Abstract
The Escherichia coli DNA damage-inducible protein DinG, a member of the superfamily 2 DNA helicases, has been implicated in the nucleotide excision repair and recombinational DNA repair pathways. Combining UV-visible absorption, EPR, and enzyme activity measurements, we demonstrate here that E. coli DinG contains a redox-active [4Fe-4S] cluster with a midpoint redox potential (Em) of –390 ± 23 mV (pH 8.0) and that reduction of the [4Fe-4S] cluster reversibly switches off the DinG helicase activity. Unlike the [4Fe-4S] cluster in E. coli dihydroxyacid dehydratase, the DinG [4Fe-4S] cluster is stable, and the enzyme remains fully active after exposure to 100-fold excess of hydrogen peroxide, indicating that DinG could be functional under oxidative stress conditions. However, the DinG [4Fe-4S] cluster can be efficiently modified by nitric oxide (NO), forming the DinG-bound dinitrosyl iron complex with the concomitant inactivation of helicase activity in vitro and in vivo. Reassembly of the [4Fe-4S] cluster in NO-modified DinG restores helicase activity, indicating that the iron-sulfur cluster in DinG is the primary target of NO cytotoxicity. The results led us to propose that the iron-sulfur cluster in DinG may act as a sensor of intracellular redox potential to modulate its helicase activity and that modification of the iron-sulfur cluster in DinG and likely in other DNA repair enzymes by NO may contribute to NO-mediated genomic instability.
The DNA damage-inducible protein DinG was initially identified from genetic screening in response to DNA-damaging agents in Escherichia coli (1–3). The sequence analysis predicted that E. coli DinG is a member of the superfamily 2 DNA helicases (4). The DNA helicase activity of E. coli DinG has since been demonstrated (5). Recent studies further showed that DinG can also unwind DNA-RNA duplexes, D-loops and R-loops, suggesting that DinG may have an important role in recombinational DNA repair and resumption of replication following DNA damage (6). Nevertheless, deletion of the gene dinG has only a mild effect on E. coli cell viability and sensitivity to UV radiation (5), likely because of redundant helicase activities in cells.
E. coli DinG is closely related to two human DNA helicases, XPD and BACH1 (7–12). XPD is a member of both the transcription initiation complex TFIIH of RNA polymerase II and the nucleotide excision repair pathway (7, 12). Inherited mutations in XPD have been linked to at least three human diseases: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy (7). BACH1 has been shown to physically interact with the BRCT motifs of BRCA1 (breast cancer 1 protein) (13). Inherited mutations in BACH1 have been implicated in deficiency of the cross-link repair pathway in Fanconi anemia patients (14). Surprisingly, recent studies have revealed that XPD homologs from Sulfolobus acidocaldarius (15) and Ferroplasma acidarmanus (16) contain an iron-sulfur cluster located between the Walker A and B motifs in the N terminus of the protein and that the iron-sulfur cluster is essential for helicase activity (15, 16). Mutations that affect iron-sulfur cluster binding or stability in XPD abolish helicase activity (15). X-ray crystallographic studies further revealed that the [4Fe-4S] cluster is located in the vicinity of the DNA-binding site of XPD (9–11). Although iron-sulfur clusters have been discovered in a large number of proteins that have interactions with DNA or RNA (17–29), the specific functions of the iron-sulfur clusters in these proteins mostly remain elusive.
E. coli DinG has ∼48% identity with human XPD in the regions of the helicase motif (5). Although DinG does not possess the corresponding conserved cysteine residues (Cys-92, Cys-113, Cys-128, and Cys-164, Thermoplasma acidophilum numbering) in XPD that host the [4Fe-4S] cluster (9–11), sequence analysis of a subset of the DinG homologs from diverse bacterial species revealed four conserved cysteine residues (Cys-120, Cys-194, Cys-199, and Cys-205, E. coli numbering) that may provide ligands for a putative iron-sulfur cluster (6, 15). In this study, we report that purified E. coli DinG contains a redox-active [4Fe-4S] cluster with a midpoint redox potential (Em) of –390 ± 23 mV (pH 8.0) and that reduction of the [4Fe-4S] cluster in DinG reversibly switches off helicase activity. Importantly, unlike the E. coli dihydroxyacid dehydratase [4Fe-4S] cluster (30), the DinG [4Fe-4S] cluster is stable, and the enzyme remains fully active after exposure to 100-fold excess of hydrogen peroxide, indicating that DinG could be functional under oxidative stress conditions. In contrast, nitric oxide (NO), a physiological free radical produced in activated macrophages and other mammalian cells (31–34), can efficiently modify the DinG [4Fe-4S] cluster, forming the DinG-bound dinitrosyl iron complex (DNIC)2 with the concomitant inactivation of helicase activity in vitro and in vivo. Combined with the previous results that NO can modify the DNA repair enzyme endonuclease III [4Fe-4S] cluster and inactivate the enzyme activity (35), we propose that modification of the iron-sulfur clusters in XPD/DinG, endonuclease III, and possibly other DNA repair enzymes by NO may contribute to the NO-mediated initiation of the carcinogenic process and genomic instability in mammalian cells (36, 37).
