Abstract
Members of the RegIII family of intestinal C-type lectins are directly antibacterial proteins that play a vital role in maintaining host-bacterial homeostasis in the mammalian gut, yet little is known about the mechanisms that regulate their biological activity. Here we show that the antibacterial activities of mouse RegIIIγ and its human ortholog, HIP/PAP, are tightly controlled by an inhibitory N-terminal prosegment that is removed by trypsin in vivo. NMR spectroscopy revealed a high degree of conformational flexibility in the HIP/PAP inhibitory prosegment, and mutation of either acidic prosegment residues or basic core protein residues disrupted prosegment inhibitory activity. NMR analyses of pro-HIP/PAP variants revealed distinctive colinear backbone amide chemical shift changes that correlated with antibacterial activity, suggesting that prosegment-HIP/PAP interactions are linked to a two-state conformational switch between biologically active and inactive protein states. These findings reveal a novel regulatory mechanism governing C-type lectin biological function and yield new insight into the control of intestinal innate immunity.
The gastrointestinal tracts of mammals are heavily colonized with vast symbiotic microbial communities and are also a major portal of entry for bacterial pathogens. To cope with these complex microbial challenges, intestinal epithelial cells produce a diverse repertoire of protein antibiotics from multiple distinct protein families (1). These proteins are secreted apically into the luminal environment of the intestine where they play a pivotal role in protecting against enteric infections (2, 3) and may also function to limit opportunistic invasion by symbiotic bacteria (4).
We previously identified lectins as a novel class of secreted antibacterial proteins in the mammalian intestine. RegIIIγ is a member of the RegIII subgroup of the C-type lectin family and is expressed in the small intestine in response to microbial cues (5), stored in epithelial cell secretory granules, and released into the small intestinal lumen (5). Similarly, HIP/PAP (hepatointestinal pancreatic/pancreatitis-associated protein; the human ortholog of RegIIIγ)6 is expressed in the human intestine (6) and is up-regulated in patients with inflammatory bowel disease (7). These proteins are produced in multiple epithelial lineages, including enterocytes and Paneth cells (5, 6). Both RegIIIγ and HIP/PAP are directly bactericidal at low micromolar concentrations for Gram-positive bacteria (5), revealing a previously unappreciated biological function for mammalian lectins. The antibacterial functions of RegIIIγ and HIP/PAP are dependent upon binding bacterial targets through interactions with peptidoglycan (5). As peptidoglycan is localized on surfaces of Gram-positive bacteria but is buried in the periplasmic space of Gram-negative bacteria, this binding activity provides a molecular explanation for the Gram-positive specific bactericidal effects of these lectins. Although the mechanism of lectin-mediated antibacterial activity remains unclear, RegIIIγ and HIP/PAP have been shown to elicit extensive damage to the cell surfaces of targeted bacteria (5).
In this study, we show that C-type lectin bactericidal activity is under stringent post-translational control. RegIIIγ and HIP/PAP each undergo in vivo proteolytic removal of a flexible anionic N-terminal prosegment that maintains the proteins in a biologically inactive state. NMR spectroscopy suggests that the prosegment functions by controlling a two-state conformational switch between the biologically active and inactive states of the protein. We propose that this regulatory mechanism allows the host to restrict expression of RegIII lectin antibacterial activity to the intestinal lumen. Together, our findings represent a unique example of post-translational control of C-type lectin biological activity, and provide novel insight into the regulation of lectin-mediated innate immunity in the mammalian intestine.
EXPERIMENTAL PROCEDURES
Purification of Endogenous RegIIIγ—Endogenous RegIIIγ was purified from the small intestines of C57BL/6 mice. Intestinal tissues were homogenized in 20% acetic acid solution containing protease inhibitors using a pre-chilled homogenization probe and lysed by sonication using a Misonix XL sonicator. The extract was dialyzed against 25 mm MES pH 5.0, 25 mm NaCl and was loaded onto a cation exchange column (SP-Sepharose, Sigma). The column was washed with 25 mm MES pH 5.0, 150 mm NaCl, and eluted with 25 mm MES pH 5.0, 500 mm NaCl. The eluate was concentrated and loaded onto a Sephacryl S-100 column (GE Healthcare). RegIIIγ-containing fractions were identified by Western blot, pooled, and purified by passage over immobilized anti-RegIIIγ (8). The eluate was concentrated, transferred to Immobilon P, and subjected to Edman degradation on an ABI494 sequencer (PE Biosystems) to determine the N-terminal sequence.
Expression and Purification of Recombinant Proteins—Recombinant pro-RegIIIγ (rpro-RegIIIγ) and rpro-HIP/PAP were expressed and purified as previously described (8). To generate the recombinant processed form of RegIIIγ, a 417-bp amplicon was generated using the rpro-RegIIIγ expression construct (pET3a-RegIIIγ) (8) as template and the specific primers 5′-ATTGCGAGGCATATGAGCAGCTGCCCCAAGGGCTCCC-3′ (forward) and 5′-CTATGGGGATCCCTAGGCCTTGAATTTGCAGACATAGGGT-3′ (reverse). The forward primer contained an NdeI restriction site (underlined) for cloning into pET3a. The reverse primer contained the native stop codon followed by an engineered BamHI site (underlined). The amplicon was digested with NdeI and BamHI and ligated into NdeI/BamHI-digested pET3a (Novagen). Similarly, the rHIP/PAP expression construct was generated using pET3a-HIP/PAPmut (8) as template and specific primers 5′-ATTGCGAGGCATATGATTCGATGTCCAAAAGGCTCCAAG-3′ (forward) and 5′-CTATGGTGATCATCAGTGAACTTTGCAGACATAGGGTAACC-3′ (reverse). The forward primer contained an NdeI site (underlined) while the reverse primer contained a BclI site (underlined) after the native stop codon. The amplicon was digested with NdeI and BclI and ligated into NdeI/BamHI-digested pET3a. Point mutations were introduced into rpro-HIP/PAP using the QuikChange II Site-directed Mutagenesis kit (Stratagene) and specific primers harboring the desired mutations. Expression and purification of recombinant wild-type and mutant proteins was carried out as previously described (8). Antimicrobial assays and peptidoglycan binding assays were carried out as previously described (5). Proteins were stored at –20 °C for a maximum of 1 week prior to assay.
