Abstract
Lactobacillus sakei is a food-borne bacterium naturally found in meat and fish products. A study was performed to examine the intraspecies diversity among 73 isolates sourced from laboratory collections in several different countries. Pulsed-field gel electrophoresis analysis demonstrated a 25% variation in genome size between isolates, ranging from 1,815 kb to 2,310 kb. The relatedness between isolates was then determined using a PCR-based method that detects the possession of 60 chromosomal genes belonging to the flexible gene pool. Ten different strain clusters were identified that had noticeable differences in their average genome size reflecting the natural population structure. The results show that many different genotypes may be isolated from similar types of meat products, suggesting a complex ecological habitat in which intraspecies diversity may be required for successful adaptation. Finally, proteomic analysis revealed a slight difference between the migration patterns of highly abundant GapA isoforms of the two prevailing L. sakei subspecies (sakei and carnosus). This analysis was used to affiliate the genotypic clusters with the corresponding subspecies. These findings reveal for the first time the extent of intraspecies genomic diversity in L. sakei. Consequently, identification of molecular subtypes may in the future prove valuable for a better understanding of microbial ecosystems in food products.
In foods, the need for microorganisms to adapt to different technological and ripening processes may result in the evolution of strain differences. Unfortunately, intraspecies genetic variations among food-borne bacteria are a largely unexplored area, so we have little understanding of the interactive mechanisms taking place within complex microbial communities existing in food ecosystems. This is particularly relevant in the case of Lactobacillus sakei, a meat-borne lactic acid bacterium potentially useful as a meat biopreservative (6, 39). L. sakei has been isolated from a range of meat and fish products, where it is the predominant Lactobacillus species (8). Ecologically, meat can be viewed as a diverse and changing environment that influences the growth potential of a variety of bacterial species during storage (27). An implication of survival in such an environment is that meat-borne bacteria may diverge genetically as they evolve mechanisms to acclimatize and compete in local microenvironments. Indeed, L. sakei strains are known to display a range of key phenotypic differences that have resulted in difficulties in their classification (23, 35), and DNA-DNA reassociation analyses have revealed very low levels of relatedness (as low as 72%) between otherwise well-characterized L. sakei strains, indicating that the species exhibits important elements of genetic heterogeneity (7). However, it is not yet known if a strong relationship exists between the niche competition properties of L. sakei in meat products and the genetic diversity between strains.
Currently, L. sakei is divided into two subspecies based on numerical analysis of randomly amplified polymorphic DNA patterns (5, 38) and total cell soluble protein content patterns (23): L. sakei subsp. sakei (type strain ATCC 15521) and L. sakei subsp. carnosus (type strain CIP105422 to CCUG31331) (24). With the sequence of the L. sakei 23K genome now available (6), it is becoming possible to study L. sakei strain diversity at a deeper genomic level, as well as performing wider searches for differences between L. sakei strains isolated naturally from various products.
In this report, we have used a combination of techniques to examine strains. These include a PCR-based method for detecting genetic markers in a pool of variable genes allowing a hierarchical clustering of strains, pulsed-field gel electrophoresis (PFGE) genome mapping, and evaluation of strain proteomes to both compare strains and assign them to each of the two subspecies. We have specifically chosen isolates from a range of laboratory collections representing a variety of geographical locations and including various sources of meat or fish products, with the expectation that such a range of undomesticated strains will better reflect the diversity found in natural L. sakei populations.
Our methods provide for the first time an integrated genome-based framework for classifying the repertoire of L. sakei molecular subtypes. The implications of our results for the understanding of the bacterium's ecology are discussed.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The L. sakei and Lactobacillus curvatus strains used in this study are described in Table 1. For most studies, strains were grown to the mid-exponential phase in MRS broth (Becton Dickinson, Sparks, MD) (11) incubated at 30°C. For proteomic studies, bacterial strains were grown in a chemically defined medium (MCD) (28) supplemented with 0.5% glucose or MRS and incubated at 30°C. Strain 332F, cured of its endogenous plasmid pRV500 (2), was prepared as described earlier (4) by electroporating the parent strain L. sakei 332 with a pRV566 plasmid carrying resistance to erythromycin, which had been derived from a pRV500 replicon (2). One erythromycin-resistant clone was selected and cultivated for 200 generations in MRS broth without antibiotic at 30°C. Several dilutions from the last culture were plated on MRS agar. Replica plating of 200 clones was performed on MRS agar with or without erythromycin (5 μg/ml), allowing us to identify erythromycin-sensitive clones. The loss of the pR566 plasmid was verified by Southern blotting (ECL enhanced chemiluminescence system direct nucleic acid labeling; Amersham Biosciences) using a probe specific for the repA gene. The corresponding erythromycin-sensitive strain was named 332F.
