Abstract
Rap1 small GTPases interact with Rap1-GTP-interacting adaptor molecule (RIAM), a member of the MRL (Mig-10/RIAM/Lamellipodin) protein family, to promote talin-dependent integrin activation. Here, we show that MRL proteins function as scaffolds that connect the membrane targeting sequences in Ras GTPases to talin, thereby recruiting talin to the plasma membrane and activating integrins. The MRL proteins bound directly to talin via short, N-terminal sequences predicted to form amphipathic helices. RIAM-induced integrin activation required both its capacity to bind to Rap1 and to talin. Moreover, we constructed a minimized 50-residue Rap-RIAM module containing the talin binding site of RIAM joined to the membrane-targeting sequence of Rap1A. This minimized Rap-RIAM module was sufficient to target talin to the plasma membrane and to mediate integrin activation, even in the absence of Rap1 activity. We identified a short talin binding sequence in Lamellipodin (Lpd), another MRL protein; talin binding Lpd sequence joined to a Rap1 membrane-targeting sequence is sufficient to recruit talin and activate integrins. These data establish the mechanism whereby MRL proteins interact with both talin and Ras GTPases to activate integrins.
Increased affinity (“activation”) of cellular integrins is central to physiological events such as cell migration, assembly of the extracellular matrix, the immune response, and hemostasis (1). Each integrin comprises a type I transmembrane α and β subunit, each of which has a large extracellular domain, a single transmembrane domain, and a cytoplasmic domain (tail). Talin binds to most integrin β cytoplasmic domains and the binding of talin to the integrin β tail initiates integrin activation (2–4). A small, PTB-like domain of talin mediates activation via a two-site interaction with integrin β tails (5), and this PTB domain is functionally masked in the intact talin molecule (6). A central question in integrin biology is how the talin-integrin interaction is regulated to control integrin activation; recent work has implicated Ras GTPases as critical signaling modules in this process (7).
Ras proteins are small monomeric GTPases that cycle between the GTP-bound active form and the GDP-bound inactive form. Guanine nucleotide exchange factors (GEFs) promote Ras activity by exchanging bound GDP for GTP, whereas GTPase-activating proteins (GAPs)3 enhance the hydrolysis of Ras-bound GTP to GDP (for review, see Ref. 8). The Ras subfamily members Rap1A and Rap1B stimulate integrin activation (9, 10). For example, expression of constitutively active Rap1 activates integrin αMβ2 in macrophage, and inhibition of Rap1 abrogated integrin activation induced by inflammatory agonists (11–13). Murine T-cells expressing constitutively active Rap1 manifest enhanced integrin dependent cell adhesion (14). In platelets, Rap1 is rapidly activated by platelet agonists (15, 16). A knock-out of Rap1B (17) or of the Rap1GEF, RasGRP2 (18), resulted in impairment of αIIbβ3-dependent platelet aggregation, highlighting the importance of Rap1 in platelet aggregation in vivo. Thus, Rap1 GTPases play important roles in the activation of several integrins in multiple biological contexts.
Several Rap1 effectors have been implicated in integrin activation (19–21). Rap1-GTP-interacting adaptor molecule (RIAM) is a Rap1 effector that is a member of the MRL (Mig-10/RIAM/Lamellipodin) family of adaptor proteins (20). RIAM contains Ras association (RA) and pleckstrin homology (PH) domains and proline-rich regions, which are defining features of the MRL protein family. In Jurkat cells, RIAM overexpression induces β1 and β2 integrin-mediated cell adhesion, and RIAM knockdown abolishes Rap1-dependent cell adhesion (20), indicating RIAM is a downstream regulator of Rap1-dependent signaling. RIAM regulates actin dynamics as RIAM expression induces cell spreading; conversely, its depletion reduces cellular F-actin content (20). Whereas RIAM is greatly enriched in hematopoietic cells, Lamellipodin (Lpd) is a paralogue present in fibroblasts and other somatic cells (22).
Recently we used forward, reverse, and synthetic genetics to engineer and order an integrin activation pathway in Chinese hamster ovary cells expressing a prototype activable integrin, platelet αIIbβ3. We found that Rap1 induced formation of an “integrin activation complex” containing RIAM and talin (23). Here, we have established the mechanism whereby Ras GTPases cooperate with MRL family proteins, RIAM and Lpd, to regulate integrin activation. We find that MRL proteins function as scaffolds that connect the membrane targeting sequences in Ras GTPases to talin, thereby recruiting talin to integrins at the plasma membrane.
EXPERIMENTAL PROCEDURES
Antibodies and Plasmid DNA Constructs—The activation-dependent mouse monoclonal antibody PAC1, activating mouse monoclonal antibody anti-LIBS6 (Ab33) against αIIbβ3, and the αIIbβ3-specificcompetitive inhibitor Ro43-5054 were previously described (23). Anti-HA mouse monoclonal antibody (MMS-101P) was purchased from Covance Research Products Inc. (Denver, PA). Anti-green fluorescent protein (GFP) rabbit polyclonal antibody was obtained from Clontech (Mountain View, CA). Anti-HA mouse monoclonal antibody (12CA5) and anti-integrin αIIb mouse monoclonal antibody (PMI-1) were also described elsewhere (24). Anti-RhoGDI rabbit polyclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Mammalian expression vector encoding RIAM has been previously described (23). GFP-Lamellipodin was a gift from Dr. F. Gertler (Massachusetts Institute of Technology, Boston, MA). cDNAs encoding full-length and truncated human RIAM or Lamellipodin were amplified by PCR and subcloned into mammalian expression vector pEGFP-C1 and bacterial expression vector pGEX4T-1, respectively. DNA constructs containing membrane-targeting sequences of Rap1A (residues 165–184) were produced by PCR-based cloning. Plasmid constructs containing designated mutations were generated by using QuikChange® II XL site-directed mutagenesis kit from Stratagene (La Jolla, CA) with mutagenic primers. Plasmids encoding for HA-Rap1A (G12V) and HA-Rap1GAP have been described previously (23). Mammalian expression constructs for HA-mouse full-length talin and its mutant (W359A) constructs were previously reported (4). Bacterial expression vector encoding His-tagged human full-length talin in pET30a was a generous gift from Dr. Liddington (Burnham Institute for Medical Research, La Jolla, CA). The authenticity of constructs was confirmed by DNA sequencing.
