Abstract
Replication protein A (RPA), the eukaryotic single-stranded DNA-binding complex, is essential for multiple processes in cellular DNA metabolism. The “canonical” RPA is composed of three subunits (RPA1, RPA2, and RPA3); however, there is a human homolog to the RPA2 subunit, called RPA4, that can substitute for RPA2 in complex formation. We demonstrate that the resulting “alternative” RPA (aRPA) complex has solution and DNA binding properties indistinguishable from the canonical RPA complex; however, aRPA is unable to support DNA replication and inhibits canonical RPA function. Two regions of RPA4, the putative L34 loop and the C terminus, are responsible for inhibiting SV40 DNA replication. Given that aRPA inhibits canonical RPA function in vitro and is found in nonproliferative tissues, these studies indicate that RPA4 expression may prevent cellular proliferation via replication inhibition while playing a role in maintaining the viability of quiescent cells.
Replication protein A (RPA)3 is a stable complex composed of three subunits (RPA1, RPA2, and RPA3) that binds single-stranded DNA (ssDNA) nonspecifically. RPA (also referred to as canonical RPA) is essential for cell viability (1), and viable missense mutations in RPA subunits can lead to defects in DNA repair pathways or show increased chromosome instability. For example, a missense change in a high affinity DNA-binding domain (DBD) was demonstrated to cause a high rate of chromosome rearrangement and lymphoid tumor development in heterozygous mice (2). RPA has also been shown to have increased expression in colon and breast cancers (3, 4). High RPA1 and RPA2 levels in cancer cells are also correlated with poor overall survival (3, 4), which is consistent with RPA having a role in efficient cell proliferation.
RPA is a highly conserved complex as all eukaryotes contain homologs of each of the three RPA subunits (1). At least some plants (e.g. rice) and some protists (e.g. Cryptosporidium parvum) contain multiple genes encoding for subunits of RPA (5, 6). In rice, there is evidence for multiple RPA complexes that are thought to perform different cellular functions (5). In contrast, only a single alternative form of RPA2, called RPA4, has been identified in humans (7). RPA4 was originally identified as a protein that interacts with RPA1 in a yeast two-hybrid screen (7). The RPA4 subunit is 63% identical/similar to RPA2. Comparison of the sequences of RPA4 and RPA2 suggests that the two proteins have a similar domain organization.4 RPA4 appears to contain a putative core DNA-binding domain (DBD G) flanked by a putative N-terminal phosphorylation domain and a C terminus containing a putative winged-helix domain (Fig. 1A). The RPA4 gene is located on the X chromosome, intronless, and found mainly in primates.4 Initial characterization of RPA4 by Keshav et al. (7) indicated that either RPA2 or RPA4, but not both simultaneously, interacts with RPA1 and RPA3 to form a complex. This analysis also showed that RPA4 is expressed in placenta and colon tissue but was either not detected or expressed at low levels in most established cell lines examined (7).
FIGURE 1.
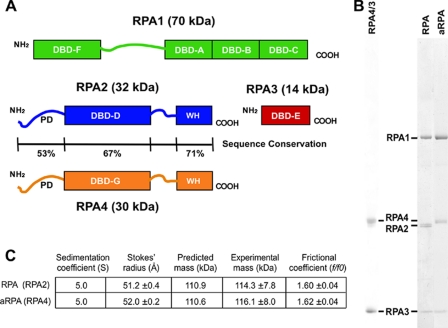
Properties of aRPA complex. A, schematic diagram of the structural and functional domains of the three subunits of RPA and (proposed for) RPA4: DNA-binding domains (DBD A-G), the phosphorylation domain (PD), winged-helix domain (WH), and linker regions (lines). The sequence similarity between RPA2 and RPA4 is indicated for each domain of the subunit. B, gel analysis of 2 μg of RPA4/3, RPA. or aRPA separated on 8-14% SDS-PAGE gels and visualized by Coomassie Blue staining. The position of each RPA subunit is indicated. C, hydrodynamic properties of aRPA and RPA complexes. The sedimentation coefficient and Stokes' radius were determined as described previously by sedimentation on a 15-35% glycerol gradient and chromatography on a Superose 6 10/300 GL column (GE Healthcare), respectively (13). Mass and frictional coefficients were calculated using the method of Siegal and Monty (8). The predicted mass was based upon the amino acid sequence derived from the DNA sequence.
These studies describe the purification and functional analysis of an alternative RPA (aRPA) complex containing RPA1, RPA3, and RPA4. The aRPA complex is a stable heterotrimeric complex similar in size and stability to the canonical RPA complex (RPA1, RPA3, and RPA2). aRPA interacts with ssDNA in a manner indistinguishable from canonical RPA; however, it does not support DNA replication in vitro. Mixing experiments demonstrate that aRPA also inhibits canonical RPA from functioning in DNA replication. Hybrid protein studies paired with structural modeling have allowed for the identification of two regions of RPA4 responsible for this inhibitory activity. Data presented here are consistent with recent analyses of RPA4 function in human cells,4 and we conclude that RPA4 has anti-proliferative properties and has the potential to play a regulatory role in human cell proliferation through the control of DNA replication.
EXPERIMENTAL PROCEDURES
Materials—HI buffers contain 30 mm HEPES (diluted from 1 m stock at pH 7.8), 1 mm dithiothreitol, 0.25 mm EDTA, 0.5% (w/v) inositol, and 0.01% (v/v) Nonidet-P40. HI was supplemented with different salt concentrations as indicated. Creatine phosphokinase from rabbit skeletal muscle and creatine phosphate disodium salt were purchased from Calbiochem. [γ-32P]ATP (250 μCi) and [α-32P]dCTP (250 μCi) were purchased from PerkinElmer Life Sciences.
