Abstract
The transient protein-protein interactions induced by guanine nucleotide-dependent conformational changes of G proteins play central roles in G protein-coupled receptor-mediated signaling systems. Leukemia-associated RhoGEF (LARG), a guanine nucleotide exchange factor for Rho, contains an RGS homology (RH) domain and Dbl homology/pleckstrin homology (DH/PH) domains and acts both as a GTPase-activating protein (GAP) and an effector for Gα13. However, the molecular mechanism of LARG activation upon Gα13 binding is not yet well understood. In this study, we analyzed the Gα13-LARG interaction using cellular and biochemical methods, including a surface plasmon resonance (SPR) analysis. The results obtained using various LARG fragments demonstrated that active Gα13 interacts with LARG through the RH domain, DH/PH domains, and C-terminal region. However, an alanine substitution at the RH domain contact position in Gα13 resulted in a large decrease in affinity. Thermodynamic analysis revealed that binding of Gα13 proceeds with a large negative heat capacity change (ΔCp°), accompanied by a positive entropy change (ΔS°). These results likely indicate that the binding of Gα13 with the RH domain triggers conformational rearrangements between Gα13 and LARG burying an exposed hydrophobic surface to create a large complementary interface, which facilitates complex formation through both GAP and effector interfaces, and activates the RhoGEF. We propose that LARG activation is regulated by an induced-fit mechanism through the GAP interface of Gα13.
Heterotrimeric G proteins3 serve as key molecular switches to transduce a large array of extracellular signals into cells by actively alternating their conformations between GDP-bound inactive and GTP-bound active forms. In the current model, the ligand-activated G protein-coupled receptors (GPCRs) catalyze the exchange of GDP for GTP on Gα subunits (1). Upon activation, three switch regions in the Gα subunit undergo significant conformational changes, followed by dissociation of the GTP-bound Gα subunit from the Gβγ subunits. Both Gα-GTP and free Gβγ interact with diverse downstream effectors to transmit intracellular signals. The Gα subunit hydrolyzes bound GTP to GDP by its intrinsic GTPase activity. This deactivation process is further accelerated by GTPase-activating proteins (GAPs) such as regulator of G protein signaling (RGS) proteins (2, 3). Gα-GDP dissociates from effectors and re-associates with Gβγ to terminate the signal.
Although this model explains the basic concept of G protein signaling, the molecular dynamics of interactions among GPCR, G protein, RGS protein, and effector during the signaling process is not well understood. It has been suggested that the GPCR signals are integrated into the intracellular signaling network at the level of G proteins (4). Accumulating evidence suggests that the Gα subunit acts as the core of the signaling complex at the membrane, which is formed through the transient protein-protein interactions of multiple signaling components (5, 6). Thus, the quantitative analysis of the dynamic molecular interactions in the GPCR signaling complex will be crucial to understanding various cellular processes.
Gα12 and Gα13 subunits have been demonstrated to regulate the activity of Rho GTPase through RhoGEFs, which contain an N-terminal RGS homology domain (RH-RhoGEFs) (7–10). RH-RhoGEFs, which consist of p115RhoGEF/Lsc, PDZ-Rho-GEF/GTRAP48, and LARG in mammalian species, directly link the activation of GPCRs by extracellular ligands to the regulation of Rho activity in cells (10–14). All three RH-RhoGEFs contain an N-terminal RH domain, which specifically recognizes the active form of Gα12 or Gα13 and central DH/PH domains characteristic of GEFs for Rho GTPases. It has been demonstrated in vitro that LARG and p115RhoGEF serve as specific GAPs for Gα12/13 through their RH domains and also as their effectors to regulate Rho GTPase activation (11–13). A structural study has demonstrated that the interface of the RH domain of p115RhoGEFs and a Gα13/i1 chimera is different from that of the RGS domain of RGS4 and Gαi1 (7). The N-terminal small element in the RH domain, which is required for GAP activity toward Gα13, contacts the switch regions and the helical domain of the Gα13/i1 chimera. The core module of the p115RhoGEF RH domain binds to the region of Gα13/i1, which is conventionally used for effector binding. These results suggest roles for the RH domain in the stimulation of GEF activity by Gα13 in addition to GAP activity. On the other hand, several studies have also indicated that regions outside of RH domain of RH-RhoGEFs, particularly the DH/PH domains, interact directly with activated Gα13 (11, 14, 15). In addition, we have demonstrated recently that p115RhoGEF interacts with distinct surfaces of Gα13 for the GAP reaction or GEF activity regulation (16). However, the molecular mechanism of LARG activation upon Gα13 binding is not clearly understood.
In this study, we have developed a quantitative method for the kinetic and thermodynamic analysis of Gα13-effector interaction using surface plasmon resonance (SPR) with sensor chips on which Gα13 was immobilized. We examined the kinetics and thermodynamics of the Gα13-LARG interaction and assessed LARG activation using both in vitro and cell-based approaches. We present evidence that, in addition to the interaction with the RH domain, the DH/PH domains and C-terminal region of LARG also interact with Gα13 to form the high affinity Gα13-LARG complex and activate RhoGEF activity. We further propose that LARG adopts the active conformation using an induced-fit mechanism through association with the GAP interface of Gα13. A similar mechanism may also be used with other Gα-effector interactions.
EXPERIMENTAL PROCEDURES
Construction of Plasmids—Human LARG cDNA (NM 015313), mouse Gα13 cDNA (NM 010303), and human p115RhoGEF (NM004706) were used in this study. Full-length LARG (FL-LARG) (aa 1–1544), LARG-RDPC (aa 307–1544), LARG-DPC (aa 617–1544), LARG-RDP (aa 307–1146), LARG-PDZ (aa 1–188), LARG-RH (aa 318–625), LARG-DH/PH (aa 607–1146), LARG-C (aa 1305–1544), PDZ-RhoGEF, FL-p115 (aa 1–912), and p115-RH (aa 1–252) were subcloned into the pcDNA-myc vector with an N-terminal myc tag (Fig. 1A). cDNAs encoding the wild-type Gα13WT and the constitutively active Gα13Q226L (Gα13QL) mutant were subcloned into the pCMV5 vector. The Gα13K204A (Gα13KA) and Gα13K204A/Q226L (Gα13KA/QL) point mutants were generated in Gα13WT or Gα13Q226L, respectively, using the QuikChange site-directed mutagenesis kit as described previously (Stratagene) (15). cDNA encoding the human V14RhoA mutant was subcloned into the pCMV5-FLAG vector and used for cell-based assays.
FIGURE 1.
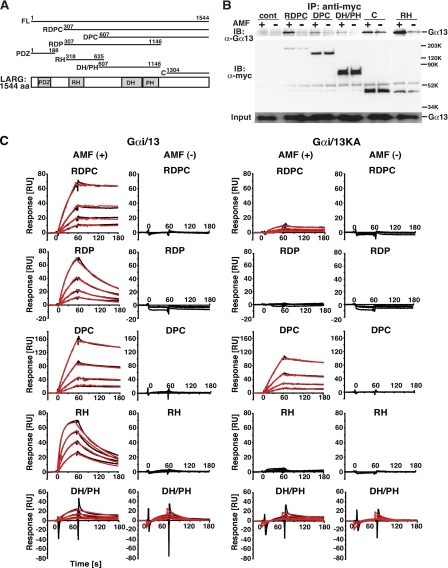
Direct interaction of LARG with Gα13 through its RH domain, DH/PH domains, and C-terminal region. A, schematic representation of LARG and its deletion constructs. The amino acid numbers encoded in each constructs are listed. PDZ, PDZ domain; RH, RGS homology domain; DH, Dbl homology domain; PH, pleckstrin homology domain. A full-length, RDPC, DPC, RDP, PDZ, RH, DH/PH, or C-terminal region of LARG is referred to as LARG-FL, -RDPC, -DPC, -RDP, -PDZ, -RH, -DH/PH, or -C, respectively. B, the binding of various LARG proteins to Gα13 in COS1 cells. COS1 cells were co-transfected with Gα13WT (0.5 μg) and the indicated myc-tagged LARG constructs: RDPC (5 μg), DPC (4 μg), DH/PH (3 μg), C(4 μg), and RH (5 μg). The LARG proteins were immunoprecipitated by anti-Myc antibody from cell lysates in the presence or absence of AMF. Immunoprecipitates were separated by SDS-PAGE, followed by Western blotting using anti-Gα13 antibody or anti-Myc antibody. C, kinetics of binding of LARG to Gαi/13 or Gαi/13KA immobilized on the SPR biosensor. Gαi/13 and Gαi/13KA proteins were immobilized on parallel channels of the Biacore sensor chip CM5 as described under “Experimental Procedures.” The association phase of the reaction between serially diluted LARG fragments and Gαi/13 was 2 min, and the dissociation phase was 1 min at 15 °C. The interactions were measured using Biacore 3000. Black lines show the experimental data. Red lines show fitting data analyzed as simultaneous ka/kd, 1:1 binding, and global fitting using the BIAevaluation program. In the absence of AMF, kinetic analyses were not performed when the responses were <1/10 of those with Gαi/13 in the presence of AMF. The concentrations of proteins were: FL, 1.1–17.5 nm; RDPC, 4.4–70 nm; RDP, 1.1–17.5 nm; DPC, 37.5–600 nm; RH, 8.8–140 nm; and DH/PH, 78.1 nm to 1.25 μm.
