Abstract
Two thioesterases are commonly found in natural product biosynthetic clusters, a type I thioesterase that is responsible for removing the final product from the biosynthetic complex and a type II thioesterase that is believed to perform housekeeping functions such as removing aberrant units from carrier domains. We present the crystal structure and the kinetic analysis of RifR, a type II thioesterase from the hybrid nonribosomal peptide synthetases/polyketide synthase rifamycin biosynthetic cluster of Amycolatopsis mediterranei. Steady-state kinetics show that RifR has a preference for the hydrolysis of acyl units from the phosphopantetheinyl arm of the acyl carrier domain over the hydrolysis of acyl units from the phosphopantetheinyl arm of acyl-CoAs as well as a modest preference for the decarboxylated substrate mimics acetyl-CoA and propionyl-CoA over malonyl-CoA and methylmalonyl-CoA. Multiple RifR conformations and structural similarities to other thioesterases suggest that movement of a helical lid controls access of substrates to the active site of RifR.
Assembly line complexes, which include modular polyketide synthases (PKS)3 and nonribosomal peptide synthetases (NRPS), are multifunctional proteins composed of modules that work in succession to synthesize secondary metabolites, many of which are precursors of potent antibiotics, immunosuppressants, anti-tumor agents, and other bioactive compounds. Rifamycin, the precursor to the anti-tuberculosis drug rifampicin, is produced by the rifamycin assembly line complex, which is an NRPS/PKS hybrid system composed of one NRPS-like and 10 PKS modules (1). Each module in an assembly line complex extends and modifies the intermediate compound before passing it on to the next module in the series (Fig. 1A). The intermediate compounds are covalently attached through a thioester linkage to the phosphopantetheine arm (Ppant) of carrier domains, one associated with each module, until they are released from the synthase, usually by a type I thioesterase (TEI) (2, 3).
FIGURE 1.
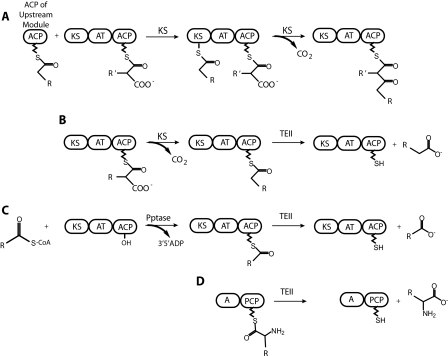
Proposed functions of thioesterase II proteins. A, chain elongation by a PKS module. The chain elongation intermediate is transferred from the ACP of the upstream module to the ketosynthase (KS) domain. The acyltransferase (AT) domain transfers an acyl group building block from CoA to the ACP within the module. The KS domain catalyzes condensation of the new building block with the intermediate, releasing CO2. B, production of a decarboxylated acyl unit by the ketosynthase domain and the subsequent hydrolysis by a TEII. C, mispriming of a PKS by transfer of an acyl-phosphopantetheine arm by a promiscuous phosphopantetheinyl transferase (Pptase) and the subsequent hydrolysis by a TEII. D, hydrolysis of an amino acid derivative by a TEII from an NRPS module comprising an adenylation domain (A) and a peptide carrier protein (PCP) domain.
TEIs are usually integrated into the final module of the assembly line complex and remove the final product through macrocyclization or hydrolysis. Occasionally, tandem type I thioesterases are integrated at the C terminus of the final module of NRPS pathways (4).
Although TEIs are covalently attached to the terminal module and generally process only the final product of an assembly line complex, type II thioesterases (TEIIs) are discrete proteins that can remove intermediates from any module in the complex. A variety of functions have been attributed to TEIIs, the most prevalent of which is a “housekeeping function,” the removal of aberrant acyl units from carrier domains. These aberrant acyl units may be due to premature decarboxylation by a PKS ketosynthase domain (5) (Fig. 1B) or to mispriming of the carrier domain by a promiscuous phosphopantetheinyl transferase (6–8) (Fig. 1C). Other proposed functions for TEIIs include the removal of intermediates from the synthase as in the case of the mammary gland rat fatty acid synthase (FAS) TEII in lactating rats, which removes medium chain C8-C12 fatty acids from the ACP domain (9) and the removal of amino acid derivatives from a carrier domain (10–13), allowing these derivatives to be incorporated into the natural product by a later module in the assembly line complex (Fig. 1D).
Disruption of the TEI function results in a complete loss of product, whereas disruption of TEII function results in a significant decrease in product yield (30–95%) (4, 14–24). Removal of the TEII from the rifamycin assembly line resulted in a 60% decrease in product yield (25). Neither TEIs nor TEIIs may rescue the disrupted function of the other (6), but a TEII from another pathway may rescue the function of a disrupted TEII (26).
Two models have been proposed for the TEII housekeeping function (5). In the high specificity model, the TEII scans the complex and efficiently removes only aberrant acyl units. In the low specificity model, the TEII removes both correct and incorrect acyl units from the Ppant arm at an inefficient rate. Correct acyl units are quickly incorporated into the growing intermediate compound. In contrast, incorrect acyl units stall the assembly line, providing a longer window of opportunity for removal by a TEII. Thus a slow, low specificity enzyme can be effective.
TEIIs from different pathways have differing specificities, but general trends include a preference for decarboxylated acyl units over carboxylated acyl units (5, 6, 27), substrates linked to a carrier domain over substrates linked to CoA or the phosphopantetheine mimic N-acetylcysteamine (7, 28), and single amino acids over di- or tri-peptides (6, 7). TEIIs are able to hydrolyze substrates attached to carrier domains from their native pathway as well as other pathways (6, 20, 28).
PKS/NRPS/FAS thioesterases belong to the α/β hydrolase family. Structures are reported for seven PKS/NRPS/FAS thioesterases: crystal structures for the TEIs from the pikromycin (PikTE) PKS (29), 6-deoxyerythronolide B (DEBSTE) PKS (30), surfactin NRPS (SrfTE) (31), fengycin NRPS (FenTE) (32), and human fatty acid synthase (hFasTE) (33) systems, and NMR structures for enterobactin TEI (34), and surfactin TEII (35). Like PKS modules, PKS TEIs are dimers. The dimer interface comprises two N-terminal helices that are unique to the PKS TEIs. NRPS TEIs are monomeric, like NRPS modules. The NRPS TEII of surfactin is also monomeric (31). Although the FAS complex is dimeric, the FAS TEI is a monomer (33). All of the TEs have an α-helical insertion after strand β5 that forms a lid over the active site. Additionally, in the PKS TEIs, the N-terminal dimer-forming helices contribute to the lid structure, forming a fixed channel that runs the length of the TE and contains the active site. In contrast, the active site pocket of monomeric NRPS TEIs and TEIIs is flexible; two conformations of the lid and active site pocket were observed in the surfactin TEI (SrfTEI) crystal structure (7), and chemical shift observations suggested greater flexibility for residues of the lid region in the surfactin TEII (SrfTEII) solution structure (35). These movements seem to be of functional importance, because a movement of a linker peptide in SrfTEI determines the shape of the active site pocket and a movement of the first lid helix appears to modulate access to the active site (31).