TABLE 1.
L. sakei and L. curvatus strains used in this study
| Laboratory collection | No. of strains | Name(s) (synonym) | Source of isolate | Source or reference(s) |
|---|---|---|---|---|
| L. sakei | ||||
| Unité Flore Lactique and Environnement Carné, INRA, Jouy-en-Josas, France | 12 | 23K, 14, 18, 21, 33, 64, 72, 112, 134, 156 | Various French-style fermented dry sausages | 5 |
| 160K | Fresh horse meat | 5 | ||
| JG3 | Fresh beef meat | 5 | ||
| Station de Recherches sur la Viande, | 11 | 195, 205, 300, 332 | Vacuum-packed beef meat | 5 |
| INRA, Theix, France | 331, 495, 504, 532, 710, 741 | Vacuum-packed pork meat | 31 | |
| L110 | Starter for French-style fermented dry sausage | 7 | ||
| Laboratoire de Génie Alimentaire, IFREMER, Nantes, France | 5 | SF770, SF771, SF841, SF842, SF843 | Smoked salmon | 20 |
| Meat Technology Centre, IRTA, Monells, Spain | 10 | CTC014, CTC041, CTC163, CTC287, CTC335, CTC427, CTC429, CTC494, CTC6469, CTC6626 | Various Spanish-style fermented dry sausages (including Chorizo) | 17, 18 |
| Meat Research Institute, ARC, Langford, | 6 | LV5, LV21, LV92 | Vacuum-packed pork meat (bacon) | 36 |
| Bristol, United Kingdom | LV52, LV59 | Vacuum-packed lamb meat | 36 | |
| LV34 | Vacuum-packed beef meat | 36 | ||
| Food Safety Group, AgResearch, Hamilton, New Zealand | 4 | AGR46, AGR48, AGR51, AGR53 | Chilled lamb meat | This study |
| Faculty of Science, Mahidol University, Bangkok, Thailand | 2 | TISTR890, TISTR911 | Nham (Thai-style fermented pork sausage) | 33, 37 |
| Institüt für Libbensmitteltechnologie, | 9 | LTH673, LTH675, LTH677, LTH5728 | Various German-style fermented moist-type sausages | 40 |
| Universität Hohenheim, Hohenheim, | LTH1764, LTH2070 | Sauerkraut | 41 | |
| Germany | LTH5588, LTH5589, LTH5590 | Human feces | 42 | |
| Institute of Meat Hygiene and Technology, Universität Berlin, Germany | 1 | CIP105422T (CCUG31331) | Raw German-style sausage | 23 |
| Federal Centre for Meat Research, Kulmbach, Germany | 1 | Lb706 | Fresh beef meat | 35 |
| Norwegian Food Research Institute, | 6 | MF1048, MF2091, MF2092 | Smoked salmon | This study |
| MATFORSK, Ås, Norway | MF2088, MF2089, MF2090 | Rakfisk (Scandinavian fermented trout) | This study | |
| Centro de Referencia para Lactobacilos (CERELA), CONICET, Tucumán, Argentina | 1 | CRL1467 | Argentinean-style fermented dry sausage | 15 |
| American Type Culture Collection (original isolate from Japan) | 1 | ATCC 15521T (DSM20017) | Spoiled moto for Saké manufacture | 22 |
| National Institute of Health, University of Tokyo, Tokyo, Japan | 5 | YMN243, YME344, YMN540, YMN557, V553 | Various fresh meat products | 32 |
| L. curvatus | ||||
| American Type Culture Collection (original isolate from Germany) | 1 | ATCC 25601T (DSM20019) | Milk | 38 |
| Meat Technology Centre, IRTA, Monells, Spain | 1 | CTC424 | Spanish-style fermented dry sausage | 17 |
Molecular techniques.
Subtractive suppressive hybridization (SSH) experiments were performed using a Clontech PCR-selected bacterial genome subtraction kit in accordance with the manufacturer's recommendations using L. sakei 23K as the driver and strain 332F as the tester. This technique resulted in the identification of 16 new genes absent from L. sakei 23K. The FGP21-0001 gene from strain L. sakei 21 was identified after sequencing a PCR product (LSA0565 to LSA566) giving an unexpected size and revealing a new bacteriocin immunity-like protein-encoding gene.