Expression and Purification of Proteins—FL-LARG, LARG-RDPC, LARG-DPC, LARG-RDP, LARG-DH/PH, LARG-RH, LARG-C, and LARG-PDZ constructs were subcloned into the pFastBacHT transfer vector with a six-histidine tag at the N terminus (Invitrogen), and their recombinant baculoviruses were generated. Recombinant Gα13 wild-type (Gα13WT), His6-geranylgeranylated RhoA (gg-RhoA), and His6-LARG proteins were purified using the Sf9-baculovirus expression system as described before (17). His6-Gαi/13 or -Gαi/13K204A chimeric protein, with the N-terminal coding region of rat Gαi1 (residues 1–28), and the backbone of Gα13WT or Gα13K204A (residues 47–377) were purified from Sf9 cells as described (9). This substitution of the N-terminal helix of Gαi1 for the corresponding region of Gα13 has been shown to facilitate the production of functional protein retaining the specific biochemical properties of wild-type Gα13. For SPR assays, the His6 tags of LARG, Gαi/13, and Gαi/13KA were removed by digestion with tobacco etch virus protease for 8 h at 4 °C.
SPR Assays—SPR measurements were carried out using a Biacore 3000 and a Biacore T100. Gαi/13 and Gαi/13KA proteins were immobilized on parallel channels of the Biacore sensor chip CM5 using amine coupling chemistry according to the manufacturer's protocol. The coupling reactions were performed at 5 μl/min at 25 °C using Running Buffer AMF (-) (10 mm Hepes, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% Surfactant P20, 4 mm MgCl2, 10 μm GDP, 1 mm dithiothreitol, 30 [micro]M AlCl3, 10 mM MgCl2, 10 mM NaF). All flow cell surfaces were activated by a 1:1 mixture of N-hydroxy-succinimide/1-ethyl-3-(3-dimethylaminopropyl) carbodiimide for 7 min, followed by injection of Gαi/13 or Gαi/13KA diluted to 10 μg/ml in Ligand Dilution Buffer (10 mm sodium acetate, pH 6.0, 1 mm MgCl2, 10 μm GDP, 1 mm dithiothreitol) to a density of ∼4000 relative units. Residual binding sites on the surface were blocked with a 7-min injection of 1 m ethanolamine.
Kinetic binding analysis was performed at 15 °C. Immobilized Gαi/13 or Gαi/13KA on a biosensor chip, which was in an inactive form in the absence of AMF (30 μm AlCl3/10 mm MgCl2/10 mm NaF), was activated by AMF in running buffer. Samples of serially diluted LARG proteins at five to six steps were injected over the inactivated or activated Gαi/13 and Gαi/13KA surfaces using Running Buffer AMF (-) or Running Buffer AMF (+) (10 mm Hepes, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% surfactant P20, 10 mm MgCl2, 10 μm GDP, 1 mm dithiothreitol, 10 mm NaF, 30 μm AlCl3) and reference flow cells at a flow rate of 10 μl/min for 1 min followed by 2-min dissociation using KINJECT mode. The sensor surfaces were regenerated by the injection of Regeneration Buffer (1 n NaCl, 1 mm MgCl2, 10 μm GDP, 1 mm dithiothreitol) after each binding cycle. All data were corrected for nonspecific binding and buffer shifts by double subtracting binding responses collected from a blank reference cell and a buffer injection over a Gαi/13 or Gαi/13KA immobilized flow cell. Data were fitted to a simultaneous ka/kd, 1:1 binding, and Global fitting using the BIAe-valuation program. Kinetic analyses were not performed when the responses were <1/10 compared with those with Gαi/13 in the presence of AMF. The experiments were performed more than four times. Thermodynamic experiments were done at a range of temperatures from 7 °C to 15 °C in 2° increments. Six different concentrations of LARG proteins were used for each temperature.
Thermodynamic Data Analysis—At an equilibrium state, the thermodynamic parameters, ΔH° (enthalpy change), ΔS° (entropy change), ΔG° (free energy change), and ΔCp° (heat capacity change) were derived from the van't Hoff equation, which states that ΔG° =ΔH° - TΔS° = RTlnKD, by measuring the temperature dependence of KD (equilibrium constant). Such parameters determined by the van't Hoff method are demonstrated to agree with those determined directly using calorimetry (18–20). If ΔH and ΔS have significant temperature dependence and ΔCp was assumed to be independent of temperature over a short temperature range, the relationship between the parameters becomes,
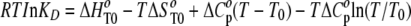 |
(Eq. 1) |
with T0 defined as the reference temperature (25 °C). The thermodynamic parameters in this study were calculated by fitting the data to Equation 1 using BiacoreT100 software.
For thermodynamic analysis of a transition state, the Eyring theory was employed (21),
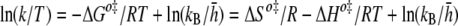 |
(Eq. 2) |
where the ‡ symbol denotes the transition state, k is the association or dissociation constant, kB is the Boltzmann constant, and [tserbian] is the Planck constant. According to Equation 2, the thermodynamic parameters at association and dissociation were calculated by linear fitting by measuring ka and kd at several temperatures using BiacoreT100 software.
Binding of LARG Proteins with His-Gαi/13 Immobilized on Ni-NTA-Agarose—COS1 cells were transfected using the Lipofectamine PLUS reagent with the indicated LARG constructs: myc-LARG-DPC (8 μg) and myc-RH (4 μg). Transfected cells were incubated at 37 °C in 10% CO2 for 24 h. Cells were lysed with 650 ml of 1.25 × lysis buffer B (50 mm Tris-HCl, pH 7.5/150 mm NaCl/1% Nonidet P-40/0.5 mm EDTA/9.8 mm 2-mercaptoethanol/10 mm β-glycerophosphate/10 mm Na3VO4/10 mm MgCl2/100 μm ATP/30 μm GDP/protease inhibitors) and centrifuged at 100,000 × g for 30 min. Equal amounts of supernatants were separated and incubated with 1.4 μg of His-Gαi/13 immobilized on 5 μl of Ni-NTA-agarose beads in 1 × lysis buffer B with or without AMF for 60 min at 4 °C. The mixtures were washed three times 1 × lysis buffer B with or without AMF. The relative amount of LARG fragments bound to the activated- or deactivated-Gαi/13 was visualized by immunoblot analysis using anti-Myc antibody.
Statistical Analysis—Data represent the mean ± S.E. of at least three independent experiments. Statistical significance of the difference was assigned based on results of Student's t test (Figs. 2C and 3D), or analysis of variance followed by Fisher's protected least significant difference test (Fig. 3b) (*, p < 0.05). Actual n and p values are provided in each figure legend.