Immunoblotting—Intestinal extracts were prepared from wild-type C57BL/6 or MMP7–/– mice (Jackson Laboratories) as previously described (5). Extracts from human intestinal tissues were prepared by extraction in acetic acid as previously described (9). 20 μg of each extract was loaded onto 15% SDS-PAGE gels and blotted onto Immobilon P (Millipore). Blots were probed with anti-RegIIIγ antiserum (8) and goat anti-rabbit-horseradish peroxidase (Amersham Biosciences), and were detected by chemiluminescence using the Pierce SuperSignal West Pico Chemiluminescent detection kit.
In Vitro Trypsin Proteolytic Processing—Recombinant pro-RegIIIγ and pro-HIP/PAP were digested with bovine pancreatic trypsin (Sigma) at a 1:200 molar ratio of trypsin:lectin at 37 °C for 2 h. Proteins were immediately analyzed by SDS-PAGE on 15% gels, and were subjected to N-terminal Edman sequencing.
NMR Spectroscopy—To prepare samples for NMR experiments, Escherichia coli BL21-CodonPlus (DE3)-RILP transformed with expression plasmids were grown in M9 minimal media containing 1 g/liter of 15NH4Cl for uniformly 15N-labeled samples, further substituting unlabeled glucose with 3 g/liter of 13C6-glucose for uniformly 15N/13C-labeled samples. RegIIIγ and HIP/PAP were purified as previously described (8). Backbone 15N, 13C, and 1H chemical shift assignments of rpro-HIP/PAP were generated from triple resonance NMR data recorded at 25 °C using a cryoprobe-equipped Varian Inova 600 MHz spectrometer. Backbone assignments were made from a 300 μm sample of 15N/13C-labeled protein in 25 mm MES pH 5.5, 25 mm NaCl using standard methods (10). All spectra were processed using NMRPipe (11) and analyzed with NMRView (12).
RESULTS
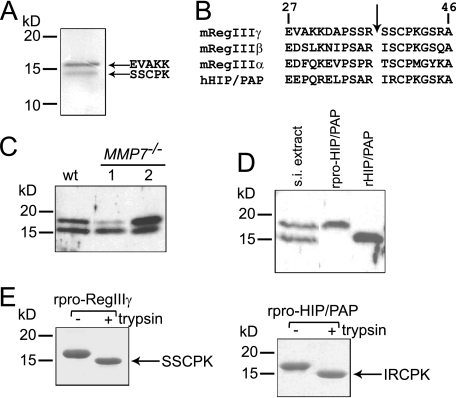
RegIIIγ Is Proteolytically Processed by Trypsin in Vivo—Two distinct forms of endogenous RegIIIγ are present in the mouse small intestine (5). The higher molecular weight form co-migrates at ∼16.5 kDa with recombinant RegIIIγ that lacks its N-terminal secretion signal, while the lower molecular weight form migrates at ∼15 kDa. The presence of the lower molecular weight form suggested that RegIIIγ might be proteolytically processed in vivo. To test this idea we purified endogenous RegIIIγ from mouse small intestine by acid extraction, gel filtration chromatography, and antibody affinity chromatography. Edman sequencing of the purified proteins verified that the higher molecular weight RegIIIγ form indeed harbors an N terminus (Glu27) that is generated by removal of the signal sequence (Fig. 1A). The N terminus of the lower molecular weight form is Ser38, corresponding to the predicted trypsin cleavage site located at Arg37–Ser38 (Fig. 1A). This site is conserved among all mouse and human RegIII family members (Fig. 1B), and is also present in other Reg subfamilies including RegI (13). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) of both RegIIIγ forms did not reveal any further modifications of the protein (data not shown). These results indicate that the N terminus of endogenous mouse RegIIIγ is proteolytically processed by trypsin or a trypsin-like protease in vivo.
FIGURE 1.
RegIIIγ is proteolytically processed by trypsin in vivo. A, purification and N-terminal sequencing of endogenous mouse RegIIIγ reveals processing at the conserved N-terminal trypsin site. B, conserved canonical trypsin site (indicated by arrow) is present near the N terminus of mouse and human RegIII family members. Residue numbers are based on the deduced sequence which includes the signal peptide. Position 1 corresponds to the initiating methionine. C, MMP-7 is dispensable for RegIIIγ processing. 20 μg of protein extract from wild-type and MMP7–/– mice were immunoblotted with anti-RegIIIγ antibody. D, evidence for in vivo proteolytic processing of HIP/PAP. 20 μg of human intestinal protein extract was immunoblotted and probed with anti-RegIIIγ antiserum. Recombinant pro-HIP/PAP (rpro-HIP/PAP; with the N-terminal signal sequence replaced by methionine) and recombinant processed HIP/PAP (rHIP/PAP; with the N-terminal tryptic fragment replaced by methionine) were included for size comparison. s.i., small intestinal E, in vitro incubation of purified recombinant pro-RegIIIγ (rpro-RegIIIγ) and rpro-HIP/PAP with bovine trypsin results in quantitative cleavage at the conserved trypsin site to yield a homogeneous product. Proteins were digested with a 1:200 molar ratio of trypsin:lectin and were analyzed by SDS-PAGE. N-terminal sequencing verified cleavage at Arg37–Ser38 and Arg37–Ile38, respectively.
