Abstract
A novel humanized mouse model of Cooley's Anemia (CA) was generated by targeted gene replacement in embryonic stem (ES) cells. Because the mouse does not have a true fetal hemoglobin, a delayed switching human γ to β0 globin gene cassette (γβ0) was inserted directly into the murine β globin locus replacing both adult mouse β globin genes. The inserted human β0 globin allele has a mutation in the splice donor site that produces the same aberrant transcripts in mice as described in human cells. No functional human β globin polypeptide chains are produced. Heterozygous γβ0 mice suffer from microcytic anemia. Unlike previously described animal models of β thalassemia major, homozygous γβ0 mice switch from mouse embryonic globin chains to human fetal γ globin during fetal life. When bred with human α globin knockin mice, homozygous CA mice survive solely upon human fetal hemoglobin at birth. This preclinical animal model of CA can be utilized to study the regulation of globin gene expression, synthesis, and switching; the reactivation of human fetal globin gene expression; and the testing of genetic and cell-based therapies for the correction of thalassemia.
β thalassemia is a common hereditary disease caused by a reduction in β globin chain production caused by mutations of the adult β globin gene. Frequently these mutations occur at splice sites that reduce (β+ thalassemia) or eliminate (β0 thalassemia) β chain production from a single mutated allele (β thalassemia minor) resulting in asymptomatic or mild anemia. Mutations of both alleles (β thalassemia major) result in severe disease that ranges from anemia requiring sporadic transfusion (thalassemia intermedia) to chronic transfusion dependence and progressive organ damage (Cooley's Anemia, CA2) (1–3). Hematological hallmarks of β thalassemia are red blood cell microcytosis, hypochromia, targeting, and anisopoikilocytosis (2, 4). In the absence of sufficient β globin chain levels for hemoglobin (Hb) tetramer formation, excess α globin chains precipitate and form inclusions that cause either the premature death of progenitors in the bone marrow (5, 6) or reduced half-life of circulating erythrocytes (7). The anemia that results from this ineffective erythropoiesis stimulates the expansion of more progenitors producing erythroid hyperplasia and extramedullary hematopoiesis.
The β globin loci of both humans and mice are controlled by a powerful regulatory region called the locus control region (LCR) that lies far upstream of the adult β globin genes and is demarcated by a series of erythroid specific developmentally stable DNaseI hypersensitive sites (8–11). In humans there are five functional β-like globin genes (ε, Gγ, Aγ, δ, and β) while in the mouse there are four genes (εY, βh1, βmaj, and βmin) located at each β globin locus. There are two β-like Hb switches in humans that occur during development (12). The first switch occurs when the major hematopoietic site shifts from primitive erythropoiesis in the yolk sac blood islands to definitive erythropoiesis in the fetal liver. Concomitant with this shift in the site of hematopoiesis is a switch from the production of embryonic ε globin chains to fetal γ globin chains. The second human Hb switch takes place around birth when the major site of hematopoiesis shifts from the fetal liver to the bone marrow. Erythrocytes that contain mostly fetal γ globin chains at birth are gradually replaced by red blood cells containing the major (β) and minor (δ) adult globins over the first year of life.
A variety of natural occurring and experimentally engineered mouse models of β thalassemia have been described (13–20). The major problem encountered when modeling human hemoglobin disorders in the mouse is that the developmental timing of Hb switching in humans and mice are different. The mouse has no fetal Hb gene equivalent (21, 22). The embryonic, z and y chains, synthesized from the βh1 and εY globin genes, respectively, are sequentially expressed and synthesized in maturing circulating primitive erythroid cells that were produced in the yolk sac blood islands. The onset of definitive erythropoiesis begins around embryonic day 12 (E12) when the major site of hematopoiesis shifts to the fetal liver in the mouse (21, 22). These definitive erythrocytes synthesize the adult β globin genes, βmaj and βmin, early in fetal development and become the sole β globin chain synthesized from E15 through adulthood. Thus, due to the lack of a fetal globin gene equivalent, βmaj and βmin globin knock-out (KO) mice die around 1 week before birth (15).
Here we report a novel CA mouse model made by targeted gene replacement of both adult mouse β globin genes with a delayed switching human γ to β0 globin gene cassette. Heterozygous γβ0 knockin (KI) mice exhibit β thalassemia intermedia. After breeding to human α globin KI mice, humanized homozygous CA mice survive solely upon human fetal Hb, HbF, during fetal life and expire due to severe anemia upon completion of the Hb switch after birth.
EXPERIMENTAL PROCEDURES
Targeting Construct and ES Cell Culture—Targeting plasmid, from 5′ to 3′, contains 1.7 kb of mouse homology upstream of the mouse βmaj globin gene (HindIII fragment), 5.7 kb human Aγ globin gene fragment (GenBank™ Accession No. U01317: 38,066–43,728), 4.1 kb human β globin gene fragment (GenBank™ Accession No. U01317: 61,320–65,426), a phosphoglycerate kinase promoter driving a hygromycin resistance gene (hyg) flanked by two loxP sites, and 7 kb of mouse homology downstream of the mouse βmin globin gene (BamHI fragment). G to A mutation was introduced into the first base of intron 1 of human β globin gene by PCR. The targeting plasmid was linearized by NotI digestion and electroporated into human γβ KI hprt “tagged” ES cells3 (23). The ES cells were plated on mitomycin C-treated mouse embryonic fibroblasts monolayer in ES cell medium (Dulbecco's modified Eagle's medium), 15% fetal bovine serum (HyClone, Logan, UT), 1× nucleosides, 2 mm l-glutamine, 1× nonessential amino acids, 50 international units/ml penicillin, 50 μg/ml streptomycin, 0.1 mm β-mercaptoethanol, and 1000 units/ml leukemia inhibitory factor). Hygromycin B (125 μg/ml) was added to the ES medium 36 h after electroporation and replenished every other day until colonies were picked. 6-thioguanine (2 μm) was added to the ES medium 4 or 5 days after electroporation to select against cells that still contain the hprt “tagged” locus and enrich for homologous recombinants. ES cell colonies that were resistant to both hygromycin B and 6-thioguanine were picked, expanded, and frozen. DNA aliquots from ES cell colonies were screened by PCR to identify human γβ0 homologous recombinants.