We report the structure and activity of recombinant RifR, the TEII of the rifamycin biosynthetic cluster. Steady-state kinetic analysis of the hydrolytic activity of RifR on a wide range of acyl-CoA and acyl-ACP substrates demonstrates that acyl-ACP substrates are preferred over the acyl-CoAs. Aberrant, decarboxylated acyl units are processed more efficiently than are the natural rifamycin building blocks. We report the crystal structure of RifR, the first for any hybrid PKS/NRPS TEII. The size and shape of the substrate chamber are variable, because one of the elements forming the chamber, an extended linker segment, is highly flexible, and different crystal forms reveal different shapes for the substrate binding site. Access to the active site is severely restricted, and structural comparisons with other thioesterases suggest that a conformational change in the lid and the flexible linker region is required for access to the substrate pocket.
EXPERIMENTAL PROCEDURES
Materials—Nonradioactive acyl-CoAs were obtained from Sigma at the highest purity available. DL-2-[methyl-14C]Methylmalonyl-CoA (54 mCi/mmol) and [malonyl-2-14C]malonyl-CoA (52 mCi/mmol) were from PerkinElmer Life Sciences, and [acetyl-1-14C]acetyl-CoA (54 mCi/mmol) and [propionyl-1-14C]propionyl-CoA (53 mCi/mmol) were from Moravek Biochemicals. Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) and restriction enzymes were from Invitrogen.
Manipulation of DNA and Strains—DNA manipulations were performed in Escherichia coli Novablue (Novagen) or DH5α using standard culture conditions (36). Polymerase chain reactions were carried out using Platinum Pfx polymerase (Invitrogen) as recommended by the manufacturer.
Construction of Expression Vectors for Wild-type and S94A RifR—PCR-based gene synthesis was used to assemble the rifR gene (GenBank™ accession number AF040570, nucleotides 96034–96813) encoding RifR from a set of 34 overlapping oligonucleotides (37). The terminal 5′- and 3′-oligonucleotides were designed to flank the synthetic gene with NdeI and XhoI restriction sites, respectively. After assembly, the gene was PCR-amplified, digested with NdeI and XhoI, and ligated to pET21 (Novagen) digested with the same enzymes to generate pMS8, an expression vector for RifR with a natural N terminus and a hexahistidine sequence appended to its C terminus. The identity of the rifR synthetic gene was confirmed by DNA sequencing. The QuikChange method (Stratagene) was used to generate the S94A mutant of RifR; the serine nucleophile of the catalytic triad was converted to alanine by mutating AGT to GCT at the appropriate location in pMS8 to give expression vector pHC2. The mutation was confirmed by sequencing.
Construction of an Expression Vector for S639A Rif M1—The natural sequence 5′-CGCGCC-3′ at nucleotides 24260–24265 (GenBank™ accession number AF040570), corresponding to the C-terminal end of Rif Module1 (M1), was chosen on the basis of an alignment of DEBS and Rif thiolation (T) domain sequences (38) for replacement with the SpeI recognition sequence 5′-ACTAGT-3′. The BsaBI-SpeI fragment encoding Rif M1 was then fused to the SpeI-EcoRI fragment encoding the DEBS TE via replacement of the BsaBI-SpeI fragment encoding DEBS M3 in pST132 (39) to give pSA10. The presence of the DEBS TE domain was undesirable for this study, so its coding sequence was eliminated by ligating the NdeI-SpeI fragment of pSA10 encoding Rif M1 to the NdeI-NheI fragment of pET25b (Novagen). This yielded pMS24, an expression vector for Rif M1 with hexahistidine appended to the C terminus. The QuikChange method (Stratagene) was used to generate Rif M1 with an inactive acyltransferase domain: the active site serine of the acyltransferase domain was converted to alanine by mutating TCG at nucleotides 21434–21436 of the original sequence to GCG to give expression vector pMS25, which was fully sequenced to confirm its identity.
Expression and Purification of Proteins—Expression plasmids were transformed into E. coli strain BL21 Star™ (DE3) (Invitrogen). One-liter cultures were grown at 37 °C in 2-liter flasks containing LB medium supplemented with 0.1 mg/ml ampicillin. Protein expression was induced with 100 μm isopropyl β-d-thiogalactopyranoside at an optical density at 600 nm of 0.8. After induction, incubation was continued for 20 h at 15 °C. The cells were then harvested by centrifugation at 2500 × g and resuspended in 50 mm sodium phosphate (pH 8.0), 300 mm NaCl, 10 mm imidazole, 1 mm MgCl2, 1 mm CaCl2, 0.1 mg/ml DNase I, 10% v/v glycerol.
All purification procedures were performed at 4 °C. The resuspended cells were disrupted by two passages through a French press at 16,000 p.s.i., and the lysate was collected by centrifugation at 47,800 × g and loaded onto a previously equilibrated Histrap HP column (1 ml; GE Healthcare). The column was washed with 10 mm imidazole in 50 mm sodium phosphate (pH 8), 300 mm NaCl, 10% v/v glycerol, and the proteins were eluted with an imidazole gradient (10–100 mm) in the same solution. For Rif M1, pooled fractions containing S639A Rif M1 were diluted with 20 mm Tris (pH 7.5), 50 mm NaCl, 1 mm EDTA, 10% v/v glycerol and loaded onto a previously equilibrated HiTrapQ HP anion exchange column (1 ml; GE Biosciences). The column was washed with 50 mm NaCl in 20 mm Tris (pH 7.5), 1 mm EDTA, 10% v/v glycerol, and S639A Rif M1 was eluted with a NaCl gradient (50–500 mm) in the same solution. Pooled fractions containing S639A Rif M1 were buffer-exchanged into 50 mm HEPES (pH 7.5), 50 mm NaCl, 1 mm EDTA, 1 mm TCEP, 10% v/v glycerol and concentrated with an Amicon Ultra-15 centrifugal filter unit (Millipore). For wild-type and S94A RifR, metal affinity column fractions containing RifR were pooled, diluted with 20 mm Tris (pH 7.5), 50 mm NaCl, 1 mm EDTA, 10% v/v glycerol, and loaded onto a previously equilibrated Mono Q 5/50 GL anion exchange column (GE Biosciences). RifR was present in the column flow through and was buffer-exchanged into 50 mm HEPES (pH 7.5), 50 mm NaCl, 1 mm EDTA, 1 mm TCEP, 10% (v/v) glycerol and concentrated with an Amicon Ultra-15 centrifugal filter unit (Millipore).
Purified proteins were flash-frozen in liquid nitrogen and stored at -80 °C. Protein concentrations were determined using the calculated extinction coefficients (40) at 280 nm: 18,450 m-1 cm-1 for RifR, and 166,840 m-1 cm-1 for S639A Rif M1. Typical 1-liter cultures yielded 10 mg of purified RifR or 4 mg of purified S639A Rif M1.
Selenomethionyl (SeMet) RifR was produced with a protocol as for RifR, modified according to Guerrero et al. (41), in which a 50-ml overnight culture was pelleted and added to minimal medium supplemented with SeMet prior to induction.