Small intestinal α-defensins are a distinct family of antimicrobial peptides that also undergo N-terminal proteolytic processing. While human α-defensins are processed by trypsin (9), the processing enzyme for mouse α-defensins is matrix metalloproteinase-7 (MMP-7) (2). To verify that MMP-7 is not the processing enzyme for mouse RegIIIγ we performed Western blots on small intestinal tissue extracts from wild-type and MMP7–/– mice. Both RegIIIγ forms were detectable in MMP7–/– mice (Fig. 1C), establishing that MMP-7 is dispensable for RegIIIγ processing. Thus, RegIIIγ and α-defensins are processed by distinct proteases in the mouse small intestine.
In the human small intestine, HIP/PAP is the predominantly expressed ortholog of RegIIIγ (6, 14) and performs a similar antibacterial function (5). Immunoblotting of human small intestinal extract revealed two forms of HIP/PAP with different molecular weights, suggesting that HIP/PAP is also expressed as a pro-protein that is proteolytically processed in vivo (Fig. 1D). Although the small amounts of HIP/PAP present in human tissue presented an obstacle to purification and sequencing, we established that the lower molecular weight form co-migrates with recombinant HIP/PAP engineered to lack the 11 amino acids N-terminal to the predicted Arg37–Ile38 trypsin cleavage site (Fig. 1D). The idea that the HIP/PAP N terminus is processed in vivo by trypsin is supported by two additional observations. First, two trypsin isoforms are produced by human small intestinal epithelia and have been shown to process other intestinal proteins in vivo (9). Second, in vitro incubation of recombinant pro-HIP/PAP (rpro-HIP/PAP) with bovine trypsin (at a 1:200 molar ratio of enzyme to lectin) resulted in quantitative cleavage at Arg37–Ile38 (Fig. 1E).
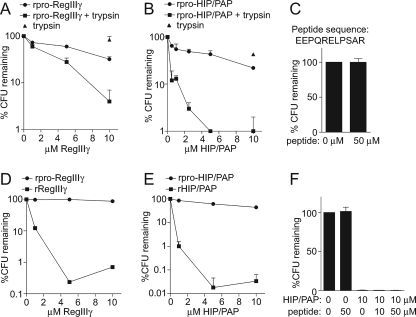
Proteolytic Processing by Trypsin Activates RegIII Lectin Antimicrobial Activity—We next investigated the biological relevance of lectin proteolytic processing. To test the hypothesis that processing regulates RegIII lectin antibacterial activity, we incubated recombinant pro-RegIIIγ (rpro-RegIIIγ) with trypsin in vitro and quantitated antimicrobial activity against Listeria monocytogenes in standard antibacterial assays (5). Mock-digested rpro-RegIIIγ exhibited limited antibacterial activity, with L. monocytogenes numbers declining by 68% in the presence of 10 μm rpro-RegIIIγ (Fig. 2A). However, trypsin-digested rpro-RegIIIγ exhibited enhanced antibacterial potency, with L. monocytogenes numbers declining by >96% in the presence of 10 μm of digested RegIIIγ (Fig. 2A). A comparison of proteolytically cleaved and uncleaved rpro-HIP/PAP yielded similar results (Fig. 2B). The enhanced activity of trypsin-cleaved HIP/PAP is not attributable to the N-terminal prosegment itself, as addition of 50 μm of synthetic N-terminal peptide to L. monocytogenes did not affect bacterial viability (Fig. 2C). These data demonstrate that removal of the N-terminal prosegment by trypsin enhances RegIIIγ and HIP/PAP antibacterial activity.
FIGURE 2.
Proteolysis of the N terminus by trypsin activates lectin antibacterial activity. A and B, trypsin proteolysis of recombinant pro-RegIIIγ (rpro-RegIIIγ) and recombinant pro-HIP/PAP (rpro-HIP/PAP) activates antibacterial activity. Purified rpro-RegIIIγ (A) and rpro-HIP/PAP (B) were digested with bovine trypsin as in Fig. 1e. L. monocytogenes was exposed to the indicated lectin concentrations at 37 °C for 2 h, and surviving bacteria were quantitated by dilution plating. An assay which included trypsin but no lectin was run as a control. C, addition of 50 μm of the HIP/PAP N-terminal peptide did not diminish L. monocytogenes viability, indicating that the prosegment alone does not exhibit antibacterial activity. D and E, antibacterial activities of recombinant mature RegIIIγ and HIP/PAP. The 11-amino acid prosegments of RegIIIγ and HIP/PAP were removed and replaced with methionine to yield rRegIIIγ and rHIP/PAP, and bactericidal activity was determined in comparison with rpro-RegIIIγ (D) and rpro-HIP/PAP (E). F, HIP/PAP N-terminal prosegment does not inhibit bactericidal activity in trans. The synthetic HIP/PAP N-terminal peptide depicted in C was added to bactericidal assays with rHIP/PAP.
