradiography. For the inhibitor experiments, cells were pretreated for 3 h with 1 μm 17-AAG, 30 min with 10 μm celastrol, or both prior to the pulse-chase. Pulse-chase experiments of endogenous PKC α followed the same protocol as described above. Densitometric analysis of scanned autoradiograms was performed using NIH Image J analysis software, and the kinetic analysis was performed using Kaleidograph software (version 4.0).
Immunoprecipitation—To examine the interaction of PKC with endogenous Hsp90, COS7 cells were transfected with either WT PKC (α, βII, δ, ζ) or the respective construct with a mutated PXXP motif. For PKC βII, the interaction of Hsp90 with the kinase-dead mutant PKC βII-K371R and PKC βII-Y446A was also assessed. Approximately 24 h after transfection, the cells were lysed in Buffer A and centrifuged at 16,000 × g for 5 min at 22 °C, and the detergent-solubilized supernatants were incubated with the appropriate PKC antibody overnight at 4 °C. The immune complexes were collected with Ultra-Link protein A/G beads, washed with Buffer A, and analyzed by SDS-PAGE and Western blotting. This method was also used to assess the interaction of PKC with endogenous Cdc37. For the PDK-1 co-immunoprecipitation experiments, COS7 cells were co-transfected with WT PKC βII, PKC βII-K371R, or PKC β-P616A/P619A and either empty vector or Myc-PDK-1. Cells were lysed as described above and incubated with a monoclonal anti-Myc antibody overnight at 4 °C to form immune complexes with Myc-PDK-1. The immune complexes were collected, washed, and analyzed as described above for the Hsp90 experiments. Densitometric analysis of scanned autoradiograms was performed using NIH Image J analysis software (version 1.40).
Analysis of 17-AAG-induced Down-regulation of PKC—COS7 cells were transiently transfected with WT PKC βII or PKC βII P616A/P619A. Approximately 24 h after transfection, the cells were treated with 1 μm 17-AAG for 0, 1, 4, 12, or 24 h. For MG-132 experiments, the cells were first pretreated with 10 μm MG-132 for ∼3 h. The cells were lysed in Buffer A, and whole cell lysates were analyzed by SDS-PAGE and Western blotting. Similar experiments were performed in HeLa, MCF-10A, and MCF7 cells lines to assess endogenous PKC levels.
Hsp90 Overlay of the Catalytic Domain of PKC βII—The catalytic domain of PKC βII (amino acids 342–673) was divided into 18-amino-acid peptides, with a 2-amino-acid shift, and synthesized using the INTAVIS MultiPep peptide synthesizer (INTAVIS Bioanalytical Instruments AG), which spotted the peptides onto an AC-S01 type amino-PEGylated membrane (INTAVIS AG). After activation of the membrane with ethanol, the membrane was blocked, washed, and then incubated overnight with 100 nm human Hsp90α recombinant protein. After incubation with the Hsp90 protein, the peptide array was analyzed by Western blot analysis with a monoclonal anti-Hsp90 antibody.
RESULTS
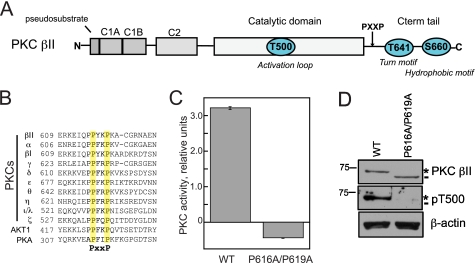
A Conserved PXXP Motif in the C-terminal Tail of PKC βII Is Necessary for Catalytic Activity—A conserved PXXP motif in the C-terminal tail of AGC kinases has been proposed to be an important site for allosteric regulation of the catalytic domain (Fig. 1, A and B) (36). To examine the biochemical role of this conserved PXXP motif in PKC βII, we mutated both Pro-616 and Pro-619 to Ala (P616A/P619A) and first assessed the catalytic activity of the construct in an in vitro kinase assay. Lysates from tsA201 cells expressing either WT PKC βII or PKC βII-P616A/P619A were assayed for activity toward a PKC peptide substrate in the presence or absence of activating cofactors; activity attributed to the transfected PKC was obtained by subtracting the endogenous activity determined from control lysates. Mutation of the PXXP motif in PKC βII abolished kinase activity; in fact, this construct behaved as a modest dominant-negative of endogenous activity (Fig. 1C). Curiously, Western blot analysis of lysates revealed that the PKC βII-P616A/P619A construct migrated faster than wild-type enzyme (Fig. 1D, dash). Previous studies have established that WT PKC βII migrates primarily as an upper band (asterisk) that represents a species quantitatively phosphorylated at the two C-terminal sites, with a minor fraction migrating as a faster mobility species (dash) that represents kinase that is unphosphorylated (44). Consistent with PKC βII-P616A/P619A migrating as the unphosphorylated species, it was not labeled by phospho-specific antibodies to the activation loop Thr-500 (Fig. 1D, middle panel), nor was it labeled by phospho-specific antibodies to the two C-terminal sites, the turn motif Thr-641, or the hydrophobic motif Ser-660 (data not shown). Thus, mutation of both Pro residues in the PXXP motif resulted in an inactive, unphosphorylated kinase.
FIGURE 1.
Mutation of a PXXP motif, conserved in all AGC kinases, in the C-terminal tail of PKC βII abolishes kinase activity. A, schematic representation of the domain composition of PKC βII showing pseudosubstrate, C1A and C1B domains, and C2 domain in the N-terminal regulatory moiety followed by the catalytic domain and C-terminal tail (Cterm tail) in the kinase moiety. The three processing phosphorylation sites, activation loop (Thr-500 (T500)), turn motif (Thr-641 (T641)), and hydrophobic motif (Ser-660 (S660)), in PKC βII are indicated by cyan circles. B, partial sequence alignment of AGC family members, PKA, Akt1, and the 10 PKC isozymes. A conserved PXXP motif in the C-terminal tail is highlighted in yellow. C, lysates from tsA201 cells expressing either WT PKC βII or PKC βII-P616A/P619A were assayed for PKC activity in the absence or presence of lipid and Ca2+. Cofactor-dependent activity attributed to transfected PKC βII (i.e. after subtraction of endogenous activity obtained from control lysates) was normalized to PKC βII expression in the lysate determined by Western blot. Data represent the mean ± S.E. from four independent experiments. D, Western blot showing the expression and phosphorylation of WT PKC βII and PKC βII-P616A/P619A used in the kinase assay with the indicated antibodies. Unphosphorylated PKC βII is labeled with a dash, and fully phosphorylated PKC βII is labeled with an asterisk. Lysates were probed for β-actin as a loading control (lower panel). pT500, pThr-500.
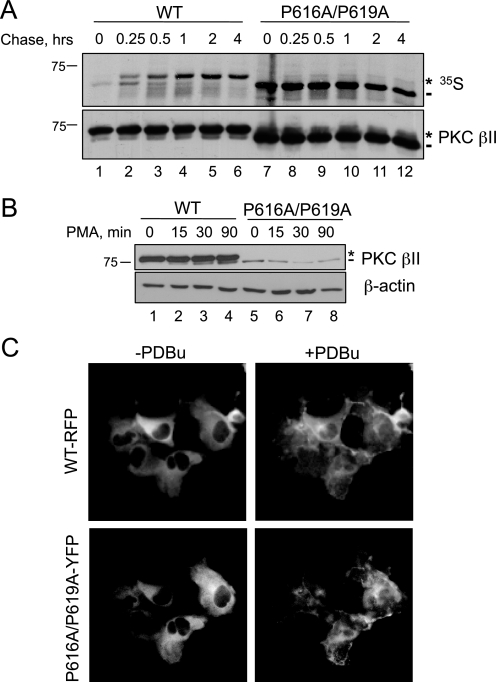
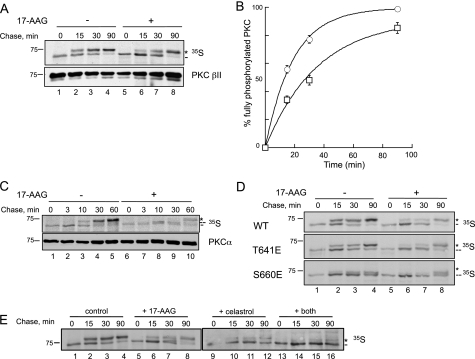
Mutation of the PXXP Motif in PKC βII Prevents the Maturation of PKC—Accumulation of unphosphorylated PKC could reflect the inability of the P616A/P619A construct to be processed by phosphorylation, or it could reflect an increased phosphatase sensitivity resulting in accumulation of matured, phosphorylated PKC that has been dephosphorylated (45). To discriminate between these two possibilities, we asked whether newly synthesized PKC matured into the phosphorylated species by pulse-chase analysis (Fig. 2A). COS7 cells transfected with WT PKC βII or PKC βII-P616A/P619A were pulse-labeled with [35S]Met/Cys and chased for up to 4 h with unlabeled media. Immunoprecipitated PKC was analyzed by autoradiography, and the processing of the pulsed pool was determined by the mobility of the radiolabeled band on SDS-PAGE. The autoradiogram in Fig. 2A shows that newly synthesized PKC βII appeared as a faster migrating species (lane 1, dash) that shifted to a slower mobility species over the course of the chase (lanes 2–6, asterisk). The mobility shift reflects the two tightly coupled processing phosphorylations at the C terminus, the turn motif (Thr-641), and the hydrophobic motif (Ser-660) (Fig. 1A). Importantly, mutation of the PXXP motif in PKC βII prevented maturation of PKC, as assessed by its co-migration with the faster-migrating, unphosphorylated species even after 4 h of chase (lanes 7–12, dash). These data reveal that mutation of both Pro residues in the PXXP motif prevents the processing of PKC βII by phosphorylation.
FIGURE 2.