FIGURE 2.
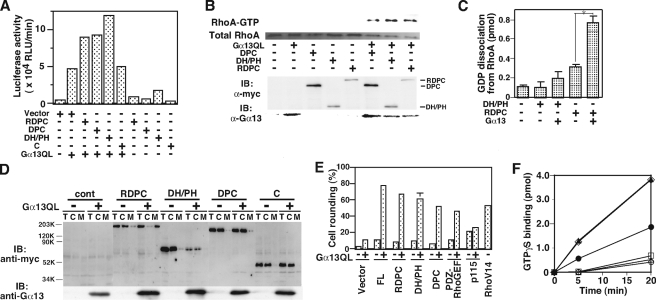
RhoGEF activation of LARG by the direct interaction of the DH/PH domains
with Gα13. A, potentiation of SRF activation by
LARG with Gα13. HeLa cells were cotransfected with 0.1 μg
of SRE-luciferase reporter plasmid and the indicated constructs:
Gα13QL (0.01 μg), myc-LARG-RDPC (0.1 μg),
myc-LARG-DPC (0.1 μg), myc-LARG-DH/PH (0.01 μg), and
myc-LARG-C (0.1 μg). SRF activities of cell lysates were measured
24 h after transfection as described in the supplemental materials.
B, RhoA activation by LARG-DH/PH with Gα13 in HeLa
cells. HeLa cells were transiently transfected with the indicated plasmids:
Gα13QL (1 μg), myc-tagged RDPC (10 μg),
myc-DPC (1 μg), and myc-DH/PH (0.5 μg). The cells were
serum-starved for 24 h after 5 h of transfection. Endogenous RhoA in the
GTP-bound form was isolated using GST-Rhotekin RBD from the cell lysates as
described in the supplemental materials. The expression of Myc-tagged LARG
constructs and Gα13 were analyzed by Western blotting using
anti-Myc and anti-Gα13 antibodies. C, in vitro
RhoGEF assays of LARG-DH/PH and -RDPC. GDP dissociation from RhoA was measured
at 20 °C after 2-min incubation in the presence of the indicated proteins:
LARG-RDPC (20 nm), LARG-DH/PH (30 nm),
 -activated Gα13WT
(100 nm). (Student's t test: n = 4, *,
p < 0.001). D, recruitment of LARG with RH domain to the
plasma membrane by constitutively active Gα13. HeLa cells
were cotransfected in the same procedure as in Fig. 2A. The
expression of LARG constructs in total cell lysate (T), crude
cytosolic (C), and membrane (M) fractions, and
Gα13 in total lysates was detected by immunoblotting using
anti-Myc antibody or anti-Gα13 antibody. E, cell
rounding induced by LARG constructs with the constitutively active
Gα13 in MDCKII cells. MDCKII cells were transiently
transfected with the indicated myc-tagged LARG constructs in the
presence or absence of Gα13QL, or FLAG-tagged V14RhoA alone.
The cells were serum-starved for 24 h after 5 h of transfection, then fixed,
and triply stained with anti-Myc antibody, anti-Gα13
antibody, and phalloidin for filamentous actin. Transfected cells were
visualized by fluorescence microscopy, identified, and scored for rounding
indicating the involvement of RhoA activation as described under the
supplemental materials. The values were calculated from four independent
experiments. The images are in supplemental Fig. S4. F,
stimulation of the RhoGEF activity of LARG by Gαi/13KA.
GTPγS binding to RhoA (200 nm) was measured at 20 °C
after 5-min incubation in the presence of the following proteins: ○,
control; ▵,
-activated Gα13WT
(100 nm). (Student's t test: n = 4, *,
p < 0.001). D, recruitment of LARG with RH domain to the
plasma membrane by constitutively active Gα13. HeLa cells
were cotransfected in the same procedure as in Fig. 2A. The
expression of LARG constructs in total cell lysate (T), crude
cytosolic (C), and membrane (M) fractions, and
Gα13 in total lysates was detected by immunoblotting using
anti-Myc antibody or anti-Gα13 antibody. E, cell
rounding induced by LARG constructs with the constitutively active
Gα13 in MDCKII cells. MDCKII cells were transiently
transfected with the indicated myc-tagged LARG constructs in the
presence or absence of Gα13QL, or FLAG-tagged V14RhoA alone.
The cells were serum-starved for 24 h after 5 h of transfection, then fixed,
and triply stained with anti-Myc antibody, anti-Gα13
antibody, and phalloidin for filamentous actin. Transfected cells were
visualized by fluorescence microscopy, identified, and scored for rounding
indicating the involvement of RhoA activation as described under the
supplemental materials. The values were calculated from four independent
experiments. The images are in supplemental Fig. S4. F,
stimulation of the RhoGEF activity of LARG by Gαi/13KA.
GTPγS binding to RhoA (200 nm) was measured at 20 °C
after 5-min incubation in the presence of the following proteins: ○,
control; ▵, 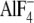 -activated
Gαi/13 (100 nm); □, AMF-activated
Gαi/13KA (100 nm); •, RDPC (10
nm); ▴, RDPC (10 nm) plus AMF-activated
Gαi/13 (100 nm); ⋄, RDPC (10 nm)
plus AMF-activated Gαi/13KA (100 nm).
-activated
Gαi/13 (100 nm); □, AMF-activated
Gαi/13KA (100 nm); •, RDPC (10
nm); ▴, RDPC (10 nm) plus AMF-activated
Gαi/13 (100 nm); ⋄, RDPC (10 nm)
plus AMF-activated Gαi/13KA (100 nm).
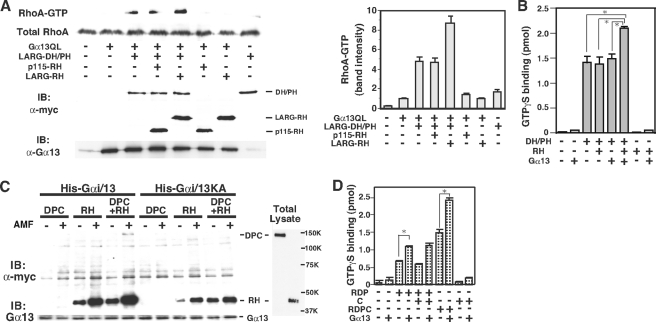
FIGURE 3.
Functional roles of the RH domain and C-terminal region for DH/PH domains-mediated GEF activation of LARG. A, Enhancement of the Gα13-stimulated RhoGEF activity of LARG-DH/PH by LARG-RH in HeLa cells. HeLa cells were transiently transfected with the indicated plasmids: Gα13QL (1 μg), myc-tagged LARG-DH/PH (1.5 μg), myc-LARG-RH (2 μg), or myc-p115-RH (0.02 μg). GTP-bound form of RhoA was isolated as described in Fig. 2B. The left of the panel is a representative result from three independent experiments. The right panel shows the mean ± S.E. of values of amount of GTP-RhoA scanned using ImageJ program. B, potentiation of the Gα13-induced RhoGEF activity of LARG-DH/PH by LARG-RH in vitro. GTPγS binding to RhoA (200 nm) was measured at 20 °C after 5-min incubation in the presence of the indicated proteins: AMF-activated Gα13 (100 nm), RH (600 nm), DH/PH (120 nm). (Fisher's protected least significant difference test: n = 3, *, p < 0.001). C, enhancement of the binding of LARG-RH and LARG-DPC with Gα13 immobilized on Ni-NTA-agarose. Equal amounts of cell lysates of COS1 cells transfected with myc-LARG-DPC (8 μg) or myc-LARG-RH (4 μg) were incubated with 1.4 μg of immobilized His-Gαi/13 or His-Gαi/13KA proteins onto Ni-NTA agarose beads in the presence or absence of AMF. The relative amount of LARG fragments bound to the activated- or deactivated-Gα13 was visualized by Western blot analysis using anti-Myc antibody. The expression of the LARG proteins in total cell lysates and the Gα13 protein in the reaction mixtures are also shown. D, the role of LARG C terminus in RhoGEF activation of LARG by Gα13. GTPγS binding to RhoA (200 nm) was measured at 20 °C after 5-min incubation in the presence of the indicated proteins: AMF-activated Gα13 (100 nm), RDP (20 nm), C (200 nm), and RDPC (20 nm) (Student's t test: n = 3, *, p = 0.0001).