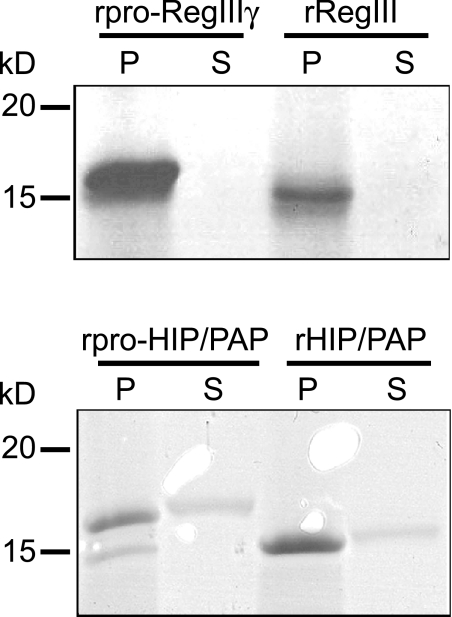
To further establish that prosegment removal activates lectin antibacterial activity, we generated recombinant forms of RegIIIγ and HIP/PAP that lacked their N-terminal prosegments. 10 μm of recombinant mature RegIIIγ (rRegIIIγ) decreased L. monocytogenes viability by over 99%, as compared with a 12% decrease for rpro-RegIIIγ (Fig. 2D). Similarly, 10 μm of recombinant mature HIP/PAP (rHIP/PAP) killed >99.9% of organisms, as compared with 56% for rpro-HIP/PAP (Fig. 2E). Addition of the HIP/PAP N-terminal prosegment in trans did not reduce the antibacterial activity of rHIP/PAP (Fig. 2F), indicating that the prosegment is inhibitory only when covalently attached (in cis). Peptidoglycan pull-down assays established that prosegment removal did not diminish the ability of either the human or mouse protein to bind peptidoglycan (Fig. 3). Collectively, these data establish that the RegIII lectin N-terminal prosegment inhibits antibacterial activity in cis, but does not affect peptidoglycan binding.
FIGURE 3.
Peptidoglycan binding activity is not altered by prosegment removal. Recombinant unprocessed and processed RegIIIγ and HIP/PAP were compared in peptidoglycan pull-down assays. 20 μg of protein was added to 50 μg of peptidoglycan and pelleted. Pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE.
We previously reported bactericidal activity for recombinant RegIIIγ and HIP/PAP that were expressed with an intact N-terminal prosegment (5). To determine why we were able to detect antibacterial activity in these protein preparations, we investigated the fate of the pro-HIP/PAP N terminus during storage. We noted that the rpro-HIP/PAP is labile and the N terminus is progressively removed over time during storage at 4 °C (supplemental Fig. S1). The presence of small amounts of mature HIP/PAP is thus likely to account for the antibacterial activity of protein preparations derived from recombinant prosegment-containing lectins. This may also explain why we detect modest antibacterial activity in rpro-RegIIIγ and rpro-HIP/PAP following mock digestion for 2 h at 37°C (Fig. 2, A and B).
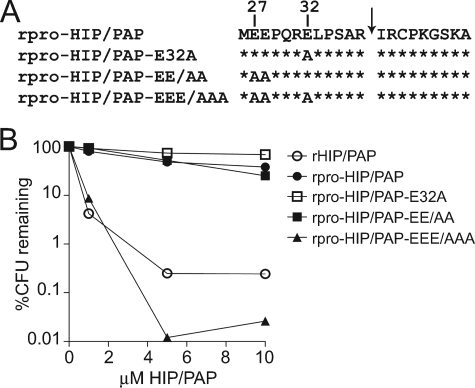
N-terminal Acidic Residues Are Essential for Prosegment Inhibitory Activity—We next sought to gain insight into the inhibitory mechanism of the RegIII lectin N-terminal prosegment. We noted the presence of either two or three acidic residues (Glu or Asp) at conserved positions in both mouse and human RegIII family members (Fig. 1B). To evaluate the functional importance of the three acidic residues contained within the HIP/PAP prosegment (Glu27, Glu28, and Glu32), we mutated combinations of these residues to Ala and assessed the mutant rpro-HIP/PAP proteins for antibacterial activity (Fig. 4A). Recombinant pro-HIP/PAP-E32A (10 μm) reduced L. monocytogenes viability by 30%, while rpro-HIP/PAP-EE/AA (harboring E27A and E28A mutations) produced a 75% decline in bacterial colony forming units (CFUs) at the same concentration (Fig. 4B). In contrast, mutation of all three acidic residues (rpro-HIP/PAP-EEE/AAA) yielded a prosegment-containing protein that produced a >99.9% reduction in the viability of L. monocytogenes at a 10 μm concentration (Fig. 4B). This demonstrates that multiple N-terminal acidic residues are essential for HIP/PAP prosegment inhibitory function, and suggests that the degree of inhibitory activity is dictated by charge.
FIGURE 4.
N-terminal acidic resides are essential for prosegment inhibitory activity. A, primary structure of the pro-HIP/PAP N terminus showing the positions of engineered mutations. B, comparison of antibacterial activity among rpro-HIP/PAP, rHIP/PAP, and rpro-HIP/PAP harboring mutations in N-terminal glutamic acid (E) residues. Antibacterial assays were performed as in Fig. 2.
Structural Analysis of the HIP/PAP-Prosegment Interaction—Although the crystal structure of pro-HIP/PAP has been determined (15), it offered limited information about the HIP/PAP-prosegment interaction. In particular, N-terminal residues 27–35 are disordered and thus absent from the structure (Fig. 5A) and residues 36–41 appear to be involved in crystal packing interactions, raising questions about the observed structure of this essential region of the protein. We therefore used nuclear magnetic resonance (NMR) spectroscopy to structurally characterize the interaction between HIP/PAP and its N-terminal prosegment. Using uniformly 15N,13C-labeled pro-HIP/PAP and standard triple resonance methods (16) we assigned the chemical shifts of 98% of the backbone resonances. We first analyzed the conformational dynamics of pro-HIP/PAP by measuring 15N relaxation rates, which are influenced by fast (ps-ns) and slow (μs-ms) motions of backbone amides throughout a protein. In particular, we measured rate constants for spin-lattice relaxation (R1), and spin-spin relaxation (R2), finding that the ratio of these values (R1/R2) was highest in residues 27–34 (Fig. 5B). This indicates a high degree of backbone conformational mobility at the N terminus. Furthermore, TALOS analyses of backbone chemical shifts (17) and 1H-1H NOEs indicated that the prosegment adopts an extended structure. Together, these data suggest that the prosegment is flexible and transiently interacts with the rest of the protein.