PCR-based detection of genes.
The PCR template was composed of 100 ng of chromosomal DNA extracted from each of the 73 L. sakei strains and two L. curvatus strains. Experiments were conducted twice to confirm the negative results. In the event of weak or spurious amplifications, PCR products were sequenced to check nucleotide polymorphism between strains, and if necessary primers were redesigned. In several cases, two or three sets of primers were designed to verify the absence/presence of genes. In cases of discrepant results between the primer sets, the corresponding genes were removed from the analysis. When negative results were obtained with the several sets, although the absence of an allelic gene with high nucleotidic polymorphism was not confirmed, we considered the gene as being a good candidate for clustering analysis. Extraction of chromosomal DNA from L. sakei and L. curvatus was performed by the method of Anderson and McKay (3). For each PCR amplification, primers were designed so that the expected product lengths were less than 2 kb (see Table S2 in the supplemental material). The PCR cycling conditions were 94°C for 4 min followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min. All PCR products were examined using 1% agarose gels and stained with ethidium bromide. The absence of one rrn copy in some strains was confirmed by long-range PCR using primers in the flanking region of the rrnAB doublet. To confirm the truncation of some genes or the products of unexpected sizes, 10 μl of the amplicons was treated with 0.1 U of shrimp alkaline phosphatase (USB Corporation) and 1 U of exonuclease I (Escherichia coli) (Biolabs) in 20 mM Tris-HCl (pH 8.0)-10 mM MgCl2 buffer for 1 h at 37°C, followed by 10 min of inactivation at 94°C. The products were then sequenced by standard technology.
PFGE experiments and I-CeuI pattern analysis.
PFGE and I-CeuI digestion pattern analyses were carried out as described earlier (13). An average of four gels was prepared for each strain. The distribution of the strains according to their genome size was examined using the HIST function and the probability DENSITY function of the R statistical package (http://www.R-project.org). A Gaussian probability distribution and a smoothing bandwidth of 30 (average standard deviation of genome size estimation) were chosen for the analyses.
Clustering of strains.
The gene contents of the strains tested were described using a two-character matrix (genes × isolates) with 0 for a gene not detected and 1 for the presence of a gene. Genes truncated by insertion (IS) elements were considered as distinct genetic identities to their wild-type counterparts. Similarities between the strains were determined using the Jacquard's correlation coefficient (19). The unsupervised hierarchical clustering was performed using the average linkage on the similarity matrix. The functions DIST, HCLUST, and DENDROGRAM of the R statistical package were used to generate the clustering dendrogram. The R package PVCLUST (R. Suzuki and H. Shimodaira; http://www.is.titech.ac.jp/∼shimo/prog/pvclust/) was used for multiscale bootstrap resampling to assess the statistical stability of each node. The number of bootstrap replicates was 1,000. Approximately unbiased P values of ≥90% and Jacquard's similarity coefficient of ≥50% were used to discriminate the possible strain clusters. The principal component analysis (PCA) for grouping of the strains was carried out using the PRINCOMP and BIPLOT functions of the R statistical package.
2D gel electrophoresis and identification of proteins by peptide mass fingerprinting.
Bacterial extract preparation and electrophoresis were performed by standard methods (21). For each strain, at least two independent cultures in MCD broth were performed for preparation of the protein samples. Each sample was analyzed twice by two-dimensional (2D) gel electrophoresis, giving a minimum of four analyses per strain. For some strains (23K, 332, 112, 64, LTH677, and JG3) from the two subspecies, up to eight runs were conducted using cells grown in different media (MRS and MCD). Gels were analyzed by Image Master software (Amersham Pharmacia Biotech). Spots were excised from Coomassie-stained gels as described earlier (29, 30), and mass spectrometry analyses were performed as previously described (16). MS-Fit (University of California—San Francisco Mass Spectrometry Facility; http://prospector.ucsf.edu) and Mascot software packages (Matrix Science, Inc., Boston, MA; http://www.matrixscience.com/search_form_select.html), installed on a local server, were used to identify proteins from peptide mass fingerprints. All searches were performed against the L. sakei 23K database (http://www.migale.jouy.inra.fr/).
Nucleotide sequence accession numbers.
The sequences of the FGP21 and FGP332 genes (the nomenclature of which indicates their placement within the flexible gene pool and their strain name origin) have been deposited in GenBank under the accession numbers given in Table 2.
TABLE 2.