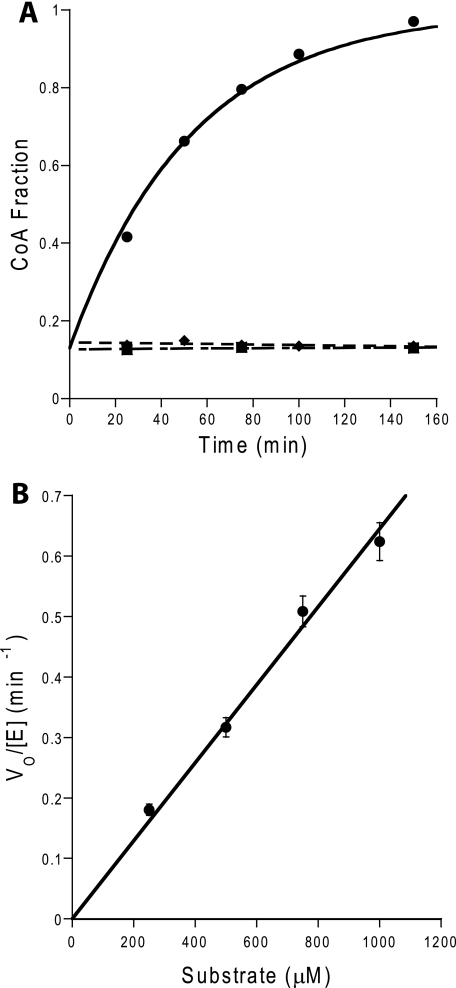
Measurement of RifR Activity toward Acyl-CoA Substrates—Starting acyl-CoA stocks contained a small amount of CoA. Acyl-CoAs (25–1000 μm) were incubated with RifR or S94A RifR (2.5–25 μm) or no enzyme in the presence of 50 mm HEPES (pH 7.5), 25 mm NaCl, 5 mm MgCl2, 1 mm TCEP, 5% v/v glycerol at 25 °C. To ensure accurately measurable hydrolysis for all acyl-CoAs over the same time frame, slower hydrolyzing acyl-CoAs (acetyl-CoA, isobutyryl-CoA, hexanoyl-CoA, malonyl-CoA, and methylmalonyl-CoA (250–1000 μm)) were incubated with 25 μm RifR, and faster hydrolyzing acyl-CoAs (butyryl-CoA, octanoyl-CoA and propionyl-CoA (25–1000 μm)) were incubated with 2.5 μm RifR. Because of its limited solubility, decanoyl-CoA was incubated at a lower concentration (25–250 μm) with RifR (2.5 μm) than were the other faster hydrolyzing substrates. At each time point, aliquots were quenched to a final concentration of 5% trichloroacetic acid, and the precipitated protein was removed by centrifugation at 20,800 × g for 5 min. The ratio of acyl-CoA to CoA in the supernatant was quantified by HPLC using a C18 reverse phase column (Altima, 5 μm, 250 × 4.6 mm) monitored by absorbance at 259 nm. Separation was performed using a modification of a published protocol (42) Briefly, a linear gradient of buffer A (75 mm potassium phosphate, pH 4.5) and buffer B (0.1% trifluoroacetic acid in acetonitrile) was used at a constant flow rate of 1.0 ml/min. Initial conditions were 96% buffer A and 4% buffer B. At 5 min, buffer B was increased to 7% over 5 min and then increased to 9% over 4 min. At 14 min, buffer B was increased to 50% over 5 min and maintained for 8 min. At 27 min, buffer B was decreased to 4% over 1 min, and the column was equilibrated at 4% buffer B for 8 min between injections. Retention times were as follows: acetyl-CoA, 18.6 min; butyryl-CoA, 20.5 min; CoA, 14.8 min; decanoyl-CoA, 22.6 min; hexanoyl-CoA, 21.4 min; isobutyryl-CoA, 20.5 min; malonyl-CoA, 13.5 min; methylmalonyl-CoA, 16.8 min; octanoyl-CoA, 22.0 min; and propionyl-CoA, 20.0 min. With the exception of isobutyryl-CoA, which was shown to saturate wild-type RifR, hydrolysis of acyl-CoAs was linearly dependent on enzyme concentration in the wild-type RifR reactions. No hydrolysis was detected in the control reactions without RifR, nor was hydrolysis observed in the S94A reactions except with isobutyryl-CoA and propionyl-CoA. Data analysis was performed using Kaleidagraph (Synergy Software). Initial velocities were extracted by fitting the hydrolysis progress plot to the equation: CoA fraction = 1 - (1 - CoA fraction0)e-tv0, where CoA fraction = [CoA]/([CoA] + [Acyl-CoA]), t = time, and v0 = initial velocity (Table 1).
TABLE 1.
Kinetic parameters for RifR hydrolysis of acyl substrates
|
Substrate
|
kcat/Km
|
Ratio
|
||||
|---|---|---|---|---|---|---|
| Wild type RifR | S94A RifR | Wild type/S94A | ACP/CoA | |||
| m–1 s–1 | ||||||
| CoA substrates | ||||||
| Decanoyl-CoA | CH3-(CH2)8-CO-S-CoA | 160 ± 18 | <0.16 | >1000 | ||
| Octanoyl-CoA | CH3-(CH2)6-CO-S-CoA | 31 ± 2.5 | <0.16 | >190 | ||
| Propionyl-CoA | CH3-CH2-CO-S-CoA | 25 ± 0.5 | 0.96 ± 0.37 | 26 | ||
| Butyryl-CoA | CH3-(CH2)2-CO-S-CoA | 13 ± 3.2 | <0.04 | >340 | ||
| Acetyl-CoA | CH3-CO-S-CoA | 11 ± 0.2 | <0.03 | >320 | ||
| Isobutyryl-CoA | (CH3)2-CH-CO-S-CoA | 9.6 ± 0.08 | 4.5 ± 0.08 | 2.1 | ||
| Hexanoyl-CoA | CH3-(CH2)4-CO-S-CoA | 5.9 ± 0.33 | <0.04 | >140 | ||
| Methylmalonyl-CoA | CO2-(CH3)CH2-CO-S-CoA | 1.8 ± 0.17 | <0.03 | >72 | ||
| Malonyl-CoA | CO2-CH2-CO-S-CoA | 1.5 ± 0.08 | <0.07 | >21 | ||
| ACP substrates | ||||||
| Propionyl-RifM1 | CH3-CH2-CO-S-ACP | 210 ± 20 | 8.4 | |||
| Acetyl-RifM1 | CH3-CO-S-ACP | 150 ± 38 | 14 | |||
| Methylmalonyl-RifM1 | CO2-(CH3)CH2-CO-S-ACP | 54 ± 6.3 | 30 | |||
Measurement of RifR Activity toward Acyl-S639A Rif M1 Substrates—To generate [14C]acyl-S639A Rif M1 substrates, [14C]acyl groups were installed on the apo T domain of S639A Rif M1 by preincubating the apo protein with [14C]acyl-CoA and the promiscuous phosphopantetheinyl transferase Sfp (43, 44). Preincubation reactions were performed at 25 °C for 60–90 min and contained 25 μm apo S639A Rif M1, 25 μm Sfp, and 25 μm [14C]acyl-CoA in 50 mm HEPES (pH 7.5), 25 mm NaCl, 5 mm MgCl2, 1 mm TCEP, 5% (v/v) glycerol. Aliquots of the preincubation reactions were then distributed into reaction tubes containing wild-type RifR, S94A RifR, or no TEII, for final reactions that consisted of varying concentrations of [14C]acyl-S639A Rif M1 (2–12 μm) and wild-type or S94A RifR (0–4 μm) in 50 mm HEPES (pH 7.5), 25 mm NaCl, 5 mm MgCl2, 1 mm TCEP, 5% v/v glycerol (plus residual Sfp and the 3′,5′-ADP product of preincubation reactions). Final reactions were incubated at 25 °C, and at desired time points 10-μl aliquots were quenched in an equal volume of 10% trichloroacetic acid. The protein precipitate was pelleted by centrifugation, washed with 150 μl of 5% trichloroacetic acid, and solubilized in 20 μl of 2% SDS, 50 mm Tris (pH 8). This solution was combined with 5 ml of liquid scintillation fluid (Ultima Gold; PerkinElmer Life Science), and the amount of [14C]acyl-S639A Rif M1 remaining at each time point was quantified by liquid scintillation counting. Disappearance of [14C]acyl-S639A Rif M1 substrates was linearly dependent on enzyme concentration in the wild-type RifR reactions, but little or no breakdown of [14C]acyl-S639A Rif M1 substrates was observed in the no-TEII and S94A RifR control reactions. Data analysis was performed using Kaleidagraph (Synergy Software), and exponential fits to the data typically gave R ≥ 0.95.