Construction of aRPA and aRPA Hybrid Expression Plasmids—To purify RPA4 in a complex with RPA1 and RPA3, a PCR fragment containing RPA4 cDNA was amplified using primers 5′-CACCTGACGTCAAAAAACCCCTCAAGACCCGTTTAGAGGCCCCAAGGGGTTATGCTATTATCAATCAGCAGACTTAAAATGCTC-3′ and 5′-TTGATGGATCCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGAGTAAGAGTGGGTTTGGGAGC-3′. This fragment was then cloned into the BamHI-AatII sites of pET16b-hSSB (9), replacing the RPA2 cDNA. Subsequently, a BsrGI-ScaI fragment containing the 3′ end of RPA3 and the entire RPA4 coding region was cloned into p11d-tRPA(10) to generate the plasmid p11d-aRPA. Plasmids for expressing RPA4 alone or with RPA3 were generated by inserting the BamHI-AatII fragment into pET-11d or pET16b-RPA32/his14 (a derivative of pET16b-hSSB in which RPA1 has been deleted), respectively. A His10 tag was added to the N terminus of RPA4 and cloned into pET11d using the same method with the primer 5′-TTGATGGATCCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGGGCCATCATCATCATCATCATCATCATCATCACAGCAGCGGCCATATCGAAGGTCGTCATATGAGTAAGAGTGGGTTTGGGAGC-3′. All plasmids were confirmed by restriction digest and DNA sequencing.
Hybrid constructs were amplified from their corresponding pEGFP plasmids4 using either primer 5′-TCTCGAGGTGGATTAATGAGTAAGAGT-3′ or 5′-CTCGAGGTGGATTAATGTGGAACAGT-3′ and 5′-AGATCCGGTGGATCCCGGGCCCGC-3′. The fragment was digested with AseI and KpnI and then cloned into the NdeI and KpnI sites of pRSF. All plasmids were confirmed by restriction analysis and DNA sequencing.
Protein Expression and Purification—RPA, aRPA, and aRPA hybrids were expressed in BL21 (DE3) cells and purified as described previously (10, 11). RPA4/3 complex was purified as described.5 When dual vectors were used, both ampicillin (120 μg/ml) and kanamycin (30 μg/ml) were used for colony selection and growth.
DNA Binding Assays—Gel mobility shift assays were carried out as described previously (11). Briefly, indicated amounts of protein and radiolabeled oligonucleotide were incubated for 20 min at 25 °C in filter binding buffer (30 mm HEPES (diluted from 1 m stock at pH 7.8), 100 mm NaC1, 5 mm MgC12, 0.5% inositol, and 1 mm dithiothreitol). Reaction mixtures were separated on a 1% agarose gel in 0.1× Tris acetate-EDTA running buffer. Bound and free DNA from gel mobility shift experiments were quantitated using a Packard Instant Imager. Apparent affinity constants were calculated by nonlinear least squares fitting of the data to the Langmuir binding equation using KaleidaGraph (Synergy Software) as described previously (13).
SV40 Replication and Elongation Assays—SV40 reactions were carried out in 25 μl. Standard reactions contained 30 mm HEPES (pH 7.5); 7 mm MgCl2;50 μm dCTP with 2.5 μCi (92.5 kBq) of [α-32P]dCTP; 100 μm each of dATP, dGTP, and dTTP; 200 μm each of CTP, GTP, and UTP; 4 mm ATP; 40 mm creatine phosphate; 2.5 μg of creatine kinase; 15 mm potassium phosphate; and 50 ng of pUC.HSO DNA template. RPA, usually 300 ng, was added as indicated. Each reaction also contained 100 μg of HeLa cell cytoplasmic extract and 0.2-0.5 μg of SV40 T-antigen. SV40 T-antigen was purified by immunoaffinity chromatography from Sf9 cells infected with a baculovirus vector containing the T-antigen gene as described previously (14). Complementation assays were carried out using a 35-65% ammonium sulfate fraction of HeLa cell extract (11). Briefly, 1 ml of complete extract was precipitated by the gradual addition of ammonium sulfate to 35%. The supernatant was further precipitated with 65% ammonium sulfate. The resulting precipitant was dissolved in one-fifth of the initial volume of 50 mm Tris-HCl, pH 7.8, 1 mm dithiothreitol, 0.1 mm EDTA, 10% glycerol and dialyzed to remove any residual ammonium sulfate. All reaction mixtures were assembled on ice and incubated at 37 °C for 2 h. The reactions were analyzed on gels as described previously (11) or quantitated by precipitation by trichloroacetic acid; reactions were quenched by the addition of 0.1 m sodium pyrophosphate to a final concentration 80 mm and precipitated with 500 μl of 10% trichloroacetic acid. The reaction mixtures were filtered through glass microfiber filters and radioactive DNA quantitated by liquid scintillation.
SV40 T-antigen dependent elongation assays (15) were done as described in the SV40 replication assay with the following modifications. Reactions were assembled as above except the [α-32P]dCTP was excluded from stage I. After incubation at 37 °C for 2 h, [α-32P]dCTP and RPA or RPA variants were added, and a stage II incubation was carried out for an additional hour at 37 °C. Products were analyzed as described above.
RESULTS
RPA4 Forms a Stable, Functional ssDNA-binding Complex—Recombinant RPA4 was produced using methodology previously described to generate recombinant canonical RPA (10). The cDNA encoding RPA4 was cloned into a bacterial expression vector either alone, with RPA3, or with RPA1 and RPA3 and expressed in Escherichia coli. Overall, RPA4 had properties that were similar to those of recombinant RPA2 (10). A His-tagged RPA4 gene expressed alone was predominantly insoluble (data not shown). When RPA4 was expressed with RPA3, both proteins were predominantly soluble and could be purified as a stable RPA4/3 complex (Fig. 1B and data not shown). When all three genes (RPA1, RPA4, RPA3) were expressed simultaneously, all three polypeptides were substantially soluble, and a complex, aRPA, could be purified to near homogeneity following the purification procedure used for canonical RPA (11). The expression of RPA4 in E. coli and the yield of aRPA complex after purification were similar to that for RPA (∼0.8 mg/liter of culture). The purified aRPA contained three intense bands of 70, 34, and 14 kDa (Fig. 1B). Although RPA4 has nine fewer amino acids than RPA2 and a predicted pI (6.07) slightly more basic than RPA2 (5.75), the RPA4 subunit consistently migrated slower in SDS-PAGE gels.