Other Procedures—Co-immunoprecipitations, SRE-luciferase assays, subcellular localization of LARG, measurement of Rho activation in cells, cell rounding assays, and in vitro Rho-GEF assays were performed as described before (9, 13). Details of these procedures are described in the supplemental information.
RESULTS
Direct Interaction of LARG with Gα13 through Its RH Domain, DH/PH Domains, and C-terminal Region—We first examined the ability of several deletion constructs of LARG to interact with Gα13 by co-immunoprecipitation using COS1 cells that were transiently transfected with myc-tagged LARG mutants and Gα13 (Fig. 1, A and B). As previously reported, LARG-RDPC and LARG-RH, which contain the RH domain, preferentially bound to AMF-activated Gα13, whose conformation resembles that of the transition state for GTP hydrolysis (15, 22). Intriguingly, Gα13 also co-immunoprecipitated with LARG constructs without the RH domain: LARG-DPC, LARG-DH/PH, and LARG-C. Although the amount of co-immunoprecipitated Gα13 was much lower than with LARG-RDPC or LARG-RH, the interaction with these LARG fragments was still dependent on activation of Gα13. We reproducibly observed a small but significant increase in the amount of precipitated Gα13 with LARG-C in response to Gα13 activation. These results suggested that LARG might also interact with activated Gα13 through the DH/PH domains and/or C-terminal region.
We thus examined the direct interaction between LARG and Gα13 using an SPR method. We have recently constructed a Gαi/13 chimera by substituting the N-terminal α1 helix of Gαi1 for the corresponding region of Gα13 and demonstrated that this Gαi/13 protein purified by an Sf9-baculovirus expression system retains the same biochemical properties as wild-type Gα13 for binding and hydrolysis of guanine nucleotides, activation of RH-RhoGEFs, and GAP response (9) (supplemental Fig. S1).
For the SPR analysis, Gαi/13 was immobilized on the biosensor chip by amine coupling. The interaction of LARG fragments with Gαi/13 on the sensor chip was analyzed by Biacore3000. An advantage of this method is that the affinity difference of an effector for the active or inactive Gα13 can be quantitatively analyzed by the addition or exclusion of AMF in buffer on the same Gαi/13 chip. For the kinetic analysis, the concentrations of LARG fragments as analytes were different for each construct. Thus, it should be noted that we cannot evaluate the binding affinity between different LARG proteins by comparing the amplitudes of the SPR responses.
Consistent with the cellular assays, the SRP assay clearly demonstrated that LARG-RH, LARG-DH/PH, and LARG-C preferentially interact with the active form of Gαi/13 (Fig. 1C and Table 1). Because FL-LARG was sensitive to degradation during the assay, we exploited LARG-RDPC instead of FL-LARG for further analysis. LARG-RDPC showed kinetic parameters similar to FL-LARG in the presence of ALF4-. For LARG-FL versus LARG-RDPC: ka (association rate constant) = 6.0 × 105 versus 4.4 × 105 m-1 s-1, kd (dissociation rate constant) = 8.1 × 10-4 versus 3.0 × 10-4 s-1, and KD (the equilibrium dissociation constant) = 1.4 × 10-9 versus 6.8 × 10-10 m.
TABLE 1.
Kinetic parameters of LARG mutants binding to Gαi/13 or Gαi/13KA ka, association rate constant; kd, dissociation rate constant; KD = kd/ka, equilibrium constant. Kinetic properties were estimated from BIAcore raw data (four to five serial dilutions for each protein) using BIAevaluation software, as indicated in Fig. 1C.
|
Protein
|
ALF
|
Gai/13
|
Gai/13KA
|
||||
|---|---|---|---|---|---|---|---|
| ka | kd | KD | ka | kd | KD | ||
| m–1 s–1 | s–1 | m | m–1 s–1 | s–1 | m | ||
| RDPC | – | NDa | ND | ND | ND | ND | ND |
| + | 4.4 × 105 | 3.0 × 10–4 | 6.8 × 10–10 | 2.9 × 105 | 6.5 × 10–3 | 2.6 × 10–8 | |
| RDP | – | ND | ND | ND | ND | ND | ND |
| + | 9.8 × 105 | 8.6 × 10–3 | 8.8 × 10–9 | ND | ND | ND | |
| DPC | – | ND | ND | ND | ND | ND | ND |
| + | 1.8 × 104 | 1.4 × 10–3 | 7.6 × 10–8 | 1.8 × 104 | 1.4 × 10–3 | 8.1 × 10–8 | |
| RH | – | ND | ND | ND | ND | ND | ND |
| + | 6.9 × 105 | 9.7 × 10–3 | 1.5 × 10–8 | ND | ND | ND | |
| DH/PH | – | 2.7 × 104 | 2.8 × 10–2 | 1.1 × 10–6 | 3.7 × 104 | 3.2 × 10–2 | 8.5 × 10–7 |
| + | 1.7 × 104 | 7.1 × 10–3 | 4.3 × 10–7 | 2.0 × 104 | 8.5 × 10–3 | 4.3 × 10–7 | |
| C | – | 2.1 × 104 | 2.0 × 10–2 | 9.6 × 10–7 | 1.3 × 104 | 1.5 × 10–2 | 1.1 × 10–6 |
| + | 2.4 × 104 | 8.6 × 10–3 | 3.6 × 10–7 | 9.4 × 103 | 9.9 × 10–3 | 1.0 × 10–6 | |
| PDZ | – | 7.2 × 103 | 1.6 × 10–2 | 2.2 × 10–6 | 8.7 × 103 | 1.4 × 10–2 | 1.6 × 10–6 |
| + | 5.0 × 103 | 1.3 × 10–2 | 2.5 × 10–6 | 3.9 × 103 | 1.1 × 10–2 | 2.9 × 10–6 | |
ND, the kinetic analysis was not done because the responses were less than 1/10 compared to those with Gαi/13 in the presence of ALF–
The affinity of LARG-RDP or LARG-RDPC for activated Gαi/13 was ∼2-fold or 20-fold higher than the RH domain alone, respectively (Table 1). This suggests that the DH/PH domains and the C-terminal region of LARG may contribute to forming the high affinity Gαi/13-LARG complex together with the RH domain. In particular, the C-terminal region of LARG greatly decreased the rate of dissociation of the Gαi/13-LARG complex. In the presence of the C-terminal region, kd was reduced by >5-fold (Table 1).
We have previously demonstrated that the mutation of lysine 204 to alanine in the switch I region of Gα13 significantly reduces its affinity for LARG and its sensitivity to the GAP activity of LARG (8, 9, 15). To further characterize the role of the RH domain in the Gα13-LARG interaction, we compared the binding kinetics of LARG fragments with Gαi/13KA and Gαi/13, which are immobilized on separate channels of the same biosensor chip (Fig. 1C and Table 1). Consistent with our previous result, the binding of LARG-RH to Gαi/13 was abolished by the K204A mutation. In contrast, the interaction of LARG-DH/PH or LARG-DPC with Gαi/13 was not affected by the K204A mutation (Table 1). These results indicate the existence of an interaction between Gα13 and the DH/PH domains. It is also important to note that the KA mutation in Gα13 significantly reduced the affinity for LARG fragments containing the RH domain but did not change the affinity for LARG-DH/PH and LARG-DPC. Thus, the initial association of the RH domain with Gα13 might be necessary to induce the proper conformational change in LARG to form a high affinity complex with Gα13.