In Vitro Protein Interaction Assay—Bacterial expression plasmids encoding glutathione S-transferase (GST)-RIAM, GST-Lpd, their mutants, or GST vector were expressed in BL21(DE3) (Novagen, Madison, WI), and recombinant proteins were purified on glutathione-Sepharose beads according to manufacturer's instructions (GE Healthcare). His-tagged full-length talin was expressed in BL21(DE3) with 0.2 mm isopropyl 1-thio-d-galactopyranoside overnight at room temperature and purified using Ni-NTA His-bind® resin (Novagen) affinity matrix. Purified talin was dialyzed overnight in a buffer (50 mm Tris-HCl (pH 7.4), 200 mm NaCl, 0.1% Triton X-100, and 1 mm dithiothreitol). Interaction of GST-RIAM proteins or GST with His-talin was conducted in a reaction buffer (50 mm Tris-HCl (pH 7.4), 200 mm NaCl, 0.1% Triton X-100, 5–10 mg/ml bovine serum albumin, 1 mm phenylmethylsulfonyl fluoride, 10 μm E-64, protease inhibitor mixture (complete mini, Roche Applied Science). 10 μg of purified GST fusion RIAM proteins on affinity matrix was mixed with 20 μg of His-tagged talin and incubated at 4 °C for 1 h. After washing the beads with reaction buffer, samples were fractionated on 4–20% SDS-PAGE gel (Invitrogen). Bound proteins were analyzed by Western blotting or Coomassie Blue staining. Peptide inhibition assay was performed in conditions described above with purified RIAM peptides (GenScript Corp.), wild type (6–30, EDIDQMFSTLLGEMDLLTQSLGVDT), or mutant (6-30-4E, EDIDQEESTEEGEMDLLTQSLGVDT).
Transient Transfection and Immunoprecipitation—A5 cells (Chinese hamster ovary cells stably expressing αIIbβ3) were cultured in Dulbecco's modified Eagle's medium supplemented with nonessential amino acids, l-glutamine, 10% fetal calf serum, and antibiotics. Lipofectamine and Plus reagents (Invitrogen) were used for transient transfection according to manufacturer's instruction. For immunoprecipitation, cell lysates were prepared in a lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.5% IGEPAL CA-630, 1 mm phenylmethylsulfonyl fluoride, protease inhibitor mixture (complete mini) (Roche Applied Science). Cell lysates were immunoprecipitated with 5 μg of anti-HA monoclonal antibody (clone 12CA5) for 4 h at 4 °C. Protein G-Sepharose (Invitrogen) was added to the reaction mixture and further incubated for 1 h at 4 °C. After three washes with lysis buffer, beads were mixed with sample buffer and subjected to SDS-PAGE. Bound proteins were detected by Western blotting.
Integrin αIIbβ3 Activation—A5 cells were transiently cotransfected with HA-tagged full-length talin and GFP-RIAM, GFP-Lpd, their mutants, or GFP control for 24 h. Three-color cytometry was employed to measure activation-specific antibody PAC1 binding to integrin αIIbβ3 as described (23). In brief, transfected cells were suspended and incubated with 0.1% PAC1 ascites, washed, and stained with R-phycoerythrin-conjugated goat anti-mouse IgM antibody (Biomeda, Foster City, CA) to detect bound PAC1. Cells were further incubated with propidium iodide (1 μg/ml), washed, and analyzed by FACScan flow cytometer (BD Biosciences). Collected data were analyzed using CellQuest software (BD Biosciences). The integrin activation index is defined as 100 × (F - F0)/(Fmax - F0), where F is the median fluorescence intensity (MFI) of PAC1 binding, F0 is MFI of PAC1 binding in the presence of 1 μm αIIbβ3-specific inhibitor Ro43-5054, and Fmax is MFI of PAC1 binding in the presence of 2 μm αIIbβ3-activating antibody anti-LIBS6. Activation index is represented as the mean ± S.E.) for n ≥ 3.
Subcellular Fractionation—A5 cells transfected for 24 h were subjected to subcellular fractionation as described (23). Briefly, cells were harvested in a lysis buffer (20 mm Tris-HCl (pH 7.4), 50 mm NaCl, 2 mm MgCl2, 5 mm KCl, 1 mm phenylmethylsulfonyl fluoride, 10 μm E-64, protease inhibitor mixture (Complete mini, Roche Applied Science), and incubated on ice for 10 min. Swollen cells were disrupted by Dounce homogenization, and a fraction (total cell lysate) was saved for analysis. The remainder lysate was centrifuged at 2000 rpm for 10 min to pellet nuclei and unbroken cells. The supernatant was further centrifuged at 14,000 rpm for 30 min to pellet the membrane fraction, which was further washed and extracted in lysis buffer including 1% IGEPAL CA-630 at 4 °C. Total lysate and cytosolic and membrane fractions were fractionated by SDS-PAGE and analyzed for protein expression by Western blotting.
Co-clustering of Integrin αIIbβ3 and Talin with RIAM or Lamellipodin—To label αIIbβ3 integrins, A5 cells transfected with mCherry-talin and various GFP-RIAM or GFP-Lpd constructs were incubated with 30 μg/ml D57-Alexa 647 (D57 monoclonal antibody directly conjugated to Alexa-Fluor 647 (Invitrogen)) for 20 min before plating on 10 μg/ml fibrinogen-coated coverslips. The cells were allowed to adhere for 30 min in complete Dulbecco's modified Eagle's medium, rinsed once in phosphate-buffered saline (PBS), and fixed with 3.7% formaldehyde in PBS. In some experiments, A5 cells were co-transfected with HA-Rap1GAP. After fixation and permeabilization in 0.1% Triton X-100 for 5 min, HA-Rap1GAP was immunostained using anti-HA monoclonal antibody (Covance Research Products) that was directly conjugated to Alexa-Fluor 350 (Invitrogen). Coverslips were subsequently mounted in Prolong Gold antifade reagent (Invitrogen) on slides. Epifluorescent images of cells were acquired with a 60× oil immersion objective on a Nikon Eclipse TE2000-U microscope equipped with the appropriate excitation and emission filter sets (Semrock Inc, Rochester, NY). Images as shown are maximal projections of deconvolved images that were acquired at 0.1-μm z-section intervals. Images were deconvolved using the 3D Blind Deconvolution algorithm of Auto-QuantX (Media Cybernetics Inc, Bethesda, MD). Additional post-acquisition processing of images were performed using ImageJ (rsb.info.nih.gov/ij) and Adobe Photoshop.