Description of L. sakei genes used for clustering analysis
| Genomic island, gene, or strain | Gene name or locus taga | Product description | GenBank accession no. |
|---|---|---|---|
| Strain 23K flexible gene pool genomic islands or genes | |||
| Island 1 | LSA0088 | Adenine deaminase | CR936503 |
| Island 2 | LSA0118 | Hypothetical protein (putative cell surface collagen-binding protein) | CR936503 |
| Island 3 | LSA0157 | Putative hydroxyl/aromatic amino acid symporter | CR936503 |
| Island 5 | LSA0165 | Putative oxidoreductase, short-chain dehydrogenase/reductase family | CR936503 |
| Island 6 | LSA0172 | CscC-type cell-surface protein with invasin/mucin-like domain and WxL domain | CR936503 |
| LSA0178 | MarR-type transcriptional regulator | CR936503 | |
| Island 7 | LSA0211/212 | CscC-type cell surface protein with adhesin-like domain and WxL domain (authentic frameshifted gene) | CR936503 |
| LSA0216 | MarR-type transcriptional regulator | CR936503 | |
| Island 8 | LSA0217 | Putative transcriptional regulator with a rhodanese-like domain, ArsR family | CR936503 |
| LSA0218 | Thioredoxin, TrxA1 | CR936503 | |
| LSA0219_b | Putative cyanate transport protein | CR936503 | |
| Independent gene | LSA0439 | Hypothetical extracellular lipase/esterase precursor | CR936503 |
| Island 12 | LSA0564_a to -_c | Putative bacteriocin-like peptides (LSA0564_ab) and cognate immunity protein (LSA0564_c) | CR936503 |
| LSA0565 to -0566 | Putative bacteriocin-like peptides | CR936503 | |
| LSA0567 to -0569_b | Putative bacteriocin-like peptides (LSA0569_ab) and cognate immunity proteins (LSA0567 and LSA0568) | CR936503 | |
| Independent gene | LSA0572 | Threonine deaminase (threonine ammonia lyase) | CR936503 |
| Island 14 | LSA0724 to -0725 | Hypothetical proteins | CR936503 |
| LSA0727 | Hypothetical cell surface precursor | CR936503 | |
| Island 15 | LSA1006 | Putative zinc-containing alcohol dehydrogenase (oxidoreductase) | CR936503 |
| Island 16 | LSA1182/1183 | Putative cytochrome P450 (authentic frameshifted gene) | CR936503 |
| Island 18 | LSA1283 | CscC-type cell-surface protein with WxL domain | CR936503 |
| Island 19 | LSA1509 | Hypothetical protein, sigma factor related | CR936503 |
| LSA1510_a to -_c | Putative teichoic acid/polysaccharide export protein complex | CR936503 | |
| LSA1510_d to -_f | Putative glycosyl transferase complex | CR936503 | |
| LSA1510_g | Putative priming glycosyl transferase | CR936503 | |
| LSA1512/1513 | Putative polysaccharide biosynthesis protein, chain length determination | CR936503 | |
| Island 20 | LSA1572 | Putative teichoic acid/polysaccharide glycosyl transferase | CR936503 |
| LSA1579/1580 | Putative teichoic acid/polysaccharide export protein complex | CR936503 | |
| LSA1581 | Putative teichoic acid-binding N-acetylmuramoyl l-alalanine amidase (cell wall hydrolase) | CR936503 | |
| LSA1584/1585 | Putative teichoic acid/polysaccharide glycosyl transferase | CR936503 | |
| Island 21 | LSA1640 | N-Acetylneuraminate lyase | CR936503 |
| LSA1641 | N-Acylglucosamine-6-phosphate 2-epimerase (N-acetylmannosamine-6-phosphate 2-epimerase) | CR936503 | |
| LSA1642 | Putative solute:Na+ symporter | CR936503 | |
| LSA1720 | Hypothetical protein (E. coli plasmidic gene) | CR936503 | |
| Island 22 | LSA1724 | MarR-type transcriptional regulator | CR936503 |
| LSA1730 | CscC-type cell surface protein with bacterial adhesin-like domain and WxL domain | CR936503 | |
| LSA1731 | CscC-type cell surface protein with hemagglutinin-like domain and WxL domain | CR936503 | |
| Island 24 | LSA1809 | Hypothetical extracellular protein precursor associated with CSC-type cluster | CR936503 |
| Island 27 | LSA1874 | MarR-type transcriptional regulator | CR936503 |
| Non-strain 23K flexible gene pool strains | |||
| Lb674 | sspT | Sakacin P ABC-transporter, ATP-binding and permease protein SspT | Z48542 |
| sspA | Bacteriocin sakacin P precursor (sakacin 674) | Z48542 | |
| 21 | FGP21-0001 | Putative bacteriocin immunity protein | EU391636 |
| KG15 | dsrB | Cell surface dextransucrase precursor (sucrose 6-glycosyltransferase) | AY697434 |
| 332F | FGP332-0001 | Putative 6-phospho-β-glucosidase | EU402602 |
| FGP332-0002 | CscC-type cell surface protein with bacterial adhesin-like domain and WxL domain | EU402603 | |