Mutation of the PXXP motif in PKC βII prevents the maturation of the kinase, a defect that cannot be rescued by translocation to membranes. A, autoradiogram from a pulse-chase analysis of COS7 cells transfected with WT PKC βII (lanes 1–6) or PKC βII-P616A/P619A (lanes 7–12). Transfected cells were labeled with [35S]Met/Cys and chased for the indicated times. The asterisk denotes the position of the mature, fully phosphorylated species of PKC, and the dash represents the newly synthesized, unphosphorylated species of PKC. Total PKC in the immunoprecipitate is labeled by an anti-PKC βII antibody. B, COS7 cells were transfected with WT PKC βII (lanes 1–4) or PKC βII-P616A/P619A (lanes 5–8) and then treated with PMA (200 nm) for the indicated times. Whole cell lysates were separated by SDS-PAGE and then analyzed for total PKC (PKC βII) or total protein content (β-actin). The asterisk denotes the position of fully phosphorylated PKC, and the dash denotes the position of unphosphorylated PKC. C, representative images of COS7 cells transfected with WT PKCβII-RFP (upper panels) and PKC βII-P616A/P619A-YFP (lower panels) before (left panels) and after (right panels) PDBu (200 nm) treatment for 15 min.
Newly synthesized PKC is loosely tethered at the membrane by its regulatory modules in an open conformation (pseudosubstrate exposed), which allows PDK-1 to phosphorylate the activation loop and initiate the maturation process (27). To test whether the lack of processing of PKC βII-P616A/P619A resulted from improper localization of the mutant to the membrane, we forced membrane association by taking advantage of the ability of phorbol esters to recruit PKC to the membrane, independently of the phosphorylation state of the enzyme. To this end, we transfected COS7 cells with either WT PKC βII or PKC βII-P616A/P619A and treated the cells with the phorbol ester PMA. The Western blot in Fig. 2B shows that the PXXP mutant continued to migrate as a faster mobility species following PMA treatment (lanes 5–8, dash). Note that PMA treatment caused a characteristic increase in the amount of unphosphorylated PKC (lanes 1–4, dash), consistent with the ability of phorbol esters to promote the dephosphorylation and eventual down-regulation of the enzyme (46). To ensure that the PXXP mutant PKC βII-P616A/P619A was properly recruited to the membrane upon phorbol ester treatment, we co-transfected COS7 cells with fluorescently tagged constructs, WT PKC βII-RFP and PKC βII-P616A/P619A-YFP, and simultaneously co-imaged them in the absence or presence of the phorbol ester PDBu. Although modest differences in localization were observed, the fluorescent images in Fig. 2C reveal that both WT PKC βII-RFP and PKC βII-P616A/P619A-YFP translocated to the membrane upon treatment with PDBu (Fig. 2C, right panels). Similar results were obtained when WT PKC βII-RFP or PKC βII-P616A/P619A-YFP were transfected separately into cells (data not shown). Therefore, these data demonstrate that the inability of PKC βII-P616A/P619A to mature is not a consequence of improper subcellular localization.
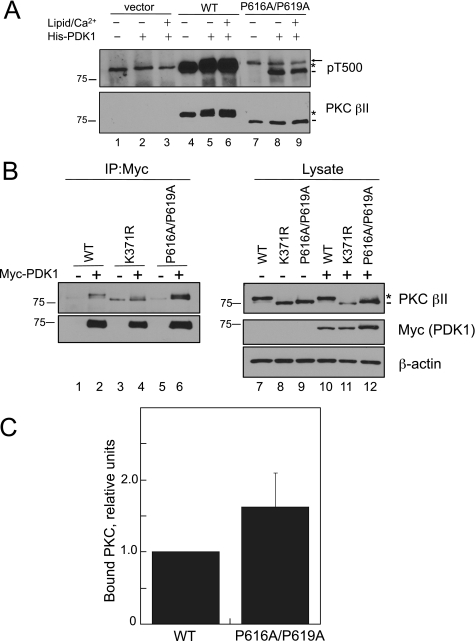
The Upstream Kinase PDK-1 Can Bind and Phosphorylate the PXXP Mutant PKC βII-P616A/P619A—The first step in the maturation process of PKC is phosphorylation of the activation loop by PDK-1. Because the PXXP mutant PKC βII-P616A/P619A has a defect in maturation (Fig. 2A), we tested whether the mutant was a suitable substrate for PDK-1. Immunoprecipitated WT PKC βII or PKC βII-P616A/P619A from detergent-solubilized lysates was incubated with purified His-PDK-1 in the absence or presence of Ca2+ and phosphatidylserine/diacylglycerol membranes (to induce the membrane-bound open conformation in which the PDK-1 site is unmasked) in an in vitro kinase assay (Fig. 3A). Activation loop phosphorylation was detected using a phospho-specific antibody to the Thr-500 activation loop site (cross-reactive with all PKC isozymes); indeed, this antibody detected endogenous PKC α in the vector lanes (Fig. 3A, lanes 1–3, arrow). WT PKC βII was robustly labeled with the pThr-500 antibody regardless of the addition of PDK-1 (Fig. 3A, lanes 4–6, asterisk), as expected for the normally matured, fully phosphorylated enzyme (43). In contrast, PKC βII-P616A/P619A was not labeled by the pThr-500 antibody in the absence of His-PDK-1 (Fig. 3A, lane 7, dash). However, the addition of His-PDK-1 resulted in the activation loop phosphorylation of the PKC βII-P616A/P619A mutant (Fig. 3A, lane 8, dash), although to a much lesser extent than WT. Additionally, PDK-1-catalyzed phosphorylation did not depend on PKC being bound to membranes (lane 9), indicating that the PKC βII-P616A/P619A mutant was in the open conformation in which the pseudosubstrate does not occupy the substrate-binding pocket, reflecting the conformation of newly synthesized PKC (27). These data reveal that the activation loop of the PKC βII-P616A/P619A is accessible and can be phosphorylated by PDK-1.
FIGURE 3.
Mutation of the PXXP motif in PKC βII does not impair phosphorylation by or binding to its upstream kinase, PDK-1. A, COS7 cells were transfected with vector (lanes 1–3), WT PKC βII (lanes 4–6), or PKC βII-P616A/P619A (lanes 7–9), and immunoprecipitated PKC βII was incubated with purified His-PDK-1 (0.5 nm), alone or together with Ca2+ and lipid, in an in vitro kinase assay. The immunoprecipitates were analyzed by Western blot for pThr-500 (pT500) and total PKC βII. The asterisk indicates the position of the fully phosphorylated PKC, the dash indicates the position of the unphosphorylated PKC, and the arrow indicates the position of endogenous PKC α. Note that phosphorylation of Thr-500 does not cause a mobility shift in WT PKC. B, COS7 cells were co-transfected with WT PKC βII (lanes 1–2), PKC βII-K371R, a catalytically inactive PKC, (lanes 3–4), or PKC βII-P616A/P619A (lanes 5–6) and either empty vector (lanes 1, 3, and 5) or Myc-PDK-1 (lanes 2, 4, and 6). Myc-PDK-1 was immunoprecipitated (IP) from detergent-solubilized lysates, and PKC interaction was assessed by Western blotting. Five percent of the detergent-solubilized lysates was analyzed by Western blotting to show the amount of Myc-PDK-1 and PKC βII present in the lysates. The asterisk denotes the position of fully phosphorylated PKC, and the dash denotes the position of unphosphorylated PKC. An anti-β-actin antibody indicates total protein loading in the lysate. C, a graph showing densitometric analysis of Western blots in B. Data represent the mean ± S.E. of 13 independent experiments.
Having established that PDK-1 can phosphorylate the activation loop of the PKC βII-P616A/P619A in vitro, we next addressed whether the phosphorylation defect observed in cells resulted from impaired binding of PDK-1 to this mutant in cells. COS7 cells were co-transfected with WT PKC βII, PKC βII-K371R (a catalytically inactive mutant that also cannot be processed by phosphorylation), or PKC βII-P616A/P619A with empty vector or Myc-PDK-1. Fig. 3B reveals that both mutants migrated as the faster mobility, unphosphorylated species (dash), whereas WT PKC βII migrated as the slower mobility, phosphorylated species (asterisk). Myc-PDK-1 was immunoprecipitated from detergent-solubilized lysates (Fig. 3B, left panel, IP), and the interaction with PKCβII was assessed by Western blot. Both WTβII and the unphosphorylated mutants, PKC βII-K371R and PKCβII-P616A/P619A, immunoprecipitated with Myc-PDK-1 (Fig. 3B, lanes 2, 4, and 6, upper panel, IP). Normalizing the amount of PKC βII bound to Myc-PDK-1 (upper panel, IP) to the amount of PKC βII in the lysate (upper panel, Lysate) indicated no significant preference of PDK-1 for the PXXP mutant when compared with WT PKC βII (Fig. 3C, n = 13, p = 0.20). Importantly, the C-terminal tail was still fully functional for binding PDK-1. Similarly, additional co-immunoprecipitation studies revealed that the binding of another C-terminal partner of PKC, Hsp70 (45, 47), was maintained with the PKC βII-P616A/P619A mutant (data not shown). Taken together with the in vitro phosphorylation experiments, these data exhibit that the defect in processing of the PKC βII-P616A/P619A mutant does not arise from defective binding of PDK-1 or inaccessibility of the activation loop site to phosphorylation by PDK-1.