RESULTS AND DISCUSSION
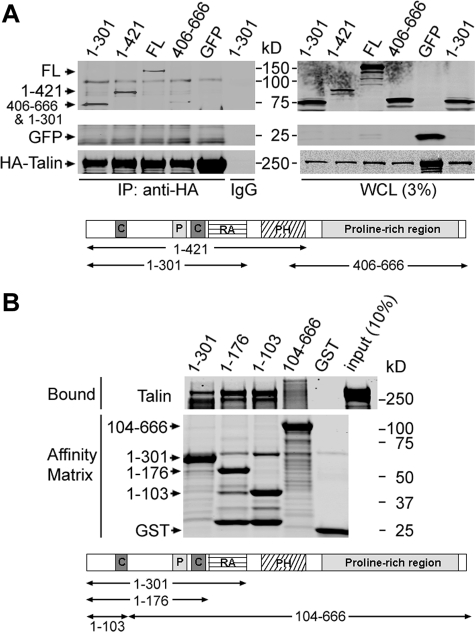
An N-terminal 103-Residue Fragment of RIAM Binds Talin—RIAM mediates Rap1-dependent integrin activation by forming a complex containing activated Rap1, talin, and the integrin (23). To investigate the mechanism by which RIAM forms this complex, we examined the interaction of various RIAM fragments with talin. cDNAs encoding influenza hemagglutinin epitope-tagged full-length talin (HA-talin) and GFP-tagged full-length RIAM or RIAM fragments were constructed and used to transfect Chinese hamster ovary cells that express recombinant integrin αIIbβ3 (A5 cells) (25). HA-talin was immunoprecipitated with anti-HA antibody, and associated GFP-RIAM fragments were detected by Western blotting with an anti-GFP antibody. As expected, full-length RIAM interacted with talin (Fig. 1A). RIAM contains several modular domains; thus, we analyzed fragments that specifically deleted one or more of these domains. The C-terminal half of RIAM, containing the principal ENA/VASP binding sites and the PH domain (20), were dispensable for talin association because fragments lacking these domains (RIAM-(1–421) and RIAM-(1–301)) associated with talin, whereas a construct containing the RIAM C terminus (406–666) associated to a much lesser extent (Fig. 1A). A talin interacting site (1–301) was, thus, localized within a RIAM fragment that contains two coiled-coil regions (63–90 and 150–182) and a RA domain (Fig. 1A). RIAM-(1–301) was well expressed as a GST fusion protein in a prokaryotic expression system, affording us the opportunity to assess the direct binding of this region of RIAM to purified recombinant talin. Purified hexahistidine-tagged full-length recombinant talin (His6-talin) was incubated with GST-tagged fragments of RIAM (GST-RIAM) immobilized on glutathione-Sepharose beads, and bound talin was detected by protein staining after SDS-PAGE fractionation. Three GST-RIAM proteins containing the N terminus residues RIAM 1–301, 1–176, and 1–103 interacted with talin, whereas GST-RIAM-(104–666) or GST alone did not (Fig. 1B). Thus, talin interacts directly with RIAM, and the interaction is mediated by an N-terminal fragment that lacks the Rap1 binding RA domain, the PH domain, and most of the ENA/VASP binding motifs.
FIGURE 1.
Talin interacts directly with RIAM. A, RIAM N-terminal fragments retain talin binding ability. A5 cells (a Chinese hamster ovary cell line expressing integrin αIIbβ3) were co-transfected with HA-tagged full-length talin and GFP-tagged RIAM constructs encompassing amino acids 1–301, 1–421, 406–666, full-length (FL) 1–666, or GFP alone, as indicated. Cell lysates were prepared and immunoprecipitated (IP) with anti-HA antibody followed by Western blotting with anti-GFP and anti-HA antibodies. A schematic of the RIAM protein indicating the amino acid positions and highlighting the following domains is shown. C, coiled-coil, P, proline-rich. B, RIAM N-terminal fragments directly bind talin. Interaction of RIAM with talin was examined by in vitro protein interaction assay. Purified recombinant His6-tagged full-length talin was incubated with GST-RIAM-(1–301), -(1–176), -(1–103), -(104–666), or GST control immobilized on glutathione-Sepharose beads, as indicated. Bound proteins were fractionated by SDS-PAGE and analyzed by staining with Coomassie Blue. WCL, whole cell lysates.
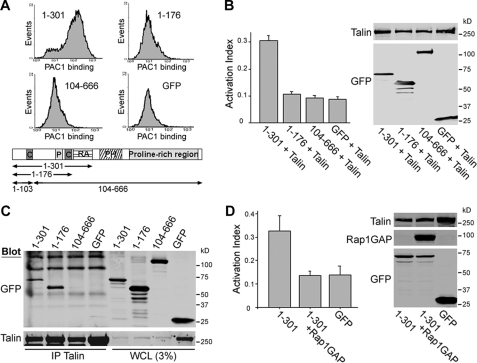
Both Talin and Rap1 Binding Regions of RIAM Are Required for Integrin Activation—As noted above, RIAM-(1–176), which lacks the Rap1 binding domain, binds directly to talin. Because the activation state of Rap1 controls RIAM-dependent integrin αIIbβ3 activation (23), we examined the capacity of this fragment to activate integrins in conjunction with talin. We first verified that there was low level spontaneous Rap1 activation in the Chinese hamster ovary cells (supplemental Fig. S1). We co-expressed GFP-RIAM fragments with HA-talin in A5 cells and used flow cytometry to assay the binding of an integrin αIIbβ3 activation-specific antibody, PAC1 (26). Cells co-transfected with HA-talin and GFP-RIAM-(1–301), which contains both the talin and Rap1 binding sites, exhibited increased PAC1 binding (Fig. 2, A and B). In contrast, no increase in PAC1 binding was observed when talin was co-transfected with either GFP-RIAM-(1–176), which lacks the RA domain, or GFP-RIAM-(104–666), which contains the RA domain but not the talin binding site (Fig. 2, A and B). In these experiments we confirmed by immunoprecipitation that GFP-RIAM-(1–176) and GFP-RIAM-(1–301) associated with HA-talin in these cells, whereas GFP-RIAM-(104–666) failed to do so (Fig. 2C). Furthermore, as with full-length RIAM (23), activation induced by RIAM-(1–301) depends on Rap1 activity, as it was blocked by co-transfection with Rap1GAP (Fig. 2D). Thus, RIAM-mediated integrin activation requires RIAM talin and RA binding domains and Rap activity.
FIGURE 2.
The RA and talin binding domains of RIAM are required for integrin αIIbβ3 activation. A, A5 cells were transiently co-transfected with plasmids encoding HA-talin and GFP-RIAM-(1–301), -(1–176), -(104–666) or GFP as indicated. Flow cytometry was used to measure the binding of PAC1, an αIIbβ3 activation-specific antibody, to GFP-expressing cells as described under “Experimental Procedures.” B, integrin activation index defined as 100 × (F - F0)/(Fmax - F0), where F is the MFI of PAC1 binding, F0 is MFI of PAC1 binding in the presence of 1 μm αIIbβ3-specific inhibitor Ro43-5054, and Fmax is the MFI of PAC1 binding in the presence of 2 μm αIIbβ3-activating antibody anti-LIBS6 calculated for the cells shown in A. Data are the means ± S.E. for n ≥ 3. Protein expression was assessed by Western blotting using anti-HA or anti-GFP antibodies. C, A5 cells were co-transfected with HA-talin and GFP-RIAM-(1–301), -(1–176), -(104–666), or GFP alone, and HA-talin was immunoprecipitated from the cell lysates with anti-HA antibody (12CA5). Co-immunoprecipitated proteins (IP talin) and protein expression in whole cell lysates (WCL) was assessed by Western blotting with antibodies against GFP for RIAM (upper panel) and against HA for talin (lower panel), as indicated. D, GFP-RIAM-(1–301) induced integrin activation was abolished upon co-transfection with Rap1GAP.