To determine the identity of the acyl products of the RifR reactions, the trichloroacetic acid supernatants of late reaction time points were analyzed by radio-HPLC. The samples were injected onto a System Gold HPLC (Beckman) equipped with an Aminex HPX-87H ion exclusion column (Bio-Rad) and a Radiomatic 150TR flow scintillation analyzer (PerkinElmer Life Science) to separate and detect 14C-labeled species. Separations were performed isocratically in 0.008 n sulfuric acid over 30 min with a flow rate of 0.6 ml/min, and flow scintillation analysis was performed on the column eluant after it was mixed with Ultima Flo liquid scintillation fluid (PerkinElmer Life Science) in a 1 to 2 ratio. As expected, [14C]acetate, [14C]propionate, and [14C]methylmalonate predominated in the trichloroacetic acid supernatants from reactions that contained [14C]acetyl-S639A Rif M1, [14C]propionyl-S639A Rif M1, and [14C]methylmalonyl-S639A Rif M1, respectively. However, significant amounts of both [14C]malonate and [14C]acetate were detected in trichloroacetic acid supernatants from [14C]malonyl-S639A Rif M1 reactions; [14C]acetate presumably results from decarboxylation of [14C]malonyl-S639A Rif M1 to [14C]acetyl-S639A Rif M1 followed by RifR-catalyzed hydrolysis during the reaction period. The concurrent decarboxylation of [14C]malonyl-S639A Rif M1 prevented us from obtaining a reliable kcat/Km value for its hydrolysis by RifR, but the accumulation of [14C]malonate over time indicates that malonyl-S639A Rif M1 is indeed a substrate.
Crystallization—RifR was crystallized by hanging drop vapor diffusion at 4 °C. Crystallization drops were set by the addition of protein stock (5–13.5 mg/ml RifR, 10 mm HEPES, pH 7.0, 2 mm dithiothreitol) to reservoir solution, (8–23% polyethylene glycol 8000, 100 mm HEPES, pH 7.0–7.6, 35–50 mm CaCl2, 2 mm dithiothreitol) in a ratio of 1:2 to 3:2. Crystallization of SeMet RifR required microseeding from native RifR crystals. Before flash freezing in liquid nitrogen, the crystals were cryoprotected by soaking 5–10 s in a solution equivalent to the reservoir solution with the addition of 10% polyethylene glycol 400.
Crystallography—X-ray diffraction data were collected at the GM/CA beamline (ID-23D) at the Advanced Photon Source (Argonne National Laboratory). A three-wavelength multi-wavelength anomalous diffraction data set was recorded from a SeMet RifR crystal for structure determination. The data were processed using the HKL2000 package (45) (Table 2). Determination of selenium atomic positions, experimental phasing, density modification phase refinement, and initial model building were performed using the programs SOLVE and RESOLVE (46, 47). Twelve of fourteen expected selenium sites were identified. Model building was carried out with Coot (48), and the model was refined using REFMAC5 in the CCP4 suite (49, 50). Rigid body motion was modeled as six translation/libration/screw groups per monomer, assigned with the aid of the TLSMD server (51). The structure was solved from monoclinic crystals with two RifR polypeptides in the asymmetric unit (P21: a = 39.5 Å, b = 94.6 Å, c = 63.2 Å, β = 90.55°). Noncrystallographic symmetry restraints were employed in refinement. Subsequent crystal forms, which were orthorhombic with a single molecule in the asymmetric unit, varied in the dimension of the long unit cell axis (82–108 Å) and were solved with molecular replacements using AMORE (52). Of the subsequent crystal forms, only one contained a fully ordered protein chain (see below) and is reported here in addition to the original crystal form. Gel filtration analysis indicates that RifR is a monomer in solution (data not shown). The final model contains residues 2–247 in both chains. The structures were validated using Mol-Probity (53), and secondary structure assignment used the Stride server (54, 55).
TABLE 2.
Crystallographic data
|
Form 1 - SeMet
|
Form 2 (native)
|
|||
|---|---|---|---|---|
| Peak | Inflection | Remote | ||
| Diffraction data | ||||
| Space group | P21 | P21 | P21 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 39.51, 94.65, 63.17 | 39.51, 94.65, 63.17 | 39.51, 94.65, 63.17 | 38.94, 62.50, 82.47 |
| α, β, γ (°) | 90, 90.55, 90 | 90, 90.55, 90 | 90, 90.55, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.97942 | 0.97959 | 0.95446 | |
| Resolution (Å) | 50.00-1.80 (1.86-1.80) | 50.00-1.90 (1.97-1.90) | 50.00-1.86 (1.94-1.86) | 50.00-1.80 (1.86-1.80) |
| Avg I/σI | 10.5 (1.5) | 19.2 (2.2) | 13.5 (2.6) | 12.5 (2.2) |
| Rsymm | 0.115 (0.540) | 0.112 (0.488) | 0.131 (0.665) | 0.069 (0.376) |
| Completeness (%) | 99.8 (87.5) | 99.0 (97.6) | 98.4 (97.1) | 96.1 (88.3) |
| Average redundancy | 3.6 (2.5) | 3.6 (3.0) | 3.5 (2.9) | 2.9 (2.5) |
| Refinement | ||||
| Resolution (Å) | 50.0-1.80 | 50.0-1.80 | ||
| No. reflections | 39950 | 17702 | ||
| Rwork/Rfree | 0.171/0.200 | 0.195/0.237 | ||
| No. atoms | ||||
| Protein | 3888 | 1926 | ||
| Ligand/ion | 2 | 14 | ||
| Water | 482 | 168 | ||
| B-factors (Å2) | ||||
| Protein | 17.2 | 22.9 | ||
| Ligand-ion | 22.7 | 48.2 | ||
| Water | 29.9 | 33.6 | ||
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.009 | 0.008 | ||
| Bond angles (°) | 1.204 | 1.181 | ||
RESULTS
We tested the ability of RifR to hydrolyze a variety of substrates from a phosphopantetheine arm delivered by both CoA (Fig. 2) and ACP carriers. In particular we tested the ability to remove carboxylated acyl units versus decarboxylated acyl units, short chain acyl units versus medium chain acyl units, and acyl units attached to a carrier domain (ACP) versus those attached to CoA (Table 1). RifR hydrolyzed all substrates tested with catalytic efficiencies over a range of 1–200 m-1 s-1. Background hydrolysis was undetectable. With the exception of isobutyryl-CoA, saturation kinetics were not observed, and individual kinetic constants could not be obtained.