EXPERIMENTAL PROCEDURES
Protein Preparation—The DNA fragment encoding the DNA damage-inducible protein DinG was amplified from E. coli genomic DNA using PCR. Two primers, DinG-1 (5′-GGTTTTCCCATGGCATTAACCGCC-3′) and DinG-2 (5′-CATCATTAAAGCTTCCGACGGCGT-3′), were used for PCR amplification. The PCR product was digested with HindIII and NcoI and ligated into expression vector pET28b+ to produce pTDinG. The cloned DNA fragment was confirmed by direct sequencing using the T7 primer (Genomic Facility, Louisiana State University). Recombinant DinG was overproduced in E. coli BL21 strain in Terrific broth and purified using a nickel-agarose column, followed by a HiTrap desalting column. The purity of purified DinG was >95% judging from the SDS-PAGE analysis followed by Coomassie blue staining. The precise molecular weight of recombinant DinG was confirmed using electrospray ionization-mass spectrometry (Chemistry Department, Louisiana State University). The protein concentration of purified DinG was measured from the absorption peak at 280 nm using an extinction coefficient of 79.0 mm–1 cm–1. The total iron content in protein samples was determined using an iron indicator, FerroZine (38). The total sulfide content in protein samples was determined according to the method of Siegel (39) as described previously (40). Site-directed mutagenesis was carried out using the QuikChange kit from Stratagene. Mutations in the gene dinG were confirmed by direct sequencing (Genomic Facility, Louisiana State University). The DinG mutant proteins were expressed and purified following the same purification procedures as described for wild-type DinG. Recombinant dihydroxyacid dehydratase (IlvD) (30) from E. coli was prepared as described (41). The specific enzyme activity of dihydroxyacid dehydratase was measured using the substrate dl-2,3-dihydroxyisovalerate, and the reaction product (keto acids) was monitored at 240 nm using an extinction coefficient of 0.19 mm–1 cm–1 (30). dl-2,3-dihydroxyisovalerate was synthesized according to the method of Cioffi et al. (42).
Helicase Activity Assay—The helicase activity of DinG was measured following the procedure described by Voloshin et al. (5) with slight modifications. Briefly, an oligonucleotide (5′-CCGTAACACTGAGTTTCGTCACCAGTACAAACTACAACGCCTGTAGCATTCCACA-3′) was labeled with [γ-32P]ATP using polynucleotide kinase (New England Biolabs). The 32P-labeled oligonucleotide (5 μm) was annealed to M13mp18 single-stranded DNA (New England Biolabs) in annealing buffer containing Tris (50 mm, pH 7.5), NaCl (50 mm), and MgCl2 (10 mm). The DNA solution was heated at 85 °C for 5 min and cooled to room temperature over 3 h. The annealed DNA duplex was purified using a gel filtration spin column (CHROMA SPIN-400, Clontech) pre-equilibrated with annealing buffer. The annealed substrate (at a final concentration of 2 nm) was incubated with DinG in 20 μl of reaction solution containing Tris (50 mm, pH 8.0), NaCl (100 mm), MgCl2 (5 mm), dithiothreitol (2 mm), glycerol (5%), and ATP (2 mm) at 30 °C for 10 min. For each experiment, two controls in which the substrate was either denatured by heating at 85 °C for 5 min or incubated at 30 °C for 10 min without any enzymes were included. The reactions were terminated by adding 4 μl of stop solution (containing 6% SDS, 60 mm EDTA, and 0.3% bromphenol blue). The reaction products (single-stranded DNA) were separated on 1% Tris acetate/EDTA-agarose gel, transferred to Nytran transfer membranes (0.45 μm; What-man), and exposed to x-ray films overnight for quantification.
Redox Titration of the DinG Iron-Sulfur Cluster—A specially designed anaerobic cuvette was used for redox titrations as described by Dutton (43). Before titration, solution containing DinG (20 μm) and safranin O (1 μm) was equilibrated with ultrapure argon gas for 50 min at room temperature. During titration, argon flow was maintained with gentle stirring using a small magnet on the bottom of the cuvette. The redox potential of the solution was adjusted by adding a small amount of freshly prepared sodium dithionite using a 10-μl gas-tight microsyringe (Hamilton, Reno, NV). The redox potential was monitored directly with a redox microelectrode (Microelectrodes Inc., Bedford, NH). Freshly prepared ZoBell solution containing potassium ferricyanide (5 mm) and potassium ferricyanide (5 mm) dissolved in Tris buffer (20 mm, pH 8.0) and NaCl (500 mm) was used as a standard (Eh = 238 mV) for calibration of the redox microelectrode.
NO Exposure of DinG and Reassembly of the Iron-Sulfur Cluster in the Protein—Purified DinG (30 μm) was incubated with the NO-releasing reagent diethylamine NONOate (0–0.5 mm; Cayman Chemicals) in buffer containing Tris (20 mm, pH 7.5) and NaCl (200 mm) anaerobically at room temperature. Diethylamine NONOate releases 1.5 eq of NO/mol of parent compound with a half-life time of 16 min at room temperature and pH 7.5. After a 20-min incubation, the protein was repurified by passage through a HiTrap desalting column to remove residual diethylamine NONOate. For in vivo NO exposure, E. coli cells containing recombinant DinG were subjected to the Silastic tubing NO delivery system (44) as described previously (41). The length of the Silastic tubing (0.025 (inner diameter) × 0.047 (outer diameter) inches) immersed in the cell culture was adjusted to such that ∼100 nm NO/s was released to the cell culture in a sealed flask under anaerobic conditions. The chosen NO release rate was comparable with reported NO production in activated polymorphonuclear leukocytes (34) or in RAW 264.7 macrophages co-cultured with arginase-deficient Helicobacter pylori (32). After the E. coli cells were subjected to NO exposure for 0, 1, 2, 4, and 10 min anaerobically, recombinant DinG was purified from cells following the procedures described above.
For reassembly of iron-sulfur clusters, NO-exposed DinG (10 μm) was incubated with freshly prepared Fe(NH4)2(SO4)2 (80 μm), l-cysteine (0.5 mm), and cysteine desulfurase IscS (1 μm) (45) in the presence of dithiothreitol (2 mm) anaerobically at 37 °C for 20 min as described (45), followed by repurification of DinG from the incubation solution. Repurified DinG was then subjected to EPR and helicase activity measurements.
EPR Measurements—The EPR spectra were recorded at X-band on a Bruker ESR-300 spectrometer using an Oxford Instruments ESR-9 flow cryostat. The routine EPR conditions were as follows: microwave frequency, 9.45 GHz; microwave power, 10 milliwatts; modulation frequency, 100 kHz; modulation amplitude, 2.0 milliteslas; sample temperature, 20 K; and receive gain, 1.0 × 105.