We examined the hydrodynamic properties of aRPA and found them to be nearly indistinguishable from those of the canonical RPA complex; the sedimentation and Stokes' radius of aRPA were determined to be 5.0 s and 52.0 Å (versus 5.0 s and 51.2 Å for canonical RPA; Fig. 1C). The mass calculated for aRPA is in close agreement to that predicted from the amino acid sequence (Fig. 1C) and indicates RPA4 is forming a heterotrimeric complex with RPA1 and RPA3. The frictional coefficients for aRPA and RPA are both consistent with an elongated shape (16), which suggests that when RPA4 is substituted for RPA2, the overall shape of the complexes in solution is similar.
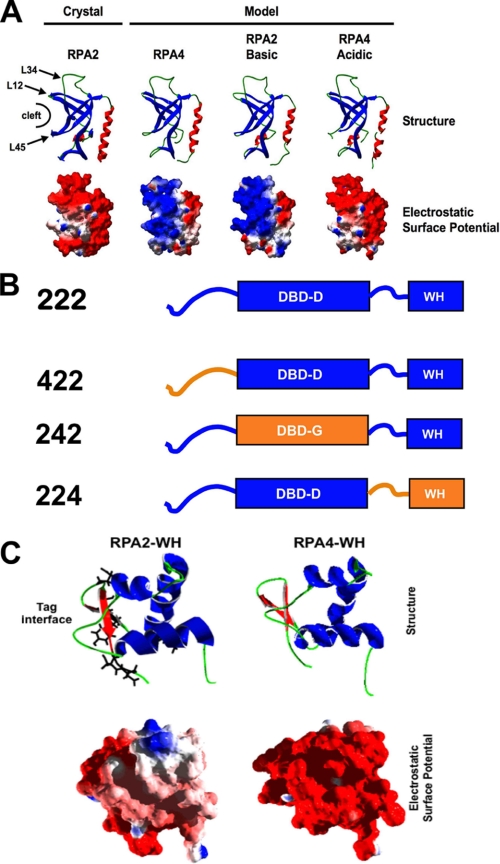
The predicted sequence of RPA4 is 63% identical/similar to RPA2.4 This similarity allows homology modeling to be used to predict the structure of the putative domains of RPA4. The known structure of the DNA-binding domain of RPA2 (DBD D; 2PI2.pdb) is shown in Fig. 2A. The shallow, putative DNA-binding cleft between the L12 and L45 loops is indicated (17). Two other prominent features of the structure are the flexible L34 loop (at the top of structure) and the C-terminal α helix, which has been shown to be part of the subunit interface of RPA2 (right side of structure) (17, 18). The known structure for DBD D of RPA2 was used to model DBD G of RPA4 (Fig. 2A). The predicted structure of DBD G is very similar to that of DBD D, suggesting that the two domains may assume similar structures (Fig. 2A). However, comparison of the predicted surface charge of the DBDs of RPA2 and RPA4 indicates that the surface of RPA2 is much more acidic than that of RPA4 (Fig. 2A, lower row, see also below).
FIGURE 2.
Structural Models of RPA2 and RPA4. A, structural models of RPA2 DBD D (2PI2.pdb) and proposed RPA4 DBD G. The proposed structure of DBD G was generated using Geno3D by modeling against DBD D. Top, the ribbon representation was generated by Swiss-PdbViewer; helices are red, β-sheet regions are blue, and coil regions are green. Putative DNA-binding cleft and important loops on DBD D are indicated. Bottom, the electrostatic surface potential of the above model are shown with regions of basic (blue), acidic (red), and neutral (white) surface potential. B, schematic of RPA2-RPA4 hybrid proteins generated. RPA2 (blue) and RPA4 (orange) domains are indicated for each hybrid. WH, winged-helix domain. C, structure of winged-helix domain. Structure of RPA2 winged-helix domain (1Z1D.pdb) and the proposed structure of the winged-helix domain of RPA4 (generated using Geno3D) are shown. Top, the ribbon representation was generated by Swiss-PdbViewer. The side chain structures of residues in the RPA2 winged helix demonstrated to interact with T antigen are shown (interface) (24). Bottom, the electrostatic surface potential of the above model is shown. All colors as in A.
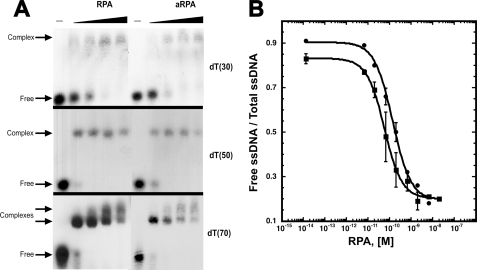
In canonical RPA, two domains in RPA1 (DBD A and B) are both necessary and sufficient for high affinity DNA binding, and RPA2 contributes little to the overall affinity of the complex for ssDNA (19-21). Therefore, aRPA, which contains RPA1, RPA4, and RPA3, was expected to bind ssDNA with high affinity. We analyzed the binding affinity of purified aRPA to (dT)30 by gel mobility shift assays. The binding of canonical RPA and aRPA is very similar; nearly equivalent concentrations of protein were needed to form a complex, and only one protein-DNA species was observed (Fig. 3A). Quantitation of the titrations demonstrated that both RPA complexes have high affinity for ssDNA; Kd equals 7.5 × 10-9 m for RPA and 20 × 10-9 m for aRPA (Fig. 3B). Binding was also examined with longer oligonucleotides, (dT)50 and (dT)70. Only one protein-DNA species was observed with (dT)50, whereas two distinct protein-DNA bands were observed with dT70 for both aRPA and RPA (Fig. 3A), suggesting that for high concentrations of both proteins, two molecules bind to dT70. Together these data indicated that the occluded binding site of aRPA is 25-35 nucleotides, which is comparable with the binding site size of RPA (13). The occluded binding site size was confirmed with stoichiometric reverse titrations monitoring changes in intrinsic protein fluorescence as described by Kim et al. (13) (data not shown). These analyses also indicate that aRPA binds with low cooperativity similar to RPA because if aRPA bound with high cooperativity, a single transition would have been observed with dT70 rather than a gradual transition between single- and double-liganded species (Fig. 3A). We conclude that aRPA has ssDNA binding properties indistinguishable from canonical RPA; it binds ssDNA with high affinity and low cooperativity.