FIGURE 5.
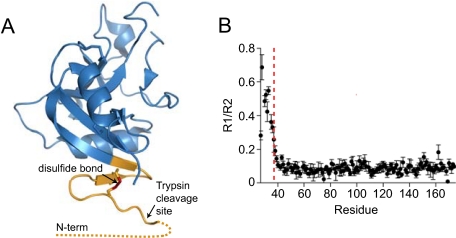
The HIP/PAP N terminus is flexible. A, ribbon diagram of the pro-HIP/PAP crystal structure (RCSB accession: 1UV0) (15). The disordered N-terminal 10 amino acids are indicated by a dotted line, and the locations of the N-terminal disulfide bond (red) and the trypsin cleavage site are indicated. B, experimental 15NR1 and R2 relaxation rates were determined for pro-HIP/PAP, and a plot of the R1/R2 ratio is shown. The location of the Arg37–Ile38 trypsin site is indicated by a dashed red line.
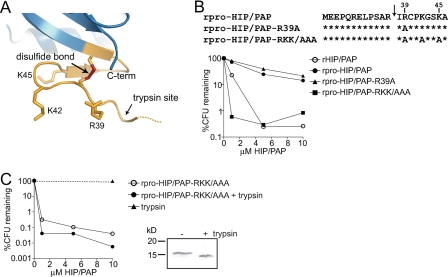
HIP/PAP Basic Residues Are Essential for Prosegment Inhibition of Antibacterial Activity—These findings suggested that the HIP/PAP N-terminal peptide may inhibit antibacterial activity through dynamic interactions with other residues in the HIP/PAP core protein. Given that acidic amino acids in the N terminus are essential for prosegment inhibitory activity, we hypothesized that interactions with nearby HIP/PAP basic residues may be required for repression of antibacterial activity. Because the prosegment is only 11 amino acids in length, we reasoned that it was likely to be constrained to local interactions with amino acids near the trypsin site. Examination of the HIP/PAP primary sequence revealed three basic residues (Arg39, Lys42, Lys45) positioned immediately C-terminal to the Arg37–Ile38 cleavage site (Figs. 1B and 6A). While introduction of a single mutation (R39A) was insufficient to activate antimicrobial activity in rpro-HIP/PAP, mutation of all three basic amino acids to alanine (rpro-HIP/PAP-RKK/AAA) yielded a prosegment-containing variant that exhibited potent antibacterial activity, producing a >99% decline in L. monocytogenes viability (Fig. 6B). Critically, the presence of these mutations had no effect on the antimicrobial activity of the mature protein lacking the prosegment (Fig. 6C). These data demonstrate that basic HIP/PAP residues, positioned C-terminal to the trypsin site, are essential for prosegment inhibitory activity but are dispensable for antibacterial function.
FIGURE 6.
HIP/PAP basic residues are essential for prosegment inhibition of antibacterial activity. A, orientation of Arg and Lys side chains near the HIP/PAP N-terminal trypsin site. The location of the trypsin cleavage site is indicated by an arrow. B, mutations of basic HIP/PAP residues yield active rpro-HIP/PAP. Comparison of antibacterial activity among rpro-HIP/PAP, rHIP/PAP, and rpro-HIP/PAP harboring the indicated mutations in basic residues. Antibacterial assays were performed as in Fig. 2. C, HIP/PAP basic residues (Arg39, Lys42, Lys45) are dispensable for antibacterial activity. Limited trypsin proteolysis was performed on rpro-HIP/PAP-RKK/AAA. SDS-PAGE analysis of the undigested and digested proteins is depicted. Digested and undigested proteins were analyzed for bactericidal activity as outlined in Fig. 2.
Structural Effects of Activating Pro-HIP/PAP Mutations—Overall, these results suggested a model in which the prosegment maintains HIP/PAP in an inactive state through transient interactions between acidic N-terminal residues and basic residues positioned C-terminal to the trypsin site. Mutation of these charged residues derepresses HIP/PAP antibacterial activity in the prosegment-containing protein, thus mimicking the derepression that occurs when the peptide is removed by proteolysis. To further examine this interaction we compared 15N/1H heteronuclear single quantum coherence (HSQC) spectra of rpro-HIP/PAP glutamic acid-to-alanine mutants with the wild-type protein (supplemental Fig. S2). We observed significant chemical shift changes (Δδ > 0.05 ppm) only in residues N-terminal to and including Ile38 (Fig. 7A). This verified that the gain-of-function phenotype produced by the EEE/AAA and RKK/AAA mutations did not result from gross protein misfolding. Furthermore, the lack of significant chemical shift changes in the main body of the protein argues against prosegment inhibitory activity being related to global changes in the core HIP/PAP structure. Notably, 15N/1H HSQC spectra of HIP/PAP with and without the wild-type prosegment showed no significant chemical shift changes aside from those at the trypsin cleavage site, confirming that proteolysis itself also does not cause a global conformational change (data not shown). Analysis of peak intensity did not reveal evidence of prosegment-modulated dimer or oligomer formation, even at the high concentration (300 μm) used for NMR spectroscopy (data not shown). However, as these spectra were collected in the absence of ligand, the possibility remains that multimer formation may occur in the presence of peptidoglycan ligands.