FIGURE 2.
Example data for hydrolysis of acyl-CoA substrates. A, deacylation of acetyl-CoA substrate by RifR under steady-state conditions. The increase of the CoA fraction during the reactions of 250 μm Acetyl-CoA with 25 μm WT RifR (•), with 25 μm S94A RifR (♦), and without RifR (▪). B, Michaelis-Menten plot for deacylation of acetyl-CoA (250–1000 μm).
Hydrolysis of Carboxylated and Decarboxylated Acyl-CoAs—The catalytic efficiency of RifR was compared directly for two natural Rif building blocks (malonyl and methylmalonyl thioesters) and their corresponding decarboxylated variants (acetyl and propionyl thioesters). RifR hydrolyzed the decarboxylated substrates, acetyl-CoA and propionyl-CoA, 7–14-fold more efficiently, respectively, than the corresponding carboxylated substrates, malonyl-CoA and methylmalonyl-CoA (Table 1). In fact, the carboxylated substrates were the poorest of all substrates tested with catalytic efficiencies of 1 m-1 s-1. Although the increased activity against decarboxylated over carboxylated substrates is suggestive of the high specificity editing model, the discrimination is modest, and the relatively slow rate of reaction is consistent with the low specificity model, in which hydrolysis of natural carboxylated building blocks occurs inefficiently and does not compete with chain elongation.
Hydrolysis of Medium Chain and Short Chain Acyl-CoAs—We tested the ability of RifR to hydrolyze acyl groups that resemble neither the natural Rif building blocks nor their decarboxylated variants. Unlike previously tested TEIIs from PKS and NRPS pathways, which had little or no activity toward acyl units of medium length (C4–C10) (6, 9, 28), RifR hydrolyzed several medium chain acyl units. Catalytic efficiency was uncorrelated with chain length: C10 > C8 > C3 > C4 ≈ C2 > C6. It was not possible to determine kinetic constants for these reactions, so we do not know whether the difference in efficiency is due to differences in kcat and/or Km values.
Hydrolysis of Acyl-ACPs—The catalytic efficiency of RifR was compared directly for acyl-ACP and acyl-CoA substrates using the natural Rif module 1 building block (methylmalonyl thioester), its decarboxylated variant (propionyl thioester), and a potential mis-acylated substrate (acetyl thioester). For these experiments, we used the ACP from Rif module 1 in the context of the full module (RifM1), a 167,000 Da multi-functional protein. PKS acyltransferase domains can possess deacylation activity toward cognate acyl-ACP domains. To avoid the possibility of deacylation by the acyltransferase in RifM1, we used an acyltransferase-inactive variant (S639A). RifR hydrolyzed all three ACP substrates (Table 1). It was not possible to obtain saturating concentrations of the acyl-ACP substrates. As for acyl-CoA substrates, RifR showed a slight (4-fold) preference for the decarboxylated substrate (propionyl-ACP) over the carboxylated (methylmalonyl-ACP) substrate. In contrast to the slight discrimination among acyl substrates, under matched reaction conditions, RifR displayed a stronger preference for acyl-ACP substrates over the corresponding acyl-CoA substrates: 8-fold for the propionyl unit, 14-fold for the acetyl unit, and 30-fold for the methylmalonyl unit (Table 1).
Hydrolysis of Acyl-CoAs by S94A RifR—Catalytic activity of wild-type RifR was compared with an active-site RifR mutant, in which the catalytic serine was substituted by alanine (S94A) (Table 1). Thioesterase activity of S94A RifR was effectively eliminated for all substrates excepting isobutyryl-CoA and propionyl-CoA. These substrates may be capable of binding in the active site such that hydroxide ion derived from water acts as the nucleophile in place of the active site serine hydroxylate, allowing hydrolysis of the acyl unit, albeit at a decreased rate.
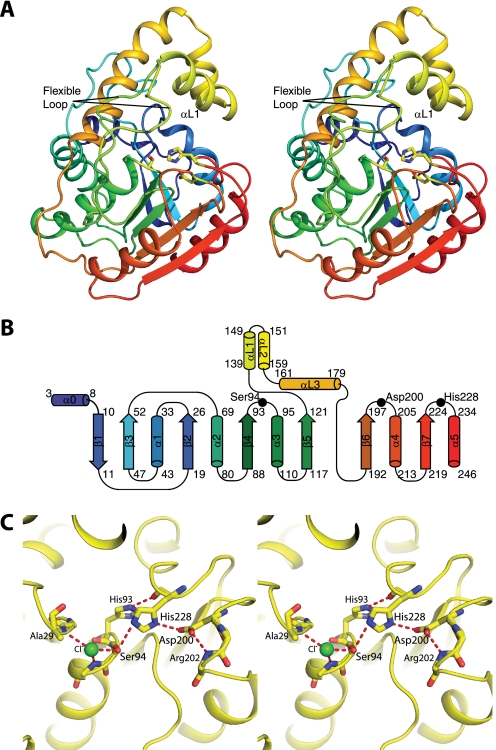
Overall Structure of RifR Type II Thioesterase—RifR is a monomeric protein (Fig. 3A) and a member of the α/β-hydrolase family, with a fold similar to the folds of an NRPS TEII (SrfTEII) (35), three NRPS TEIs (SrfTEI (31), FenTE (32) and EntTE (34)), two PKS TEIs (DEBS TEI (30) and Pik TEI (29)), and the human fatty acid synthase thioesterase (hFAS TE) (33). The α/β-hydrolase core fold is a predominantly parallel β-sheet surrounded by α-helices. The hydrolytic active site is a triad of amino acids located on loops at the C-terminal edge of the core β-sheet. Members of the diverse family differ in the location of some triad residues and in the number and location of helices that decorate the core fold. RifR contains a small subdomain (residues 130–180) that forms a three α-helix “lid” inserted between strands β5 and β6 of the α/β-hydrolase fold (Fig. 3B). The first two helices of the lid (αL1 and αL2) form a short hairpin structure comprising the top of the lid, and the third helix (αL3) forms the back of the lid.
FIGURE 3.