Stimulation of the RhoGEF Activity of LARG through Direct Interaction between the DH/PH Domains and Gα13—We next examined whether the interaction of Gα13 with the DH/PH domains of LARG induces RhoGEF activation. First, we co-expressed LARG mutants lacking the RH domain with a constitutively active, GTPase-deficient mutant of Gα13 (Gα13Q226L) in HeLa cells and assayed SRF activation of the cell lysates as an indicator of Rho activation. The SRF activity of cell lysates expressing LARG-DH/PH or LARG-DPC was further stimulated in the presence of Gα13QL similar to LARG-RDPC (Fig. 2A). The immunoblot of the transfected cell lysates showed similar expression levels of LARG constructs (Fig. 2D). However, the co-expression of Gα13WT didn't synergistically potentiate the SRF activity stimulated by LARG-DPC or LARG-RDPC (supplemental Fig. S2). We also examined the amount of GTP-bound Rho in cells using GST-RBD pulldown assays (13, 23). Consistent with Fig. 2A, GTP-bound Rho in cells expressing LARG-DH/PH, LARG-DPC, or LARG-RDPC increased with the co-expression of Gα13QL (Fig. 2B).
We then examined the stimulation of the GEF activity of the DH/PH domains by in vitro reconstitution assays using recombinant wild-type Gαi/13 (Gα13WT) and LARG proteins (13). AMF-activated Gαi/13WT stimulated the RhoGEF activity of LARG-RDPC (Fig. 2C). However, in contrast to the results from cellular assays, Gαi/13WT did not significantly stimulate the activity of LARG-DH/PH (Fig. 2, B and C). The lower binding affinity of LARG-DH/PH for Gαi/13 compared with LARG-DPC (KD: 4.3 × 10-7 m versus 7.6 × 10-8 m, respectively) may be responsible for this discrepancy.
The effect of co-expression of Gα13QL on the subcellular distribution of LARG mutants was also examined under the same conditions as for the SRF assays in Fig. 2 (A and D). All of the LARG constructs were distributed mainly in cytosolic fractions in the absence of Gα13QL. In the presence of Gα13QL, LARG-RDPC, the only construct with a RH domain, was redistributed to the membrane fraction. LARG-RH was recruited to the membrane fraction in the presence of Gα13QL, but not Gα13WT or Gα13KA (supplemental Fig. S3). These data suggest that the interaction of LARG-RH with activated Gα13 is likely to be important for inducing membrane distribution of LARG similar to the case of p115RhoGEF (5). Thus, it is suggested that Gα13 may regulate LARG activity through two distinct mechanisms, by inducing its membrane translocation through the RH domain and by stimulating its GEF activity through the DH/PH domains.
To confirm these results in a more physiological context, we performed cell rounding assays as an indicator of Rho activation (24). MDCKII cells were co-transfected with myc-tagged RH-RhoGEF constructs and Gα13QL. In the presence of Gα13QL, LARG-DH/PH or LARG-DPC induced cell rounding to a level similar to that observed with LARG-RDPC (Fig. 2E and supplemental Fig. S4). We further examined the role of lysine 204 in stimulating the GEF activity of LARG constructs. Unexpectedly, the GEF activity of LARG-RDPC was still stimulated by ALF4--activated Gαi/13KA in the reconstitution assay (Fig. 2F). These results further support the existence of a direct interaction of Gα13 with the DH/PH domains of LARG for Rho-GEF activation.
Functional Roles of the RH Domain and C-terminal Region in RhoGEF Activation Mediated by the DH/PH Domains—We next examined the effect of the isolated RH domain or C-terminal region on the Gα13-stimulated RhoGEF activity of DH/PH domains. GST-RBD pulldown assays demonstrated that co-expression of LARG-RH, but not p115RhoGEF-RH, potentiated the RhoGEF activity of LARG-DH/PH stimulated by Gα13QL (Fig. 3A). In the reconstitution assay, the Gαι/13-stimulated RhoGEF activity of LARG-DH/PH was also significantly potentiated by the addition of LARG-RH to the reaction (Fig. 3B). However, RhoGEF activity of LARG-DH/PH was not stimulated by Gα13 without the RH domain even at a higher concentration (120 nm). These results and the higher affinity of LARG-RDP versus LARG-RH or LARG-DH/PH for Gαi/13 (Table 1) suggest that, even though the effect of these separated domains is less than that of the combined molecule, the RH-Gα13 interaction increases the affinity of the DH/PH domains for Gα13.
We then examined the binding of Myc-tagged LARG-RH and LARG-DPC expressed
in COS1 cells to His-Gαi/13 immobilized on Ni-NTA-agarose
resin. The binding of LARG-DPC to
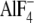 -activated Gαi/13
was dependent on the presence of LARG-RH
(Fig. 3C). In
addition, although the amount of bound LARG-DPC was much lower than that of
LARG-RH, the presence of LARG-DPC and LARG-RH enhanced their binding to
Gαi/13. With Gαi/13KA-resin, reduced binding
of LARG-RH without LARG-DPC was observed, which is consistent with the results
of the SPR analysis (Table 1).
These results suggest that the RH domain and DH/PH domains simultaneously bind
to Gα13 through separate interfaces and that the binding of
the RH domain with Gα13 would enhance the affinity of the
DH/PH domains for Gα13.
-activated Gαi/13
was dependent on the presence of LARG-RH
(Fig. 3C). In
addition, although the amount of bound LARG-DPC was much lower than that of
LARG-RH, the presence of LARG-DPC and LARG-RH enhanced their binding to
Gαi/13. With Gαi/13KA-resin, reduced binding
of LARG-RH without LARG-DPC was observed, which is consistent with the results
of the SPR analysis (Table 1).
These results suggest that the RH domain and DH/PH domains simultaneously bind
to Gα13 through separate interfaces and that the binding of
the RH domain with Gα13 would enhance the affinity of the
DH/PH domains for Gα13.
We also investigated how the interaction of the C-terminal region of LARG with Gα13 affects the RhoGEF activity using in vitro reconstitution assays (Fig. 3D). The addition of LARG-C fragment did not potentiate the GEF activity of LARG-RDP stimulated by Gαi/13. Nevertheless, the RhoGEF activity of LARG-RDPC was potentiated 2.5-fold compared with that of LARG-RDP, suggesting that the interaction of the C-terminal region with Gα13 might also be involved in the conformational change of the LARG molecule to activate the RhoGEF activity.
In summary, these results strongly indicate that the multiple interactions of LARG with Gα13 through its RH domain, DH/PH domains, and the C-terminal region coordinate together to form the active Gα13-LARG signaling complex. The association of the RH domain with the Gα13 surface, including Lys-204 likely induces the conformational change of the DH/PH domains of LARG necessary to stimulate RhoGEF activity.
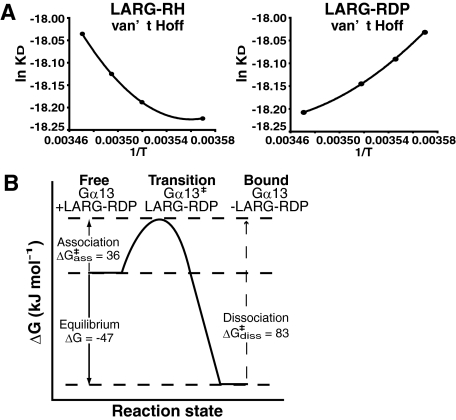
Thermodynamic Analysis of the Interaction between Gα13 and LARG—To further elucidate the dynamics of the conformational changes induced by Gα13-LARG binding, especially the contribution of the RH domain or the DH/PH domains of LARG to the interaction with Gα13, we determined the thermodynamic parameters of LARG-RH/Gα13 and LARG-RDP/Gα13 interactions by SPR analysis. The SPR analysis at several different temperatures from 7 °C to 15 °C was performed with a chip on which Gαi/13 was immobilized using BiacoreT100 as described under “Experimental Procedures.” Thermodynamic parameter (free energy G°, enthalpy H°, entropy S°, or heat capacity Cp°) changes at equilibrium state were calculated from the temperature dependence of KD using the van't Hoff equation (Table 2). The thermodynamic parameters at equilibrium estimated from SPR analysis have been consistent with those determined directly using isothermal titration calorimetry (18–20). The parameters for the transition state of the interactions were calculated by the Eyring theory using the temperature dependence of association rate constant (ka) or dissociation rate constant (kd) as described under “Experimental Procedures” (21). The temperature dependence of KD, ka, and kd from SPR analysis fit well with the theoretical equations (Fig. 4A and supplemental Fig. S5A).