Chimera Production and Breeding—Correctly targeted human γβ0 KI ES cells were injected into 3.5-day-old C57BL/6J (The Jackson Laboratory, Bar Harbor, ME) blastocysts that were transferred into the uteri of outbred CD1 (Charles River Laboratories, Inc., Wilmington, MA) pseudopregnant recipient mice for the production of chimeras (supplemental Fig. S1A). Chimeric mice were bred to C57BL/6J to produce F1 heterozygous γβ0 KI offspring (supplemental Fig. S1B). The hygromycin marker gene was deleted by breeding to CMV-Cre transgenic mice (24). Homozygous γβ0 KI mice were produced by breeding two heterozygous KI mice. All procedures were approved by the University of Alabama at Birmingham's Institutional Animal Care and Use Committee.
RNA Splice Site Determination—Total RNA was isolated from the peripheral blood of γβ0 KI mice using TRIzol LS reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Total peripheral blood RNA was digested by RNase-free DNaseI (New England Biolab) before further usage. RT-PCR was performed on the RNA using random primers and MLV-reverse transcriptase (Promega, Madison, WI) according to the manufacturer's protocol. Two primers were used to simultaneously amplify all the splicing variants by PCR: hβ32 For (5′-ACGAAGCTTACTAGCAACCTCAAACAGACACCA-3′) and hβ162 Rev (5′-ACGAAGCTTTCCAAGGGTAGACCACCAGCA-3′). PCR amplification products were digested by HindIII endonuclease and resolved by PAGE on an 8% polyacrylamide/bis-acrylamide (29:1) gel. All the bands were eluted and subcloned into the HindIII site of pBluescript SK phagemid (Stratagene, La Jolla, CA). The nucleotide sequence of the plasmid inserts was determined at the UAB Heflin Center for Human Genetics DNA Sequencing core facility.
Hematological Indices and Histopathology—Peripheral blood was collected from anesthetized mice into Microtainer® EDTA collection tubes (Becton Dickinson, Franklin Lakes, NJ). Red blood cell counts and red blood cell distribution widths (RDW) were measured on a HemaVet® 1700 (Drew Scientific, Waterbury, CT) hematology analyzer. Packed cell volume (PCV) was measured in a JorVet J503 (Jorgenson Laboratories Systems, Loveland, CO) micro-hematocrit centrifuge. Hb concentrations were determined after conversion to cyanmethemoglobin by lysing red blood cells in Drabkin's Reagent (Sigma), removal of insoluble erythrocyte membranes by centrifugation, measuring the absorbance at 540 nm on a spectrophotometer, and comparison to Hb standards. The mean corpuscular volume (MCV), hemoglobin (MCH), and hemoglobin concentration (MCHC) were calculated from the red blood cell count, Hb concentration, and PCV measurements. Reticulocyte counts were determined by flow cytometry after staining with thiazole orange (25). Tissues were fixed in 70% alcoholic formalin, embedded in paraffin, sectioned, and stained with hematoxylin-eosin by standard methods at the UAB Comparative Pathology Laboratory. Transmission electron microscopy was performed at the UAB High Resolution Imaging Facility. In brief, peripheral blood was fixed in buffered glutaraldehyde, post-fixed in osmium tetroxide, embedded in Epon 812, sections were post-stained with uranyl acetate and lead citrate, viewed with a Tecnai T12 Spirit Twin (FEI Company, Hillboro, OR) at 60kV, and images were captured with a AMT XR60B digital camera (Danvers, MA).
HPLC Analysis of Hemoglobin Chains—Hemolysates are prepared by lysing washed red blood cells in hemolysate buffer (5 mm phosphate, 0.5 mm EDTA, pH 7.4), adding NaCl to 1%, and removal of membranes by centrifugation. A linear gradient of increasing acetonitrile with 0.1% trifluoroacetic acid at a 1.0 ml/min flow rate was used to separate human and mouse globin chains on a reverse phase C4 column (Vydac, Inc., Hesperia, CA) on a Surveyor HPLC instrument (Thermo Scientific, Waltham, MA). Blood from humanized γβA KI mice3 is used for a control in Fig. 5.
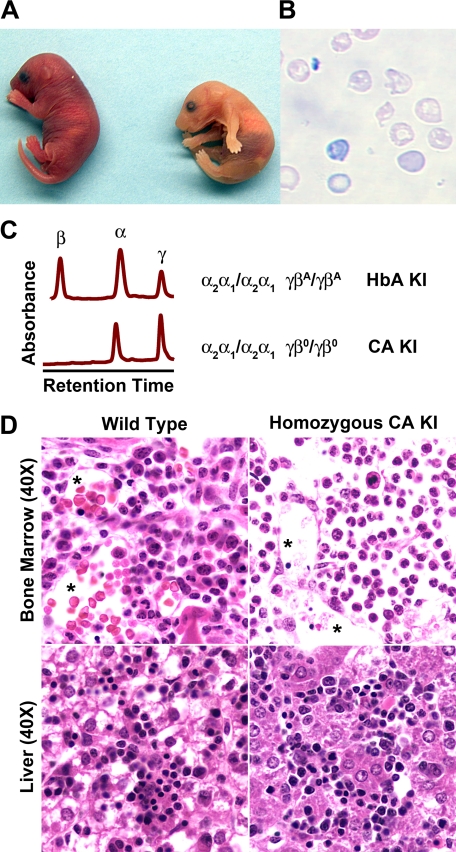
FIGURE 5.
Humanized CA mice survive to birth with only human fetal hemoglobin in their red blood cells. A, homozygous newborn CA mice (right) are anemic as evidenced by the pale skin color compared with the heterozygous littermate control (left). B, peripheral blood smear of homozygous CA mice show a reduction in the absolute number of red blood cells that exhibit hypochromia, polychromatophilia, and anisopoikilocytosis all indicative of their severe anemia (100× magnification). C, HPLC chromatograms of peripheral blood hemolysates from newborn humanized mice. Control human HbA KI mice have peaks for human α globin and both human γ and β globin chains at birth (upper chromatogram). Homozygous CA KI mice have only human α and γ globin chains present in their red blood cells at birth confirming that the CA mouse survive solely upon human fetal Hb (lower chromatogram). D, histological sections of bone marrow and liver of wild type and homozygous CA newborn mice. The vascular lumens (asterisks) in the control bone marrow are filled with red blood cells while there is a paucity of red blood cells in the lumens of anemic CA mice.