FIGURE 3.
DNA binding properties of RPA complexes. A, gel mobility shift assays were carried out as described previously (11). Autoradiograms of representative gel mobility shift assays of aRPA and RPA using radiolabeled dT30, dT50, and dT70 are shown. Radiolabeled dT30 (0.2 fmol), dT50 (2 fmol), or dT70 (2 fmol) was incubated with various amounts of protein (dT30: 0, 0.0067, 0.02, 0.067, 0.2 nm; dT50 and dT70: 0, 0.067, 0.2, 0.67, 2.0 nm) as described under “Experimental Procedures.” The positions of free DNA and shifted protein-DNA complex are indicated. B, representative binding isotherms for aRPA and RPA binding dT30 determined as described under “Experimental Procedures.” Binding data for RPA (circles) and aRPA (open triangles) and best-fit curves are shown.
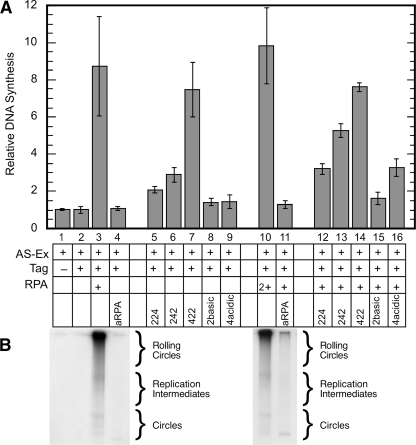
aRPA Function in SV40 Replication—RPA was originally identified as a protein essential for simian virus 40 (SV40) DNA replication (22); therefore, we examined whether aRPA could support SV40 DNA replication. Cell extracts derived from human tissue culture cells contain all of the cellular proteins required for SV40 replication, except the viral protein large T antigen (Tag) (23). RPA is required for SV40 replication and is present in the cell extracts (22); however, the extracts can be depleted of RPA using ammonium sulfate fractionation, making the DNA synthesis dependent on both Tag and RPA (11). RPA-depleted, ammonium sulfate-fractionated extract (AS-Ex) is unable to support DNA synthesis in the presence of Tag unless the reaction is also supplemented with RPA (Fig. 4A, bars 1-3). A complete reaction with canonical RPA gives robust DNA synthesis (Fig. 4A, bar 3). In contrast, supplementation with aRPA results in only background levels of DNA synthesis (Fig. 4A, bar 4). Background synthesis was also observed when purified RPA4/3 complex was added in place of RPA (data not shown). Replication of the SV40 origin containing DNA occurs by two mechanisms in these reactions: circle-to-circle and rolling circle. These mechanisms produce different products, circles and long linear DNA, respectively (Fig. 4B) (15). Analysis of the products by gel electrophoresis showed that aRPA did not support the formation of either type of product (Fig. 4B, left panel). We conclude that although aRPA binds ssDNA with high affinity, it is unable to support SV40 DNA replication.
FIGURE 4.
SV40 DNA replication with various forms of RPA. A plus sign in the table indicates an addition of the indicated component: 300 ng RPA forms, 0.2-0.5 μg SV40 large T antigen (Tag), 100 μg of HeLa cytosolic extract (AS-Ex). The 2+ indicates 600 ng of RPA. The amount of DNA synthesis after 2 h of incubation at 37 ° was quantitated by scintillation counting or analyzed by agarose gel electrophoresis. A, summary of quantitative analysis of replication. Five independent experiments with duplicate points at each condition were completed. The data from each experiment were normalized to the minus Tag and RPA control (bar 1) for that experiment, averaged, and plotted. Maximal DNA synthesis for individual experiments ranged from 23 to 60 pmol. Error bars represent standard deviation for the combined data. B, autoradiograph of the products of a representative reactions (containing the indicated components) after separation on a 1% agarose gel. The mobility of various DNA forms is indicated.
Interestingly, the addition of aRPA to unfractionated extracts also showed only background levels of synthesis (data not shown). This is surprising because canonical RPA is present in these unfractionated extracts and normally supports replication. Reactions containing both purified aRPA and purified RPA were analyzed. Additional canonical RPA (double the normal amount) in the reaction results in a modest increase in DNA synthesis (Fig. 4A, bar 10). When aRPA was added in the presence of equal amounts of RPA, no DNA synthesis was observed (Fig. 4A, bar 11). This demonstrates that aRPA has a dominant negative effect on the function of canonical RPA in SV40 DNA replication. This effect does not appear to be caused by dissociation of the aRPA complex or subunit exchange because the addition of the purified RPA4/3 complex had no effect on a complete SV40 replication containing canonical RPA (data not shown).