TABLE 2.
Thermodynamic characterization of the interaction between LARG-RH or LARG-RDP and Gα13 at equilibrium state and in transition states using Biacore T100
| Interaction phase | Thermodynamic parameter | RH | RDP |
|---|---|---|---|
| Equilibrium | ΔG° [kJ mol–1] | –43 ± 0.019 | –47 ± 0.2 |
| ΔH° [kJ mol–1] | –85 ± 0.8 | –23 ± 8.6 | |
| ΔS° [kJ mol–1 K–1] | –0.14 ± 0.0027 | 0.079 ± 0.029 | |
| TΔS° [kJ mol–1] | –43 ± 0.82 | 24 ± 8.8 | |
| ΔCp° [kJ mol–1 K–1] | –5 ± 0.056 | –3.8 ± 0.61 | |
| Association | ΔG°† [kJ mol–1] | 38 ± 0.19 | 36 ± 0.11 |
| ΔH°‡ [kJ mol–1] | 35 ± 4 | 61 ± 2.2 | |
| ΔS°‡ [kJ mol–1 K–1] | –0.01 ± 0.014 | 0.084 ± 0.0077 | |
| TΔS°‡ [kJ mol–1] | –3 ± 4.2 | 25 ± 2.3 | |
| Dissociation | ΔG°‡ [kJ mol–1] | 82 ± 0.33 | 83 ± 0.24 |
| ΔH°‡ [kJ mol–1] | 50 ± 6.8 | 32 ± 4.6 | |
| ΔS°‡ [kJ mol–1 K–1] | –0.11 ± 0.024 | –0.17 ± 0.016 | |
| TΔS°‡ [kJ mol–1] | –32 ± 7.2 | –52 ± 4.8 |
Thermodynamic parameters of the interactions were derived from the van't Hoff plots and Eyring plots in Fig. 4 and supplemental Fig. S5
FIGURE 4.
Thermodynamic analysis of Gα13-LARG interaction. A, thermodynamic analysis of the Gα13-LARG complex formation and dissociation through its RH and DH domain. van't Hoff plots and Eyring plots of the experimental data are shown. The thermodynamic parameters at an equilibrium state and at a transition state were estimated from van't Hoff plots and Eyring plots as described under “Experimental Procedures” (Table 2 and supplemental Fig. S3). B, schematic reaction profile of the thermodynamic energies at the different states of Gα13-LARG interaction.
The LARG-RDP/Gα13 interaction showed more negative free energy change (ΔG°) than the LARG-RH/Gα13 interaction, which is consistent with the higher affinity of LARG-RDP versus LARG-RH (Table 2). In addition, thermodynamic parameters demonstrated the essential differences between the Gα13-RH interaction and Gα13-RDP interaction (Table 2). The binding of LARG-RH to Gα13 is enthalpy-driven and entropically unfavorable. In contrast, the binding of LARG-RDP to Gα13 is less favorable than that of LARG-RH from the enthalpy term but is entropy-driven with a positive ΔS°. In general, a large negative heat capacity change (ΔCp°) in a protein-protein interaction indicates the removal of water-accessible hydrophobic surface area coupled to conformational changes (25, 26). ΔCp° of 1 kJ mol-1 K-1 corresponds to the removal of ∼990 Å2 of exposed hydrophobic surface area (25, 26). Thus, a large negative ΔCp° of -3.8 kJ mol-1 K-1 together with the positive ΔS° (0.084 kJ mol-1 K-1) suggests that >3500 Å2 of hydrophobic surface is buried in the interface between Gα13 and the region between the RH domain and the DH/PH domains upon complex formation. Such extensive hydrophobic interactions would certainly stabilize the Gα13-LARG complex.
Furthermore, the free energy changes at the association and dissociation phases calculated by the Eyring theory were suitable for the equation ΔG° =ΔG°‡ass - ΔG°‡diss. A free energy barrier at the transition state of association between Gα13 and LARG-RDP was calculated as ΔG°‡ass of 36 kJ mol-1. The interaction between Gα13 and LARG-RDP will be driven by this binding free energy (Fig. 4B).
DISCUSSION
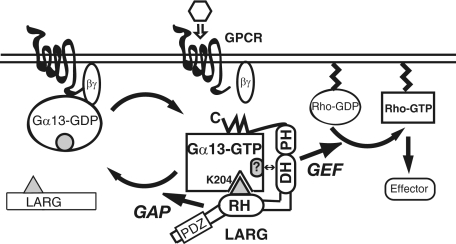
Dynamics of the Interaction between Gα13 and LARG with Large Conformational Rearrangements at the Interface—In this study, we have characterized the molecular mechanism of Gα13-LARG interaction by cellular and biochemical assays, including quantitative SPR analysis using a biosensor chip on which Gα13 was immobilized. Our results from the kinetic and thermodynamic analysis demonstrate that the simultaneous interaction of Gα13 through the RH domain, DH/PH domains, and C-terminal region of LARG facilitates the formation of the high affinity active Gα13/LARG complex. Furthermore, a large hydrophobic surface spanning >3500 Å2 is created at the interface between Gα13 and the region between the RH domain and the DH/PH domains. It has been demonstrated by Spolar and Record (26) that the formation of a such large hydrophobic surface at the interface cannot be accounted for by association between pre-existing surfaces, but rather is coupled to conformational rearrangements between the proteins, most likely by induced fit. Thus, it appears unlikely that the binding of Gα13 to the RH domain uncovers a large pre-existing surface on the DH/PH domains that contacts Gα13 via hydrophobic interactions. The K204A mutation at the RH domain contact position in Gα13 resulted in large decreases in affinity for LARG fragments containing the RH domain (Fig. 1C and Table 1). This fact strongly supports the notion that lysine 204 is in the interface between Gα13 and LARG-RH and could be a hotspot for triggering significant conformational rearrangements in the binding site, which means that the interaction is most likely driven by an induced-fit mechanism. These results indicate that the interaction between Gα13 and LARG is induced through the binding of Gα13 with the RH domain and proceeds by burying an exposed hydrophobic surface to create a large complementary interface. Thus, both GAP and effector interfaces facilitate complex formation and activation of LARG (Figs. 4 and 5). A structural study has indicated that the N-terminal small region of the DH domain of LARG is necessary to exert GEF activity for Rho (27). Interestingly, this region has been shown to act as an activation “switch” for another RhoGEF, Vav. Therefore, it is possible that the association of the RH domain with Gα13 driven by an induced-fit mechanism directly or indirectly affects this region, and enables LARG to adopt the active conformation. Further x-ray crystal structure analysis of LARG or its complex with Gα13 will be necessary to clarify these points.
FIGURE 5.
A proposed model of Rho activation through Gα13-LARG interaction. The interaction of LARG-RH with Gα13 activated by agonist-bound GPCR will induce conformational changes of DH/PH domains and C-terminal region to form an active Gα13-LARG complex.
We have recently reported that p115RhoGEF interacts with Gα13 through separate surfaces for the GAP reaction and GEF activity regulation (16). In agreement with this finding, the K204A mutation does not affect the affinity of LARG-DH/PH or -DPC for Gαi/13 (Fig. 1C), or stimulation of LARG's RhoGEF activity in reconstitution assays (Fig. 2F) strongly suggesting the existence of an additional interface between Gα13 and the DH/PH domains. Compared with a lock and key interaction, which proceeds via pre-existing complementary surfaces such as seen in the antigen-antibody interaction, the induced-fit mechanism will provide Gα subunits with much more flexibility to interact with various downstream effectors. Similar to this finding, it was demonstrated recently that the Gβγ subunit interacts with a variety of downstream effectors using homologous interfaces containing hot spots (29–32). It is thus possible that other Gα-effector interactions may be regulated by a similar mechanism.