Quantitative Real-time RT-PCR (QPCR) Expression Analysis of Globin Chains—cDNA was generated using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) with random hexamers and 500 ng of total peripheral blood RNA. Real-time PCR reactions contained ABI Taqman Universal Mastermix, 900 nm primers, 200 nm probe, and typically 2 μl of 1000-fold diluted cDNA in a 20-μl total volume. Reactions were run on a 7900HT Real-Time PCR System (Applied Biosystems). Sequences of primers and probes used are listed in Table 1. Mouse α, mouse β, and human α globin primer and probe sets measure total expression from both alleles of their respective adult genes. Experiments were performed in triplicate with a 384-well optical plate, and the standard error of the mean from three to ten biological replicates is calculated. Expression was calculated using measured efficiencies (26).
TABLE 1.
Primer and probe sequences used in QPCR assay
| Gene | Forward primer | Probe | Reverse primer |
|---|---|---|---|
| mβh1 | GGTACTTGTGGGACAGAGCATTG | CCACTCCAATCACCAGCTTCTGCCA | CCAAGGAATTCACCCCAGAG |
| mεY | ATCACCAGCAACCTCCCAGAC | TGCCATCATGGTGAACTTTACTGCTGAGG | TACTCCACAGGCCATTGATGAG |
| mβ | GCTGCTGGTTGTCTACCCTTG | CCAGCGGTACTTTGATAGCTTTGGAGACC | CCCATGATAGCAGAGGCAGAG |
| hγ | GTGGAAGATGCTGGAGGAGAAA | AGGCTCCTGGTTGTCTACCCATGGACC | TGCCAAAGCTGTCAAAGAACCT |
| hβ | CGTGCTGGTCTGTGTGCTG | CCCATCACTTTGGCAAAGAATTCACCC | CTTGTGGGCCAGGGCATTAG |
| mζ | CTCCTGTCCCACTGTCTGCTG | TCACAATGGCCGCACGCTTTCC | CAGGCTTCGTGGACCTCAG |
| mα | AGCTGAAGCCCTGGAAAGGAT | TGCTAGCTTCCCCACCACCAAGACC | GCCGTGGCTTACATCAAAGTG |
| hα | CAGACTCAGAGAGAACCCACCAT | TGCTGTCTCCTGCCGACAAGACCAA | GCCTCCGCACCATACTCG |
RESULTS
Generation of the CA Mouse Model—Humans with CA are healthy at birth because of high HbF levels present in newborn circulating erythrocytes. To mimic this condition in the mouse, we introduced a delayed switching human γ to β0 globin gene cassette directly into the mouse β globin locus (Fig. 1A). The β0 globin allele used contains a single G to A nucleotide transition in the first base of intervening sequence 1 (IVS1.1 G to A) that accounts for 13% of β0 thalassemia mutations in the Mediterranean population (27). DNA from these γβ0 KI mice was sequenced to confirm the mutation in the human β globin gene (supplemental Fig. S1C). Analyses of the murine β globin locus by Southern blotting confirmed that the delayed switching human γβ0 cassette was inserted in place of the adult mouse β globin genes (Fig. 1, B and C).
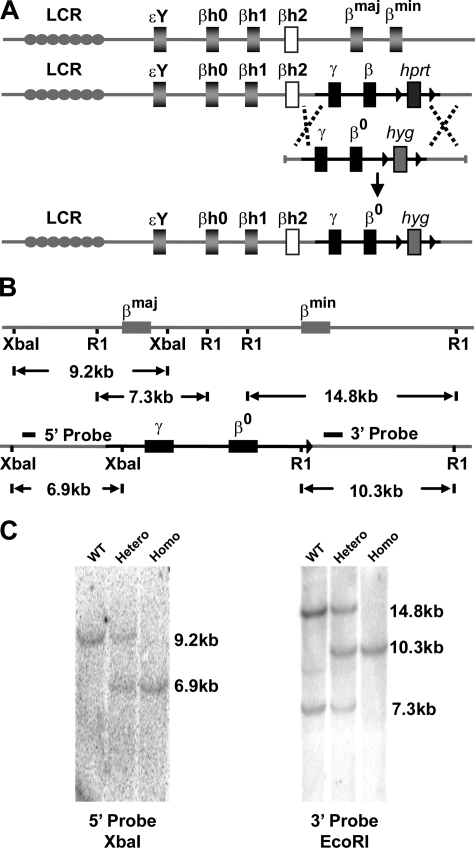
FIGURE 1.
Targeted gene replacement of adult mouse β globin genes with a human γ to β0 globin switching cassette. A, schematic demonstration of gene replacement in the mouse β globin locus tagged with human γβ globin and hprt genes with γβ0 globin and hyg genes. B, schematic demonstration of the wild-type mouse β globin locus and γβ0 KI allele after marker gene removal with labeled restriction endonuclease sites that confirm correct homologous recombination. The 5′ probe anneals to a 9.2-kb XbaI fragment from the wild-type allele and a 6.9-kb XbaI fragment from the human γβ0 globin KI allele. The 3′-probe derived from part of the βmin globin gene, anneals to a 14.8-kb EcoRI fragment from the βmin globin gene, a 7.3-kb fragment from the βmaj globin gene, and a 10.3-kb fragment from the γβ0 globin KI allele. C, Southern blot confirmation of mouse liver DNA of correct 5′ and 3′ homologous recombination. Lane 1, wild type control; lane 2, heterozygous γβ0 globin KI; lane 3, homozygous γβ0 globin KI.
The Mutant Human β0 Globin Gene Produces Aberrantly Spliced Transcripts in Vivo—Treisman et al. reported that transfected cell lines containing a β globin gene with an IVS1.1 G to A splice donor site mutation produces aberrantly spliced mRNAs (28). Loss of the normal GT dinucleotide splice donor resulted in the activation of three cryptic splice donor sites, two upstream in the first exon and one downstream in the first intron. Use of these cryptic splice sites produces altered mRNA transcripts that result in no functional β globin polypeptide synthesis from this allele. In order to determine how the mutant human β0 globin gene transcripts were spliced in the mouse, peripheral blood RNA from γβ0 KI mice were amplified by PCR using human β globin-specific primers, hBeta32 For and hBeta162 Rev, that flank the first exon and most of the second exon (Fig. 2C). Amplified PCR products were resolved by PAGE (Fig. 2A, lane 4), excised, subcloned, and sequenced. Three splice variants were identified that utilized cryptic GT splice donor sites (Fig. 2A, lanes 1–3). This demonstrates that the G to A splice donor site mutation in the human β0 globin KI allele prevents normal splicing, and the murine splicing machinery uses alternative cryptic splice donor sites in vivo.