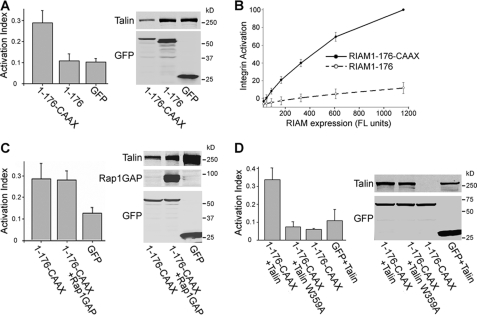
Membrane-targeting Sequences of Ras Proteins Bypass the Requirement of Rap1 Activity for RIAM-dependent Integrin Activation—As noted above, the RA domain and Rap1 activity were needed for RIAM-(1–301) to promote integrin activation. Ras family proteins such as Rap1 contain a C-terminal CAAX box that specifies C-terminal prenylation, proteolytic cleavage, and carboxymethylation (27). In combination with nearby polybasic sequences and/or acylated Cys residues, this region of Ras family proteins plays a crucial role in the localization of these proteins to cellular membranes and to particular microdomains within these membranes (28). To test the role of the membrane localization region of Rap1A in integrin activation, we joined the C-terminal 20 residues of Rap1A, which contains the CAAX box in combination with a Lys-rich region, with RIAM-(1–176), a fragment that contains the talin binding region but not the RA domain. Expression of GFP-RIAM-(1–176)-CAAX in combination with talin dramatically increased PAC1 binding to αIIbβ3, whereas as before, GFP-RIAM-(1–176) failed to do so (Fig. 3A). Because each RIAM construct was GFP-tagged, we were able to directly compare expression level with effect on integrin activation. The addition of the Rap1 membrane-localization sequence resulted in a >10-fold increase in the capacity of RIAM-(1–176) to activate integrin αIIbβ3 (Fig. 3B). Furthermore, integrin activation induced by GFP-RIAM-(1–176)-CAAX did not require Rap1 activity as it was insensitive to inhibition by Rap1GAP (Fig. 3C), indicating that RIAM-(1–176)-CAAX expression bypasses the requirement for Rap1 activity. The effect of RIAM-(1–176)-CAAX on integrin activation is dependent on the interaction between talin and integrin β3 cytoplasmic tail because GFP-RIAM-(1–176)-CAAX was not able to activate integrins when co-expressed with a talin mutant (talin(W359A)) that is deficient in binding to integrin β cytoplasmic domain (29) (Fig. 3D). These results strongly suggest that one important effect of Rap1 interaction with RIAM is to drive recruitment of a RIAM-talin complex to the plasma membrane.
FIGURE 3.
Tethering membrane targeting sequences of Rap1 to RIAM-(1–176) bypasses Rap1 requirement for integrin activation. A5 cells were co-transfected with HA-talin and GFP-tagged RIAM-(1–176) constructs with or without Rap1GAP, as indicated. Integrin activation was assayed by PAC1 binding and quantified using flow cytometry as described in Fig. 2. A, fusion of the membrane targeting sequences of Rap1 with RIAM-(1–176) to form RIAM-(1–176)-CAAX induced integrin activation. B, the extent of integrin activation (Activation index), calculated as described in legend to Fig. 2B, was plotted against expression of RIAM-(1–176) or RIAM-(1–176)-CAAX as assessed by GFP fluorescence intensity. C, integrin activation induced by RIAM-(1–176)-CAAX was insensitive to inhibition by Rap1GAP co-expression. D, integrin activation depends on talin binding to the integrin because it is abolished with HA-talin (W359A), a mutant talin that is deficient in binding to the integrin β3 cytoplasmic tail. Integrin activation indices were calculated as described in Fig. 2 and are the mean ± S.E. for n ≥ 3(A–D). Transfected protein expression levels were assessed by Western blotting (right panels) with antibodies against GFP or HA for detection of GFP-RIAM, HA-talin, or HA-Rap1GAP, as indicated (A–D).
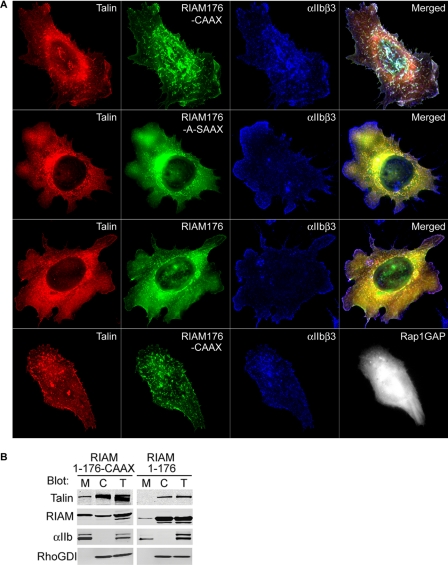
RIAM-(1–176)-CAAX Promotes Rap1-independent Talin Association with Integrins at the Plasma Membrane—Based on the observation that RIAM-(1–176)-CAAX activates integrin αIIbβ3 in a Rap1-independent manner, we hypothesized that membrane recruitment of talin, mediated by the Rap1 membrane targeting domain, leads to integrin activation. To test this idea, we examined the localization of αIIbβ3 integrins, RIAM-(1–176)-CAAX and talin. Expression of GFP-RIAM-(1–176)-CAAX promoted formation of cell surface clusters exhibiting extensive co-localization of talin and αIIbβ3 integrins (Fig. 4A, first row). In contrast, expression of GFP-RIAM-(1–176) resulted in a diffuse, predominantly cytosolic distribution of RIAM-(1–176) and talin that did not co-localize with αIIbβ3 integrins (Fig. 4A, third row). Similar results were obtained in biochemical subcellular fractionation assays. Expression of RIAM-(1–176)-CAAX led to a marked increase in membrane-associated talin (Fig. 4B). In contrast, RIAM-(1–176), which lacks a membrane-targeting sequence, failed to induce talin localization to the membrane fraction, remaining essentially cytosolic (Fig. 4B). To specifically test the role of membrane targeting by the Rap1 CAAX sequence, we expressed GFP-RIAM-(1–176)-A-SAAX, a prenylation-deficient variant in which the polybasic Lys tract within the Rap1 sequence is replaced with Ala residues. Expression of this construct produced a phenotype identical to that observed with GFP-RIAM-(1–176) (Fig. 4A, second and third rows), thus demonstrating an essential role of Rap1-mediated membrane targeting for formation of the talin-, RIAM-, and integrin-containing clusters. To quantify the extent of co-localization between talin, RIAM, and αIIbβ3 integrins, we compared the distribution of correlated pixels as a two-color component scatter plot and calculated Pearson's correlation coefficients, shown in supplemental Fig. S2. These quantitative analyses showed that talin and all of the RIAM-(1–176) constructs exhibited extensive co-localization regardless of the presence of the Rap1 CAAX sequence. However, only the RIAM-(1–176)-CAAX led to a significant correlation between the distribution of integrin αIIbβ3 with talin and RIAM (supplemental Fig. S2). In addition, maximal RIAM-induced integrin activation required both the polybasic Lys residues and the capacity to be prenylated, as combined mutation of the polylysine tract and the CAAX box in the Rap1A membrane-targeting sequence abolished its ability to support integrin activation (supplemental Fig. S3). Rap1 activity is required for full-length RIAM to induce formation of a complex containing talin and integrin αIIbβ3 (23). In sharp contrast, inhibition of Rap activity by expression of Rap1GAP had no effect on the association of talin and integrin αIIbβ3 with GFP-RIAM-(1–176)-CAAX (Fig. 4A, fourth row and supplemental Fig. S2). Thus, joining a talin binding fragment of RIAM to the membrane targeting sequence of Rap1A is sufficient to induce talin recruitment to the plasma membrane, association with integrin αIIbβ3, and integrin activation even when Rap1 activity is inhibited.