RPA2 and RPA4 are both composed of three distinct functional domains: the phosphorylation domain, a DBD, and a winged-helix domain (Fig. 1A). To determine what region(s) of RPA4 is responsible for the properties of aRPA in DNA replication, three hybrid proteins were generated in which the phosphorylation domain, the DBD, or the C terminus of RPA2 was replaced with the corresponding domain of RPA4, named RPA2 (422), RPA2 (242), and RPA2 (224), respectively (Fig. 2B). These domain hybrid proteins were expressed with RPA1 and RPA3, and the resulting complexes were purified. All three complexes purified with a yield similar to RPA and bound (dT)30 with an affinity equivalent to wild-type RPA (data not shown). When the trimeric complexes, RPA·2 (422), RPA·2 (242), and RPA·2 (224), were examined for the ability to support DNA synthesis, only the RPA·2 (422) hybrid complex was able to support wild-type levels of DNA synthesis (Fig. 4A, bars 5-7). RPA·2 (242) and RPA·2 (224) both supported levels of synthesis that were slightly above background and aRPA levels. We conclude that the phosphorylation domain of RPA4 is not responsible for the phenotype observed with aRPA. These data also indicate that both the DBD and the winged-helix domains of RPA2 are necessary for RPA function in SV40 DNA replication and that both of these domains of RPA4 are contributing to the aRPA phenotype.
Mixing experiments were also carried out with the RPA2-RPA4 hybrids. RPA·2 (422) did not inhibit the function of RPA and showed levels of synthesis comparable with that of RPA alone. Both RPA·2 (242) and RPA·2 (224) showed levels of DNA synthesis that were significantly reduced from that of RPA (t test; p < 0.005 and p < 0.001, respectively) but greater than that of aRPA (Fig. 4A, bars 12-14). RPA·2 (224) consistently showed more inhibition than RPA·2 (242), suggesting that the two domains may have different effects on SV40 DNA replication. We conclude that both the DBD and the winged-helix domain of aRPA are contributing to the inhibitory effect of RPA4.
Mechanism of aRPA Inhibition of SV40 Replication—RPA is required for both initiation and elongation phases of DNA replication. To examine which phase of replication is being affected by aRPA, two-stage elongation assays were carried out. Time course experiments have shown that in the SV40 replication reaction, initiation predominantly occurs during early times (stage I), and at later times (stage II), only elongation synthesis on rolling-circle intermediates is occurring (15). It is therefore possible to examine aRPA function in elongation in a two-stage reaction. Stage I contains all the components necessary for initiation and elongation of SV40 origin-containing DNA except for the radioactive dCTP tracer. This stage is incubated for 2 h at 37 °C, during which normal initiation and elongation occur, but the DNA synthesized is not labeled. In stage II, [α-32P]dCTP and various forms of RPA are added, the incubation is continued for 1 h, and the elongation DNA synthesis is quantitated. This assay measures DNA synthesis occurring during the elongation phase and is independent of the initiation processes (15).
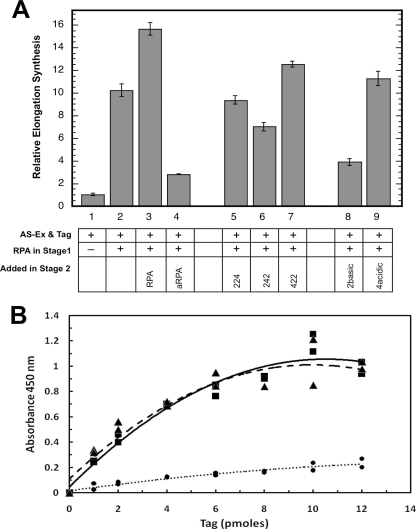
Substantial elongation synthesis was observed in the stage II elongation phase (Fig. 5A, bar 2). This synthesis was dependent on the presence of RPA from the start of the reaction and could be stimulated by additional RPA at the beginning of stage II (Fig. 5A, bars 1-3). aRPA strongly inhibits elongation synthesis, demonstrating that aRPA inhibits the normal function of canonical RPA at the pre-existing replication fork (Fig. 5A, bar 4). This strong inhibition was not observed with the hybrid subunits (Fig. 5A, bars 5-7). RPA·2 (422) causes a slight increase in elongation synthesis similar to the addition of canonical RPA (t test; p < 0.0005) and consistent with its ability to promote replication. RPA·2 (224) had no effect on elongation synthesis (t test; p < 0.11), whereas RPA·2 (242) showed slightly reduced levels of DNA synthesis (t test; p < 0.001). Together these experiments indicate that the putative phosphorylation domain of RPA4 has no role in inhibiting elongation synthesis, whereas DBD G of RPA4 inhibits elongation synthesis. Interestingly, the putative winged helix of RPA4 appears to have a separation of function phenotype. Although this region results in inhibition of the complete SV40 DNA synthesis, it does not affect elongation synthesis. This suggests that the putative winged helix-containing C terminus of RPA4 is defective for replication initiation only.
FIGURE 5.
Function in elongation ad Tag interactions. A, SV40 elongation assay. Two-stage SV40 elongation assays were carried out as described under “Experimental Procedures.” A plus sign in the table indicates the addition of 300 ng RPA in stage I or 300 ng of the indicated form of RPA in stage II. 100 μg of HeLa cytosolic extract (AS-Ex) and 0.2-0.5 μg of SV40 large T antigen (Tag) were added to all reactions. Three independent experiments with all reactions done in duplicate were completed. The data from each experiment were normalized to the minus Tag and RPA control (bar 1) for that experiment, averaged, and plotted. Maximal DNA synthesis for individual experiments ranged from 128 to 135 pmol. Error bars represent standard deviation for the combined data. B, interactions of aRPA or RPA with Tag monitored by enzyme-linked immunosorbent assay (11). The lines indicate the average of two independent experiments: aRPA (closed triangle), RPA (closed square), and bovine serum albumin (closed circle). Microtiter plate wells were coated with 1 μg of indicated protein for 1 h, blocked, and washed. The indicated quantities of SV40 large T antigen (Tag) were then incubated in each well for 1 h. After washing, wells were incubated sequentially with Pab419 SV40 Tag antibody (12) and peroxidase-conjugated secondary antibody each for 1 h. After the final incubation, the wells were washed and developed using 200 μl of 0.8 mg/ml o-phenylenediamine in a 0.50 m phosphate-citrate buffer, and absorbance at 450 nm was determined. All steps were carried out at room temperature.