FIGURE 7.
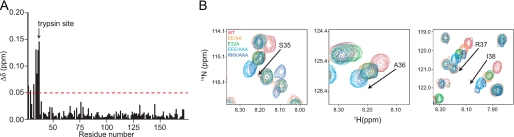
Structural effects of activating pro-HIP/PAP mutations. A, chemical shift changes in the 15N/1H HSQC spectra of rpro-HIP/PAP and the activated mutant rpro-HIP/PAP-EEE/AAA are plotted as a function of residue number. Chemical shift changes >0.05 ppm are indicated with the red line. The trypsin cleavage site is indicated. B, superimposed 15N/1H HSQC spectra of 15N-labeled rpro-HIP/PAP and activating rpro-HIP/PAP mutations reveal colinear chemical shift perturbations among four residues surrounding the trypsin cleavage site at Arg37/Ser38. Arrows indicate the direction of larger chemical shift changes from wild-type and progression toward enhanced HIP/PAP killing activity.
We noted a striking co-linear pattern of change in key N-terminal backbone amide chemical shifts when comparing the 15N/1H HSQC spectra of rpro-HIP/PAP and single, double or triple glutamic acid-to-alanine mutants. Peaks from Ser35, Ala36, Arg37, and Ile38 shifted in the same direction in each of the rpro-HIP/PAP mutants (Fig. 7B), with the triple EEE/AAA mutant exhibiting chemical shift changes that are close to the additive sum of those observed for the single E32A and double E27A/E28A mutants (Table 1) (18). Such a co-linear chemical shift pattern signifies a molecule that is in conformational equilibrium between two states, with intermediate chemical shifts deriving from a population-weighted average of these states (16). Furthermore, the additivity of these chemical shift changes indicates a simple, non-cooperative interaction between the charges on the prosegment and the rest of the HIP/PAP protein. Notably, similar chemical shift changes for the Ser35–Ile38 segment were observed in the activated rpro-HIP/PAP-RKK/AAA mutant, suggesting that the same conformational change is being triggered from the “opposite” side of the putative charge/charge interaction (Fig. 7B). Close inspection of the 15N/1H HSQC spectrum for this mutant shows that peaks for residues Ser35–Ile38 are doubled on this vector, indicative of further conformational exchange occurring on a slow time scale. Collectively, these data support a model in which perturbation of the interaction between HIP/PAP and its prosegment drives a shift in the conformational equilibrium toward a biologically active state (Fig. 8). Our data further suggest that the degree to which the prosegment shifts the HIP/PAP population to the inactive state is simply determined by the number of intramolecular charge-charge interactions rather than any specific ordering of the prosegment against the core HIP/PAP structure.
TABLE 1.
Simple additivity of chemical shift differences in HIP/PAP E/A mutants
| S35 | A36 | R37 | I38 | |
|---|---|---|---|---|
| ppm | ppm | ppm | ppm | |
| E/Aa (E32A) | 0.565 | 0.265 | 0.543 | 0.399 |
| EE/AAa (E27A/E28A) | 0.542 | 0.268 | 0.318 | 0.510 |
| EEE/AAAa (E27A/E28A/E32A) | 1.017 | 0.511 | 0.859 | 0.872 |
| Calculated EEE/AAAb (E/A + EE/AA) | 1.107 | 0.533 | 0.861 | 0.909 |
Chemical shift differences (ΔδTOT) were obtained using the following equation: ΔδTOT = [Δδ1H)2 + (χ × Δδ15N)2]1/2 (normalized for proton with the scale factor χ = 0.17, established from estimates of atom-specific chemical shift ranges in a protein environment) (14).
Calculated chemical shift differences were determined by ΔδTOT = ΔδE/A + ΔδEE/AA.
FIGURE 8.
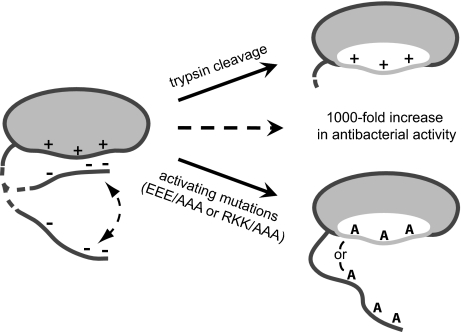
Model of HIP/PAP prosegment inhibition of antibacterial activity. The HIP/PAP hinge region (dashed line) undergoes a two-state conformational shift between a closed (inhibited) and an open (active) form. Maintenance of the closed, inactive state depends on transient interactions between negatively charged prosegment residues and positively charged residues on the HIP/PAP core protein. Inhibition is relieved either by cleavage at a conserved trypsin site in the hinge region, or by mutation of either charged region, leading to a ∼1000-fold increase in antibacterial activity. Note that the hinge region in the activating mutants is in a constitutively open form, as demonstrated by NMR. We propose that disruption of prosegment-core protein interactions alleviates inhibition by unmasking a region (shown in white) necessary for bacterial cell surface damage or for multimerization that may be required for bactericidal activity.
DISCUSSION
The RegIII lectins constitute a vital component of the intestinal epithelial antibacterial arsenal which protect the host against pathogens (19) and maintain homeostasis with symbiotic bacteria (5). However, little is known about how RegIII antibacterial activity is regulated. In this report we show that RegIII lectins are subject to repression by an inhibitory N-terminal prosegment that is removed in vivo by trypsin. This represents a novel regulatory mechanism governing C-type lectin biological activity. The inhibitory activity of the N-terminal segment depends on charge-charge interactions with the main body of the protein. Derepression of antibacterial activity occurs when these interactions are perturbed, either through proteolytic removal of the prosegment or by mutation of the charged residues (Fig. 8).