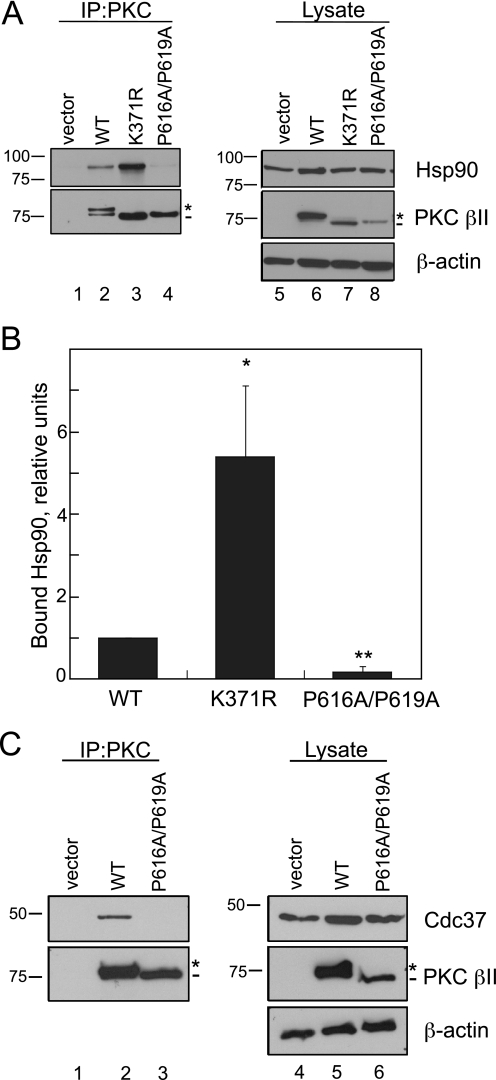
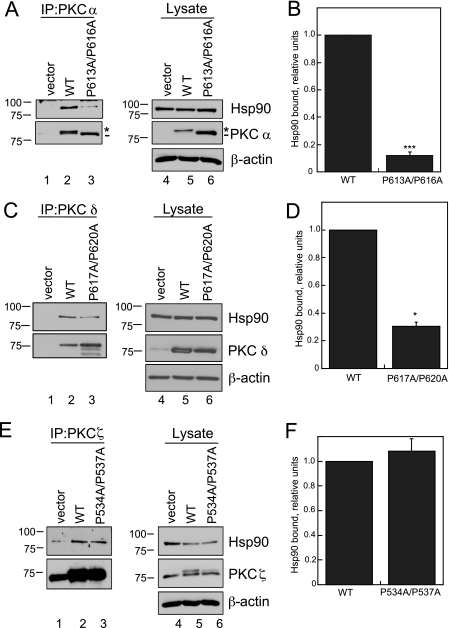
Mutation of the PXXP Motif in PKC βII Disrupts the Interaction with Hsp90 and Its Cochaperone, Cdc37—Recent studies with other AGC kinase family members, Akt and PDK-1, have described the role of heat shock proteins, particularly Hsp90, in the maturation and stabilization of these kinases (11–13). Because the PKC βII-P616A/ P619A mutant has a clear defect in maturation (Fig. 2A), we explored whether Hsp90 might play a role in this process. WT PKC βII, PKC βII-K371R, or PKC βII-P616A/P619A was expressed in COS7 cells and immunoprecipitated from detergent-solubilized lysates. The amount of endogenous Hsp90 bound was detected on Western blots (Fig. 4A). As in previous experiments, the phospho-defective mutants migrated as the faster mobility, unphosphorylated species (middle panel, dash), whereas WT PKC βII migrated primarily as the mature, phosphorylated species (middle panel, asterisk). Hsp90 bound to WT PKC βII, an interaction that was markedly enhanced with the unphosphorylated kinase-dead construct PKC βII-K371R. In marked contrast, there was no detectable binding of Hsp90 to the PXXP mutant PKC βII-P616A/P619A (Fig. 4A, upper panel, lane 4). Quantitative analysis of the ratio of bound Hsp90 to PKC in the immunoprecipitate demonstrated that mutation of the PXXP motif in PKC βII decreased the interaction with Hsp90 a striking 5-fold relative to WT PKC βII (Fig. 4B, p < 0.05, n = 4). Conversely, 5-fold more Hsp90 bound the PKC βII-K371R construct when compared with WT PKC βII (Fig. 4B, p < 0.01, n = 4). Thus, although both constructs are completely unphosphorylated and inactive, Hsp90 discriminated between an intact PXXP motif (in the PKC βII-K371R mutant) and a mutated PXXP motif. Consistent with this result, another mutant PKC that cannot be processed by phosphorylation, PKC βII-T634A/T641A/S654A (44), bound Hsp90 significantly better than WT PKC βII (data not shown). These data show that Hsp90 preferentially recognizes the unphosphorylated form of PKC via a mechanism that is driven by the PXXP motif.
FIGURE 4.
Mutation of the PXXP motif in PKC βII decreases binding to the chaperones Hsp90 and Cdc37. A, COS7 cells were transfected with empty vector (lanes 1 and 5), WT PKCβII (lanes 2 and 6), PKCβII-K371R (lanes 3 and 7), or PKC βII-P616A/P619A (lanes 4 and 8). PKC was immunoprecipitated (IP) from detergent-solubilized lysates, and endogenous Hsp90 interaction was assessed by Western blot. Five percent of the detergent-solubilized lysates was analyzed to show the amount of Hsp90 and PKCβII present in the lysate. The asterisk denotes the position of fully phosphorylated PKC, and the dash denotes the position of unphosphorylated PKC. B, a graph representing densitometric analysis of the Western blots shown in A. Data represent the mean ± S.E. of six independent experiments. *, p < 0.05 versus WT, and **, p < 0.01 versus WT by Student's t test. C, COS7 cells were transfected with vector (lanes 1 and 4), WT PKC βII (lanes 2 and 5), or PKC βII-P616A/P619A (lanes 3 and 6). Endogenous Cdc37 interaction was assessed as described in A. The asterisk denotes the position of fully phosphorylated PKC, and the dash denotes the position of unphosphorylated PKC.
Hsp90 functions as a complex in cells with other cochaperones such as Cdc37 (16, 17). To test whether the PXXP motif mediates the interaction with other cochaperones in the Hsp90 complex, we checked whether Cdc37 was present in immune complexes of PKC (Fig. 4C). WT PKC βII or PKC βII-P616A/P619A was immunoprecipitated from detergent-solubilized lysates (Fig. 4C, middle panel, IP), and bound endogenous Cdc37 was assessed by Western blot analysis (upper panel, IP). This cochaperone was readily detected in immunoprecipitates of WT PKC βII (IP, lane 2) but was not detectable in immunoprecipitates of the PKC βII-P616A/P619A mutant (lane 3). Thus, the PXXP motif mediates the interaction of PKC with both Hsp90 and its cochaperone Cdc37.
Inhibition of Hsp90 Slows the Maturation of PKC—Because the PXXP mutant PKC βII-P616A/P619A cannot be processed by phosphorylation and, furthermore, has reduced interaction with the Hsp90-Cdc37 chaperones, we next addressed whether Hsp90 activity is required for the maturation of PKC. Specifically, we tested how the Hsp90 inhibitor 17-AAG affected the rate of maturation of PKC as assessed by pulse-chase analysis (Fig. 5A). This potent inhibitor is a derivative of geldanamycin, an antibiotic of the ansamycin class produced by Streptomyces hygroscopicus, which binds to the N-terminal ATP-binding pocket of Hsp90 and prevents ATP binding and hydrolysis that is essential for protein folding processes (6). COS7 cells transfected with WT PKC βII were pretreated with 17-AAG prior to pulse-chase analysis (Fig. 5A). The autoradiogram in Fig. 5A shows that newly synthesized PKC migrated as a single, faster mobility band (dash) and then shifted to a slower mobility band (asterisk) over the course of the chase. The addition of 17-AAG slowed the maturation of PKC (compare lane 2 and lane 7). Quantitative analysis revealed that in the absence of the inhibitor, PKC was processed with a half-time of 13.2 ± 0.8 min (Fig. 5B, circles); however, in the presence of the inhibitor, the rate of processing of PKC was slowed 2-fold, with the half-time of processing increasing to 25 ± 4 min (squares). These data reveal that inhibition of endogenous Hsp90 activity decreases the rate of processing of PKC.
FIGURE 5.
Hsp90 activity facilitates the maturation of PKC. A, autoradiogram from a pulse-chase analysis of COS7 cells transfected with WT PKC βII in the absence (lanes 1–4) or presence (lanes 5–8) of 17-AAG (1 μm). Transfected cells were labeled with [35S]Met/Cys and then chased for the indicated times. PKC was immunoprecipitated from detergent-solubilized lysates and analyzed by autoradiography. The asterisk denotes the position of mature, fully phosphorylated PKC, and the dash denotes the position of newly synthesized, unphosphorylated PKC. An anti-PKC βII antibody indicates the amount of PKC in the immunoprecipitates. B, a graph representing the data shown in the autoradiogram in A. This graph shows the relative amount of fully phosphorylated PKC (asterisk) as a percentage of the total PKC protein in the absence (circles) or presence (squares) of 17-AAG (1 μm). Data are representative of three independent experiments. C, autoradiogram from a pulse-chase analysis of endogenous PKC α in COS7 cells in the absence (lanes 1–5) or presence (lanes 6–10) of 17-AAG (1 μm). Cells were labeled with [35S]Met/Cys and then chased for the indicated times. PKC was immunoprecipitated from detergent-solubilized lysates and analyzed by autoradiography. The asterisk denotes the position of mature, fully phosphorylated PKC (phosphorylated at both C-terminal sites); the double dash denotes the position of PKC phosphorylated at only one C-terminal site; and the single dash denotes the position of newly synthesized, unphosphorylated PKC. An anti-PKC α antibody indicates the amount of PKC present in the immunoprecipitates. D, autoradiogram from a pulse-chase analysis of COS7 cells transfected with WT PKC βII, PKC βII-T641E, or PKC βII S660E in the absence (lanes 1–4) and presence (lanes 5–8) of 17-AAG (1 μm). Transfected cells were pulse-labeled and analyzed as shown in A. The asterisk denotes the fully phosphorylated, mature PKC; the double dash denotes the position of phosphorylation at Thr-641; and the dash denotes the newly synthesized, unphosphorylated PKC. E, autoradiogram from a pulse-chase analysis of COS7 cells transfected with WT PKC βII and treated with dimethyl sulfoxide (DMSO) (lanes 1–4), 17-AAG (1 μm, lanes 5–8), celastrol (10 μm, lanes 9–12), or both 17-AAG and celastrol (lanes 13–16). Transfected cells were pulse-labeled and analyzed as shown in A. The asterisk denotes the fully phosphorylated, mature PKC, and the dash denotes the newly synthesized, unphosphorylated PKC.