Structural Basis of RPA4 Inhibition—The DNA-binding domains of RPA2 and RPA4 are predicted to have similar structures but very different electrostatic surface potentials (Fig. 2A). Because the solution structure of the C-terminal region of RPA2 is known (24), we used homology modeling to predict the structure of the C terminus of RPA4. Fig. 2C shows that the predicted structure for the winged helix of RPA4 is very similar to the known structure of the RPA2 winged helix. The predicted surface potential is predominantly acidic for both winged-helix domains; however, the N terminus of the predicted winged helix of RPA4 is much more acidic than the equivalent region of RPA2 (Fig. 2C). The inhibition studies discussed above suggest that the putative winged-helix domain of RPA4 is inhibiting initiation; RPA·2 (224) inhibits the complete reaction but has no effect on elongation synthesis. This is consistent with a previous analysis that described an important role for the winged-helix domain of RPA2 in initiation of SV40 replication (24). In contrast, RPA·2 (242) inhibits both the complete SV40 replication reaction and the elongation reaction. This suggests that DBD G of RPA4 (Fig. 2A) is affecting an RPA function (or functions) normally required for both phases of replication.
Initiation of SV40 replication requires binding of the origin of replication by SV40 large T antigen (Tag) and specific interactions between RPA and T antigen to promote unwinding of the origin sequence and loading of DNA polymerase α/primase complex (25-27). Protein interaction assays were carried out to determine whether aRPA interacts with T antigen. aRPA interacts with SV40 T antigen to the same extent as RPA (Fig. 5B). It has been shown that T antigen interacts with both the core DNA-binding domain of RPA1 and the C terminus (winged helix) of RPA2 (key residues Glu-252, Tyr-256, Ser-257, Asp-261, Thr-267, Asp-268) (24, 28). This region of the winged helix of RPA2 is partially conserved in RPA4, with 3 of the 6 key residues differing between RPA2 and RPA4 (Glu-252, Tyr-256, Pro-257*, Arg-261*, Ala-267*, Asp-268; asterisks indicate non-conserved residues). The finding that aRPA interacts strongly with T antigen (Fig. 5B) suggests either that the interaction is primarily mediated through RPA1 or that the partial conservation of the C terminus of the winged helix in RPA4 is sufficient for interaction with Tag.
The inability of aRPA to support SV40 DNA replication indicates either that aRPA is forming a nonfunctional initiation complex with Tag or that aRPA is inhibiting another part of the initiation reaction. T antigen has origin-dependent helicase activity, which can be stimulated nonspecifically by RPA or other single-stranded DNA-binding proteins (22, 29). We found that aRPA stimulated T antigen-dependent unwinding at levels comparable with canonical RPA (data not shown). This indicates that aRPA does not inhibit T antigen helicase and that the defects in replication are more likely to be in subsequent steps of initiation such as loading of DNA polymerase α/primase complex or primer synthesis by DNA polymerase α/primase complex.
Construction of a Dominant Negative Form of RPA2—DBD G has a more basic surface charge than DBD D and is capable of inhibiting the function of the canonical RPA complex. This suggests that electrostatic interactions may be responsible for the altered function of DBD G. A comparison of the sequences of RPA2 and RPA4 identified one region that was very poorly conserved between RPA2 and RPA4, amino acids 108-123 in RPA2, referred to as the L34 loop (Fig. 2A). These residues are acidic in DBD D (5/17 acidic and 0/17 basic residues) and basic in DBD G (1/16 acidic and 3/16 basic residues). To test whether this region is responsible for the difference in activity between RPA4 and RPA2, the acidic region of RPA2 and the basic region of RPA4 were exchanged for one another. This resulted in two mutated subunits: an RPA2 subunit that has the basic L34 loop of DBD G (RPA2Basic) and an RPA4 subunit with the acidic L34 loop of DBD D (RPA4Acidic). These structures were again modeled against the crystal structure of DBD D (2PI2.pdb; Ref. 17), and the electrostatic surface potential was displayed for each (Fig. 2A). The electrostatic surface potential shows that RPA2Basic has a predicted surface potential similar to RPA4 and that RPA4Acidic has a predicted surface potential similar to RPA2.
The RPA2Basic and RPA4Acidic complexes were expressed in E. coli and purified. Both complexes bound (dT)30 with an affinity equivalent to wild-type RPA (data not shown). Each was then tested for the ability to support SV40 DNA replication as described above. Neither trimeric complex, RPA·2Basic nor RPA·4Acidic, was able to support DNA synthesis in the SV40 system (Fig. 4A, bars 8-9). Mixing experiments containing equal amounts of RPA, and RPA·2Basic had background levels of DNA synthesis (Fig. 4A, bars 8 and 15), indicating that RPA·2Basic is strongly inhibitory of RPA activity in DNA replication. This is similar to that observed for aRPA. In contrast, mixing RPA and RPA·4Acidic resulted in intermediate levels of synthesis that were similar to those obtained with RPA·2 (224) (Fig. 4A, bars 9, 12, and 16). This suggests that removing the basic L34 loop from DBD G reduces the inhibitory activity of this domain. In elongation assays, RPA·2Basic inhibited DNA synthesis almost as well as aRPA, whereas RPA·4Acidic had no inhibitory effect on elongation synthesis (Fig. 5A). These findings indicate that the L34 loop of RPA4 is both necessary and sufficient for the inhibitory activity of DBD G. In HeLa cells studies, RPA·2Basic is defective in chromosomal DNA replication and has a dominant negative effect.4 Therefore, replication inhibition (both viral and cellular) appears to be a general property of this short amino acid stretch of RPA4.