This regulatory mechanism may have evolved to give the host control over the timing and location of the expression of RegIII antibacterial activity. As RegIII lectin antibacterial activity involves damage to microbial cell surfaces (5), it is possible that these proteins could also damage host cell membranes. This would suggest a need to maintain RegIII lectins in an inactive state prior to their luminal release. Two trypsin isozymes are expressed in human gut epithelial cells and are stored as inactive zymogens that are activated by proteolysis in the intestinal lumen (9). It thus seems likely that trypsin-mediated activation of RegIII proteins occurs only after they are secreted. Given the fact that we were able to detect processed RegIIIγ and HIP/PAP in small intestinal tissue extracts, we cannot completely rule out the possibility of some intracellular cleavage. However, it is likely that this reflects extracellular processing of proteins that are trapped in the mucus layer that overlies the intestinal epithelium. Thus, we propose that this regulatory mechanism evolved to ensure that lectin bactericidal activity is inhibited during intracellular storage, and activated upon secretion into the gut lumen.
In the small intestine, members of the α-defensin family of antimicrobial peptides also require activation through proteolytic removal of an N-terminal prosegment by specific intestinal proteases (2, 9). While trypsin is the processing enzyme for at least one member of the human α-defensin family (9), mouse α-defensins undergo processing by matrix metalloproteinase-7 (matrilysin; MMP-7) (2). Our findings demonstrate that trypsin coordinately activates multiple innate immune effector proteins in both mice and humans, suggesting that this protease may function as a central regulator of diverse intestinal antibacterial responses in multiple host species.
Our results indicate that HIP/PAP prosegment inhibitory activity is governed by interactions between acidic prosegment residues and cationic residues on the remainder of the protein. The prosegments of key intestinal α-defensins confer inhibitory activity through a similar charge-charge interaction (20). However, our results suggest that the functional outcome of this interaction is quite different in the two protein families. The α-defensin prosegment neutralizes cationic amino acids that bind to negatively charged bacterial phospholipids and are thus essential for antibacterial function (21). In contrast, while HIP/PAP cationic amino acids are required for peptide inhibitory activity, they are dispensable for antibacterial function, indicating that the HIP/PAP prosegment represses antibacterial activity through a distinct mechanism. This is consistent with our finding that RegIIIγ and HIP/PAP bind to bacterial surfaces through specific recognition of peptidoglycan, and that this binding interaction is not modulated by the prosegment. Collectively, these observations suggest that the RegIII lectins may mediate their antibacterial functions via mechanisms that differ from the α-defensins.
NMR spectroscopy has provided structural insight into the HIP/PAP-prosegment interaction. Our analysis revealed a distinctive colinear correspondence between backbone amide shifts as point mutations were made to either side of a putative charge-charge interaction pair. Notably, point mutants with derepressed killing activity demonstrated the greatest degree of chemical shift changes. Such correspondence between structural and functional changes is distinctive, and has previously been observed in several other systems (22–24). The linearity of this behavior further suggests that intramolecular interactions between HIP/PAP and its N-terminal prosegment govern a shift between two states with differing activities. Although the physical basis for this shift is not yet clear, we envision at least three possibilities. One is that disruption of HIP/PAP-prosegment interactions results in allosteric changes in residues that are physically distant from the N terminus. This seems very unlikely given the limited chemical shift changes observed outside the N-terminal region in comparisons of the wildtype and EEE/AAA 15N/1H HSQC spectra. A second possibility is that the activating mutations increase accessibility of the trypsin site to cleavage. However, the lack of HIP/PAP proteolysis in our in vitro antibacterial assays suggests that this is unlikely to explain the enhanced bactericidal activity of the activated pro-HIP/PAP mutants (supplemental Fig. S3). A third possibility is that disruption of the HIP/PAP-prosegment interaction unmasks surfaces on the HIP/PAP core structure that are required for bactericidal activity, and that our chemical shift changes reflect a hinging motion of the prosegment away from the rest of the protein. Such movement could allow the HIP/PAP molecule to adopt an optimal conformation to damage microbial cell surfaces, or could induce formation of multimers that may be required for bactericidal activity (Fig. 8). The latter of these two mechanisms has been observed in other systems in which bactericidal activity requires formation of protein oligomers (25–27). Although our NMR spectra did not show evidence of HIP/PAP multimerization at high concentration (300 μm) in the absence of ligand, it is possible that such association requires both the presence of peptidoglycan ligands and the disruption of HIP/PAP-prosegment interactions.
In summary, we have uncovered a novel mechanism controlling C-type lectin biological activity. We propose that impaired regulation of RegIII antibacterial activity in human populations could result in compromised intestinal immunity and a predisposition to enteric infections and inflammation. Furthermore, the structural insights from these studies could aid in the design of novel antimicrobial therapeutics.
Supplementary Material
Author's Choice—Final version full access.
This work was supported, in whole or in part, by National Institutes of Health Grants DK070855 (to L. V. H.), GM081875 (to K. H. G.), and CA130441 (to C. L. P.). This work was also supported by the Burroughs Wellcome Foundation (New Investigators in the Pathogenesis of Infectious Diseases Award (to L. V. H.), and the Robert A. Welch Foundation (I-1659, to L. V. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: HIP/PAP, hepatointestinal pancreatic/pancreatitis-associated protein; MES, 4-morpholineethanesulfonic acid; MMP, matrix metalloproteinase.