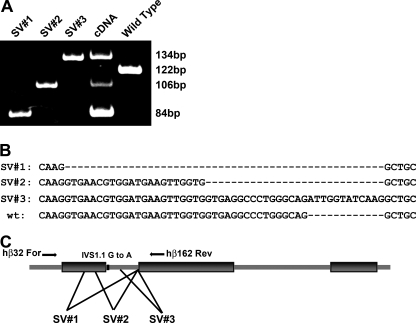
FIGURE 2.
Determination of the mRNA splice variants (SV) from the human β0 globin KI allele caused by the IVS1.1 G to A mutation. A, alternatively spliced human β0 globin mRNA products were amplified by RT-PCR from peripheral blood RNA and resolved by PAGE. Lanes 1, 2, and 3 are the PCR products amplified from plasmid DNA from individually subcloned SV. Lane 4 is the PCR products amplified from γβ0 globin KI mouse. Lane 5 is the PCR product of wild-type human β globin. B, DNA sequence of each subcloned human β0 globin SV and wild type human β globin control. Two cryptic splice donor sites in the first exon (SV1 and SV2) and one cryptic splice donor site in the first intron (SV3) were used to process the mutant human β0 globin transcripts. C, schematic of the human β globin gene primary transcript illustrating exon/intron boundaries, locations of the primers used to amplify the SV, the G to A mutation of the first base of IVS1, and the approximate location of the cryptic splice donor sites.
Humanized CA Mice Switch from Mouse Embryonic to Human Fetal Hb during Development—The human γβ0 KI mice were analyzed to determine whether the human globin genes would be expressed at high levels in a temporally regulated way when inserted 30-kb downstream of the LCR in the murine β globin locus. Expression of both human and mouse globin genes were studied at the protein and RNA levels in developing γβ0 globin KI mice. Fetal blood RNA and hemolysates were prepared from γβ0 globin KI and wild-type littermate controls from embryonic day 10.5 (E10.5) through adulthood (Fig. 3).
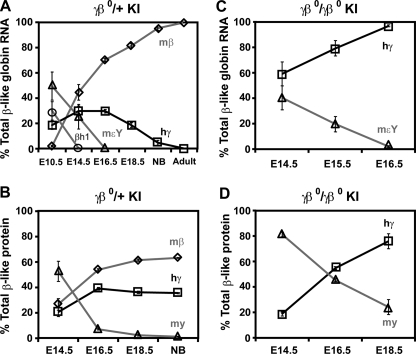
FIGURE 3.
The humanγβ0 globin KI mice switch from mouse embryonic to human γ globin gene expression during fetal development. Circulating blood RNA from heterozygous (A) and homozygous (C)γβ0 globin KI embryos were analyzed by QPCR to quantify the level of β-like globin gene expression per gene copy during fetal development. Mouse embryonic βh1 (○) and εY (▵) globins are silenced by E14.5 and E16.5, respectively. Human γ globin (□) expression persists through fetal life becoming the predominant β-like globin chain produced in homozygous CA mice after E16.5. Already high at E14.5, mouse β globin (⋄) expression continues to rise into adulthood in heterozygous mice. Globin protein chains produced in heterozygous (B) and homozygous (D) γβ0 globin KI embryos during fetal development were measured by HPLC and expressed as a percentage of total β-like chains per gene copy. High levels of human γ globin chains are present throughout fetal life. n ≥ 3 for each time point in A–D. mβ is the average level of combined mouse βmaj and βmin globin gene RNA and protein levels. mεY and my are the mouse embryonic εY globin gene RNA and protein levels, respectively. hγ is the human γ globin gene RNA and protein levels. The low levels of short-lived hβ0 transcripts were left out of panels A and C for clarity, but may be found in supplemental Table S1.
Heterozygous γβ0 globin KI mice express high levels of human γ globin during development (Fig. 3A). In E10.5 primitive erythrocytes human γ globin levels represent about 19% of the total β-like globin gene mRNA per gene copy. During definitive hematopoiesis in the fetal liver, human γ globin levels rise to 30% of total β-like globin mRNA from E14.5 through E16.5 (Fig. 3A). As the human fetal gene switches to the nonfunctional human β0 allele the levels drop to 5% at birth and to less than 1% by weaning age of the adult mouse β globin genes (Fig. 3A). The aberrantly spliced mRNA variants produced from the mutant human β0 allele are detectable, but very short lived representing only a fraction of a percent (0.5%) of total β-like mRNA at birth (supplemental Table S1, Part A). The human β0 globin transcripts increase slightly in adult blood reticulocytes (0.6%) to levels that are 3-fold higher than the final human γ globin expression level (0.2%, supplemental Table S1, Part A). Higher levels of human β0 globin mRNA are found in erythroblasts in the adult bone marrow (1.8%, supplemental Table S1, Part A).
The expression of the murine embryonic globin genes, βh1 and εY, are silenced during development in the heterozygous γβ0 globin KI mice (Fig. 3A). The murine βh1 globin genes are expressed at high levels in primitive erythrocytes at E10.5 (30%), but are silenced by E14.5 (0.4%). Likewise, the expression of the murine εY globin genes are 50% of total β-like globin mRNA in primitive erythrocytes at E10.5, but are silenced by E16.5 (0.1%). The predominant β-like globin mRNA present from E14.5 into adulthood is transcribed from the endogenous adult mouse β globin genes from the wild-type non-targeted β globin locus. The adult murine β globin transcripts increase throughout fetal life and represent over 99% of total β-like globin chains in adult heterozygous γβ0 globin KI mice.