Structure of RifR. A, stereodiagram of RifR TEII. In this ribbon diagram, the polypeptide is colored as a rainbow from blue at the N terminus to red at the C terminus. The lid domain is colored yellow. Active site triad residues (Ser94, Asp200, and His228) are shown as sticks. Two conformations (from different crystal forms) are shown for the flexible linker region between strand β5 and the lid domain. B, topology of RifR. C, stereodiagram of the active site. The catalytic triad (Ser94, Asp200, and His228) and surrounding residues are shown as sticks. A chloride ion (green) occupies the oxyanion hole formed by backbone amides of residues Ala29 and Met95 (side chain not shown). The atomic colors are used in A and C for stick figures with yellow as carbon, red as oxygen, blue as nitrogen, and green as chlorine.
Catlytic Triad—The active site of RifR is a classic catalytic triad comprising residues Ser94, Asp200, and His228 (Fig. 3C). The triad residues of RifR, like those of other TEIIs (supplemental Fig. S1), follow strands β4 (Ser), β6 (Asp), and β7 (His) of the α/β-hydrolase fold (Fig. 3). The active site serine, found within the signature sequence Gly92-His93-Ser94-Xaa95-Gly96, is between strand β4 and helix α3 and has the constrained geometry typical of a nucleophilic elbow, a hallmark of the α/β-hydrolase family. A number of hydrogen bonds position residues in the catalytic center. Notably, His93 of the signature sequence forms a hydrogen bond with the backbone carbonyl of the active site His228, stabilizing its alignment within the triad (Fig. 3C). The oxyanion hole, which stabilizes the tetrahedral intermediate, is formed by the backbone amides of Met95 and Ala29 and contains a single chloride ion in the crystal structures (Fig. 3C). The aspartate of the catalytic triad follows strand β6, in contrast to the PKS, NRPS, and FAS TEIs, where it follows strand β5 (supplemental Fig. S1).
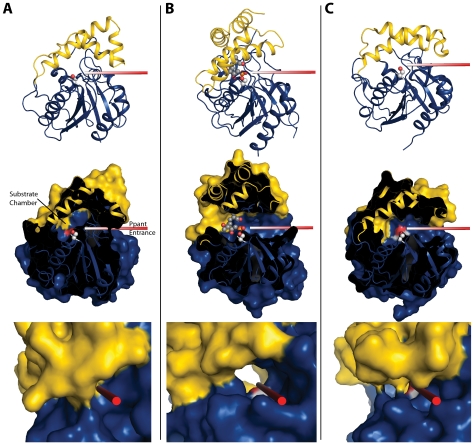
Flexible Lid Subdomain—The lid subdomain of RifR covers the active site like similar lids in other PKS, NRPS, and FAS TEs. In each TE, the lid is centered over the catalytic triad and defines a “Ppant entrance” on one side of the triad and a “substrate chamber” on the other side (Fig. 4). Several lines of evidence are consistent with these functional assignments. The substrate of each TE is delivered to the active site on the Ppant arm of a carrier domain. The Ppant entrance is inferred from the position of the TE N terminus, where the carrier domains are fused to PKS TEIs (30), and from a solution structure of EntTE in complex with its cognate ACP domain (34). The substrate chamber is inferred from the structures of substrate-analog affinity-labeled PKS TEIs (56) and of an inhibitor complex of hFAS TE (33). The size, shape, and character of the substrate chamber determine which substrates can be accommodated and whether the TE hydrolyzes the thioester to a linear product or, like many PKS TEIs, forms a macrolactone.
FIGURE 4.
Cartoon and surface diagrams in equivalent orientations for RifR TEII (A), Pik TEI (Protein Data Bank code 2HFJ) with affinity label (B), and SrfTEI (Protein Data Bank code 1JMK) (C). The rods show the Ppant entrance path to the catalytic serine (shown in spheres). The middle panels show a cut-away surface diagram in the same orientation as the top panels. Access to the active site for RifR TEII is blocked by the lid helices (yellow). The bottom panels are close-up views along the Ppant entrances, showing the closed entrance in RifR, the tunnel-like entrance characteristic of dimeric PKS TEIs (Pik TEI) and trough-like entrance of the monomeric NRPS TEI (SrfTEI in the open form).
All of the TE lids are helical; however, they differ in the number and disposition of helices and in their flexibility. The RifR lid is similar to the lids of the monomeric NRPS TEIs and TEII, which are continuous in sequence and flexible. In contrast to the RifR lid, the lids of the dimeric PKS TEIs lack flexibility and contain four nonconsecutive α-helices, two of which are an N-terminal extension of the sequence and form the dimer interface (supplemental Fig. S1).
Variation among RifR crystal structures provides evidence for lid flexibility. RifR crystallized in a range of related forms with similar crystal packing along two shorter unit cell axes of ∼39 and ∼64 Å. The longer unit-cell axis displayed remarkable variation from 82 to 109 Å. The lid subdomain participated in a crystal lattice contact along the direction of this long unit-cell edge. The various crystal forms captured different solution conformations of the lid. The structures fall into three distinct classes, which differ in the conformation of a flexible “lid loop” (residues 122–138) that is an integral part of the substrate chamber and links strand β5 of the α/β-hydrolase core to the first helix of the lid domain (αL1). In “Form 1” crystals (long axis, 94–99Å), the lid loop is positioned toward the active site; in “Form 2” structures (long axis, ∼82 Å), the lid loop lies along αL3; and in other crystal forms (long axies, 88–92 and 108–109 Å), the lid loop is disordered. The atomic mobility (B) factors are higher, and the electron density for the lid loop is poorer in Form 2 than in Form 1 (supplemental Fig. S2, A and B). Additionally, αL1 is shifted toward the α/β-hydrolase core and rotated inward in Form 2 with respect to Form 1 (supplemental Fig. S2C).
Movement of the lid helices and the flexible lid loop has dramatic effects on the size and shape of the substrate chamber (supplemental Fig. 2, D and E). This flexibility in the substrate chamber is consistent with the modest substrate preferences and wide substrate range exhibited by RifR (Table 1). The inside of the RifR Ppant entrance port contains residues that are conserved across TEIIs, but, consistent with our kinetic results, neither the entrance port nor the substrate chamber contains any obvious structural features that would confer exclusive preference for decarboxylated substrates over carboxylated ones, (Table 1).
The lid movements also affect access to the catalytic triad from the presumed Ppant entrance. The substrate entrance of RifR is bounded by helix αL1 of the lid and helix α1 of the α/β hydrolase core. In all crystal forms, the Ppant entrance is blocked by contact of these α-helices (Fig. 4). In part for this reason we think that movement of helix αL1 is required to open the binding site for the Ppant arm.