References
- 1.Mukherjee, S., Vaishnava, S., and Hooper, L. V. (2008) Cell Mol. Life Sci. 65 3019–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson, C. L., Ouellette, A. J., Satchell, D. P., Ayabe, T., Lopez-Boado, Y. S., Stratman, J. L., Hultgren, S. J., Matrisian, L. M., and Parks, W. C. (1999) Science 286 113–117 [DOI] [PubMed] [Google Scholar]
- 3.Salzman, N. H., Ghosh, D., Huttner, K. M., Paterson, Y., and Bevins, C. L. (2003) Nature 422 522–526 [DOI] [PubMed] [Google Scholar]
- 4.Salzman, N. H., Underwood, M. A., and Bevins, C. L. (2007) Semin. Immunol. 19 70–83 [DOI] [PubMed] [Google Scholar]
- 5.Cash, H. L., Whitham, C. V., Behrendt, C. L., and Hooper, L. V. (2006) Science 313 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christa, L., Carnot, F., Simon, M. T., Levavasseur, F., Stinnakre, M. G., Lasserre, C., Thepot, D., Clement, B., Devinoy, E., and Brechot, C. (1996) Am. J. Physiol. 271 G993–G1002 [DOI] [PubMed] [Google Scholar]
- 7.Ogawa, H., Fukushima, K., Naito, H., Funayama, Y., Unno, M., Takahashi, K., Kitayama, T., Matsuno, S., Ohtani, H., Takasawa, S., Okamoto, H., and Sasaki, I. (2003) Inflamm. Bowel Dis. 9 162–170 [DOI] [PubMed] [Google Scholar]
- 8.Cash, H. L., Whitham, C. V., and Hooper, L. V. (2006) Protein Expr. Purif. 48 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh, D., Porter, E., Shen, B., Lee, S. K., Wilk, D., Drazba, J., Yadav, S. P., Crabb, J. W., Ganz, T., and Bevins, C. L. (2002) Nat. Immunol. 3 583–590 [DOI] [PubMed] [Google Scholar]
- 10.Sattler, M., Schleucher, J., and Griesinger, C. (1999) Prog. NMR Spectrosc. 34 93–158 [Google Scholar]
- 11.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277–293 [DOI] [PubMed] [Google Scholar]
- 12.Johnson, B. A. (2004) Methods Mol. Biol. 278 313–352 [DOI] [PubMed] [Google Scholar]
- 13.Graf, R., Schiesser, M., Scheele, G. A., Marquardt, K., Frick, T. W., Ammann, R. W., and Bimmler, D. (2001) J. Biol. Chem. 276 21028–21038 [DOI] [PubMed] [Google Scholar]
- 14.Ogawa, H., Fukushima, K., Sasaki, I., and Matsuno, S. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 279 G492–499 [DOI] [PubMed] [Google Scholar]
- 15.Abergel, C., Chenivesse, S., Stinnakre, M. G., Guasco, S., Brechot, C., Claverie, J. M., Devinoy, E., and Christa, L. (1999) Acta Crystallogr. D Biol. Crystallogr. 55 1487–1489 [DOI] [PubMed] [Google Scholar]
- 16.Cavanagh, J., Fairbrother, W. J., Palmer, A. G., Rance, M., and Skelton, N. J. (2006) Protein NMR Spectroscopy: Principles and Practice, Academic Press, San Diego, CA
- 17.Cornilescu, G., Delaglio, F., and Bax, A. (1999) J. Biomol. NMR 13 289–302 [DOI] [PubMed] [Google Scholar]
- 18.Farmer, B. T., 2nd, Constantine, K. L., Goldfarb, V., Friedrichs, M. S., Wittekind, M., Yanchunas, J., Jr., Robertson, J. G., and Mueller, L. (1996) Nat. Struct. Biol. 3 995–997 [DOI] [PubMed] [Google Scholar]
- 19.Brandl, K., Plitas, G., Schnabl, B., Dematteo, R. P., and Pamer, E. G. (2007) J. Exp. Med. 204 1891–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weeks, C. S., Tanabe, H., Cummings, J. E., Crampton, S. P., Sheynis, T., Jelinek, R., Vanderlick, T. K., Cocco, M. J., and Ouellette, A. J. (2006) J. Biol. Chem. 281 28932–28942 [DOI] [PubMed] [Google Scholar]
- 21.Tanabe, H., Qu, X., Weeks, C. S., Cummings, J. E., Kolusheva, S., Walsh, K. B., Jelinek, R., Vanderlick, T. K., Selsted, M. E., and Ouellette, A. J. (2004) J. Biol. Chem. 279 11976–11983 [DOI] [PubMed] [Google Scholar]
- 22.Pufall, M. A., Lee, G. M., Nelson, M. L., Kang, H. S., Velyvis, A., Kay, L. E., McIntosh, L. P., and Graves, B. J. (2005) Science 309 142–145 [DOI] [PubMed] [Google Scholar]
- 23.Volkman, B. F., Lipson, D., Wemmer, D. E., and Kern, D. (2001) Science 291 2429–2433 [DOI] [PubMed] [Google Scholar]
- 24.Li, P., Martins, I. R., Amarasinghe, G. K., and Rosen, M. K. (2008) Nat. Struct. Mol. Biol. 15 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoover, D. M., Rajashankar, K. R., Blumenthal, R., Puri, A., Oppenheim, J. J., Chertov, O., and Lubkowski, J. (2000) J. Biol. Chem. 275 32911–32918 [DOI] [PubMed] [Google Scholar]
- 26.Clayberger, C., and Krensky, A. M. (2003) Curr. Opin. Immunol. 15 560–565 [DOI] [PubMed] [Google Scholar]
- 27.Hadders, M. A., Beringer, D. X., and Gros, P. (2007) Science 317 1552–1554 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.