The individual globin polypeptide chain levels in the heterozygous γβ0 globin KI mice mirror the expression data described above; however, they lag the mRNA levels by about 2 days of development (Fig. 3B). The mouse embryonic y chain synthesized from the εY globin gene is the major chain present at E14.5, but decreases to background level of total β-like globin chains per gene copy by birth. Human γ globin chains reach their peak level of 39% on E16.5 and then slowly decrease to 36% of total β-like globin protein in newborn mice. Thus, heterozygous human γβ0 globin KI mice maintain high levels of human γ globin chains during fetal life that represent approximately one-third of total chains at birth.
Fetal blood RNA and hemolysates from homozygous CA embryos (γβ0/γβ0) were also analyzed by QPCR and HPLC (Fig. 3, C and D). The data demonstrate the completion of a mouse εY globin to human γ globin switch from E14.5 to E16.5 (Fig. 3C). The mouse εY globin gene mRNA levels drop from 40% on E14.5 to 2% on E16.5. The human γ globin gene mRNA levels are the predominant β-like globin gene expressed during fetal life increasing from 60% at E14.5 to 96% by E16.5. While the human γ globin mRNA levels represent the majority of all β-like globin gene expression during late fetal life, the percentage of total mouse α-like globin to total β-like globin mRNA increases during this period as the γ globin genes switch to the nonfunctional human β0 globin genes that have a relatively short-lived mRNA (supplemental Table S1, Part B). Thus, erythroid cells become increasingly thalassemic during late fetal life in the homozygous γβ0 KI fetuses.
At the protein level there is a clear Hb switch from mouse embryonic globin chains to human fetal chains during development in homozygous γβ0 KI mice (Fig. 3D). As the murine embryonic y chains decrease from E14.5 to E18.5 there is a reciprocal rise in the human γ globin chain levels. The human γ globin chains increase from 19% in E14.5 fetuses to the predominant β-like chain present in fetal blood at E16.5 and to 76% of β-like chains at E18.5 shortly before their death (Fig. 3D). At their time of death these CA mice must survive upon mouse α/human γ hybrid Hb, mα2hγ2, which has an extremely high oxygen affinity (29).
Humanized CA Mice Have a Severe β Thalassemic Phenotype—Heterozygous adult human γβ0 globin KI mice are anemic and exhibit β thalassemia intermedia phenotypes due to the replacement of their wild-type β globin alleles with the nonfunctional human β0 globin gene. Peripheral blood smears shows a marked anisopoikilocytosis with numerous targeted and hypochromic red blood cells (Fig. 4A). Excess α globin chains form inclusions that are easily seen after vital staining (supplemental Fig. S2) or by electron microscopy (Fig. 4B). Red blood cell indices demonstrate that the γβ0 KI mice have a microcytic anemia with significant reductions in the PCV and Hb levels compared with wild-type controls (Table 2). This anemia causes an increase in the production and early release of nascent macrocytic red blood cells, reticulocytes, from hematopoietic tissues into the circulation that results in a 9-fold increase in the reticulocyte count above normal levels. Overall there is a 14% reduction in circulating red blood cell MCV as reticuloendothelial cells remove excess α globin chain inclusions by “pitting” (30). This simultaneous mixture of macrocytic reticulocytes and mature microcytic red blood cells causes a doubling in the measured RDW. Other changes observed include significant decreases in the red blood cell count, MCH, and MCHC levels (Table 2).
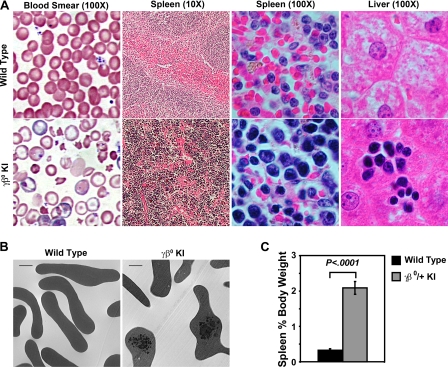
FIGURE 4.
A, histology of blood, spleen, and liver, from heterozygous human γβ0 globin KI mice shows β thalassemic phenotypes. Wright-Giemsa stained peripheral blood smears of wild-type control and heterozygous γβ0 globin KI mice. Typical red blood cell targeting, hypochromia, and anisopoikilocytosis are observed in thalassemic human γβ0 globin KI mice. The normal red (erythroid) and white (lymphoid) pulp sections seen in the wild-type spleen were replaced by an expanded erythroid compartment in the γβ0 globin KI mice compared with the wild-type control. Livers of thalassemic γβ0 globin KI mice contain clusters of erythroid cells indicative of extramedullary hematopoiesis. Image magnifications are shown in parentheses. Slides were analyzed on Nikon Eclipse E800 or TE2000 microscopes (Nikon, Tokyo, Japan). Image magnifications are shown in parentheses. B, electron microscopy of circulating red blood cells from wild type and γβ0 globin KI adult mice demonstrating α globin chain inclusions (scale bar, 1 μm). C, comparison of spleen size as a percentage of body weight of wild type and γβ0 globin KI mice show a significant splenomegaly in the thalassemic mice (n ≥ 8 in each group).
TABLE 2.
Heterozygous human γβ0 globin KI mice have β thalassemic red blood cell indices
Male and female mice were analyzed 8 to 10 weeks after birth. Values represent mean ± S.E. Statistical significances were determined for the γβ0/+ KI mice compared to the wild-type control mice. p values were calculated by the two tailed Student's t test. RBC, red blood cell; PCV, packed cell volume; Hb, hemoglobin; MCV, mean corpuscular volume; MCH, mean corpuscular Hb; MCHC, mean corpuscular Hb concentration; Retic, reticulocyte; RDW, red blood cell distribution width.
| Mice | n | RBC | PCV | Hb | MCV | MCH | MCHC | Retic | RDW |
|---|---|---|---|---|---|---|---|---|---|
| 106/μl | % | g/dl | fl | pg | g/dl | % | % | ||
| Wild type | 8 | 8.8 ± 0.2 | 43.8 ± 0.8 | 13.8 ± 0.4 | 50.3 ± 0.8 | 15.6 ± 0.4 | 29.2 ± 0.6 | 2.1 ± 0.2 | 16.1 ± 0.2 |
| γβ0/+ KI | 11 | 7.3 ± 0.2a | 31.3 ± 0.7a | 7.9 ± 0.2a | 43.3 ± 1.0a | 10.9 ± 0.2a | 25.2 ± 0.4a | 19.6 ± 2.1a | 34.1 ± 0.7a |
p < .0001.