DISCUSSION
We have shown that aRPA and RPA have similar biochemical properties but not similar functions. Both complexes have similar solution structures and DNA binding activity, but aRPA is unable to support in vitro SV40 DNA replication. Analysis of the mechanism of aRPA action indicates that aRPA inhibits the function of canonical RPA in the initiation and elongation phases of DNA replication (Fig. 6). Recent findings suggest that RPA4 also does not support chromosomal replication in the absence of RPA2, suggesting that these properties identified in vitro also hold true for cellular replication.4 Our findings suggest a model by which RPA4 levels could regulate DNA replication in the cell. At low concentrations of RPA4, aRPA complex formation is also low, and efficient DNA replication will occur, utilizing canonical RPA. When RPA4 is expressed at higher levels, aRPA forms and exists at a level that can inhibit the replication activity of canonical RPA. RPA4 is expressed in some human tissues (7), suggesting that cell viability is maintained in the presence of RPA4. Thus, we would predict that in cells that need to perform genome maintenance, but not genome duplication (i.e. quiescent cells), aRPA might be able to substitute for canonical RPA. Alternatively, it has been recently shown that there is another single-stranded binding protein (hSSB1) in human cells that may have a role in DNA repair (30). This protein could help maintain viability in RPA4-expressing cells. It will be important to determine whether aRPA and/or hSSB1 can support at least some basal processes normally performed by RPA, such as DNA repair. Multiple protein-protein interactions are important for RPA function in SV40 DNA replication. These include interactions with SV40 Tag, DNA polymerase α, and topoi-somerase I during initiation (21, 31) and interactions with RF-C, DNA polymerase α, and polymerase δ in elongation (32). Because aRPA has ssDNA binding properties similar to RPA, it is most likely that altered protein interactions are responsible for the inability of aRPA to function in replication. aRPA interacts with SV40 Tag and can stimulate Tag DNA unwinding at the same level as RPA, suggesting that the inhibitory properties of aRPA result from aRPA either forming nonfunctional complexes with the replication machinery or being unable to participate in a subset of essential protein interactions.
FIGURE 6.
Schematic showing observed properties for RPA2, RPA4, and RPA2 hybrids. PD, phosphorylation domain; WH, winged-helix domain. See “Discussion” for details.
Two regions of RPA4 have been identified to be involved in its activity: the basic L34 loop of DBD G and the winged-helix domain (Fig. 6). Analysis of RPA4Acidic and RPA2Basic indicated that the L34 loop in RPA4 is both necessary and sufficient for inhibition of SV40 DNA replication. The RPA4Acidic complex, which contains RPA4 with the L34 loop from RPA2, has properties similar to RPA·2 (224) in both DNA replication and elongation. In contrast, the RPA2Basic complex, which contains RPA2 with the L34 loop of RPA4, strongly inhibits all replication reactions, identical to the full aRPA complex. Recent analysis of RPA4 function in human cells indicates that the L34 loop also inhibits cellular chromosomal replication,4 suggesting that this is a general property of this loop. The importance of the L34 loop in RPA function has not been previously identified. Modeling of the electrostatic surface potential of DBD G (Fig. 2A) indicates that the basic loop from RPA4 has a large influence on the surface potential of this domain. It seems likely that this change in surface potential is causing the inhibitory activity of this domain.
The second domain of RPA4, the C terminus containing a putative winged helix, also affects SV40 DNA replication. RPA·2 (224) strongly inhibited a complete replication reaction but had minimal effects on elongation, suggesting that the putative winged-helix domain is only playing a critical role in the initiation of SV40 replication. Structure-function analysis of RPA2 previously mapped the T antigen interaction domain to the C terminus of the winged-helix domain and showed that this interaction was important for initiation of SV40 replication (24). Our analysis of hybrid RPA2 complexes demonstrates that this interaction is either only necessary for initiation or can also occur with RPA4. In contrast, the DNA-binding domain of RPA4 is inhibitory in both complete and elongation assays. Furthermore, previous studies have shown that the DNA-binding domain is the only domain of RPA2 essential for life in yeast (33).5 Additional analysis is necessary to understand the complete function of the winged helix-containing domains of RPA2 and RPA4.
It has recently been demonstrated that RPA4 expression in human cells does not allow the cell to replicate its genome nor proceed through the cell cycle.4 In addition, the original analysis of RPA4 by Keshav et al. (7) showed that RPA4 expression occurs in predominantly quiescent cells and not in cell lines, which are by definition proliferative. Our detailed biochemical characterization of purified alternative RPA complex (containing RPA1, RPA3, and RPA4) provides definitive evidence that not only does aRPA prevent DNA replication, it does so in the presence of canonical RPA. The fact that RPA4 is expressed in at least some tissues suggests that it may have an active role in preventing cell proliferation and promoting quiescence. Canonical RPA is crucial for maintenance of the genome. aRPA has similar solution properties and DNA binding activity, so it seems likely that aRPA can function in at least some cell maintenance processes normally carried out by RPA. These findings suggest that RPA4 has potential functions as a therapeutic agent and/or target not only in preventing cell proliferation (i.e. cancer) but also as a potential antiviral agent (i.e. through prevention of viral duplication).
Acknowledgments
We thank Anindya Dutta for providing RPA4 plasmid. We also thank the members of the Wold laboratory for evaluation of data and critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Research Grant (GM44721). This work was also supported by grants from the Center for Aging and Cancer at the University of Iowa (CA103672). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RPA, human replication protein A; aRPA, alternative RPA; RPA1, 70-kDa subunit of RPA; RPA2, 32-kDa subunit of RPA; RPA3, 14-kDa subunit of RPA; RPA4, product of the RPA4 gene; ssDNA, single strand DNA; DBD, DNA-binding domain; AS-Ex, ammonium sulfate fractionated extract; hSSB 1, human single-stranded binding protein 1.