Our result using 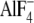 -activated
Gα13 represents the transition state of GTP hydrolysis. We
have found that LARG-DH/PH has a higher affinity for Gα13QL,
which mimics the GTP-bound state, than for
-activated
Gα13 represents the transition state of GTP hydrolysis. We
have found that LARG-DH/PH has a higher affinity for Gα13QL,
which mimics the GTP-bound state, than for
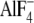 -activated Gα13
(data not shown). In contrast, LARG-RH demonstrated the opposite affinity as
previously demonstrated. The discrepancy between activation of LARG-DH/PH by
Gα13 in cell-based assays and reconstitution assays may
result from the affinity difference of LARG-DH/PH for Gα13QL
and
-activated Gα13
(data not shown). In contrast, LARG-RH demonstrated the opposite affinity as
previously demonstrated. The discrepancy between activation of LARG-DH/PH by
Gα13 in cell-based assays and reconstitution assays may
result from the affinity difference of LARG-DH/PH for Gα13QL
and 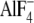 -activated
Gα13.
-activated
Gα13.
Gα/GAP/Effector Signaling Complex—We have demonstrated that the interaction of Gα13 with LARG through the RH domain (a GAP interface) and the DH/PH domains (an effector interface) could coordinate together to stimulate the RhoGEF activity of LARG (Fig. 3, A and B). The spatial and kinetic connection of GAP and GEF activities within a LARG molecule may help to regulate amplification of G protein signaling, increase the temporal resolution of the response, and increase the specificity of the signal output. Because Gα12/13 have characteristically slow rates of nucleotide exchange and GTP hydrolysis, this mechanism could be a rational system for Gα12/13 signaling to regulate multiple important cellular functions with fast responses as well as long term processes (33, 34). Cooperative regulation of GAP and catalytic activities in one single effector molecule may also apply to other Gα effectors such as PLCβ (35).
The simultaneous interaction of GAP and effector with activated Gα has been clearly demonstrated with the crystal structure of the Gαt-RGS9-PDEγ complex (36). The structure explains the mechanism by which PDEγ potentiates the GAP activity of RGS9-1: by increasing the affinity of RGS9-1 for Gαt to enhance the fidelity and temporal resolution of visual signal transduction (37, 38). Furthermore, the crystal structures of Gαq-p63RhoGEF-RhoA and Gαq-GRK2-Gβγ, where the switch I region appears available for the simultaneous binding of RGS proteins, suggest that Gα can bind an effector and a GAP at the same time (8, 39). Thus, simultaneous and non-overlapping binding of a GAP and an effector with Gα may be a conserved mechanism to help efficiently regulate G protein-mediated signaling output.
It is also interesting to note that the Gα13-RH interaction through the GAP interface may confer specificity on the Gα13-LARG interaction through its effector interface (Fig. 3A). The result is analogous to the selective potentiation of the GAP activity of RGS9 for Gαt by PDEγ (40–42). The interaction of the RH domain with Gα13 outside of the switch regions might contribute to determining the specificity of GEF activation by Gα13 (7, 9). These results suggest that RGS proteins may play another important role in GPCR signaling pathways by specifying the effector molecule for Gα. We also demonstrated that activation of Gα13 contributes to the subcellular redistribution of LARG from the cytosol to the plasma membrane through the interaction with its RH domain (Fig. 2D), which is consistent with the case of p115RhoGEF (5). This translocation will help LARG closely interact with its target, RhoA at the membrane.
Role of the C-terminal Region of LARG—Our kinetic studies indicate that the C-terminal region of LARG significantly affects the interaction with Gαi/13, especially the dissociation of LARG from Gα13 (Fig. 1C and Table 1), and regulates the Gα13-stimulated RhoGEF activity of LARG (Fig. 3D). Our preliminary thermodynamic analysis of the LARG-RDPC/Gα13 interaction could not produce an appropriate fitting to the theoretical curve, which suggests that the C-terminal region might be unstable, and some additional interacting protein may be required to stabilize the conformation upon binding to Gα13 (supplemental Fig. 3B). Recently, the presence of homo- or hetero-oligomerization of RH-RhoGEFs through their C-terminal regions has been reported (43–45). However, the physiological function of this oligomerization process in cells has not yet been clearly understood. Further study is required to understand the functional role of the C-terminal region in the regulation of Gα13-LARG signaling.
CONCLUSION
In this study we have characterized the molecular dynamics of Gα13-effector interaction by applying both kinetic and thermodynamic analyses. The Gα12/13-RhoGEF-Rho signaling pathway has been shown to participate in a variety of disease conditions, such as leukemia, cancer invasion, and hypertension (28, 46, 47). Detailed characterization of the mechanism of regulation of RH-RhoGEFs by Gα13 as demonstrated in this study will contribute to the screening of novel drugs to control these diseases. X-ray crystallography structures of Gα12/13 and the complex with target molecules has made progress toward understanding Gα12/13-mediated signaling pathways (7, 9). However, it is essential to understand the molecular dynamics of the system to provide an active view of the protein-protein interactions during the signaling process. The combined efforts of the dynamic-interaction analysis such as SPR presented in this study together with the high resolution x-ray crystallography approach will be critical to further understand the molecular mechanism of this Gα12/13-mediated signaling pathway.
Supplementary Material
Acknowledgments
We thank Yoshikazu Tanaka for help with thermodynamic analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant GM61454 (to T. K.). This work was also supported by the American Heart Association (to T. K.), by Translational Systems Biology and Medicine Initiative from the Ministry of Education, Culture, Sports, Science and Technology of Japan, by the Program of Fundamental Studies in Health Science of the National Institute of Biomedical Innovation in Japan, and by the NFAT project of the New Energy and Industrial Technology Development Organization in Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Figs. S1–S5, and references.
Footnotes
The abbreviations used are: G protein, guanosine nucleotide-binding regulatory protein; GPCR, G protein-coupled receptor; GEF, guanine nucleotide exchange factor; LARG, leukemia-associated RhoGEF; RGS, regulator of G protein signaling; RH, RGS homology; DH, Dbl homology; PH, pleckstrin homology; GAP, GTPase-activating protein; SPR, surface plasmon resonance; SRE, serum response element; GTPγS, guanosine 5′-3-O-(thio)triphosphate; PLC, phospholipase C; PDEγ, the γ subunit of cGMP phosphodiesterase; aa, amino acid(s); WT, wild type; GST, glutathione S-transferase; Ni-NTA, nickel-nitrilotriacetic acid; RBD, Rhotekin Rho-binding domain.