Erythroid hyperplasia and extramedullary hematopoiesis, consequences of the anemia of β thalassemia are easily observed in the adult heterozygous CA mice. The femoral bone marrow cavities are enlarged and filled with erythroid progenitors in the heterozygous γβ0 globin KI mice (supplemental Fig. S2). Splenomegaly, measured as a percentage of the adult animal's body weight, is increased 6-fold as normal splenic architecture of red and white pulp is disorganized by expansion of erythroid progenitor cells in response to their severe anemia (Fig. 4, A and C). Extramedullary hematopoiesis is present as clusters of maturing erythroid precursors in the liver of heterozygous γβ0 globin KI mice (Fig. 4A). Lastly, the early destruction of erythroid cells leads to an increased deposition of iron in the erythroid tissues of the bone marrow, liver, and spleen (supplemental Fig. S2 and data not shown).
Heterozygous CA mice were bred together to generate homozygous γβ0 KI mice to examine their viability through embryonic development. Pregnancies were terminated at various gestational times and the fetuses were removed for genotyping. There was no significant loss of either heterozygous or homozygous γβ0 KI mice up to embryonic day 18.5 in 14 litters analyzed (supplemental Table S2). Additionally, though heterozygous γβ0 globin KI mice suffer from β thalassemia intermedia, no significant survival disadvantage was observed in the heterozygotes by 3 weeks of age at the time of weaning in 15 litters analyzed (supplemental Table S2). Longer term survival studies will need to be performed to determine if the adult heterozygous CA mice have a shortened lifespan. Homozygous γβ0 KI mice did not survive to birth. Although homozygous fetuses express high levels of human γ globin late in fetal life, these chains must dimerize with mouse α globin to form mouse/human hybrid dimers and Hb tetramers. This mouse/human hybrid Hb, mα2/hγ2, may be unstable and has an extremely high oxygen affinity that may affect the viability of homozygous CA fetuses (29). Alternatively, because human γ globin is in the process of switching to the linked nonfunctional human β0 globin allele the expression level of human γ globin may not be high enough for survival to birth. Thus, homozygous γβ0 KI mice die around E18.5 presumably due to a lethal anemia.
Fully Humanized CA Mice Live to Birth—If the homozygous γβ0 KI fetuses die around E18.5 due to an unstable or high oxygen affinity mα2/hγ2 hybrid Hb, then we should be able to rescue them by replacing their mouse α globin chains with human α globin. The γβ0 KI mice were bred to human α2α1 globin KI mice3 that have a targeted gene replacement of both endogenous adult mouse α globin genes with human adult α2 and α1 globin genes to generate mice that synthesize only human Hb in their definitive erythrocytes. Similar experiments that incorporate functional human γβA globin alleles into the murine β globin locus produce humanized mice that switch from mouse embryonic to human fetal to human adult globin genes during development.3 These humanized HbA KI mice have high γ globin chain levels at birth (38%) and complete their fetal to adult Hb switch by 3 weeks of age when their γ chains levels decline to less than 1%3.
After multiple rounds of intercrossing with human α2α1 globin KI mice, fully humanized CA mice were produced that survived to birth. These newborn CA mice are extremely pale compared with newborn control mice (Fig. 5A). Their peripheral blood smears exhibited hypochromic and polychromatic cells with an overall reduction in absolute red blood cell number indicating a severe anemia (Fig. 5B). HPLC analyses of newborn hemolysates confirmed that these humanized CA mice were surviving solely upon human HbF at birth (Fig. 5C). Similar to fetal life the majority of erythroid production at birth is found in the liver. Similar numbers of erythropoietic islands are seen in the livers of control and homozygous CA newborns (Fig. 5D). The percentage of cells in the newborn liver that express the erythroid-specific surface antigen Ter119 were the same in control, heterozygous, and homozygous γβ0 KI mice, showing a lack of erythroid hyperplasia in newborn CA mice (supplemental Fig. S3A). Mouse β globin KO mice (mβthal3/+) also lack any erythroid hyperplasia in their liver or bone marrow at birth (supplemental Fig. S3B), even though their β thalassemia and anemia begins much earlier in fetal life at the time of the primitive to definitive shift in erythropoiesis (15, 16). Thus, while there are numerous myeloid cells present in the bone marrow of control, CA, and mouse β globin KO newborns there was no indication of erythroid hyperplasia (Fig. 5D and supplemental Fig. S3B). However the marked anemia in the CA mice is clearly evident by the paucity of red blood cells in the vasculature of the bone marrow of the homozygous CA mouse compared with control (Fig. 5D). Analysis of blood RNA in the newborn homozygous CA mice demonstrates a severe imbalance in expression levels with the ratio of human γ to human α globin transcripts decreased to less than 9% (supplemental Table S1, Part C). Similar to CA patients, without treatment these fully humanized CA mice expire early in life as the Hb switch from human γ to the nonfunctional β0 globin is completed.
DISCUSSION
Several murine models of β thalassemia have been described. The first was a naturally occurring 3.3 kb deletion of one (βmaj) of the two adult β globin genes (13). Because the remaining βmin globin gene was still expressed this early model was characterized as a β+ thalassemia. Genotypically the second murine β thalassemia model was similar to the first, a deletion of βmaj globin generated by targeted deletion in ES cells, but the phenotypes were different (14). The marker gene that was inserted into the locus reduced the remaining βmin globin gene expression resulting in perinatal lethality of homozygotes. The first β0 thalassemia mouse model was developed by deletion of both adult β globin genes in ES cells (15). A similar β0 thalassemia mouse model was produced by a two step process referred to as the “plug and socket” (16) that was also used to make the first human splicing mutant KI model (17). All these homozygous β0 thalassemia models died early in fetal development due to the lack of any functional adult β globin chains. A non-heritable β0 thalassemia model was made by transplantation of fetal liver cells from homozygous β globin KO mouse embryos into lethally radiated adult wild type mice (18). β0 thalassemic symptoms develop 6 weeks after transplant and recipients die from severe anemia after 7 to 9 weeks. More recently a bacterial artificial chromosome (BAC) transgenic β0 thalassemic mouse model was described (19, 20). Similar to the earlier β0 models, these BAC transgenic mice die during fetal development when homozygous for the mouse β globin gene KO.