S. J. Haring, T. D. Humphreys, and M. S. Wold, submitted for publication.
A. M. Dickson, Y. Krasikova, P. Pestryakov, O. Lavrik, and M. S. Wold, submitted for publication.
References
- 1.Wold, M. S. (1997) Annu. Rev. Biochem. 66 61-92 [DOI] [PubMed] [Google Scholar]
- 2.Wang, Y., Putnam, C. D., Kane, M. F., Zhang, W., Edelmann, L., Russell, R., Carrion, D. V., Chin, L., Kucherlapati, R., Kolodner, R. D., and Edelmann, W. (2005) Nat. Genet. 37 750-755 [DOI] [PubMed] [Google Scholar]
- 3.Givalos, N., Gakiopoulou, H., Skliri, M., Bousboukea, K., Konstantinidou, A. E., Korkolopoulou, P., Lelouda, M., Kouraklis, G., Patsouris, E., and Karatzas, G. (2007) Mod. Pathol. 20 159-166 [DOI] [PubMed] [Google Scholar]
- 4.Tomkiel, J. E., Alansari, H., Tang, N., Virgin, J. B., Yang, X., VandeVord, P., Karvonen, R. L., Granda, J. L., Kraut, M. J., Ensley, J. F., and Fernandez-Madrid, F. (2002) Clin. Cancer Res. 8 752-758 [PMC free article] [PubMed] [Google Scholar]
- 5.Ishibashi, T., Kimura, S., and Sakaguchi, K. (2006) J. Biochem. (Tokyo) 139 99-104 [DOI] [PubMed] [Google Scholar]
- 6.Rider, S. D., Jr., Cai, X., Sullivan, W. J., Jr., Smith, A. T., Radke, J., White, M., and Zhu, G. (2005) J. Biol. Chem. 280 31460-31469 [DOI] [PubMed] [Google Scholar]
- 7.Keshav, K. F., Chen, C., and Dutta, A. (1995) Mol. Cell. Biol. 15 3119-3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegal, L. M., and Monty, K. J. (1966) Biochim. Biophys. Acta 112 346-362 [DOI] [PubMed] [Google Scholar]
- 9.Zhang, D., Frappier, L., Gibbs, E., Hurwitz, J., and O'Donnell, M. (1998) Nucleic Acids Res. 26 631-637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henricksen, L. A., Umbricht, C. B., and Wold, M. S. (1994) J. Biol. Chem. 269 11121-11132 [PubMed] [Google Scholar]
- 11.Binz, S. K., Dickson, A. M., Haring, S. J., and Wold, M. S. (2006) Methods Enzymol. 409 11-38 [DOI] [PubMed] [Google Scholar]
- 12.Harlow, E., Crawford, L. V., Pim, D. C., and Williamson, N. M. (1981) J. Virol. 39 861-869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, C., Paulus, B. F., and Wold, M. S. (1994) Biochemistry 33 14197-14206 [DOI] [PubMed] [Google Scholar]
- 14.Brush, G. S., Kelly, T. J., and Stillman, B. (1995) Methods Enzymol. 262 522-548 [DOI] [PubMed] [Google Scholar]
- 15.Walther, A. P., Bjerke, M. P., and Wold, M. S. (1999) Nucleic Acids Res. 27 656-664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomes, X. V., and Wold, M. S. (1996) Biochemistry 35 10558-10568 [DOI] [PubMed] [Google Scholar]
- 17.Deng, X., Habel, J. E., Kabaleeswaran, V., Snell, E. H., Wold, M. S., and Borgstahl, G. E. (2007) J. Mol. Biol. 374 865-876 [DOI] [PubMed] [Google Scholar]
- 18.Bochkareva, E., Korolev, S., Lees-Miller, S. P., and Bochkarev, A. (2002) EMBO J. 21 1855-1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walther, A. P., Gomes, X. V., Lao, Y., Lee, C. G., and Wold, M. S. (1999) Biochemistry 38 3963-3973 [DOI] [PubMed] [Google Scholar]
- 20.Sibenaller, Z. A., Sorensen, B. R., and Wold, M. S. (1998) Biochemistry 37 12496-12506 [DOI] [PubMed] [Google Scholar]
- 21.Fanning, E., Klimovich, V., and Nager, A. R. (2006) Nucleic Acids Res. 34 4126-4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wold, M. S., and Kelly, T. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 2523-2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly, T. J. (1988) J. Biol. Chem. 263 17889-17892 [PubMed] [Google Scholar]
- 24.Arunkumar, A. I., Klimovich, V., Jiang, X., Ott, R. D., Mizoue, L., Fanning, E., and Chazin, W. J. (2005) Nat. Struct. Mol. Biol. 12 332-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melendy, T., and Stillman, B. (1993) J. Biol. Chem. 268 3389-3395 [PubMed] [Google Scholar]
- 26.Collins, K. L., and Kelly, T. J. (1991) Mol. Cell. Biol. 11 2108-2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murakami, Y., Eki, T., and Hurwitz, J. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 952-956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun, K. A., Lao, Y., He, Z., Ingles, C. J., and Wold, M. S. (1997) Biochemistry 36 8443-8454 [DOI] [PubMed] [Google Scholar]
- 29.Kenny, M. K., Lee, S.-H., and Hurwitz, J. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 9757-9761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard, D. J., Bolderson, E., Cubeddu, L., Wadsworth, R. I., Savage, K., Sharma, G. G., Nicolette, M. L., Tsvetanov, S., McIlwraith, M. J., Pandita, R. K., Takeda, S., Hay, R. T., Gautier, J., West, S. C., Paull, T. T., Pandita, T. K., White, M. F., and Khanna, K. K. (2008) Nature 453 677-681 [DOI] [PubMed] [Google Scholar]
- 31.Simmons, D. T., Gai, D., Parsons, R., Debes, A., and Roy, R. (2004) Nucleic Acids Res. 32 1103-1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuzhakov, A., Kelman, Z., Hurwitz, J., and O'Donnell, M. (1999) EMBO J. 18 6189-6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philipova, D., Mullen, J. R., Maniar, H. S., Gu, C., and Brill, S. J. (1996) Genes Dev. 10 2222-2233 [DOI] [PubMed] [Google Scholar]