We next examined the effect of inhibiting Hsp90 on the processing of endogenous PKC α, another conventional PKC isozyme, by pulse-chase analysis (Fig. 5C). The autoradiogram in Fig. 5C shows that newly synthesized PKC α migrated as a faster mobility band (dash) and shifted to a slower mobility band (asterisk) over the course of the chase with a half-time of ∼10 min (lane 3) in the absence of the Hsp90 inhibitor. However, in the presence of the Hsp90 inhibitor, the half-time to shift to the fully phosphorylated, slower mobility species (asterisk) was ∼4-fold slower (half-time ∼45 min). Interestingly, a band of intermediate mobility (double dash) was detected at 10 min of chase (Fig. 5C, lane 8) in the sample treated with the Hsp90 inhibitor. This band represents a species of PKC that is phosphorylated at only one of the two C-terminal sites (44). The half-time for phosphorylation at this site was similar to that for full phosphorylation in the absence of inhibitor (the ratio of upper (asterisk or double dash) and lower bands (dash) at the 10-min chase time points are similar with and without inhibitor (Fig. 5C, lanes 3 and 8)). These data suggest that Hsp90 inhibition impairs the processing of one of the two C-terminal sites.
To explore which of the two C-terminal sites Hsp90 regulates, we investigated the rate of processing of phosphorylation site constructs of PKC βII in which C-terminal phosphorylation site residues were individually replaced with Glu, the phospho-mimetics PKC βII-T641E (turn motif mutant) and PKC βII S660E (hydrophobic motif mutant) (Fig. 5D). Both mutants were processed by phosphorylation with half-times only slightly slower from those of WT PKCβII; analysis of the ratio of upper to lower mobility species indicated that ∼70% of WT PKC, 50% of T641E, and 40% of S660E were processed following a 30-min chase (Fig. 5D, lane 3). Note that the mobility shift for the T641E mutant reflects phosphorylation of Ser-660, which causes a readily detectable shift, and that the mobility shift for the S660E mutant reflects phosphorylation of Thr-641, a modification that causes a smaller mobility shift. Importantly, 17-AAG treatment slowed the processing of WT PKC βII and PKC βII-T641E ∼2-fold (compare the ratios of the upper and lower mobility species at the 30-min time point, lanes 3 and 7) but did not affect the rate of processing of the PKC βII-S660E mutant (the ratio of upper to lower mobility species was ∼1 at the 90-min time point both in the absence and in the presence of 17-AAG (lanes 4 and 8)). The lack of sensitivity of PKC βII-S660E phosphorylation at Thr-641 to Hsp90 inhibition suggests that Hsp90 controls the final phosphorylation step of PKC, that of Ser-660.
The Hsp90 inhibitor, 17-AAG, used in the preceding pulse-chase experiments, specifically targets the ATPase activity of Hsp90 and would not affect potential regulation of PKC by Cdc37. To investigate the effect of disrupting the interaction of Hsp90 with its cochaperones on PKC maturation, we took advantage of a recently described Hsp90 inhibitor, celastrol, that not only inhibits ATP binding to Hsp90 but also inhibits the interaction of Hsp90 with Cdc37 (21, 48). COS7 cells transfected with WT PKC βII were pretreated with 17-AAG, celastrol, or both prior to pulse-labeling with [35S]Met/Cys (Fig. 5E). The autoradiogram in Fig. 5E shows that the processing of PKC was slowed ∼2-fold by 17-AAG, as observed previously in Fig. 5A. The amount of unphosphorylated lower band (dash) and phosphorylated upper band (asterisk) was approximately equal at the 15-min (lane 2) and 30-min (lane 7) time points in the absence or presence of 17-AAG, respectively. Celastrol had a much more striking effect on the maturation of PKC, with PKC migrating primarily as the unphosphorylated species even after a 90-min chase. In the presence of both 17-AAG and celastrol, processing of PKC was completely inhibited. These data underscore the importance of both Hsp90 activity and the Hsp90-Cdc37 interaction for the proper maturation of PKC.
The PXXP Motif Also Mediates the Interaction of Other Conventional and Novel PKC Isozymes with Hsp90—Given the conservation of the hydrophobic motif among PKC isozymes, we addressed whether other isozymes of PKC are controlled by Hsp90. We mutated the PXXP motif to AXXA for another conventional PKC, PKC α (PKC α-P613A/P616A), a novel PKC, PKC δ (PKC δ-P617A/P620A), and an atypical PKC, PKC ζ (PKC ζ-P534A/P537A), and tested the interaction of each mutant with Hsp90 (Fig. 6). COS7 cells were transfected with empty vector (lanes 1 and 4), WT PKC isozymes (lanes 2 and 5), or the corresponding PXXP mutant (lanes 3 and 6). Specific PKC isozymes were immunoprecipitated from detergent-solubilized lysates, and bound endogenous Hsp90 was assessed by Western blot analysis. Hsp90 was detected in the immune complexes of all three wild-type isozymes as well as the corresponding AXXA mutants (Fig. 6, A, C, and E, lanes 2 and 3). However, quantitative analysis of the amount of Hsp90 bound to PKC (IP, upper panel), normalized to the amount of Hsp90 in the lysate (Lysate, upper panel), indicated striking isozyme-specific effects of perturbing the PXXP motif. Notably, mutation of the PXXP motif in the conventional PKC α dramatically reduced the interaction with Hsp90 to ∼10% of the level bound to WT PKC α (Fig. 6B, ***, p < 0.001, n = 3). This protein migrated with a faster mobility than wild-type PKC α on SDS-PAGE (lane 6, dash), indicating that it, like its counterpart in PKC βII, was not processed by phosphorylation. This result is consistent with Hsp90 inhibition slowing the maturation of endogenous PKC α in COS7 cells (Fig. 5C). Similarly, mutation of the PXXP motif in the novel PKC δ reduced the interaction with Hsp90; ∼70% less Hsp90 was observed in the immune complex of the mutant when compared with that of WT PKC δ (Fig. 6D, *, p < 0.05, n = 3). Indeed, similar to that of the conventional PKCs, the maturation of PKC δ-P617A/P620A is impaired, and it has reduced phosphorylation at both the turn and the hydrophobic motif sites (data not shown), although the mobility of the mutant is not detectably different from that of WT PKC δ (Fig. 6C, middle panel). Pulse-chase analysis of another novel PKC isozyme, PKC ε, revealed that the rate of processing was also slowed 2-fold by 17-AAG (data not shown). In contrast to mutation of the conventional and novel isozymes, mutation of the PXXP motif in the atypical PKC ζ did not affect the interaction with Hsp90 (Fig. 6F, n = 3). It is noteworthy that PKC ζ differs from the conventional and novel isozymes in that it has a Glu at the hydrophobic motif phosphorylation site. These data are consistent with Hsp90 binding and facilitating the phosphorylation of conventional and novel PKC isozymes via the conserved PXXP motif.
FIGURE 6.
Mutation of the PXXP motif in conventional and novel PKC isozymes decreases the interaction with Hsp90. A, COS7 cells were transiently transfected with empty vector (lanes 1 and 4), WT PKC α (lanes 2 and 5), or PKC α-P613A/P616A (lanes 3 and 6). PKC was immunoprecipitated (IP) from detergent-solubilized lysates, and endogenous Hsp90 interaction was assessed by Western blot. Five percent of the detergent-solubilized lysate was analyzed by Western blot to show the amount of PKC and Hsp90 present in the lysate. The asterisk denotes the position of fully phosphorylated PKC, and the dash denotes the position of unphosphorylated PKC. Total protein loading is indicated by an anti-β-actin antibody. B, a graph representing densitometric analysis of the Western blots shown in A. Data represent the mean ± S.E. of three independent experiments. ***, p < 0.001 versus WT by Student's t test. C, COS7 cells were transiently transfected with empty vector (lanes 1 and 4), WT PKC δ (lanes 2 and 5), or PKC δ-P617A/P620A (lanes 3 and 6). PKC was immunoprecipitated, and endogenous Hsp90 interaction was analyzed as described in A. D, a graph representing densitometric analysis of the Western blots shown in C. Data represent the mean ± S.E. of three independent experiments. *, p < 0.05 versus WT by Student's t test. E, COS7 cells were transiently transfected with empty vector (lanes 1 and 4), WT PKC ζ (lanes 2 and 5), or PKC ζ-P534A/P537A (lanes 3 and 6). PKC was immunoprecipitated, and endogenous Hsp90 interaction was analyzed as described in A. F, a graph representing densitometric analysis of Western blots shown in E. Data represent the mean ± S.E. of three independent experiments.
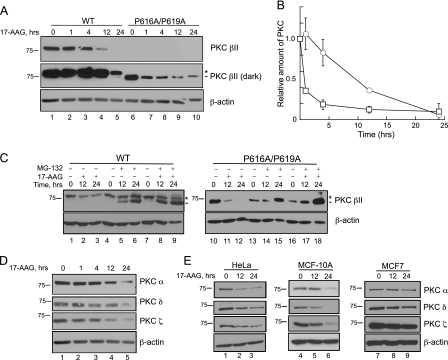
Inhibition of Hsp90 Activity Results in the Down-regulation of PKC via a Proteasome-mediated Pathway—Chaperones, such as Hsp90, not only facilitate the proper folding and maturation of kinases but also stabilize vulnerable structural conformations that are susceptible to degradation mechanisms (6, 8). Therefore, we next addressed whether Hsp90 serves an additional stabilizing role for PKC. COS7 cells were transfected with either WT PKC βII or PKC βII P616A/P619A and treated with 17-AAG for increasing time; protein levels in whole cell lysates were analyzed by Western blot (Fig. 7A). The top panel in Fig. 7A shows that WT PKC βII was significantly depleted by 12 h (lane 4) and absent by 24 h (lane 5) with inhibitor treatment. 17-AAG also down-regulated PKC βII-P616A/P619A (lanes 6–10; darker exposure). Quantitative analysis of the PKC levels normalized to actin (bottom panel) revealed that PKC βII-P616A/P619A was ∼10-fold more sensitive to Hsp90 inhibition than WT PKC βII (Fig. 7B, half-time for depletion of PKC βII-P616A/P619A was ∼1 h when compared with 10 h for WT PKC βII). These data indicate that Hsp90 activity not only controls the processing of PKC but also the degradation of PKC.