RESULTS
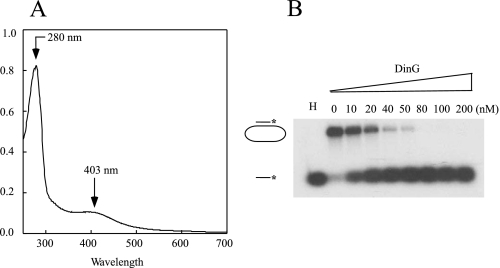
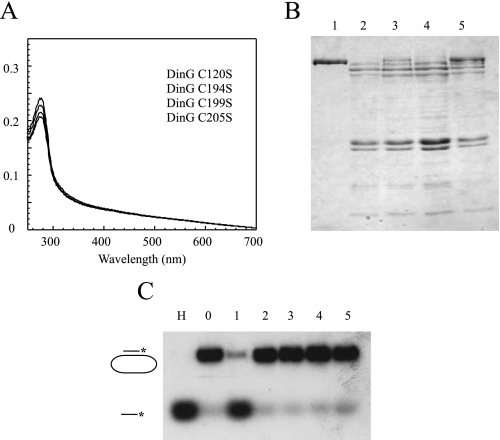
E. coli DinG Contains an Iron-Sulfur Cluster Essential for Protein Stability and Helicase Activity—Fig. 1A shows the UV-visible absorption spectrum of purified E. coli DinG. The major peak at ∼403 nm represents a typical absorption of iron-sulfur clusters in proteins. The overall spectrum of purified DinG was similar to that of the purified XPD homolog [4Fe-4S] cluster from S. acidocaldarius (15) and the endonuclease III [4Fe-4S] cluster from E. coli (35). The total iron and sulfur content analyses of purified DinG showed that each DinG monomer contained ∼3.1 ± 0.5 iron and 2.8 ± 0.8 sulfide (n = 3), indicating that DinG contains one [4Fe-4S] cluster per monomer. Purified DinG was further analyzed for its DNA helicase activity. Following the procedures described by Voloshin et al. (5), we demonstrated that as-purified DinG was able to unwind double-stranded DNA in an ATP-dependent reaction (Fig. 1B). E. coli DinG has 11 cysteine residues; four of them (Cys-120, Cys-194, Cys-199, and Cys-205) are conserved among a subset of DinG proteins from diverse bacteria (6, 15). To test whether the conserved cysteine residues are required for iron-sulfur cluster binding, we substituted each of these four cysteine residues in DinG with serine using site-directed mutagenesis as described under “Experimental Procedures.” All four mutant proteins (C120S, C194S, C199S, and C205S), purified using the same procedure as wild-type DinG, had no absorption peak at 403 nm (Fig. 2A). The SDS-PAGE analyses revealed that unlike wild-type DinG, the DinG mutants expressed in E. coli cells were largely degraded (Fig. 2B) and had no detectable helicase activity (Fig. 2C). Thus, these conserved cysteine residues appear to be essential for the iron-sulfur cluster binding in DinG and protein stability.
FIGURE 1.
Purified E. coli DinG contains an iron-sulfur cluster. A, UV-visible absorption spectrum of purified E. coli DinG. The protein concentration was ∼10 μm. B, the helicase activity of purified E. coli DinG. Purified DinG (at a final concentration of 0–200 nm) was incubated with the 32P-radioactively labeled substrate in the presence of ATP (2 mm) at 30 °C for 10 min. The reaction product (single-stranded DNA) was separated by agarose gel (1%) electrophoresis as described under “Experimental Procedures.” In lane H, the sample was heated at 85 °C for 5 min. The concentration of DinG in the reaction solution is indicated at the top of each lane.
FIGURE 2.
The conserved cysteine residues in DinG are required for iron-sulfur cluster binding, protein stability, and helicase activity. The DinG mutants (C120S, C194S, C199S, and C205S) were constructed and purified as described under “Experimental Procedures.” A, UV-visible absorption spectra of purified DinG mutants. The protein concentrations were ∼3 μm. B, SDS-PAGE analysis of purified wild-type DinG and DinG mutants. The same amount of cells containing either wild-type DinG or the DinG mutants was used for protein purification. Equal amounts of purified proteins were analyzed on the SDS-polyacrylamide gel. Lanes 1–5, wild-type DinG and mutants C120S, C194S, C199S, and C205S, respectively. C, the helicase activity of purified wild-type DinG and the DinG mutants. Purified proteins (200 nm) were incubated with the 32P-radioactively labeled substrate in the presence of ATP (2 mm) at 30 °C for 10 min. The reaction product (single-stranded DNA) was separated by agarose gel (1%) electrophoresis as described under “Experimental Procedures.” Lane H, sample heated at 85 °C for 5 min; lane 0, no enzyme added; lanes 1–5, wild-type DinG and mutants C120S, C194S, C199S, and C205S, respectively.
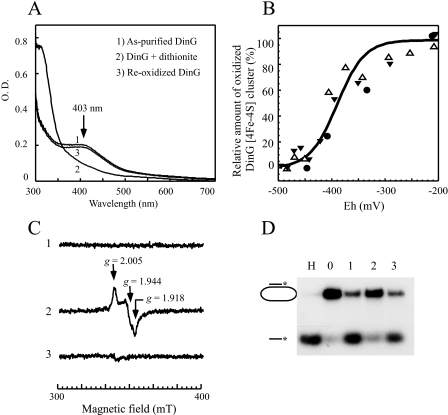
Redox State of the Iron-Sulfur Cluster in DinG Controls Helicase Activity—Although the iron-sulfur cluster in wild-type DinG was stable under aerobic conditions, addition of sodium dithionite quickly bleached the absorption peak at 403 nm (Fig. 3A). Nevertheless, when reduced DinG was reoxidized by exposure to air or by the oxidant potassium ferricyanide, the absorption peak at 403 nm of DinG reappeared (Fig. 3A), suggesting that the iron-sulfur cluster in DinG can be reversibly reduced by sodium dithionite.