References
- 1.Hepler, J. R., and Gilman, A. G. (1992) Trends Biochem. Sci. 17 383-387 [DOI] [PubMed] [Google Scholar]
- 2.Ross, E. M., and Wilkie, T. M. (2000) Annu. Rev. Biochem. 69 795-827 [DOI] [PubMed] [Google Scholar]
- 3.Zerangue, N., and Jan, L. Y. (1998) Curr. Biol. 8 R313-316 [DOI] [PubMed] [Google Scholar]
- 4.Ross, E. M. (1989) Neuron 3 141-152 [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharyya, R., and Wedegaertner, P. B. (2003) Biochem. J. 371 709-720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells, C. D., Gutowski, S., Bollag, G., and Sternweis, P. C. (2001) J. Biol. Chem. 276 28897-28905 [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z., Singer, W. D., Sternweis, P. C., and Sprang, S. R. (2005) Nat. Struct. Mol. Biol. 12 191-197 [DOI] [PubMed] [Google Scholar]
- 8.Tesmer, V. M., Kawano, T., Shankaranarayanan, A., Kozasa, T., and Tesmer, J. J. (2005) Science 310 1686-1690 [DOI] [PubMed] [Google Scholar]
- 9.Kreutz, B., Yau, D. M., Nance, M. R., Tanabe, S., Tesmer, J. J., and Kozasa, T. (2006) Biochemistry 45 167-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuhara, S., Murga, C., Zohar, M., Igishi, T., and Gutkind, J. S. (1999) J. Biol. Chem. 274 5868-5879 [DOI] [PubMed] [Google Scholar]
- 11.Hart, M. J., Jiang, X., Kozasa, T., Roscoe, W., Singer, W. D., Gilman, A. G., Sternweis, P. C., and Bollag, G. (1998) Science 280 2112-2114 [DOI] [PubMed] [Google Scholar]
- 12.Kozasa, T., Jiang, X., Hart, M. J., Sternweis, P. M., Singer, W. D., Gilman, A. G., Bollag, G., and Sternweis, P. C. (1998) Science 280 2109-2111 [DOI] [PubMed] [Google Scholar]
- 13.Suzuki, N., Nakamura, S., Mano, H., and Kozasa, T. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 733-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wells, C. D., Liu, M. Y., Jackson, M., Gutowski, S., Sternweis, P. M., Rothstein, J. D., Kozasa, T., and Sternweis, P. C. (2002) J. Biol. Chem. 277 1174-1181 [DOI] [PubMed] [Google Scholar]
- 15.Nakamura, S., Kreutz, B., Tanabe, S., Suzuki, N., and Kozasa, T. (2004) Mol. Pharmacol. 66 1029-1034 [DOI] [PubMed] [Google Scholar]
- 16.Kreutz, B., Hajicek, N., Yau, D. M., Nakamura, S., and Kozasa, T. (2007) Cell Signal. 19 1681-1689 [DOI] [PubMed] [Google Scholar]
- 17.Tanabe, S., Kreutz, B., Suzuki, N., and Kozasa, T. (2004) Methods Enzymol. 390 285-294 [DOI] [PubMed] [Google Scholar]
- 18.Horn, J. R., Russell, D., Lewis, E. A., and Murphy, K. P. (2001) Biochemistry 40 1774-1778 [DOI] [PubMed] [Google Scholar]
- 19.Krogsgaard, M., Prado, N., Adams, E. J., He, X. L., Chow, D. C., Wilson, D. B., Garcia, K. C., and Davis, M. M. (2003) Mol. Cell 12 1367-1378 [DOI] [PubMed] [Google Scholar]
- 20.Wear, M. A., and Walkinshaw, M. D. (2006) Anal. Biochem. 359 285-287 [DOI] [PubMed] [Google Scholar]
- 21.Roos, H., Karlsson, R., Nilshans, H., and Persson, A. (1998) J. Mol. Recognit. 11 204-210 [DOI] [PubMed] [Google Scholar]
- 22.Coleman, D. E., Berghuis, A. M., Lee, E., Linder, M. E., Gilman, A. G., and Sprang, S. R. (1994) Science 265 1405-1412 [DOI] [PubMed] [Google Scholar]
- 23.Ren, X. D., and Schwartz, M. A. (2000) Methods Enzymol. 325 264-272 [DOI] [PubMed] [Google Scholar]
- 24.Aragay, A. M., Collins, L. R., Post, G. R., Watson, A. J., Feramisco, J. R., Brown, J. H., and Simon, M. I. (1995) J. Biol. Chem. 270 20073-20077 [DOI] [PubMed] [Google Scholar]
- 25.Murphy, K. P., Bhakuni, V., Xie, D., and Freire, E. (1992) J. Mol. Biol. 227 293-306 [DOI] [PubMed] [Google Scholar]
- 26.Spolar, R. S., and Record, M. T., Jr. (1994) Science 263 777-784 [DOI] [PubMed] [Google Scholar]
- 27.Kristelly, R., Gao, G., and Tesmer, J. J. (2004) J. Biol. Chem. 279 47352-47362 [DOI] [PubMed] [Google Scholar]
- 28.Wirth, A., Benyo, Z., Lukasova, M., Leutgeb, B., Wettschureck, N., Gorbey, S., Orsy, P., Horvath, B., Maser-Gluth, C., Greiner, E., Lemmer, B., Schutz, G., Gutkind, J. S., and Offermanns, S. (2008) Nat. Med. 14 64-68 [DOI] [PubMed] [Google Scholar]
- 29.Bonacci, T. M., Mathews, J. L., Yuan, C., Lehmann, D. M., Malik, S., Wu, D., Font, J. L., Bidlack, J. M., and Smrcka, A. V. (2006) Science 312 443-446 [DOI] [PubMed] [Google Scholar]
- 30.Ghosh, M., Peterson, Y. K., Lanier, S. M., and Smrcka, A. V. (2003) J. Biol. Chem. 278 34747-34750 [DOI] [PubMed] [Google Scholar]
- 31.Johnston, C. A., Willard, F. S., Jezyk, M. R., Fredericks, Z., Bodor, E. T., Jones, M. B., Blaesius, R., Watts, V. J., Harden, T. K., Sondek, J., Ramer, J. K., and Siderovski, D. P. (2005) Structure 13 1069-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott, J. K., Huang, S. F., Gangadhar, B. P., Samoriski, G. M., Clapp, P., Gross, R. A., Taussig, R., and Smrcka, A. V. t. (2001) EMBO J. 20 767-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozasa, T., and Gilman, A. G. (1995) J. Biol. Chem. 270 1734-1741 [DOI] [PubMed] [Google Scholar]
- 34.Singer, W. D., Miller, R. T., and Sternweis, P. C. (1994) J. Biol. Chem. 269 19796-19802 [PubMed] [Google Scholar]
- 35.Berstein, G., Blank, J. L., Jhon, D. Y., Exton, J. H., Rhee, S. G., and Ross, E. M. (1992) Cell 70 411-418 [DOI] [PubMed] [Google Scholar]
- 36.Slep, K. C., Kercher, M. A., He, W., Cowan, C. W., Wensel, T. G., and Sigler, P. B. (2001) Nature 409 1071-1077 [DOI] [PubMed] [Google Scholar]
- 37.He, W., Cowan, C. W., and Wensel, T. G. (1998) Neuron 20 95-102 [DOI] [PubMed] [Google Scholar]
- 38.Skiba, N. P., Hopp, J. A., and Arshavsky, V. Y. (2000) J. Biol. Chem. 275 32716-32720 [DOI] [PubMed] [Google Scholar]
- 39.Lutz, S., Shankaranarayanan, A., Coco, C., Ridilla, M., Nance, M. R., Vettel, C., Baltus, D., Evelyn, C. R., Neubig, R. R., Wieland, T., and Tesmer, J. J. (2007) Science 318 1923-1927 [DOI] [PubMed] [Google Scholar]
- 40.McEntaffer, R. L., Natochin, M., and Artemyev, N. O. (1999) Biochemistry 38 4931-4937 [DOI] [PubMed] [Google Scholar]
- 41.Skiba, N. P., Yang, C. S., Huang, T., Bae, H., and Hamm, H. E. (1999) J. Biol. Chem. 274 8770-8778 [DOI] [PubMed] [Google Scholar]
- 42.Sowa, M. E., He, W., Wensel, T. G., and Lichtarge, O. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1483-1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisenhaure, T. M., Francis, S. A., Willison, L. D., Coughlin, S. R., and Lerner, D. J. (2003) J. Biol. Chem. 278 30975-30984 [DOI] [PubMed] [Google Scholar]
- 44.Chikumi, H., Barac, A., Behbahani, B., Gao, Y., Teramoto, H., Zheng, Y., and Gutkind, J. S. (2004) Oncogene 23 233-240 [DOI] [PubMed] [Google Scholar]
- 45.Grabocka, E., and Wedegaertner, P. B. (2007) Mol. Pharmacol. 72 993-1002 [DOI] [PubMed] [Google Scholar]
- 46.Kelly, P., Moeller, B. J., Juneja, J., Booden, M. A., Der, C. J., Daaka, Y., Dewhirst, M. W., Fields, T. A., and Casey, P. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8173-8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kourlas, P. J., Strout, M. P., Becknell, B., Veronese, M. L., Croce, C. M., Theil, K. S., Krahe, R., Ruutu, T., Knuutila, S., Bloomfield, C. D., and Caligiuri, M. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 2145-2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.