FIGURE 7.
Inhibition of Hsp90 down-regulates PKC through a proteasome-dependent mechanism in a cell-type-dependent manner. A, COS7 cells were transiently transfected with WT PKC βII (lanes 1–5) or PKC βII-616A/P619A (lanes 6–10). Cells were treated with 17-AAG (1 μm) for the indicated times. Whole cell lysates were analyzed by Western blotting for total PKC (PKC βII) and total protein content (β-actin). The asterisk denotes the position of fully phosphorylated PKC, and the dash denotes the position of unphosphorylated PKC. B, a graph representing densitometric analysis of Western blots shown in A. The relative amounts of WT PKC βII (white circles) and PKC βII-P616A/P619A (white squares) are indicated. Data are representative of three independent experiments. C, COS7 cells were transiently transfected with WT PKC βII (left) or PKC βII-P616A/P619A (right). Cells were then treated with 17-AAG (1 μm, lanes 2–3, 11–12), MG-132 (10 μm, lanes 5–6, 14–15), or both (lanes 8–9, 17–18) for the times indicated. Whole cell lysates were separated by SDS-PAGE and analyzed by Western blotting. PKC βII and β-actin were detected with the indicated antibodies. The asterisk denotes the position of fully phosphorylated PKC, and the dash denotes the position of unphosphorylated PKC. Data are representative of three independent experiments. D, COS7 cells were treated with 17-AAG (1 μm) for the indicated times. Whole cell lysates were analyzed by Western blotting for endogenous PKC α, PKC δ, PKC ζ, and β-actin for total protein loading. Data are representative of three independent experiments. E, HeLa cells (lanes 1–3), MCF-10A cells (lanes 4–6), and MCF7 cells (lanes 7–9) were treated with 17-AAG (1 μm) for the indicated times. Whole cell lysates were analyzed by Western blotting for endogenous PKCα, PKCδ, PKCζ, andβ-actin for total protein loading. Data are representative of three independent experiments.
Studies have shown that Hsp90 inhibition leads to down-regulation via ubiquitination and degradation by the proteasome (29, 49). To determine whether the down-regulation we observed with 17-AAG occurred via a proteasome-mediated pathway, COS7 cells were transfected with WT PKC βII or PKC βII-P616A/P619A, pretreated with the proteasome inhibitor MG-132, and then treated with 17-AAG for 12 or 24 h (Fig. 7C). The Western blot in Fig. 7C reveals that the steady-state levels of both WT PKC βII and PKC βII-P616A/P619A were reduced following 17-AAG treatment, as reported above (Fig. 7C, lanes 2–3 and lanes 11–12). Pretreatment with MG-132 completely protected the PKC βII-P616A/P619A construct from degradation both in the absence (lanes 14–15) as well as in the presence 17-AAG (lanes 17–18); in fact, inhibition of the proteasome dramatically increased the steady-state levels of this construct. The proteasome inhibitor also prevented the 17-AAG-triggered depletion of WT PKC βII (lanes 8–9). Note that the proteasome inhibitor resulted in the accumulation of dephosphorylated PKC (dash); this is consistent with the dephosphorylated species of PKC being degraded by proteasomal pathways. These data demonstrate that Hsp90 protects PKC from proteasomal degradation, as reported previously for other client proteins.
To determine whether long term Hsp90 inhibition down-regulated other PKC isozymes, we examined the levels of endogenous PKC α, PKC δ, and PKC ζ in COS7 cells upon treatment with 17-AAG (Fig. 7D). Hsp90 inhibition resulted in a significant loss of each of these isozymes (lane 5). Although Hsp90 regulation of the processing of PKC is an isozyme-specific process, these data illustrate that Hsp90 control of the degradation of PKC is not isozyme-specific. However, because a previous study reported that PKC is insensitive to 17-AAG-induced down-regulation in MCF7 cells (11), we investigated whether the 17-AAG-induced degradation depended on cell type. To this end, we treated HeLa cells (Fig. 7E, lanes 1–3), MCF-10A cells (lanes 4–6), or MCF7 cells (lanes 7–9) with 17-AAG for 0, 12, or 24 h and monitored endogenous PKC levels. PKC α, PKC δ, and PKC ζ levels were dramatically reduced in both HeLa and MCF-10A (a normal breast cell line) cells (Fig. 7E, lanes 1–6, top three panels). In marked contrast, 17-AAG caused only a very modest degradation of PKC isozymes in MCF7 cells (a breast cancer cell line), even after 24 h treatment (Fig. 7E, lanes 7–9, top three panels). Therefore, the efficiency of Hsp90-mediated stabilization of PKC isozymes depends on cell type.
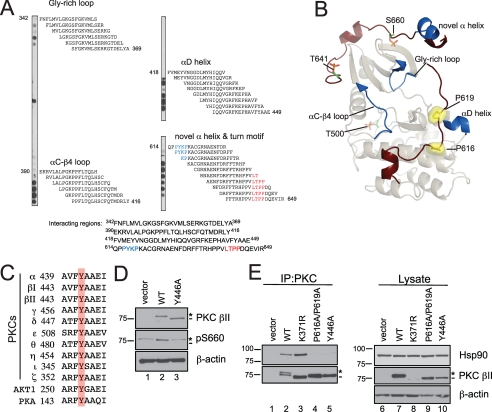
Mutation of a Conserved Tyr That Interacts with the PXXP Motif Recapitulates the Defect of Mutating the PXXP Motif—To determine whether the interaction of Hsp90 with PKC was mediated directly by the PXXP motif, or whether, instead, the PXXP motif indirectly controls the binding of Hsp90, we performed a peptide walk of the PKC sequence and asked which segments bind Hsp90. Specifically, 18-mer peptides derived from the catalytic domain of PKC βII (amino acids 342–673), moving down the sequence 2 residues at a time, were spotted onto a membrane, which was then incubated with purified Hsp90 protein. Hsp90 specifically bound clusters of sequences; these are shown in Fig. 8A, with the sequence of the spotted peptide shown to the right of the peptide spot. Four interacting regions were detected in the array: the Gly-rich loop, the αC-β4 loop, the αD-helix, and the novel α helix and turn motif (highlighted in red) sequences of PKC βII (Fig. 8A) (50). Interestingly, Hsp90 did not significantly bind the peptide sequences that contained the PXXP motif (highlighted in blue in the novel α helix and turn motif segment). These data show that Hsp90 directly binds peptide segments that are proximal to the PXXP motif but not the PXXP motif itself. These binding surfaces are mapped onto the structure of PKC βII (Fig. 8B, dark blue). Note that the αC-β4 loop and the αD-helix, two of the strongest Hsp90-binding regions, flank the N and C-terminal ends of the PXXP motif, respectively.
FIGURE 8.
Mutation of a conserved Tyr in the αE-helix of the catalytic domain of PKC βII mimics the defect of the PXXP mutant, PKC βII-P616A/P619A. A, peptide overlay of the catalytic domain of PKC βII with purified Hsp90α. Eighteen-residue peptides covering the catalytic domain (amino acids 342–673) of PKC βII and staggered by two amino acids were spotted onto a membrane and overlaid with 100 nm Hsp90α, and Hsp90 binding was detected with an anti-Hsp90 antibody. The residues highlighted in blue are the PXXP motif, and the residues highlighted in red are the residues delineating the turn motif phosphorylation site. B, ribbon diagram representation of the catalytic domain of PKC βII. A homology model of the PKC βII was built using the crystal structure of PKC θ (Protein Data Bank number: 1XJD) as the template. The C-terminal tail is highlighted in dark red with the Pro of the PXXP motif as yellow surface representations. The processing phosphorylation sites (activation loop (Thr-500), turn motif (Thr-641), and hydrophobic motif (Ser-660)) are shown as stick representations (orange and green). The regions identified in the peptide overlay are mapped onto the catalytic domain and are highlighted in dark blue. C, partial sequence alignment of part of the conserved αE-helix in the AGC kinases, the PKCs, Akt1, and PKA. A conserved Tyr is highlighted in red. D, COS7 cells were transiently transfected with empty vector (lane 1), WT PKC βII (lane 2), or PKC βII-Y446A (lane 3). Whole cell lysates were analyzed for total PKC (PKC βII), phosphorylated PKC (pSer-660), and total protein content (β-actin) by Western blotting. The asterisk denotes the position of phosphorylated PKC, and the dash denotes the position of unphosphorylated PKC. E, COS7 cells were transfected with empty vector (lanes 1 and 6), WT PKC βII (lanes 2 and 7), PKC βII-K371R (lanes 3 and 8), PKC βII-P616A/P619A (lanes 4 and 9), or PKC βII-Y446A (lanes 5 and 10). PKC was immunoprecipitated (IP) from detergent-solubilized lysates, and endogenous Hsp90 interaction was assessed by Western blot. Five percent of the detergent-solubilized lysates was analyzed to show the amount of Hsp90 and PKC βII present in the lysate. The asterisk denotes the position of fully phosphorylated PKC, and the dash denotes the position of unphosphorylated PKC. Data are representative of three independent experiments.