FIGURE 3.
Redox state of the iron-sulfur cluster in DinG regulates its helicase activity. A, UV-visible absorption spectra of reduced and oxidized DinG. Purified DinG (25 μm; spectrum 1) was reduced with sodium dithionite (200 μm) under anaerobic conditions (spectrum 2) and then reoxidized by exposure to air for 30 min (spectrum 3). The absorption peak at 403 nm reflects the oxidized DinG [4Fe-4S] cluster. B, redox titration of the DinG [4Fe-4S] cluster. Purified DinG (25 μm) in buffer containing Tris (50 mm, pH 8.0) and NaCl (500 mm) was supplemented with safranin O (1 μm) as a redox mediator. Redox titration was performed in an anaerobic redox cuvette as described under “Experimental Procedures.” The x axis shows the redox potentials measured with a redox microelectrode. The y axis shows the relative absorbance at 403 nm, normalized to 0 or 100% for a fully reduced or oxidized DinG [4Fe-4S] cluster, respectively. The solid line drawn through the data points represents the best fit to the Nernst equation (n = 1) with Em of –390 ± 23 mV. Data were from three independent experiments represented with three different symbols. C, EPR spectra of purified DinG. Purified DinG (500 μm) (spectrum 1) was reduced with sodium dithionite (2 mm; spectrum 2) or reoxidized with potassium ferricyanide (2 mm; spectrum 3). D, the helicase activity of DinG under different redox potentials. Purified DinG (100 nm) was either reduced with sodium dithionite or reoxidized with potassium ferricyanide before the 32P-radioactively labeled DNA substrate was added to the incubation solutions. After a 5-min incubation at 30 °C, the reaction was terminated, and the product (single-stranded DNA) was separated by agarose gel (1%) electrophoresis as described under “Experimental Procedures.” Lane H, sample heated at 85 °C for 5 min; lane 0, no enzyme added; lane 1, purified DinG; lane 2, purified DinG reduced with dithionite (1.0 mm); lane 3, dithionite-reduced DinG reoxidized with potassium ferricyanide (2 mm).
The absorption peak at 403 nm was then used to determine the Em of the DinG iron-sulfur cluster. Purified DinG was dissolved in buffer containing the redox mediator safranin O under anaerobic conditions. The redox potential of the solution was adjusted by adding freshly prepared sodium dithionite and directly monitored using a microelectrode as described by Dutton (43). The UV-visible absorption spectra were taken at different redox potentials, and the absorption peak at 403 nm of the DinG [4Fe-4S] cluster was plotted as a function of the poised redox potentials (Fig. 3B). The data from three experiments were fitted to a Nernst equation (n = 1) with a midpoint redox potential of –390 ± 23 mV, a value close to that of the intracellular redox potential in E. coli (17).
EPR spectroscopy was further used to explore the redox state of the DinG iron-sulfur cluster. As shown in Fig. 3C, purified DinG had no EPR signal at around the g = 2.0 region under the experimental conditions (spectrum 1). However, when freshly prepared sodium dithionite was added to purified DinG, a rhombic EPR signal with gx = 1.918, gy = 1.944, and gz = 2.005, indicative of a reduced [4Fe-4S] cluster, appeared (spectrum 2). Spin quantification revealed that there was ∼0.8–0.9 spin per each iron-sulfur cluster in the dithionite-reduced DinG. The observed g-values were comparable with those of the reduced [4Fe-4S]+ cluster observed in other proteins (46). The relatively small Δg could reflect the unique property of the [4Fe-4S] cluster in DinG. The rhombic EPR signal was completely eliminated when dithionite-reduced DinG was reoxidized by exposure to air or by potassium ferricyanide (spectrum 3), confirming that the DinG [4Fe-4S] cluster can be reversibly reduced by sodium dithionite. It is worth mentioning that no EPR signal at g = 2.018 of the [3Fe-4S] cluster (47) was observed when purified DinG was treated with potassium ferricyanide (data not shown), suggesting that the DinG [4Fe-4S] cluster is resistant to oxidation.
Because the [4Fe-4S] cluster in the archaeal XPD homologs is located in the vicinity of the DNA-binding site of the enzyme (9–11), we speculated that the redox state of the iron-sulfur cluster in DinG may modulate helicase activity. To test this idea, we compared the helicase activity of DinG when its iron-sulfur cluster was either reduced or oxidized. Fig. 3D shows that the helicase activity of DinG was greatly diminished when the iron-sulfur cluster was reduced with dithionite and largely restored once the reduced iron-sulfur cluster was reoxidized, demonstrating that the reduction of the iron-sulfur cluster in DinG can reversibly switch off helicase activity at least in vitro.
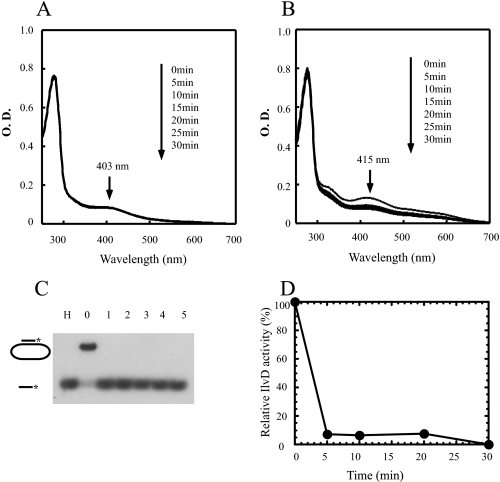
Iron-Sulfur Cluster in DinG Is Resistant to Hydrogen Peroxide—As a DNA damage-inducible protein, it is somewhat surprising that DinG contains a [4Fe-4S] cluster that is presumably susceptible to reactive oxygen species (30). To determine the sensitivity of the DinG [4Fe-4S] cluster to reactive oxygen species, we incubated purified DinG with 100-fold excess of H2O2 at 25 °C for 30 min and found that the absorption peak at 403 nm of the DinG [4Fe-4S] cluster (Fig. 4A) and helicase activity (Fig. 4C) remained essentially unchanged before and after incubation. In contrast, when the purified E. coli dihydroxyacid dehydratase [4Fe-4S] cluster (30) was incubated with 50-fold excess of H2O2 at 25 °C, both the absorption peak at 415 nm of the dihydroxyacid dehydratase [4Fe-4S] cluster (Fig. 4B) and its enzyme activity (Fig. 4D) were abolished as reported previously (30). Thus, unlike the dihydroxyacid dehydratase [4Fe-4S] cluster, the DinG [4Fe-4S] cluster is stable, and its helicase activity remains fully active after exposure to 100-fold excess of hydrogen peroxide.