FIGURE 4.
Expression of RIAM-(1–176)-CAAX targets talin to integrins in the plasma membrane. A, A5 cells transiently expressing mCherry-talin and GFP-tagged RIAM-(1–176)-CAAX, RIAM-(1–176)-A-SAAX, or RIAM-(1–176) (top three panels), or with RIAM-(1–176)-CAAX and HA-Rap1GAP (bottom panel) were adhered to fibrinogen-coated coverslips and stained to visualize integrin αIIbβ3 and Rap1GAP. Depicted are the localization of αIIbβ3(blue), talin (red), RIAM (green), or Rap1GAP (white). Images shown are maximal projections of deconvoluted 0.1-μm z-section images of the entire cell volume. B, A5 cells expressing HA-talin and GFP-tagged RIAM-(1–176)-CAAX or RIAM-(1–176) were Dounce-homogenized, fractionated, and analyzed by Western blotting for the distribution of HA-talin and GFP-RIAM in membrane (M), cytosol (C), and total (T) fractions with the corresponding anti-HA or anti-GFP antibodies. In addition, integrin αIIb and RhoGDI were immunoblotted to validate separation of the membrane and cytosol fractions, respectively.
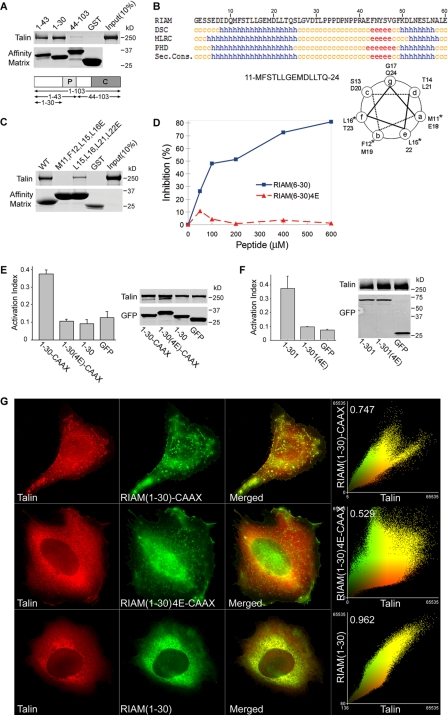
Talin Binding to RIAM Is Required for Integrin Activation—The foregoing studies show that the talin binding function localizes to the N terminus of RIAM-(1–103). To further narrow down the talin binding sequence, we expressed additional truncation mutants of RIAM as GST-tagged proteins and assessed their ability to bind purified recombinant talin. RIAM-(1–30)- and RIAM-(1–43)-bound talin, whereas RIAM-(44–103) did not (Fig. 5A). Analysis of the secondary structure predicted an ∼20-residue amphipathic α-helix within this region (Fig. 5B). Because hydrophobic interactions of amphipathic helices are crucial in many protein-protein interactions such as that of talin with vinculin (30), we mutated the hydrophobic residues predicted to form one face of the helix to charged glutamic acids. Specifically, we made these substitutions in RIAM residues Met-11, Phe-12, Leu-15, and Leu-16 to form RIAM-(1–30)-4E. GST-RIAM-(1–30)-4E failed to bind talin (Fig. 5C). In contrast, another control mutant GST-RIAM(L15E,L16E,L21E,L22E) (1–30) retained partial binding to talin, suggesting the first two turns of the predicted amphipathic helix are most important for talin binding. Furthermore, synthetic peptides containing RIAM-(6–30) sequence but not those containing the mutant RIAM-(6–30)-4E sequence inhibited the binding of full-length RIAM to talin (Fig. 5D). Thus, RIAM-(6–30) contains a minimal binding site for talin, indicating that the N-terminal 30 residues of RIAM are both sufficient and necessary for talin binding (Fig. 5D).
FIGURE 5.