Previous analysis of AGC kinase sequences and structure had identified key residues in the αE-helix of the catalytic domain that serve as a docking site for the PXXP motif (36). In particular, a conserved tyrosine (Tyr-446) in the αE-helix maximally packs up against Pro-616 in the PXXP motif to simulate a molecular clamp (Fig. 9). Sequence alignment portrays that this Tyr is conserved in several AGC family members, including all PKC isozymes, Akt, and PKA (Fig. 8C, red). To test whether mutation of this conserved Tyr in PKC βII (Tyr-446) to an Ala would recapitulate the processing defect of the PKC βII-P616A/P619A mutant, COS7 cells were transfected with empty vector, WT PKC βII, or PKC βII-Y446A, and whole cell lysates were analyzed by Western blot. The Western blot in Fig. 8D indicates that the Y446A construct migrated as the faster mobility, unphosphorylated form (dash). Consistent with this result, a phospho-specific antibody to the hydrophobic motif site (pSer-660) did not detect any phosphorylation of this mutant (middle panel, asterisk; note that the upper band labeled with the pSer-660 antibody in the vector and Y446A lanes represents endogenous PKC α). Furthermore, the Y446A mutant was not labeled by phospho-specific antibodies to the other two sites, Thr-500 and Thr-641 (data not shown). Thus, perturbation of the intramolecular clamp between the αE helix and the PXXP segment by mutation of Tyr-446 recapitulated the processing defect observed upon mutation of the PXXP motif.
FIGURE 9.
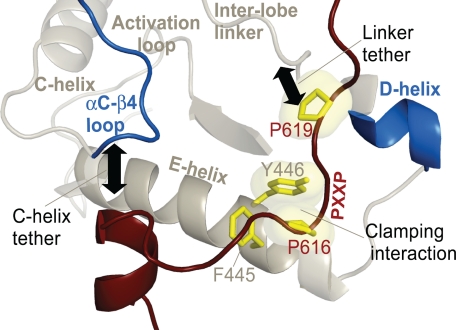
Structural representation of the residues that comprise the molecular clamp between the PXXP motif and catalytic core. The PXXP motif in the C-terminal tail (highlighted in dark red) and the determinants in the αE-helix of the catalytic domain are highlighted in yellow. Other key regions, such as the αC-β4 loop, C-helix, D-helix, interlobe linker, and the activation loop, of the catalytic domain are also represented. The tethering of theαC-β4 loop and the interlobe linker by the C-terminal tail is highlighted by black arrows.
Lastly, we wanted to determine whether mutation of Tyr-446, similar to mutation of the PXXP motif that abrogated maturation, abolished Hsp90 binding. COS7 cells were transfected with vector, WT PKC βII, PKC βII-K371R, PKC βII-P616A/P619A, or PKC βII-Y446A, the PKC was immunoprecipitated, and bound endogenous Hsp90 was detected by Western blot (Fig. 8E, upper panel, IP). As reported above, Hsp90 bound both WT PKC βII and PKC βII-K371R (upper panel, IP, lanes 2–3). However, Hsp90 failed to bind either the PKC βII-P616A/P619A mutant or the PKC βII-Y446A mutant (upper panel, IP, lanes 4–5, n = 3). These data are consistent with Hsp90 binding a surface that is structured by the PXXP motif and αE helix.
DISCUSSION
The foregoing data demonstrate that the molecular chaperones, Hsp90 and Cdc37, control the processing by phosphorylation of conventional and novel PKC isozymes. Binding of these chaperones is dependent upon a molecular clamp that is formed between a conserved PXXP motif in the C-terminal tail of PKC and determinants in the αE helix of the catalytic domain. Perturbation of this structural motif by mutation or pharmacological inhibition of the chaperones results in an abrogation of the maturation process. Of the two C-terminal phosphorylation sites on PKC, only that of the hydrophobic motif is sensitive to Hsp90 inhibition. In addition, we show that Hsp90 has a second role in stabilizing conventional, novel, and atypical PKC isozymes. Thus, the Hsp90-Cdc37 complex plays an essential role in allowing the maturation of isozymes that require hydrophobic motif phosphorylation and, additionally, in stabilizing all subclasses of this family of kinases.
The PXXP Motif Forms an Intramolecular Clamp—Our findings show that the PXXP motif is critical for the maturation and proper catalytic function of PKC. Specifically, the PXXP motif anchors the flexible C-terminal tail to the catalytic core by forming a molecular clamp with residues in the αE-helix. A number of residues in this clamp, including the sequence FYAAE, are conserved across all AGC kinases. Mutation of the conserved Tyr (Tyr-446 in PKC βII) in this sequence mimics the processing defect of mutating the PXXP motif in PKC βII. Thus, mutation of either portion of this clamp, i.e. the PXXP motif or the conserved Tyr of the αE-helix, perturbs the structure of the catalytic domain, preventing Hsp90-Cdc37 from binding and thus inhibiting the maturation of PKC by phosphorylation. In support of this model, mutation of the adjacent conserved Phe, Phe-445 in PKC βII, also mimics the processing defect of mutating the PXXP motif (data not shown). Thus, the PXXP motif serves as a folding nucleus to modulate events necessary for proper catalytic function by promoting interaction with the Hsp90-Cdc37 complex.
The PXXP motif is conserved across all AGC kinases and may also serve a common function. Our data indicate that Hsp90 facilitates maturation for both conventional and novel PKC isozymes. Studies with Akt have shown that this PXXP motif is necessary for proper localization in T-cells as well as activation (51, 52). Indeed, the Akt1 PXXP mutant P424A/P427A is unphosphorylated, catalytically inactive, and migrates as a faster mobility species (52). Inhibition of Hsp90 blocks the activation and maturation of Akt (12). Whether the PXXP motif controls Akt maturation in the same manner as it does for PKC remains to be established.
PKC Is a Client Kinase of the Hsp90-Cdc37 Chaperone Complex—The Hsp90-Cdc37 complex has over 100 known client kinases involved in a wide array of signaling pathways (53). PDK-1 and Akt, both members of the AGC kinase superfamily, have been previously established as clients of these chaperones (11–13). The identification of PKC as another client of the Hsp90-Cdc37 chaperone complex underscores the importance of this chaperone in kinase regulation. In the case of PKC, the chaperoning activity of Hsp90-Cdc37 serves two purposes: 1) facilitating the maturation of PKC by processing phosphorylations and 2) regulating the stability of PKC.
An intact PXXP motif is required for the selective binding of Hsp90 to unphosphorylated, unprocessed forms of PKC when compared with phosphorylated PKC. The tight binding of Hsp90 to mutants of PKC that cannot be phosphorylated (e.g. kinase-inactive mutant PKC βII-K371R or the turn motif mutant PKC βII-T641AAA (data not shown (38)) is abolished upon mutation of the PXXP motif. Thus, the PXXP motif critically regulates the binding of Hsp90 to unphosphorylated PKC.
In order for PKC to mature into a catalytically competent enzyme, it must first undergo three processing phosphorylations on the activation loop, the turn motif, and the hydrophobic motif. The activation loop step is dependent upon the upstream kinase PDK-1, whereas the latter two phosphorylation steps rely on accessory protein complexes and autophosphorylation. Hsp90 does not appear to be necessary for the first phosphorylation step by PDK-1 because the PKC βII-P616A/P619A mutant was phosphorylated by PDK-1 in vitro and bound PDK-1 in cells, although it was unable to complete the maturation process in cells. Recent studies have shown that Hsp90 is necessary to stabilize mutants in Akt and PKC that lack turn motif phosphorylation, which is promoted by the mTORC2 complex (29, 30). However, our data reveal that phosphorylation at the turn motif site is not sensitive to Hsp90 inhibition. The rate of processing of a mutant with a constitutive negative charge at the hydrophobic motif (PKC βII S660E) is not slowed by Hsp90 inhibition, whereas the rate of processing of a mutant with a constitutive negative charge at the turn motif (PKC βII T641E) is slowed. These data suggest that Hsp90-Cdc37 facilitates the hydrophobic motif phosphorylation step. Consistent with this model, the processing of atypical PKC isozymes, which have a Glu at the phospho-acceptor position of the hydrophobic motif, is not impaired by mutation of the PXXP motif.
How Hsp90 facilitates the phosphorylation of the hydrophobic motif is not clear. Enzymological studies with pure protein have demonstrated that, in the case of PKC βII, this site is autophosphorylated by an intramolecular mechanism (54). This result would suggest that this segment of the C-terminal tail must access the active site during the maturation of PKC. Thus, one possibility is that Hsp90 facilitates conformational transitions that allow this to occur. It is noteworthy that mutation of the hydrophobic motif site to Ala in either PKC α or PKC βII does not prevent the maturation of PKC. Although constructs with Ala at this position are less stable than phosphorylated wild-type enzyme, they are nonetheless phosphorylated at the PDK-1 site and turn motif and retain catalytic activity (38, 55). Thus, Hsp90 not only facilitates the phosphorylation of the hydrophobic motif, but it likely plays an additional role in the maturation of PKC.
Once phosphorylated, the hydrophobic motif phosphorylation site serves as an important site of regulation in AGC kinases. Phosphorylation at the hydrophobic motif promotes a stable, active, and phosphatase-resistant kinase; dephosphorylation of this site in PKC promotes its ubiquitination and down-regulation (56). Structural studies have shown that hydrophobic motif phosphorylation in AGC kinases stabilizes the N-lobe of the kinase domain by docking into a hydrophobic groove. This interaction leads to an ordering of the αC helix that allows optimal binding to ATP and also stabilizes a closed, active conformation by interacting with the activation loop phosphate (57, 58). Studies have shown that Hsp90-Cdc37 can bind to the N-lobe and αC helix of the catalytic domain of various client kinases, which are important regions involved in kinase dynamics (8, 59). Because hydrophobic motif phosphorylation is necessary for generation of a stable, active kinase for PKC, it is plausible that the role of Hsp90-Cdc37 in the maturation process is to stabilize these regions as PKC completes its maturation.