FIGURE 4.
The iron-sulfur cluster in DinG is resistant to hydrogen peroxide. A, effect of H2O2 on the DinG [4Fe-4S] cluster. Purified DinG (10 μm) was incubated with H2O2 (1 mm) at 25 °C. UV-visible spectra were taken every 5 min after addition of H2O2 for 30 min. B, effect of H2O2 on the dihydroxyacid dehydratase [4Fe-4S] cluster. Purified E. coli dihydroxyacid dehydratase (20 μm) was incubated with hydrogen peroxide (1 mm) at 25 °C. UV-visible spectra were taken every 5 min after addition of H2O2 for 30 min. C, effect of H2O2 on the DinG helicase activity. After incubation with H2O2 for the indicated time, DinG (at a final concentration of 100 nm) was used for the helicase activity assay. Lane H, sample heated at 85 °C for 5 min; lane 0, no enzyme added; lanes 1–5, purified DinG after incubation with H2O2 for 0, 5, 10, 20, and 30 min, respectively. The reaction product (single-stranded DNA) was separated by agarose gel (1%) electrophoresis as described under “Experimental Procedures.” D, effect of H2O2 on the enzyme activity of dihydroxyacid dehydratase. The relative enzyme activity of dihydroxyacid dehydratase after incubation with H2O2 (1 mm) was measured as described under “Experimental Procedures” and plotted as a function of incubation time with H2O2.
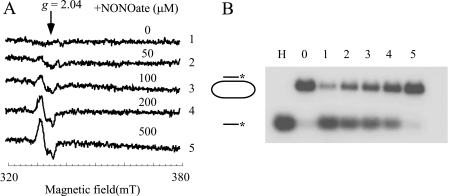
Iron-Sulfur Cluster in DinG Can Be Efficiently Modified by NO—NO is a physiological free radical that acts as a signal molecule (31) as well as a powerful weapon to kill pathogenic bacteria and tumor cells (32–34). Chronic exposure to NO has also been attributed to the initiation of the carcinogenic process and genomic instability (36, 37). Among cellular components, iron-sulfur proteins are considered the primary targets of NO cytotoxicity (41, 48). In vitro and in vivo studies have shown that NO can readily modify iron-sulfur clusters in proteins, forming the protein-bound DNIC (35, 41, 47, 49–52). To test whether the DinG [4Fe-4S] cluster can also be modified by NO, purified DinG was exposed to NO using the NO-releasing reagent diethylamine NONOate under anaerobic conditions. Fig. 5A shows that when purified DinG was incubated with an increasing amount of diethylamine NONOate (0–500 μm), the DinG [4Fe-4S] cluster was gradually modified by NO, forming the DinG-bound DNIC with a typical EPR signal at g = 2.04 as previously reported for other iron-sulfur proteins (35, 47, 49–52). Parallel helicase activity measurements showed that DinG was progressively inactivated by NO exposure (Fig. 5B). Thus, NO can effectively modify the DinG [4Fe-4S] cluster and inactivate helicase activity in vitro.
FIGURE 5.
The DinG iron-sulfur cluster is sensitive to NO. A, modification of the DinG [4Fe-4S] cluster by NO. Purified DinG (30 μm) was incubated with different amounts of diethylamine NONOate in buffer containing Tris (20 mm, pH 7.5) and NaCl (200 mm) at room temperature under anaerobic conditions. After a 20-min incubation, protein was repurified by passage through a HiTrap desalting column. Spectra 1–5, purified DinG incubated with 0, 50, 100, 200, and 500 μm NONOate under anaerobic conditions. The protein concentrations of repurified DinG were ∼4 μm. mT, milliteslas. B, inactivation of DinG helicase activity by NO. After incubation with different amounts of NONOate, repurified DinG (at a final concentration of 100 nm) was used for the helicase activity assay. Lane H, sample heated at 85 °C for 5 min; lane 0, no enzyme added; lanes 1–5, repurified DinG after incubation with 0, 50, 100, 200, and 500 μm NONOate. The reaction product (single-stranded DNA) was separated by agarose gel (1%) electrophoresis as described under “Experimental Procedures.”
To further explore the sensitivity of the DinG [4Fe-4S] cluster to NO in vivo, we exposed E. coli cells containing recombinant DinG to pure NO gas using the Silastic tubing NO delivery system (44) as described previously (41). A releasing rate of 100 nm NO/s was chosen to emulate NO production in activated polymorphonuclear leukocytes (34) or macrophages (32). Recombinant DinG was then purified from E. coli cells after exposure with different amounts of NO. EPR measurements of purified DinG showed that the DinG-bound DNIC was gradually increased with the concomitant inactivation of helicase activity when the E. coli cells were exposed to increasing amounts of NO (data not shown). About 4 min of NO exposure at a rate of 100 nm NO/s was sufficient to completely modify the recombinant DinG [4Fe-4S] cluster and inactivate helicase activity in E. coli cells. These results suggested that the DinG [4Fe-4S] cluster can be efficiently modified with the concomitant inactivation of its helicase activity in E. coli cells by NO.