DISCUSSION
RifR displayed broad substrate specificity, hydrolyzing carboxylated and decarboxylated acyl thioesters, as well as short, medium, and branched chain substrates. Despite the broad substrate range, RifR preferentially hydrolyzed aberrant decarboxylated acyl thioesters over natural Rif building blocks, consistent with its function as a scavenger of aberrant acyl groups. However, the preference for decarboxylated over carboxylated substrates (4–14-fold) was modest (Table 1). Methylmalonate is the building block for most modules in the Rif pathway, so the decarboxylated variant of methylmalonyl-ACP (propionyl-ACP) should be a primary target of any editing enzyme. RifR had a modest preference for propionyl-ACP over methylmalonyl-ACP (4-fold; Table 1). Our results are consistent with the low specificity model for TEII editing in which both aberrant and natural acyl thioesters are hydrolyzed from carrier domains more slowly than the assembly line pathway processes the natural building blocks. The rate of Rif pathway throughput is unknown, as is the catalytic efficiency of individual ketosynthase condensing domains, so it is not possible to compare throughput and editing rates. Nevertheless RifR is a rather slow enzyme with efficiencies between 1 and 200 m-1 s-1.
The structural variability of the RifR substrate chamber matches the observed broad specificity of the enzyme. The chamber is malleable because of the flexibility of the lid loop (residues 122–138) and loop helix αL1. The plasticity of the substrate chamber likely allows it to accommodate a variety of acyl groups, accounting in part for the broad substrate specificity. The crystal structures captured two variations of the substrate chamber, as well as a highly open chamber in which the lid loop is disordered. These variants likely represent a small subset of substrate chamber shapes that are accessible to the protein in solution. In addition to plasticity, the interior surface of the substrate chamber appears able to accommodate a variety of substrates. The surface of the substrate chamber is hydrophilic in both crystal forms and appears unable to distinguish between charged and uncharged substrates, or short, medium, and branched acyl thioesters. The chamber is accessible to bulk solvent in all crystal forms, also consistent with the broad substrate specificity.
A closed Ppant entrance was observed in all crystal forms of RifR, but differences among these crystal structures provided evidence of lid motion. A substantially populated closed lid form of RifR in solution could account for the observed slow turnover of the enzyme. Lid flexibility is a hallmark of monomeric PKS and NRPS TEs. The SrfTEI crystallized with two independent molecules, one with an open Ppant entrance, the other closed (31). Solution (NMR) structures of EntTEI (34) and SrfTEII (35) also suggest movement in the lid region. In fact, the flexible lid of SrfTEII was reported in an extremely open conformation with no contacts to the α/β hydrolase core. In contrast, no flexibility has been observed for lid α-helices or lid loops in the dimeric PKS TEIs. The extra N-terminal helices, which comprise the dimerization domain in the Pik TEI and DEBS TEI lids, likely stabilize the lid loop region.
The 8–30-fold preference of RifR for substrates carried by ACP over those carried by CoA is consistent with an editing function for RifR. If RifR is a scavenger of aberrant acyl units that stall the Rif pathway, then it should have poor or no activity with CoA substrates. The observed carrier preference could be due to either favorable interactions of RifR with Rif ACP or unfavorable interactions with CoA. The Ppant arm, common to ACP and CoA carriers, is long enough to reach the catalytic triad from the enzyme surface at the Ppant entry. Therefore the RifR carrier preference must be specified on the enzyme surface. The RifR surface surrounding the Ppant entrance is neither strongly hydrophobic nor strongly electronegative and thus lacks features that could lead to unfavorable electrostatic or van der Waals' interactions with CoA. It seems more likely that favorable protein-protein interactions with Rif ACP account for the carrier preference.
Our working model for RifR editing invokes the dynamic property of the lid. The lid must be open for acylated Ppant to reach the active site, and any RifR molecules in a closed lid form are temporarily unavailable for catalysis. Evidence of lid motion comes from differences in RifR crystal forms (supplemental Fig. S2) and from larger scale motions observed or implied in structures of SrfTEI and SrfTEII. We propose that helix αL1 moves to allow proper substrate binding. In this manner, lid dynamics could be a strategy to prevent wasteful hydrolysis of CoA substrates. If Rif ACPs interact preferentially with an open lid form of RifR and if CoA and non-Rif ACPs have no such preference, then Rif ACPs would be the preferred RifR substrate carriers. Thus the “correct” carrier increases editing efficiency by facilitating lid opening. Most characterized editing TEs have weak or no acyl group specificity. It may be a general feature of editing thioesterases that interaction with appropriate ACP-linked substrates stimulates a relatively modest level of activity. Control of thioesterase activity in this manner would help to limit improper hydrolysis of metabolically important acyl-CoA or acyl-ACP substrates by an otherwise promiscuous enzyme.
Supplementary Material
Acknowledgments
We thank Jamie Razelun for assistance with crystallization. We are indebted to the staff of the GM/CA beamlines (supported by the NIGMS and NCI, National Institutes of Health) at the Advanced Photon Source (supported by the United States Department of Energy).
The atomic coordinates and structure factors (codes 3FLA and 3FLB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant DK42303 (to J. L. S.). This work was also supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund (to S. J. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: PKS, polyketide synthase; NRPS, nonribosomal peptide synthetase; TEI, type I thioesterase; TEII, type II thioesterase; Ppant, phosphopantetheinyl arm; FAS, fatty acid synthase; TCEP, Tris(2-carboxyethyl)phosphine hydrochloride; SeMet, selenomethionyl; HPLC, high pressure liquid chromatography; ACP, acyl carrier protein.