In addition to its role in facilitating maturation, the Hsp90-Cdc37 complex also stabilizes PKC. Long term treatment with Hsp90 inhibitors promotes the down-regulation of PKC isozymes by a proteasomal-dependent mechanism. Interestingly, we also observed rapid down-regulation of PKC βII-P616A/P619A, suggesting that any residual interaction occurring with Hsp90 was sensitized to Hsp90 inhibition. This 17-AAG-induced down-regulation was observed in all PKC isozyme families. Indeed, Hsp90 inhibition leads to the degradation of other client kinases (8, 53). However, this effect is cell type-dependent. A previous study reported that PKC was not a client of Hsp90 by assessing 17-AAG-induced down-regulation in MCF7 cells (11). Consistent with this, we see decreased PKC levels in not only COS7 cells but also HeLa and MCF-10A cells, but we did not observe as striking a decrease in MCF7 cells. These findings indicate that Hsp90 regulation of PKC is cell-type dependent and may reflect abnormalities within signaling pathways of different cell environments.
PKC, Hsp90, and Cdc37 in Cancer—Altered expression levels of both PKC βII and the molecular chaperones Hsp90 and Cdc37 have been observed in various cancers (21, 24, 25, 60). These chaperones are unique as oncogenes because they promote tumorigenicity by stabilizing overexpressed or mutated oncogenic proteins (6). Whether PKC levels are elevated in cancer because of defective interaction with Hsp90-Cdc37 remains to be established. However, it is noteworthy that a recent study of the genes mutated in glioblastoma multiforme identified a mutation in PKC α, P613S, as a cancer-driving mutation (5). This residue in PKC α corresponds to the first Pro of the PXXP motif (Fig. 1B). Based on our data, it is tempting to speculate that the tumorigenic phenotype could arise from the lack of processing of PKC α due to mutation of the PXXP motif. Interestingly, a recent study has shown that PKC α protein expression is necessary for glioma cell proliferation and not necessarily its kinase activity (61).
Hsp90 has become an appealing drug target in cancer therapeutics. A recent study showed that the Hsp90 inhibitor 17-AAG potentiated the activity of the PKC βII-specific inhibitor, enzastaurin, in malignant glioma (62). Although Akt activity is up-regulated in this cancer pathway (62), the efficacy of these two inhibitors could also be explained by Hsp90 regulation of PKC. The identification of PKC as a client of the Hsp90-Cdc37 chaperone complex suggests that Hsp90 and PKC inhibitors could serve as potential therapeutics in treating pathologies that have associated PKC mutations or elevated levels of PKC.
Summary—Our data reveal that PKC is a client kinase of the Hsp90-Cdc37 chaperone complex. Interaction with these chaperones depends on a molecular clamp created by a conserved PXXP motif in the C-terminal tail and determinants in the αE helix of the catalytic domain (Fig. 9). Hsp90-Cdc37 serves two functions for PKC. 1) It facilitates maturation into a signaling-competent enzyme, and 2) it stabilizes the mature enzyme. These data identify new molecular mechanisms that regulate the function and stability of PKC. Knowledge of these mechanisms could be helpful in the design of therapies to target pathologies that have aberrant PKC signaling.
Supplementary Material
Acknowledgments
We thank Charles C. King for purifying His-PDK-1, C. J. Allison for synthesizing the peptide array, Robert Romano for technical advice, and the Newton laboratory members for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant Grants 2T32 GM-07752 (to C. G.) and by National Institutes of Health Grants GM-43154 and P01 DK54441. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: Hsp, heat shock protein; PKC, protein kinase C; PKA, cAMP-dependent protein kinase; AGC, c-AMP-dependent protein kinase/protein kinase G/protein kinase C; PDK-1, phosphoinositide-dependent kinase 1; Cdc37, cell division cycle 37; mTOR, mammalian target of rapamycin; RFP, red fluorescent protein; YFP, yellow fluorescent protein; PDBu, phorbol-12,13-dibutyrate; PMA, phorbol 12-myrstate 13-acetate; 17-AAG, 17-(allylamino)-17-demethoxygeldanamycin; MG-132, cabobenzoxyl-l-leucyl-l-leucyl-leucinal; DMEM, Dulbecco's modified Eagle's medium; WT, wild type.
References
- 1.Manning, G., Whyte, D. B., Martinez, R., Hunter, T., and Sudarsanam, S. (2002) Science 298 1912–1934 [DOI] [PubMed] [Google Scholar]
- 2.Torkamani, A., and Schork, N. J. (2008) Cancer Res. 68 1675–1682 [DOI] [PubMed] [Google Scholar]
- 3.Majumder, P. K., and Sellers, W. R. (2005) Oncogene 24 7465–7474 [DOI] [PubMed] [Google Scholar]
- 4.Greenman, C., Stephens, P., Smith, R., Dalgliesh, G. L., Hunter, C., Bignell, G., Davies, H., Teague, J., Butler, A., Stevens, C., Edkins, S., O'Meara, S., Vastrik, I., Schmidt, E. E., Avis, T., Barthorpe, S., Bhamra, G., Buck, G., Choudhury, B., Clements, J., Cole, J., Dicks, E., Forbes, S., Gray, K., Halliday, K., Harrison, R., Hills, K., Hinton, J., Jenkinson, A., Jones, D., Menzies, A., Mironenko, T., Perry, J., Raine, K., Richardson, D., Shepherd, R., Small, A., Tofts, C., Varian, J., Webb, T., West, S., Widaa, S., Yates, A., Cahill, D. P., Louis, D. N., Goldstraw, P., Nicholson, A. G., Brasseur, F., Looijenga, L., Weber, B. L., Chiew, Y. E., DeFazio, A., Greaves, M. F., Green, A. R., Campbell, P., Birney, E., Easton, D. F., Chenevix-Trench, G., Tan, M. H., Khoo, S. K., Teh, B. T., Yuen, S. T., Leung, S. Y., Wooster, R., Futreal, P. A., and Stratton, M. R. (2007) Nature 446 153–15817344846 [Google Scholar]
- 5.McLendon, R., Friedman, A., Bigner, D., Van Meir, E. G., Brat, D. J., Mastrogianakis, M., Olson, J. J., Mikkelsen, T., Lehman, N., Aldape, K., Alfred Yung, W. K., Bogler, O., Vandenberg, S., Berger, M., Prados, M., Muzny, D., Morgan, M., Scherer, S., Sabo, A., Nazareth, L., Lewis, L., Hall, O., Zhu, Y., Ren, Y., Alvi, O., Yao, J., Hawes, A., Jhangiani, S., Fowler, G., San Lucas, A., Kovar, C., Cree, A., Dinh, H., Santibanez, J., Joshi, V., Gonzalez-Garay, M. L., Miller, C. A., Milosavljevic, A., Donehower, L., Wheeler, D. A., Gibbs, R. A., Cibulskis, K., Sougnez, C., Fennell, T., Mahan, S., Wilkinson, J., Ziaugra, L., Onofrio, R., Bloom, T., Nicol, R., Ardlie, K., Baldwin, J., Gabriel, S., Lander, E. S., Ding, L., Fulton, R. S., McLellan, M. D., Wallis, J., Larson, D. E., Shi, X., Abbott, R., Fulton, L., Chen, K., Koboldt, D. C., Wendl, M. C., Meyer, R., Tang, Y., Lin, L., Osborne, J. R., Dunford-Shore, B. H., Miner, T. L., Delehaunty, K., Markovic, C., Swift, G., Courtney, W., Pohl, C., Abbott, S., Hawkins, A., Leong, S., Haipek, C., Schmidt, H., Wiechert, M., Vickery, T., Scott, S., Dooling, D. J., Chinwalla, A., Weinstock, G. M., Mardis, E. R., Wilson, R. K., Getz, G., Winckler, W., Verhaak, R. G., Lawrence, M. S., O'Kelly, M., Robinson, J., Alexe, G., Beroukhim, R., Carter, S., Chiang, D., Gould, J., Gupta, S., Korn, J., Mermel, C., Mesirov, J., Monti, S., Nguyen, H., Parkin, M., Reich, M., Stransky, N., Weir, B. A., Garraway, L., Golub, T., Meyerson, M., Chin, L., Protopopov, A., Zhang, J., Perna, I., Aronson, S., Sathiamoorthy, N., Ren, G., Wiedemeyer, W. R., Kim, H., Won Kong, S., Xiao, Y., Kohane, I. S., Seidman, J., Park, P. J., Kucherlapati, R., Laird, P. W., Cope, L., Herman, J. G., Weisenberger, D. J., Pan, F., Van Den Berg, D., Van Neste, L., Mi Yi, J., Schuebel, K. E., Baylin, S. B., Absher, D. M., Li, J. Z., Southwick, A., Brady, S., Aggarwal, A., Chung, T., Sherlock, G., Brooks, J. D., Myers, R. M., Spellman, P. T., Purdom, E., Jakkula, L. R., Lapuk, A. V., Marr, H., Dorton, S., Gi Choi, Y., Han, J., Ray, A., Wang, V., Durinck, S., Robinson, M., Wang, N. J., Vranizan, K., Peng, V., Van Name, E., Fontenay, G. V., Ngai, J., Conboy, J. G., Parvin, B., Feiler, H. S., Speed, T. P., Gray, J. W., Brennan, C., Socci, N. D., Olshen, A., Taylor, B. S., Lash, A., Schultz, N., Reva, B., Antipin, Y., Stukalov, A., Gross, B., Cerami, E., Qing Wang, W., Qin, L. X., Seshan, V. E., Villafania, L., Cavatore, M., Borsu, L., Viale, A., Gerald, W., Sander, C., Ladanyi, M., Perou, C. M., Neil Hayes, D., Topal, M. D., Hoadley, K. A., Qi, Y., Balu, S., Shi, Y., Wu, J., Penny, R., Bittner, M., Shelton, T., Lenkiewicz, E., Morris, S., Beasley, D., Sanders, S., Kahn, A., Sfeir, R., Chen, J., Nassau, D., Feng, L., Hickey, E., Weinstein, J. N., Barker, A., Gerhard, D. S., Vockley, J., Compton, C., Vaught, J., Fielding, P., Ferguson, M. L., Schaefer, C., Madhavan, S., Buetow, K. H., Collins, F., Good, P., Guyer, M., Ozenberger, B., Peterson, J., and Thomson, E. (2008) Nature 455 1061–106818772890 [Google Scholar]