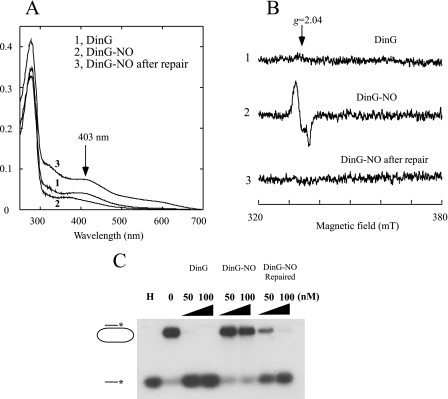
We then attempted to reassemble the iron-sulfur cluster in NO-modified DinG using the iron-sulfur cluster repair system (l-cysteine, cysteine desulfurase IscS, ferrous iron, and dithiothreitol) in vitro as described previously (35). After incubation with the repair system at 37 °C for 30 min, the iron-sulfur cluster was reassembled in NO-modified DinG (Fig. 6A), the DinG-bound DNIC was decomposed (Fig. 6B), and the helicase activity was largely restored (Fig. 6C). Thus, the iron-sulfur cluster in DinG, like other iron-sulfur proteins (41), could be the primary target of NO cytotoxicity.
FIGURE 6.
Reactivation of NO-modified DinG by reassembly of iron-sulfur clusters. Purified DinG (30 μm) was exposed to NO (0.5 mm NONOate) under anaerobic conditions, followed by repair using the iron-sulfur cluster repair system as described under “Experimental Procedures.” A, UV-visible absorption spectra of DinG. Spectrum 1, purified DinG before NO exposure (DinG); spectrum 2, purified DinG after NO exposure (DinG-NO); spectrum 3, NO-exposed DinG repaired with the iron-sulfur cluster repair system (DinG-NO repaired). The protein concentrations of DinG were ∼5 μm. B, EPR spectra of DinG. Spectrum 1, purified DinG before NO exposure; spectrum 2, purified DinG after NO exposure; spectrum 3, NO-exposed DinG repaired with the iron-sulfur cluster assembly system. The protein concentrations of DinG were ∼5 μm. mT, milliteslas. C, reversible inactivation of DinG by NO. Two concentrations of DinG (50 and 100 nm) were used for the helicase activity assay. In lane H, the sample was heated at 85 °C for 5 min. In lane 0, no enzyme was added. The reaction product (single-stranded DNA) was separated by agarose gel (1%) electrophoresis as described under “Experimental Procedures.”
DISCUSSION
In this study, we have reported that the E. coli DNA damage-inducible protein DinG helicase contains a redox-active [4Fe-4S] cluster with Em of approximately –390 ± 23 mV (pH 8.0) and that reduction of the iron-sulfur cluster reversibly switches off the helicase activity of DinG. Although the iron-sulfur cluster in DinG is stable in the presence of oxygen or hydrogen peroxide, it can be efficiently modified by NO, forming the DinG-bound DNIC with the concomitant inactivation of helicase activity in vitro and in vivo. The results led us to propose that DinG helicase activity can be modulated by intracellular redox potential and by NO via its iron-sulfur cluster.
In the past decade, a large number of iron-sulfur proteins that have specific interactions with DNA or RNA have been reported. According to their functions, these iron-sulfur proteins may be divided into two groups. The first group includes the transcription or translation regulators that directly bind to DNA or RNA. Some well characterized examples are the redox transcription factor SoxR [2Fe-2S] cluster (17, 18), the anaerobic growth factor FNR (Fumarate and nitrate reductase regulator) [4Fe-4S] cluster (19), the repressor IscR [2Fe-2S] cluster that regulates iron-sulfur cluster biosynthesis (20), and the IRP-1 (iron regulatory protein-1) [4Fe-4S] cluster that controls the post-translational control of intracellular iron contents in mammalian cells (21). In this group of proteins, iron-sulfur clusters generally act as sensors of specific signals and modulate the subtle interactions between the protein and DNA or RNA. The second group includes the iron-sulfur enzymes that chemically modify RNA or DNA molecules. The ribosomal RNA methyltransferase (the RumA [4Fe-4S] cluster) (22) and the bifunctional radical S-adenosylmethionine enzyme MiaB [4Fe-4S] cluster (23) are two examples of RNA-modifying enzymes. More recently, the p58 subunit of human DNA primase has been shown to contain a [4Fe-4S] cluster (28, 29). The iron-sulfur enzymes that chemically modify DNA are mostly the DNA repair enzymes such as the endonuclease III [4Fe-4S] cluster (24, 25), the MutY [4Fe-4S] cluster (26), the family 4 uracil-DNA glycosylase [4Fe-4S] cluster (27), the DNA helicase XPD [4Fe-4S] clusters (9–11), and the E. coli DinG helicase [4Fe-4S] cluster (Refs. 5 and 6 and this study). Evidently, the function of iron-sulfur clusters in these diverse DNA/RNA-modifying enzymes could not be readily generalized. Here, we have shown that the [4Fe-4S] cluster in DinG is stable in the presence of oxygen and hydrogen peroxide (Fig. 4), a feature that could be important for helicase activity in repairing DNA damage under oxidative stress conditions. More importantly, we have demonstrated that the [4Fe-4S] cluster in DinG is redox-active with a midpoint redox potential of –390 ± 23 mV (pH 8.0) and that reduction of the [4Fe-4S] cluster in DinG reversibly switches off helicase activity (Fig. 3). We postulate that reduction of the [4Fe-4S] cluster in DinG, like that of the redox transcription factor SoxR [2Fe-2S] cluster (17), may modulate the overall structure of the catalytic center and thus inactivate helicase activity. It should be pointed out that DinG homologs in some other bacteria do not have the conserved cysteine residues and therefore no iron-sulfur clusters (9). Whether there are other means to regulate the activity of these DinG helicases remains to be investigated.