The creation of a mouse model of CA that accurately mimics the disorder in humans has been difficult to achieve because the mouse lacks a true fetal Hb. By incorporating a delayed human γ to β0 globin switching cassette and replacing all of the adult murine globin genes with human genes, the β thalassemia major model in this report is the first to synthesize functional human HbF throughout fetal life at levels high enough to survive until birth. Similar to CA infants upon completion of their Hb switch to a nonfunctional β0 globin KI allele, CA mice require therapeutic intervention for life after birth.
Modeling human Hb switching in transgenic mice has been studied extensively. When a single human globin gene is linked in cis to LCR sequences and introduced into the mouse embryo, these transgenes are expressed at high levels during both primitive and definitive erythropoiesis; that is, the temporal control of these genes is lost (9, 11, 31–35). However, when multiple temporally regulated human globin genes are linked together in cis to the LCR, temporal control is restored as the individual genes compete for interaction with the LCR for expression (34–39). Cosmid, BAC, and yeast artificial chromosome transgenic mice that contain some or all of the human β globin loci correctly undergo a human γ to β globin switch during development (34, 36, 37, 40, 41). However, the timing of the switch mimics the pattern of the endogenous mouse genes; hence, the fetal to adult Hb switch is completed by E15. Transgenes that contain the human LCR linked in cis to γ and β globin genes with much of the intergenic region between them deleted exhibit a delay in the fetal to adult Hb switch (38, 39). Indeed, the delay in the Hb switch from human γ to β globin with an LCRγβ transgene was instrumental in the production of a viable knock-out-transgenic mouse model of sickle cell anemia by keeping sickle Hb levels low and HbF levels high throughout fetal life (38).
In this report, a human γ to β globin switching cassette was directly targeted to the murine β globin locus without any human LCR sequences linked in cis. Rather than having multiple copy, head to tail arrays that are randomly inserted in the genome and vary from one founder line to the next as is the case in transgenic mouse experiments, this human γβ0 KI is single copy and precisely located in the genome. We show that the human γ globin gene is expressed at high levels, presumably by interacting with the murine LCR located far upstream. We also show that similar to the human β globin locus there are now two switches at the murine β globin locus, mouse embryonic (εY) to human fetal (γ) and human fetal to a nonfunctional adult human β. It remains for future experimentation to determine if the human γ globin KI allele would autonomously silence when inserted into the murine β locus in the absence of any linked adult globin gene.
This humanized CA model should be useful for studying the regulation of human globin gene expression, synthesis, and switching. In particular the epigenetic modifications to the chromatin surrounding the human genes can be analyzed through development as the human γ to β globin switch is completed. Additionally, any maturational switch from embryonic to fetal to adult globin gene expression in both the primitive and definitive erythroid precursors awaits further study in this KI model of Hb switching. Furthermore, this model is ideal for experiments designed to reactivate the silenced human γ globin gene by transacting factors or small molecules because successful reactivation simultaneously cures a preclinical disease model. New transfusion strategies and chelation therapies could also be tested in this β thalassemia major model. These mice should also be useful for the study of iron overload and the regulation of iron homeostasis in disease.
Homozygous humanized CA mice survive less than 24 h after birth. Their short viability can make this model experimentally challenging to work with because therapies designed to cure or ameliorate the anemia of CA must be administered during a relatively short perinatal period. While this foreshortened therapeutic window may have its drawbacks for some experimental protocols it could also be a blessing for others. These newborn CA pups may be an ideal model system for transplantation and cell-based therapies because the incoming donor hematopoietic cells would have a tremendous survival advantage over the thalassemia major cells in the recipient.
Future improvements of this work could focus on ways to extend the neonatal life of the CA pups. Expanding the window of time for therapeutic interventions designed to rescue the dying newborns could be achieved by increasing the human γ globin expression levels by further delaying the switch from γ to β0 globin. Incorporating γ globin promoter mutations that result in the hereditary persistence of fetal Hb in humans could extend the postnatal life of this model. Alternatively, incorporating the human δ globin gene into the KI construct used to make these humanized CA mice would provide some minor adult human HbA2 similar to human patients. In summary, this humanized mouse model of CA has the following characteristics: has a single human γ and mutant β0 globin KI allele at each murine β globin locus; expresses 100% human fetal Hb in adult red blood cells; completes the human γ to β globin gene switch after birth; synthesizes no functional adult β globin chains after birth; and is genetically heritable.
Supplementary Material
Acknowledgments
We thank Dr. Trenton Schoeb of the UAB Comparative Pathology Laboratory and C. Leigh Millican of the UAB High Resolution Imaging Facility for help with experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL072351 (to T. M. R.) and R01 HL073440 (to T. M. R.); UAB CMB Training Grant T32 GM008111 (to S. C. M.); and the Carmichael Scholarship (to S. C. M. and T.-T. Z.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
Footnotes
The abbreviations used are: CA, Cooley's Anemia; ES, embryonic stem; γβ0, delayed switching human γ to β0 globin gene cassette; Hb, hemoglobin; LCR, locus control region; E12, embryonic day 12; KO, knockout; KI, knockin; HbF, fetal hemoglobin; RDW, red blood cell distribution width; PCV, packed cell volume; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; IVS1.1, first base of intervening sequence 1; QPCR, quantitative real-time RT-PCR; SV, splice variant; BAC, bacterial artificial chromosome.