References
- 1.Yu, T.-W., Shen, Y., Doi-Katayama, Y., Tang, L., Park, C., Moore, B. S., Richard Hutchinson, C., and Floss, H. G. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9051-9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weissman, K. J. (2004) Philos. Transact. A Math Phys. Eng. Sci. 362 2671-2690 [DOI] [PubMed] [Google Scholar]
- 3.Hutchinson, C. R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3010-3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roongsawang, N., Washio, K., and Morikawa, M. (2007) ChemBioChem 8 501-512 [DOI] [PubMed] [Google Scholar]
- 5.Heathcote, M. L., Staunton, J., and Leadlay, P. F. (2001) Chem. Biol. 8 207-220 [DOI] [PubMed] [Google Scholar]
- 6.Schwarzer, D., Mootz, H. D., Linne, U., and Marahiel, M. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14083-14088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeh, E., Kohli, R. M., Bruner, S. D., and Walsh, C. T. (2004) ChemBioChem 5 1290-1293 [DOI] [PubMed] [Google Scholar]
- 8.Linne, U., Schwarzer, D., Schroeder, G. N., and Marahiel, M. A. (2004) Eur. J. Biochem. 271 1536-1545 [DOI] [PubMed] [Google Scholar]
- 9.Libertini, L. J., and Smith, S. (1978) J. Biol. Chem. 253 1393-1401 [PubMed] [Google Scholar]
- 10.Chen, H., Hubbard, B. K., O'Connor, S. E., and Walsh, C. T. (2002) Chem. Biol. 9 103-112 [DOI] [PubMed] [Google Scholar]
- 11.Chen, H., and Walsh, C. T. (2001) Chem. Biol. 8 301-312 [DOI] [PubMed] [Google Scholar]
- 12.Fujimori, D. G., Hrvatin, S., Neumann, C. S., Strieker, M., Marahiel, M. A., and Walsh, C. T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 16498-16503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel, J., Hoyt, J. C., and Parry, R. J. (1998) Tetrahedron 54 15927-15936 [Google Scholar]
- 14.Butler, A. R., Bate, N., and Cundliffe, E. (1999) Chem. Biol. 6 287-292 [DOI] [PubMed] [Google Scholar]
- 15.Xue, Y., Zhao, L., Liu, H. W., and Sherman, D. H. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 12111-12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider, A., and Marahiel, M. A. (1998) Arch. Microbiol. 169 404-410 [DOI] [PubMed] [Google Scholar]
- 17.Chen, S., Roberts, J. B., Xue, Y., Sherman, D. H., and Reynolds, K. A. (2001) Gene (Amst.) 263 255-264 [DOI] [PubMed] [Google Scholar]
- 18.Wertheimer, A. M., Verweij, W., Chen, Q., Crosa, L. M., Nagasawa, M., Tolmasky, M. E., Actis, L. A., and Crosa, J. H. (1999) Infect. Immun. 67 6496-6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geoffroy, V. A., Fetherston, J. D., and Perry, R. D. (2000) Infect. Immun. 68 4452-4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, Z., Pfeifer, B. A., Chao, E., Murli, S., Kealey, J., Carney, J. R., Ashley, G., Khosla, C., and Hutchinson, C. R. (2003) Microbiology 149 2213-2225 [DOI] [PubMed] [Google Scholar]
- 21.Reimmann, C., Patel, H. M., Walsh, C. T., and Haas, D. (2004) J. Bacteriol. 186 6367-6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cosmina, P., Rodriguez, F., de Ferra, F., Grandi, G., Perego, M., Venema, G., and van Sinderen, D. (1993) Mol. Microbiol. 8 821-831 [DOI] [PubMed] [Google Scholar]
- 23.Roberts, G. A., Staunton, J., and Leadlay, P. F. (1993) Eur. J. Biochem. 214 305-311 [DOI] [PubMed] [Google Scholar]
- 24.Yu, F.-M., Qiao, B., Zhu, F., Wu, J.-C., and Yuan, Y.-J. (2006) Appl. Biochem. Biotechnol. 135 145-158 [DOI] [PubMed] [Google Scholar]
- 25.Doi-Katayama, Y., Yoon, Y. J., Choi, C.-Y., Yu, T.-W., Floss, H. G., and Hutchinson, C. R. (2000) J. Antibiot. 53 484-495 [DOI] [PubMed] [Google Scholar]
- 26.Kotowska, M., Pawlik, K., Butler, A. R., Cundliffe, E., Takano, E., and Kuczek, K. (2002) Microbiology 148 1777-1783 [DOI] [PubMed] [Google Scholar]
- 27.Kim, B. S., Cropp, T. A., Beck, B. J., Sherman, D. H., and Reynolds, K. A. (2002) J. Biol. Chem. 277 48028-48034 [DOI] [PubMed] [Google Scholar]
- 28.Tang, Y., Koppisch, A. T., and Khosla, C. (2004) Biochemistry 43 9546-9555 [DOI] [PubMed] [Google Scholar]
- 29.Tsai, S. C., Lu, H., Cane, D. E., Khosla, C., and Stroud, R. M. (2002) Biochemistry 41 12598-12606 [DOI] [PubMed] [Google Scholar]
- 30.Tsai, S.-C., Miercke, L. J. W., Krucinski, J., Gokhale, R., Chen, J. C. H., Foster, P. G., Cane, D. E., Khosla, C., and Stroud, R. M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14808-14813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruner, S. D., Weber, T., Kohli, R. M., Schwarzer, D., Marahiel, M. A., Walsh, C. T., and Stubbs, M. T. (2002) Structure 10 301-310 [DOI] [PubMed] [Google Scholar]
- 32.Samel, S. A., Wagner, B., Marahiel, M. A., and Essen, L.-O. (2006) J. Mol. Biol. 359 876-889 [DOI] [PubMed] [Google Scholar]
- 33.Chakravarty, B., Gu, Z., Chirala, S. S., Wakil, S. J., and Quiocho, F. A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15567-15572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frueh, D. P., Arthanari, H., Koglin, A., Vosburg, D. A., Bennett, A. E., Walsh, C. T., and Wagner, G. (2008) Nature 454 903-906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koglin, A., Lohr, F., Bernhard, F., Rogov, V. V., Frueh, D. P., Strieter, E. R., Mofid, M. R., Guntert, P., Wagner, G., Walsh, C. T., Marahiel, M. A., and Dotsch, V. (2008) Nature 454 907-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., Fritsch, E. F., and and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 37.Hoover, D. M., and Lubkowski, J. (2002) Nucleic Acids Res. 30 e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gokhale, R. S., Tsuji, S. Y., Cane, D. E., and Khosla, C. (1999) Science 284 482-485 [DOI] [PubMed] [Google Scholar]
- 39.Tsuji, S. Y., Cane, D. E., and Khosla, C. (2001) Biochemistry 40 2326-2331 [DOI] [PubMed] [Google Scholar]
- 40.Gasteiger, E., Hoogland, C., Gattiker, A., Durand, S., Wilkins, M. R., Appel, R. D., and Bairoch, A. (2005) in The Proteomics Protocols Handbook (Walker, J. M., ed) pp. 571-607, Humana Press, Totowa, NJ
- 41.Guerrero, S. A., Hecht, H. J., Hofmann, B., Biebl, H., and Singh, M. (2001) Appl. Microbiol. Biotechnol. 56 718-723 [DOI] [PubMed] [Google Scholar]
- 42.Deutsch, J., Rapoport, S. I., and Rosenberger, T. A. (2002) Neurochem. Res. 27 1577-1582 [DOI] [PubMed] [Google Scholar]
- 43.Lambalot, R. H., Gehring, A. M., Flugel, R. S., Zuber, P., LaCelle, M., Marahiel, M. A., Reid, R., Khosla, C., and Walsh, C. T. (1996) Chem. Biol. 3 923-936 [DOI] [PubMed] [Google Scholar]
- 44.Quadri, L. E., Weinreb, P. H., Lei, M., Nakano, M. M., Zuber, P., and Walsh, C. T. (1998) Biochemistry 37 1585-1595 [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski, Z., and Minor, V. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 46.Terwilliger, T. C., and Berendzen, J. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 849-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger, T. C. (2003) Acta Crystallogr. Sect. D Biol. Crystallogr. 59 38-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 49.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 50.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D50 760-763 [Google Scholar]
- 51.Painter, J., and Merritt, E. (2006) J. Appl. Crystallogr. 39 109-111 [Google Scholar]
- 52.Navaza, J. (1994) Acta Crystallogr. Sect. A Found. Crystallogr. 50 157-163 [Google Scholar]
- 53.Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B., III, Snoeyink, J., Richardson, J. S., and Richardson, D. C. (2007) Nucleic Acids Res. 35 375-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frishman, D., and Argos, P. (1995) Proteins 23 566-579 [DOI] [PubMed] [Google Scholar]
- 55.Heinig, M., and Frishman, D. (2004) Nucleic Acids Res. 32 500-502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akey, D. L., Kittendorf, J. D., Giraldes, J. W., Fecik, R. A., Sherman, D. H., and Smith, J. L. (2006) Nat. Chem. Biol. 2 537-542 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.