A minimized Rap-RIAM module is sufficient for integrin activation. A, minimal talin binding sequence of RIAM is in the N-terminal 30 residues. Purified His6-talin was incubated with GST-RIAM-(1–103), -(1–43), or -(1–30) or GST immobilized on glutathione-Sepharose. Bound proteins were fractionated by SDS-PAGE and analyzed by Coomassie staining. B, talin binding fragment of RIAM-(1–30) contains a putative amphipathicα-helix. RIAM-(1–30) was analyzed using a secondary structure prediction program (PBIL) and predicted helical residues (h) were identified with DSC, MLRC, and PHD programs. Symbols represent α-helix (h), random coil (c), and extended strand (e). Sec. Cons., secondary consensus residues. Helical wheel analysis was performed on RIAM-(11–24) containing the predicted α-helical region, and the asterisk denotes hydrophobic amino acid residues that were mutated to charged Glu residues (see Fig. 5C). C, disruption of hydrophobic face of the predicted helix abolishes talin binding. Hydrophobic amino acid residues aligned along one side of the predicted amphipathic α-helix in RIAM-(1–30) were mutated to glutamic acids and expressed as GST-tagged proteins immobilized on glutathione-Sepharose beads. Purified recombinant His6-talin was incubated with GST-RIAM-(1–30) and the corresponding mutants M11E,F12E,L15E,L16E (RIAM-(6–30)-4E) or L15E,L16E,L21E,L22E. Bound proteins were fractionated by SDS-PAGE followed by Coomassie Blue staining for detection. D, interaction of RIAM with talin is inhibited by a short RIAM wild type peptide but not by mutant peptide. The complex of GST-RIAM-(1–301) was incubated with full-length recombinant talin, and binding was performed as described in Fig. 1B in the presence of increasing amounts of peptides containing sequences from RIAM; that is, wild type peptides spanning (6–30) or a mutant peptide (6–30)-4E. 4E denotes the M11E,F12E,L15E,L16E mutant. Bound talin was quantified by densitometry of Coomassie Blue-stained bands, and percent inhibition was calculated as 100 × (B0 - B)/B0, where B0 = binding in the absence of peptide, and B = binding in the presence of peptide. E, Rap1 membrane targeting sequence fused to RIAM-(1–30) induce integrin activation that is abrogated by mutations that abolish talin binding. A5 cells expressing HA-talin in combination with the indicated GFP-RIAM-(1–30) proteins or GFP control were assessed for PAC1 binding using flow cytometry to measure activation of αIIbβ3 (data are the mean ± S.E. of independent experiments; n ≥ 3). Transfected protein expression was verified by Western blotting. F, mutations that perturb talin binding abolishes integrin activation induced by RIAM-(1–301). A5 cells expressing HA-talin and GFP-tagged RIAM-(1–301), RIAM-(1–301)-4E. or GFP vector control were assessed for their ability to bind PAC1 using flow cytometry (data are the mean ± S.E. of independent experiments; n ≥ 3). 4E denotes the M11E,F12E,L15E,L16E mutant. Transfected protein expression was assessed by Western blotting. G, RIAM-(1–30)-CAAX recruits talin to clusters at the membrane. A5 cells expressing mCherry-talin and GFP-tagged RIAM-(1–30)-CAAX, RIAM-(1–30)-4E-CAAX, or RIAM-(1–30) were adhered to fibrinogen-coated coverslips and imaged to show talin (red) and RIAM (green). Epifluorescent images as shown are maximal projections of deconvoluted 0.1-μm z-section images of the entire cell volume. The right panels show a two-color component scatter plot comparing the distribution of correlated pixels for the indicated labeled proteins and the calculated Pearson's correlation coefficient.
Having identified a short talin binding sequence and mutations that disrupt talin binding, we tested the importance of the talin binding activity in integrin activation by creating a minimized Rap1-RIAM module. We joined the short talin binding sequence, RIAM-(1–30), to the Rap1 membrane targeting sequence to form RIAM-(1–30)-CAAX. Co-transfection of this 50-amino acid chimera with talin promoted activation of integrin αIIbβ3 (Fig. 5E). In sharp contrast, RIAM-(1–30)-4E-CAAX had no effect on the activation of αIIbβ3 (Fig. 5E). Furthermore, the introduction of the same four Glu mutations into RIAM-(1–301) abolished talin binding (not shown) and consequently blocked integrin activation (Fig. 5F). When expressed in A5 cells, GFP-RIAM-(1–30)-CAAX co-localized with talin in cell surface clusters (first row, Fig. 5G). In contrast, GFP-RIAM-(1–30) co-localized with talin throughout the cytoplasm and failed to form the cell surface clusters (third row, Fig. 5G). Finally, the talin binding-deficient RIAM-(1–30)-4E-CAAX failed to recruit talin to the membrane (second row, Fig. 5G). Thus, the RIAM talin binding site is within a short sequence that contains an amphipathic α-helix. Furthermore, the capacity of RIAM to bind to talin is required for its ability to promote integrin activation.
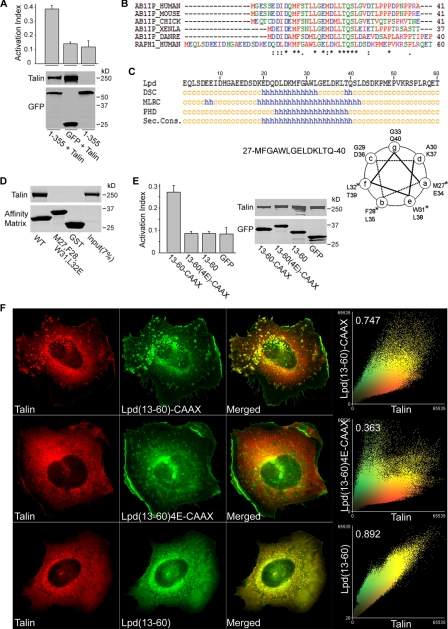
A Shared Amphipathic Helical Motif Mediates Lamellipodin Binding to Talin and Integrin Activation—RIAM is a member of the MRL protein family that includes its closest paralogue, Lpd (22). Lpd can induce both talin recruitment and integrin activation (31); however, Lpd overexpression can decrease the adhesion of HEK 293 cells to fibronectin (32). Nevertheless, we found that expression of full-length Lpd (not shown) or Lpd(1–355), which contains both the talin binding site and an RA domain, in conjunction with talin activated αIIbβ3 integrins (Fig. 6A). The N-terminal fragment of Lpd, Lpd-(13–60), contains a region (17–46) that is ∼47% identical to the talin binding site of RIAM-(1–30) (Fig. 6B) and is predicted to contain an amphipathic α-helix (Fig. 6C) similar to that contained within RIAM-(1–30). Indeed, Lpd-(13–60) bound to purified talin (Fig. 6D). In contrast, Lpd(M27E,F28E,W31E,L32E)-(13–60) (abbreviated Lpd-(13–60)-4E)), incorporating mutations predicted to disrupt the hydrophobic face of the helix, did not bind talin (Fig. 6D). As noted above, RIAM requires the ability to bind to both Rap1 and talin to mediate integrin activation. Nevertheless, Lpd has been reported not to bind to Rap1 even though it contains an RA domain (22). Because other Ras proteins might bind to Lpd and serve to provide membrane localization, we asked whether the membrane targeting sequences of other Ras GTPases might be able to recruit talin appropriately to induce integrin activation. Attachment of the membrane targeting sequences from the N-, K-, R-, or H-Ras GTPases to RIAM-(1–176) all increased integrin activation (supplemental Fig. S4). Based on these results, we fused a CAAX-containing membrane-targeting sequence to GFP-Lpd-(13–60) and found that it could promote activation of integrin αIIbβ3 in conjunction with talin (Fig. 6E). As with RIAM, both talin binding and membrane targeting were required, as the talin binding-deficient mutant Lpd-(13–60)-4E-CAAX and membrane targeting-deficient Lpd-(13–60) did not promote activation (Fig. 6E). To investigate if integrin activation by the Lpd fragment leads to integrin cluster formation, we co-expressed talin with Lpd-(13–60)-CAAX, the talin binding-defective mutant Lpd-(13–60)-4E-CAAX, or Lpd-(13–60). Indeed, GFP-Lpd-(13–60)-CAAX led to co-clustering with talin (first row, Fig. 6F) and integrins (not shown). In contrast, the talin binding-deficient GFP-Lpd-(13–60)-4E-CAAX failed to recruit talin to the membrane (second row, Fig. 6F), whereas GFP-Lpd-(13–60) co-localized with talin throughout the cytoplasm but failed to form the cell surface clusters (third row, Fig. 6F). Thus, both RIAM and Lpd can mediate integrin activation through a common scaffolding mechanism; they each contain amphipathic helices that mediate direct binding to talin and RA domains that bind Ras superfamily GTPases. MRL proteins thereby act as scaffolds that mediate the membrane recruitment of talin where it interacts with and activates integrins.