T. M. Ryan, unpublished data.
References
- 1.Bank, A. (1985) Curr. Top. Hematol. 5 1–23 [PubMed] [Google Scholar]
- 2.Bunn, H., and Forget, B. (1986) Hemoglobin: Molecular, Genetic, and Clinical Aspects, pp. 333–343, W. B. Saunders, Philadelphia
- 3.Kazazian, H. H., Jr., Ginder, G. D., Snyder, P. G., Van Beneden, R. J., and Woodhead, A. P. (1975) Proc. Natl. Acad. Sci. U. S. A. 72 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinberg, M. H. (1988) Am. J. Med. Sci. 296 308–321 [DOI] [PubMed] [Google Scholar]
- 5.Mathias, L. A., Fisher, T. C., Zeng, L., Meiselman, H. J., Weinberg, K. I., Hiti, A. L., and Malik, P. (2000) Exp. Hematol. 28 1343–1353 [DOI] [PubMed] [Google Scholar]
- 6.Yuan, J., Angelucci, E., Lucarelli, G., Aljurf, M., Snyder, L. M., Kiefer, C. R., Ma, L., and Schrier, S. L. (1993) Blood 82 374–377 [PubMed] [Google Scholar]
- 7.Kean, L. S., Brown, L. E., Nichols, J. W., Mohandas, N., Archer, D. R., and Hsu, L. L. (2002) Exp. Hematol. 30 394–402 [DOI] [PubMed] [Google Scholar]
- 8.Forrester, W. C., Thompson, C., Elder, J. T., and Groudine, M. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 1359–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosveld, F., van Assendelft, G. B., Greaves, D. R., and Kollias, G. (1987) Cell 51 975–985 [DOI] [PubMed] [Google Scholar]
- 10.Tuan, D., and London, I. M. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 2718–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan, T. M., Behringer, R. R., Martin, N. C., Townes, T. M., Palmiter, R. D., and Brinster, R. L. (1989) Genes Dev. 3 314–323 [DOI] [PubMed] [Google Scholar]
- 12.Nienhuis, A. W., and Stamatoyannopoulos, G. (1978) Cell 15 307–315 [DOI] [PubMed] [Google Scholar]
- 13.Skow, L. C., Burkhart, B. A., Johnson, F. M., Popp, R. A., Popp, D. M., Goldberg, S. Z., Anderson, W. F., Barnett, L. B., and Lewis, S. E. (1983) Cell 34 1043–1052 [DOI] [PubMed] [Google Scholar]
- 14.Shehee, W. R., Oliver, P., and Smithies, O. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 3177–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciavatta, D. J., Ryan, T. M., Farmer, S. C., and Townes, T. M. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 9259–9263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang, B., Kirby, S., Lewis, J., Detloff, P. J., Maeda, N., and Smithies, O. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 11608–11612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis, J., Yang, B., Kim, R., Sierakowska, H., Kole, R., Smithies, O., and Maeda, N. (1998) Blood 91 2152–2156 [PubMed] [Google Scholar]
- 18.Rivella, S., May, C., Chadburn, A., Riviere, I., and Sadelain, M. (2003) Blood 101 2932–2939 [DOI] [PubMed] [Google Scholar]
- 19.Jamsai, D., Zaibak, F., Khongnium, W., Vadolas, J., Voullaire, L., Fowler, K. J., Gazeas, S., Fucharoen, S., Williamson, R., and Ioannou, P. A. (2005) Genomics 85 453–461 [DOI] [PubMed] [Google Scholar]
- 20.Vadolas, J., Nefedov, M., Wardan, H., Mansooriderakshan, S., Voullaire, L., Jamsai, D., Williamson, R., and Ioannou, P. A. (2006) J. Biol. Chem. 281 7399–7405 [DOI] [PubMed] [Google Scholar]
- 21.Kingsley, P. D., Malik, J., Emerson, R. L., Bushnell, T. P., McGrath, K. E., Bloedorn, L. A., Bulger, M., and Palis, J. (2006) Blood 107 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitelaw, E., Tsai, S. F., Hogben, P., and Orkin, S. H. (1990) Mol. Cell. Biol. 10 6596–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu, L. C., Sun, C. W., Ryan, T. M., Pawlik, K. M., Ren, J., and Townes, T. M. (2006) Blood 108 1183–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nozaki, M., Ohishi, K., Yamada, N., Kinoshita, T., Nagy, A., and Takeda, J. (1999) Lab. Investig. 79 293–299 [PubMed] [Google Scholar]
- 25.Ferguson, D. J., Lee, S. F., and Gordon, P. A. (1990) Am. J. Hematol. 33 13–17 [DOI] [PubMed] [Google Scholar]
- 26.Pfaffl, M. W. (2001) Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kollia, P., Karababa, P. H., Sinopoulou, K., Voskaridou, E., Boussiou, M., Papadakis, M., and Loukopoulos, D. (1992) Gene Geogr. 6 59–70 [PubMed] [Google Scholar]
- 28.Treisman, R., Orkin, S. H., and Maniatis, T. (1983) Nature 302 591–596 [DOI] [PubMed] [Google Scholar]
- 29.Nishino, T., Cao, H., Stamatoyannopoulos, G., and Emery, D. W. (2006) Br. J. Haematol. 134 100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forget, B. G., and Nathan, D. G. (1975) Annu. Rev. Med. 26 345–351 [DOI] [PubMed] [Google Scholar]
- 31.Behringer, R. R., Ryan, T. M., Reilly, M. P., Asakura, T., Palmiter, R. D., Brinster, R. L., and Townes, T. M. (1989) Science 245 971–973 [DOI] [PubMed] [Google Scholar]
- 32.Ryan, T. M., Behringer, R. R., Townes, T. M., Palmiter, R. D., and Brinster, R. L. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 37–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan, T. M., Townes, T. M., Reilly, M. P., Asakura, T., Palmiter, R. D., Brinster, R. L., and Behringer, R. R. (1990) Science 247 566–568 [DOI] [PubMed] [Google Scholar]
- 34.Behringer, R. R., Ryan, T. M., Palmiter, R. D., Brinster, R. L., and Townes, T. M. (1990) Genes Dev. 4 380–389 [DOI] [PubMed] [Google Scholar]
- 35.Enver, T., Raich, N., Ebens, A. J., Papayannopoulou, T., Costantini, F., and Stamatoyannopoulos, G. (1990) Nature 344 309–313 [DOI] [PubMed] [Google Scholar]
- 36.Strouboulis, J., Dillon, N., and Grosveld, F. (1992) Genes Dev. 6 1857–1864 [DOI] [PubMed] [Google Scholar]
- 37.Peterson, K. R., Clegg, C. H., Huxley, C., Josephson, B. M., Haugen, H. S., Furukawa, T., and Stamatoyannopoulos, G. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 7593–7597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan, T. M., Ciavatta, D. J., and Townes, T. M. (1997) Science 278 873–876 [DOI] [PubMed] [Google Scholar]
- 39.Ryan, T. M., Sun, C. W., Ren, J., and Townes, T. M. (2000) Nucleic Acids Res. 28 2736–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaensler, K. M., Kitamura, M., and Kan, Y. W. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 11381–11385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaufman, R. M., Pham, C. T., and Ley, T. J. (1999) Blood 94 3178–3184 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.