The observed Em of the DinG [4Fe-4S] cluster (–390 ± 23 mV at pH 8.0) (Fig. 3) is close to that of the intracellular redox potential in E. coli (17). Several attempts were made to observe the redox state of the recombinant DinG [4Fe-4S] cluster in E. coli cells. Unfortunately, no EPR signal of the reduced DinG [4Fe-4S] cluster was observed in vivo (data not shown), likely because of insufficient amounts of recombinant DinG expressed in E. coli cells. It has been reported that DNA binding shifts the Em of the endonuclease III [4Fe-4S] cluster toward oxidation, converting the redox-inactive endonuclease III [4Fe-4S] cluster to a typical high-potential [4Fe-4S] protein (25). Here, we have found that unlike the endonuclease III [4Fe-4S] cluster, the DinG [4Fe-4S] cluster is redox-active even without any DNA binding (Fig. 3). Whether DNA binding will change the redox property of the DinG [4Fe-4S] cluster remains to be investigated. Nevertheless, on the basis of the results presented in this study, we propose that the DinG [4Fe-4S] cluster may be partially oxidized in E. coli cells under normal growth conditions. When cells are subjected to oxidative stresses, the reduced [4Fe-4S] cluster is oxidized, and DinG becomes fully active to repair the inflicted DNA damage and resume DNA replication.
NO is a physiological free radical involved in signal transduction in neuronal and cardiovascular systems (31). Excessive production of NO in activated macrophages and other mammalian cells can also act as a powerful weapon to kill pathogenic bacteria and tumor cells (32, 33). In some studies, chronic NO exposure has been linked to the carcinogenic process and genomic instability (36, 37). Nevertheless, the etiology of NO cytotoxicity has not been fully understood. Here, we have reported that the DinG [4Fe-4S] cluster can be modified by NO forming the DinG-bound DNIC with the concomitant inactivation of helicase activity in vitro and in vivo. Because genetic defects in the human XPD gene (ERCC2) have been associated with the increase of cancer incidence and aging phenotypes (7, 9, 10, 15), it is plausible that chronic NO exposure may inactivate the iron-sulfur cluster-containing DNA repair enzymes such as DinG/XPD and contribute to the initiation of the carcinogenic process and genomic instability (36, 37).
Acknowledgments
We thank P. Leslie Dutton for inspiring discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA107494 from the United States Public Health Service. This work was also supported by National Science Foundation Grant MCB-0416537 (to H. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DNIC, dinitrosyl iron complex; Em, midpoint redox potential; NONOate, dinitric oxide.
References
- 1.Lewis, L. K., Jenkins, M. E., and Mount, D. W. (1992) J. Bacteriol. 174 3377–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Dyk, T. K., DeRose, E. J., and Gonye, G. E. (2001) J. Bacteriol. 183 5496–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez De Henestrosa, A. R., Ogi, T., Aoyagi, S., Chafin, D., Hayes, J. J., Ohmori, H., and Woodgate, R. (2000) Mol. Microbiol. 35 1560–1572 [DOI] [PubMed] [Google Scholar]
- 4.Koonin, E. V. (1993) Nucleic Acids Res. 21 1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voloshin, O. N., Vanevski, F., Khil, P. P., and Camerini-Otero, R. D. (2003) J. Biol. Chem. 278 28284–28293 [DOI] [PubMed] [Google Scholar]
- 6.Voloshin, O. N., and Camerini-Otero, R. D. (2007) J. Biol. Chem. 282 18437–18447 [DOI] [PubMed] [Google Scholar]
- 7.Lehmann, A. R. (2001) Genes Dev. 15 15–23 [DOI] [PubMed] [Google Scholar]
- 8.Cantor, S. B., Bell, D. W., Ganesan, S., Kass, E. M., Drapkin, R., Grossman, S., Wahrer, D. C., Sgroi, D. C., Lane, W. S., Haber, D. A., and Livingston, D. M. (2001) Cell 105 149–160 [DOI] [PubMed] [Google Scholar]
- 9.Liu, H., Rudolf, J., Johnson, K. A., McMahon, S. A., Oke, M., Carter, L., McRobbie, A. M., Brown, S. E., Naismith, J. H., and White, M. F. (2008) Cell 133 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, L., Fuss, J. O., Cheng, Q. J., Arvai, A. S., Hammel, M., Roberts, V. A., Cooper, P. K., and Tainer, J. A. (2008) Cell 133 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolski, S. C., Kuper, J., Hanzelmann, P., Truglio, J. J., Croteau, D. L., Van Houten, B., and Kisker, C. (2008) PLoS Biol. 6 e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coin, F., Oksenych, V., and Egly, J. M. (2007) Mol. Cell 26 245–256 [DOI] [PubMed] [Google Scholar]
- 13.Cantor, S., Drapkin, R., Zhang, F., Lin, Y., Han, J., Pamidi, S., and Livingston, D. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2357–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litman, R., Peng, M., Jin, Z., Zhang, F., Zhang, J., Powell, S., Andreassen, P. R., and Cantor, S. B. (2005) Cancer Cell 8 255–265 [DOI] [PubMed] [Google Scholar]
- 15.Rudolf, J., Makrantoni, V., Ingledew, W. J., Stark, M. J., and White, M. F. (2006) Mol. Cell 23 801–808 [DOI] [PubMed] [Google Scholar]
- 16.Pugh, R. A., Honda, M., Leesley, H., Thomas, A., Lin, Y., Nilges, M. J., Cann, I. K., and Spies, M. (2008) J. Biol. Chem. 283 1732–1743 [DOI] [PubMed] [Google Scholar]