FIGURE 6.
Lamellipodin binds talin and activates integrin αIIbβ3. A, overexpression of Lpd activates integrin αIIbβ3. A5 cells were transfected with GFP-Lpd(1–355), a fragment that aligns with RIAM-(1–301), or co-transfected with talin in combination with GFP-Lpd(1–355), or with GFP only and were assessed for integrin activation by flow cytometry. Data are the mean ± S.E. of independent experiments, calculated as described in Fig. 2. Protein expression was examined by Western blotting. B, sequence alignment of RIAM (AB1IP) talin binding sites in different species with Lamellipodin (RAPH1). asterisk, identical; colon, conservative substitution; dot, semi-conservative substitution. C, Lpd-(13–60) encodes a predicted amphipathic α-helix. Lpd-(13–60) sequences was analyzed using a secondary structure prediction program (PBIL) and analyzed with DSC, MLRC, and PHD programs. Helical wheel analysis was performed on Lpd-(27–40) containing a predicted α-helical region, and the asterisk denotes amino acid residues that were mutated to Glu residues. See below in D. D, Lpd N-terminal fragment 13–60 directly binds talin. Purified recombinant His6-talin was incubated with GST-tagged Lpd-(13–60), Lpd-(13–60)-4E, or GST. Bound proteins were fractionated by SDS-PAGE followed by Coomassie Blue staining for analysis. Lpd-(13–60)-4E contains M27E,F28E,L31E,L32E mutations that align with those of RIAM-4E mutations shown in Fig. 5F. WT, wild type. E, linking Rap membrane-targeting sequences to Lpd talin binding fragment leads to integrin activation. A5 cells were co-transfected with HA-talin and GFP-tagged Lpd-(13–60)-CAAX, Lpd-(13–60)-4E-CAAX, Lpd-(13–60), or GFP vector control and were assessed for their ability to bind PAC1 using flow cytometry (data are the mean ± S.E. of independent experiments; n ≥ 3). Western blotting was used to verify protein expression. F, Lpd-(13–60)-CAAX recruits talin to clusters at the membrane. A5 cells expressing mCherry-talin and GFP-tagged Lpd-(13–60)-CAAX, Lpd-(13–60)-4E-CAAX, or Lpd-(13–60) were adhered on fibrinogen-coated coverslips and imaged to show talin (red) and Lpd (green). Epifluorescent images as shown are maximal projections of deconvoluted 0.1-μm z-section images of the entire cell volume. The right panels show a two-color component scatter plot comparing the distribution of correlated pixels for the indicated labeled proteins and the calculated Pearson's correlation coefficient.
In platelets, agonist stimulation leads to talin redistribution from the cytoplasm to the plasma membrane (33). The work described here provides a detailed biochemical explanation for the redistribution of talin. Specifically, as noted above, Rap1B is activated in response to platelet agonists, and RIAM mediates the Rap-dependent relocalization of talin to the plasma membrane (23). We now show that, RIAM can bind to both Rap1 and talin, thereby forming a scaffold that uses the Rap1 membrane-targeting sequence to induce talin relocalization. Furthermore, the minimized Rap-RIAM module that contains only talin binding and membrane targeting sequences was sufficient to replace both RIAM and Rap activity for talin redistribution to the plasma membrane and integrin activation. Thus, these studies show that talin recruitment to the plasma membrane is important for integrin activation and provide a molecular mechanism for this recruitment by MRL proteins.
MRL proteins contain multiple protein and lipid binding domains, yet only talin binding and Ras binding domains were required for integrin activation in our studies. These proteins contain PH domains that can interact with inositol phospholipids. Thus, it is possible that these PH domains could play a regulatory role in MRL protein function either by controlling localization or by influencing the conformation of the MRL protein. It is noteworthy that phosphatidylinositol 3-kinase has been implicated in activation of integrin αIIbβ3 (34); it will be of interest to explore the potential role of modulation of MRL proteins as a potential mechanism for this effect of phosphatidylinositol 3-kinase. Similarly, the ENA/VASP and profilin binding sites of RIAM and Lpd were dispensable for integrin activation in our experiments, yet these interactions are likely to make an important contribution to both RIAM and Lpd capacity to promote actin polymerization (20, 22); the capacity of the MRL proteins to bind talin through a conserved motif suggests that RIAM could also serve as a link between activated integrins, talin, and polymerizing actin.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL 078784, AR27214, and HL 31950. This work was also supported by Cell Migration Consortium Grant U54 GM06434. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: GAP, GTPase-activating protein; RIAM, Rap1-GTP-interacting adaptor molecule; MRL, Mig-10/RIAM/Lamellipodin; RA, Ras association; PH, pleckstrin homology; HA, hemagglutinin; GFP, green fluorescent protein; GST, glutathione S-transferase; MFI, median fluorescence intensity; Lpd, Lamellipodin.