- 6.Whitesell, L., and Lindquist, S. L. (2005) Nat. Rev. Cancer 5 761–772 [DOI] [PubMed] [Google Scholar]
- 7.Hartl, F. U., and Hayer-Hartl, M. (2002) Science 295 1852–1858 [DOI] [PubMed] [Google Scholar]
- 8.Caplan, A. J., Mandal, A. K., and Theodoraki, M. A. (2007) Trends Cell Biol. 17 87–92 [DOI] [PubMed] [Google Scholar]
- 9.Pearl, L. H., and Prodromou, C. (2006) Annu. Rev. Biochem. 75 271–294 [DOI] [PubMed] [Google Scholar]
- 10.Xu, Y., Singer, M. A., and Lindquist, S. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basso, A. D., Solit, D. B., Chiosis, G., Giri, B., Tsichlis, P., and Rosen, N. (2002) J. Biol. Chem. 277 39858–39866 [DOI] [PubMed] [Google Scholar]
- 12.Sato, S., Fujita, N., and Tsuruo, T. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 10832–10837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita, N., Sato, S., Ishida, A., and Tsuruo, T. (2002) J. Biol. Chem. 277 10346–10353 [DOI] [PubMed] [Google Scholar]
- 14.Xu, W., Mimnaugh, E., Rosser, M. F., Nicchitta, C., Marcu, M., Yarden, Y., and Neckers, L. (2001) J. Biol. Chem. 276 3702–3708 [DOI] [PubMed] [Google Scholar]
- 15.Xu, W., Mimnaugh, E. G., Kim, J. S., Trepel, J. B., and Neckers, L. M. (2002) Cell Stress Chaperones 7 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearl, L. H. (2005) Curr. Opin. Genet. Dev. 15 55–61 [DOI] [PubMed] [Google Scholar]
- 17.Karnitz, L. M., and Felts, S. J. (2007) Science's STKE 2007 pe22. [DOI] [PubMed] [Google Scholar]
- 18.Vaughan, C. K., Gohlke, U., Sobott, F., Good, V. M., Ali, M. M., Prodromou, C., Robinson, C. V., Saibil, H. R., and Pearl, L. H. (2006) Mol. Cell 23 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed, S. I. (1980) Genetics 95 561–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacLean, M., and Picard, D. (2003) Cell Stress Chaperones 8 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray, P. J., Jr., Prince, T., Cheng, J., Stevenson, M. A., and Calderwood, S. K. (2008) Nat. Rev. Cancer 8 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamal, A., Boehm, M. F., and Burrows, F. J. (2004) Trends Mol. Med. 10 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray, P. J., Jr., Stevenson, M. A., and Calderwood, S. K. (2007) Cancer Res. 67 11942–11950 [DOI] [PubMed] [Google Scholar]
- 24.Pick, E., Kluger, Y., Giltnane, J. M., Moeder, C., Camp, R. L., Rimm, D. L., and Kluger, H. M. (2007) Cancer Res. 67 2932–2937 [DOI] [PubMed] [Google Scholar]
- 25.McCarthy, M. M., Pick, E., Kluger, Y., Gould-Rothberg, B., Lazova, R., Camp, R. L., Rimm, D. L., and Kluger, H. M. (2008) Ann. Oncol. 19 590–594 [DOI] [PubMed] [Google Scholar]
- 26.Newton, A. C. (2003) Biochem. J. 370 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutil, E. M., and Newton, A. C. (2000) J. Biol. Chem. 275 10697–10701 [DOI] [PubMed] [Google Scholar]
- 28.Gao, T., Toker, A., and Newton, A. C. (2001) J. Biol. Chem. 276 19588–19596 [DOI] [PubMed] [Google Scholar]
- 29.Facchinetti, V., Ouyang, W., Wei, H., Soto, N., Lazorchak, A., Gould, C., Lowry, C., Newton, A. C., Mao, Y., Miao, R. Q., Sessa, W. C., Qin, J., Zhang, P., Su, B., and Jacinto, E. (2008) EMBO J. 27 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikenoue, T., Inoki, K., Yang, Q., Zhou, X., and Guan, K. L. (2008) EMBO J. 27 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keranen, L. M., Dutil, E. M., and Newton, A. C. (1995) Curr. Biol. 5 1394–1403 [DOI] [PubMed] [Google Scholar]
- 32.Orr, J. W., Keranen, L. M., and Newton, A. C. (1992) J. Biol. Chem. 267 15263–15266 [PubMed] [Google Scholar]
- 33.Ziegler, W. H., Parekh, D. B., Le Good, J. A., Whelan, R. D., Kelly, J. J., Frech, M., Hemmings, B. A., and Parker, P. J. (1999) Curr. Biol. 9 522–529 [DOI] [PubMed] [Google Scholar]
- 34.Freeley, M., Volkov, Y., Kelleher, D., and Long, A. (2005) Biochem. Biophys. Res. Commun. 334 619–630 [DOI] [PubMed] [Google Scholar]
- 35.Standaert, M. L., Bandyopadhyay, G., Kanoh, Y., Sajan, M. P., and Farese, R. V. (2001) Biochemistry 40 249–255 [DOI] [PubMed] [Google Scholar]
- 36.Kannan, N., Haste, N., Taylor, S. S., and Neuwald, A. F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1272–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeong, S. S., Zhu, Y., Smith, D., Verma, C., Lim, W. G., Tan, B. J., Li, Q. T., Cheung, N. S., Cai, M., Zhu, Y. Z., Zhou, S. F., Tan, S. L., and Duan, W. (2006) J. Biol. Chem. 281 30768–30781 [DOI] [PubMed] [Google Scholar]
- 38.Edwards, A. S., Faux, M. C., Scott, J. D., and Newton, A. C. (1999) J. Biol. Chem. 274 6461–6468 [DOI] [PubMed] [Google Scholar]
- 39.Edwards, A. S., and Newton, A. C. (1997) J. Biol. Chem. 272 18382–18390 [DOI] [PubMed] [Google Scholar]
- 40.Chou, M. M., Hou, W., Johnson, J., Graham, L. K., Lee, M. H., Chen, C. S., Newton, A. C., Schaffhausen, B. S., and Toker, A. (1998) Curr. Biol. 8 1069–1077 [DOI] [PubMed] [Google Scholar]
- 41.Dries, D. R., Gallegos, L. L., and Newton, A. C. (2007) J. Biol. Chem. 282 826–830 [DOI] [PubMed] [Google Scholar]
- 42.Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dutil, E. M., Toker, A., and Newton, A. C. (1998) Curr. Biol. 8 1366–1375 [DOI] [PubMed] [Google Scholar]
- 44.Sonnenburg, E. D., Gao, T., and Newton, A. C. (2001) J. Biol. Chem. 276 45289–45297 [DOI] [PubMed] [Google Scholar]
- 45.Gao, T., and Newton, A. C. (2006) J. Biol. Chem. 281 32461–32468 [DOI] [PubMed] [Google Scholar]
- 46.Gould, C. M., and Newton, A. C. (2008) Curr. Drug Targets 9 614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao, T., and Newton, A. C. (2002) J. Biol. Chem. 277 31585–31592 [DOI] [PubMed] [Google Scholar]
- 48.Zhang, T., Hamza, A., Cao, X., Wang, B., Yu, S., Zhan, C. G., and Sun, D. (2008) Mol. Cancer Ther. 7 162–170 [DOI] [PubMed] [Google Scholar]
- 49.Moriwaki, Y., Kim, Y. J., Ido, Y., Misawa, H., Kawashima, K., Endo, S., and Takahashi, R. (2008) Neurosci. Res. 61 43–48 [DOI] [PubMed] [Google Scholar]
- 50.Grodsky, N., Li, Y., Bouzida, D., Love, R., Jensen, J., Nodes, B., Nonomiya, J., and Grant, S. (2006) Biochemistry 45 13970–13981 [DOI] [PubMed] [Google Scholar]
- 51.Kane, L. P., Mollenauer, M. N., and Weiss, A. (2004) J. Immunol. 172 5441–5449 [DOI] [PubMed] [Google Scholar]
- 52.Jiang, T., and Qiu, Y. (2003) J. Biol. Chem. 278 15789–15793 [DOI] [PubMed] [Google Scholar]
- 53.Citri, A., Harari, D., Shohat, G., Ramakrishnan, P., Gan, J., Lavi, S., Eisenstein, M., Kimchi, A., Wallach, D., Pietrokovski, S., and Yarden, Y. (2006) J. Biol. Chem. 281 14361–14369 [DOI] [PubMed] [Google Scholar]
- 54.Behn-Krappa, A., and Newton, A. C. (1999) Curr. Biol. 9 728–737 [DOI] [PubMed] [Google Scholar]
- 55.Bornancin, F., and Parker, P. J. (1997) J. Biol. Chem. 272 3544–3549 [DOI] [PubMed] [Google Scholar]
- 56.Gao, T., Brognard, J., and Newton, A. C. (2008) J. Biol. Chem. 283 6300–6311 [DOI] [PubMed] [Google Scholar]
- 57.Yang, J., Cron, P., Thompson, V., Good, V. M., Hess, D., Hemmings, B. A., and Barford, D. (2002) Mol. Cell 9 1227–1240 [DOI] [PubMed] [Google Scholar]
- 58.Frodin, M., Antal, T. L., Dummler, B. A., Jensen, C. J., Deak, M., Gammeltoft, S., and Biondi, R. M. (2002) EMBO J. 21 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huse, M., and Kuriyan, J. (2002) Cell 109 275–282 [DOI] [PubMed] [Google Scholar]
- 60.Fields, A. P., and Gustafson, W. C. (2003) Methods Mol. Biol. 233 519–537 [DOI] [PubMed] [Google Scholar]
- 61.Cameron, A. J., Procyk, K. J., Leitges, M., and Parker, P. J. (2008) Int. J. Cancer 123 769–779 [DOI] [PubMed] [Google Scholar]
- 62.Jane, E. P., and Pollack, I. F. (2008) Cancer Lett. 268 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.