- 17.Ding, H., Hidalgo, E., and Demple, B. (1996) J. Biol. Chem. 271 33173–33175 [DOI] [PubMed] [Google Scholar]
- 18.Gaudu, P., and Weiss, B. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 10094–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mettert, E. L., and Kiley, P. J. (2007) J. Bacteriol. 189 3036–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz, C. J., Giel, J. L., Patschkowski, T., Luther, C., Ruzicka, F. J., Beinert, H., and Kiley, P. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14895–14900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paraskeva, E., and Hentze, M. W. (1996) FEBS Lett. 389 40–43 [DOI] [PubMed] [Google Scholar]
- 22.Agarwalla, S., Stroud, R. M., and Gaffney, B. J. (2004) J. Biol. Chem. 279 34123–34129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez, H. L., Pierrel, F., Elleingand, E., Garcia-Serres, R., Huynh, B. H., Johnson, M. K., Fontecave, M., and Atta, M. (2007) Biochemistry 46 5140–5147 [DOI] [PubMed] [Google Scholar]
- 24.Cunningham, R. P., Asahara, H., Bank, J. F., Scholes, C. P., Salerno, J. C., Surerus, K., Munck, E., McCracken, J., Peisach, J., and Emptage, M. H. (1989) Biochemistry 28 4450–4455 [DOI] [PubMed] [Google Scholar]
- 25.Boal, A. K., Yavin, E., Lukianova, O. A., O'Shea, V. L., David, S. S., and Barton, J. K. (2005) Biochemistry 44 8397–8407 [DOI] [PubMed] [Google Scholar]
- 26.Porello, S. L., Cannon, M. J., and David, S. S. (1998) Biochemistry 37 6465–6475 [DOI] [PubMed] [Google Scholar]
- 27.Hinks, J. A., Evans, M. C., De Miguel, Y., Sartori, A. A., Jiricny, J., and Pearl, L. H. (2002) J. Biol. Chem. 277 16936–16940 [DOI] [PubMed] [Google Scholar]
- 28.Weiner, B. E., Huang, H., Dattilo, B. M., Nilges, M. J., Fanning, E., and Chazin, W. J. (2007) J. Biol. Chem. 282 33444–33451 [DOI] [PubMed] [Google Scholar]
- 29.Klinge, S., Hirst, J., Maman, J. D., Krude, T., and Pellegrini, L. (2007) Nat. Struct. Mol. Biol. 14 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flint, D. H., Emptage, M. H., Finnegan, M. G., Fu, W., and Johnson, M. K. (1993) J. Biol. Chem. 268 14732–14742 [PubMed] [Google Scholar]
- 31.Ignarro, L. J. (1999) Biosci. Rep. 19 51–71 [DOI] [PubMed] [Google Scholar]
- 32.Gobert, A. P., McGee, D. J., Akhtar, M., Mendz, G. L., Newton, J. C., Cheng, Y., Mobley, H. L., and Wilson, K. T. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13844–13849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacMicking, J., Xie, Q. W., and Nathan, C. (1997) Annu. Rev. Immunol. 15 323–350 [DOI] [PubMed] [Google Scholar]
- 34.Krieglstein, C. F., Cerwinka, W. H., Laroux, F. S., Salter, J. W., Russell, J. M., Schuermann, G., Grisham, M. B., Ross, C. R., and Granger, D. N. (2001) J. Exp. Med. 194 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers, P. A., Eide, L., Klungland, A., and Ding, H. (2003) DNA Repair 2 809–817 [DOI] [PubMed] [Google Scholar]
- 36.Gal, A., and Wogan, G. N. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 15102–15107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, C. Q., Trudel, L. J., and Wogan, G. N. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 10364–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowart, R. E., Singleton, F. L., and Hind, J. S. (1993) Anal. Biochem. 211 151–155 [DOI] [PubMed] [Google Scholar]
- 39.Siegel, L. M. (1965) Anal. Biochem. 11 126–132 [DOI] [PubMed] [Google Scholar]
- 40.Yang, J., Bitoun, J. P., and Ding, H. (2006) J. Biol. Chem. 281 27956–27963 [DOI] [PubMed] [Google Scholar]
- 41.Ren, B., Zhang, N., Yang, J., and Ding, H. (2008) Mol. Microbiol. 70 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cioffi, E. A., Shaw, K. J., Bailey, W. F., and Berg, C. M. (1980) Anal. Biochem. 104 485–488 [DOI] [PubMed] [Google Scholar]
- 43.Dutton, P. L. (1978) Methods Enzymol. 54 411–435 [DOI] [PubMed] [Google Scholar]
- 44.Tamir, S., Lewis, R. S., de Rojas Walker, T., Deen, W. M., Wishnok, J. S., and Tannenbaum, S. R. (1993) Chem. Res. Toxicol. 6 895–899 [DOI] [PubMed] [Google Scholar]
- 45.Yang, W., Rogers, P. A., and Ding, H. (2002) J. Biol. Chem. 277 12868–12873 [DOI] [PubMed] [Google Scholar]
- 46.McDevitt, C. A., Hanson, G. R., Noble, C. J., Cheesman, M. R., and McEwan, A. G. (2002) Biochemistry 41 15234–15244 [DOI] [PubMed] [Google Scholar]
- 47.Kennedy, M. C., Antholine, W. E., and Beinert, H. (1997) J. Biol. Chem. 272 20340–20347 [DOI] [PubMed] [Google Scholar]
- 48.Spiro, S. (2007) FEMS Microbiol. Rev. 31 193–211 [DOI] [PubMed] [Google Scholar]
- 49.Drapier, J. C. (1997) Methods (San Diego) 11 319–329 [DOI] [PubMed] [Google Scholar]
- 50.Foster, M. W., and Cowan, J. A. (1999) J. Am. Chem. Soc. 121 4093–4100 [Google Scholar]
- 51.Ding, H., and Demple, B. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5146–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cruz-Ramos, H., Crack, J., Wu, G., Hughes, M. N., Scott, C., Thomson, A. J., Green, J., and Poole, R. K. (2002) EMBO J. 21 3235–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]