References
- 1.Hynes, R. O. (2002) Cell 110 673-687 [DOI] [PubMed] [Google Scholar]
- 2.Petrich, B. G., Fogelstrand, P., Partridge, A. W., Yousefi, N., Ablooglu, A. J., Shattil, S. J., and Ginsberg, M. H. (2007) J. Clin. Investig. 117 2250-2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrich, B. G., Marchese, P., Ruggeri, Z. M., Spiess, S., Weichert, R. A., Ye, F., Tiedt, R., Skoda, R. C., Monkley, S. J., Critchley, D. R., and Ginsberg, M. H. (2007) J. Exp. Med. 204 3103-3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tadokoro, S., Shattil, S. J., Eto, K., Tai, V., Liddington, R. C., de Pereda, J. M., Ginsberg, M. H., and Calderwood, D. A. (2003) Science 302 103-106 [DOI] [PubMed] [Google Scholar]
- 5.Wegener, K. L., Partridge, A. W., Han, J., Pickford, A. R., Liddington, R. C., Ginsberg, M. H., and Campbell, I. D. (2007) Cell 128 171-182 [DOI] [PubMed] [Google Scholar]
- 6.Yan, B., Calderwood, D. A., Yaspan, B., and Ginsberg, M. H. (2001) J. Biol. Chem. 276 28164-28170 [DOI] [PubMed] [Google Scholar]
- 7.Kinbara, K., Goldfinger, L. E., Hansen, M., Chou, F. L., and Ginsberg, M. H. (2003) Nat. Rev. Mol. Cell Biol. 4 767-776 [DOI] [PubMed] [Google Scholar]
- 8.Marshall, C. J. (1996) Curr. Opin. Cell Biol. 8 197-204 [DOI] [PubMed] [Google Scholar]
- 9.Bos, J. L. (2005) Curr. Opin. Cell Biol. 17 123-128 [DOI] [PubMed] [Google Scholar]
- 10.Kooistra, M. R., Dube, N., and Bos, J. L. (2007) J. Cell Sci. 120 17-22 [DOI] [PubMed] [Google Scholar]
- 11.Bertoni, A., Tadokoro, S., Eto, K., Pampori, N., Parise, L. V., White, G. C., and Shattil, S. J. (2002) J. Biol. Chem. 277 25715-25721 [DOI] [PubMed] [Google Scholar]
- 12.Caron, E., Self, A. J., and Hall, A. (2000) Curr. Biol. 10 974-978 [DOI] [PubMed] [Google Scholar]
- 13.Reedquist, K. A., Ross, E., Koop, E. A., Wolthuis, R. M., Zwartkruis, F. J., van Kooyk, Y., Salmon, M., Buckley, C. D., and Bos, J. L. (2000) J. Cell Biol. 148 1151-1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebzda, E., Bracke, M., Tugal, T., Hogg, N., and Cantrell, D. A. (2002) Nat. Immunol. 3 251-258 [DOI] [PubMed] [Google Scholar]
- 15.Franke, B., Akkerman, J. W., and Bos, J. L. (1997) EMBO J. 16 252-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franke, B., van Triest, M., de Bruijn, K. M., van Willigen, G., Nieuwenhuis, H. K., Negrier, C., Akkerman, J. W., and Bos, J. L. (2000) Mol. Cell. Biol. 20 779-785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chrzanowska-Wodnicka, M., Smyth, S. S., Schoenwaelder, S. M., Fischer, T. H., and White, G. C., Jr. (2005) J. Clin. Investig. 115 680-687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergmeier, W., Goerge, T., Wang, H. W., Crittenden, J. R., Baldwin, A. C., Cifuni, S. M., Housman, D. E., Graybiel, A. M., and Wagner, D. D. (2007) J. Clin. Investig. 117 1699-1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katagiri, K., Maeda, A., Shimonaka, M., and Kinashi, T. (2003) Nat. Immunol. 4 741-748 [DOI] [PubMed] [Google Scholar]
- 20.Lafuente, E. M., van Puijenbroek, A. A., Krause, M., Carman, C. V., Freeman, G. J., Berezovskaya, A., Constantine, E., Springer, T. A., Gertler, F. B., and Boussiotis, V. A. (2004) Dev. Cell 7 585-595 [DOI] [PubMed] [Google Scholar]
- 21.Zhang, Z., Rehmann, H., Price, L. S., Riedl, J., and Bos, J. L. (2005) J. Biol. Chem. 280 33200-33205 [DOI] [PubMed] [Google Scholar]
- 22.Krause, M., Leslie, J. D., Stewart, M., Lafuente, E. M., Valderrama, F., Jagannathan, R., Strasser, G. A., Rubinson, D. A., Liu, H., Way, M., Yaffe, M. B., Boussiotis, V. A., and Gertler, F. B. (2004) Dev. Cell 7 571-583 [DOI] [PubMed] [Google Scholar]
- 23.Han, J., Lim, C. J., Watanabe, N., Soriani, A., Ratnikov, B., Calderwood, D. A., Puzon-McLaughlin, W., Lafuente, E. M., Boussiotis, V. A., Shattil, S. J., and Ginsberg, M. H. (2006) Curr. Biol. 16 1796-1806 [DOI] [PubMed] [Google Scholar]
- 24.Shadle, P. J., Ginsberg, M. H., Plow, E. F., and Barondes, S. H. (1984) J. Cell Biol. 99 2056-2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loftus, J. C., O'Toole, T. E., Plow, E. F., Glass, A., Frelinger, A. L., III, and Ginsberg, M. H. (1990) Science 249 915-918 [DOI] [PubMed] [Google Scholar]
- 26.Shattil, S. J., Cunningham, M., and Hoxie, J. A. (1987) Blood 70 307-315 [PubMed] [Google Scholar]
- 27.Gutierrez, L., Magee, A. I., Marshall, C. J., and Hancock, J. F. (1989) EMBO J. 8 1093-1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock, J. F. (2003) Nat. Rev. Mol. Cell Biol. 4 373-384 [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Alvarez, B., de Pereda, J. M., Calderwood, D. A., Ulmer, T. S., Critchley, D., Campbell, I. D., Ginsberg, M. H., and Liddington, R. C. (2003) Mol. Cell 11 49-58 [DOI] [PubMed] [Google Scholar]
- 30.Izard, T., Evans, G., Borgon, R. A., Rush, C. L., Bricogne, G., and Bois, P. R. (2004) Nature 427 171-175 [DOI] [PubMed] [Google Scholar]
- 31.Watanabe, N., Bodin, L., Pandey, M., Krause, M., Coughlin, S., Boussiotis, V. A., Ginsberg, M. H., and Shattil, S. J. (2008) J. Cell Biol. 181 1211-1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinn, C. C., Pfeil, D. S., Chen, E., Stovall, E. L., Harden, M. V., Gavin, M. K., Forrester, W. C., Ryder, E. F., Soto, M. C., and Wadsworth, W. G. (2006) Curr. Biol. 16 845-853 [DOI] [PubMed] [Google Scholar]
- 33.Beckerle, M. C., Miller, D. E., Bertagnolli, M. E., and Locke, S. J. (1989) J. Cell Biol. 109 3333-3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kovacsovics, T. J., Bachelot, C., Toker, A., Vlahos, C. J., Duckworth, B., Cantley, L. C., and Hartwig, J. H. (1995) J. Biol. Chem. 270 11358-11366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.