| FGP332-0003 | Hypothetical cell surface protein | EU402604 | |
| FGP332-0005 | Putative pyridine nucleotide-disulfide oxidoreductase | EU402605 | |
| FGP332-0006 | Putative ferritin-like DNA-binding protein (oxidative damage protectant; Dps type) | EU402605 | |
| lacC | Putative tagatose-6-phosphate kinase | EU402605 | |
| lacG | Putative 6-phospho-β-galactosidase | EU402605 | |
| FGP332-0007 | Putative autotransporter protein | EU914886 | |
| FGP332-0008 | Hypothetical protein | EU914887 | |
| FGP332-0009 | Hypothetical protein | EU914888 | |
| FGP332-0010 | Hypothetical protein | EU914889 | |
| FGP332-0011 | Putative transcriptional regulator, LysR family | EU914890 | |
| FGP332-0012 | Putative quinine oxidoreductase | EU914891 | |
| FGP332-0013 | Putative asparagine synthase | EU914892 | |
| FGP332-0014 | CscC-type cell surface protein with WxL domain | EU914893 | |
| FGP332-0015 | Putative glycine/betaine reductase | EU914894 |
For “FGP” entries (e.g., FGP21-001), the format is “FGP” (flexible gene pool) name-CDS number.
RESULTS
Selection of L. sakei strains.
To represent the range of ecosystems in which L. sakei populations are found, strains were examined from a variety of meat and fish products (raw or fermented) and from other sources, including human feces and sauerkraut. Additionally, because sampling biases may exist in individual laboratory bacterial collections due to isolation procedures or the type of food materials analyzed, the L. sakei strains were selected from 14 different laboratory collections geographically scattered across Europe, Asia, Argentina, and New Zealand. We took care to discard from the analysis strains known to be identical but belonging to different laboratory collections (and often renamed). In total, 73 L. sakei strains were selected and analyzed (Table 1), as well as two strains of Lactobacillus curvatus, a close relative of L. sakei included as an external species reference (outgroup reference).
Identification of the main L. sakei molecular subtypes by PCR-based detection of the flexible gene pool.
Analysis of the flexible gene pool (genes that are often associated with horizontal transfer and assumed to vary between strains) is a method commonly used to perform intraspecies strain clustering (9). To identify genes belonging to the L. sakei flexible gene pool, an in silico analysis of the L. sakei 23K chromosome using codon bias and atypical phylogenetic protein profiles was carried out and revealed a gene pool comprising 27 genomic islands and 57 independent genes (our experimental unpublished data). We decided to verify by conventional PCR the presence or, on the contrary, lack of detection of this pool of genes for clustering analysis of the L. sakei isolates (see Materials and Methods). This strategy was first tested using a preliminary PCR experiment on a set of 20 strains to demonstrate intraspecies variation for the selected genes (data not shown). The results indicated that only five islands were highly conserved, and these were accordingly removed from the analysis. To avoid clustering disturbance due to highly laterally transferable mobile elements (IS sequences or phage) or elements easily mobilized in lateral transfer (restriction/modification systems), genes encoding such elements were also removed from the analysis. Additionally, we noticed that most genes inside each genomic island usually showed similar patterns of variation (i.e., the whole island was usually present or not detected), so to avoid a bias from the large genomic clusters (containing more genes than the smaller ones), a maximum of four genes were selected for analysis from each cluster, representing those that eventually demonstrated different patterns of variation. Finally, we incorporated in our analysis 20 chromosomally encoded genes from other L. sakei strains that were absent from the L. sakei 23K chromosome. These genes were partly chosen from previously published clusters and partly taken from subtractive hybridization experiments carried out with strain 332F, known to be distantly related to strain 23K (5). Our selection process resulted in 40 genes originating from L. sakei 23K (representing 20 genomic islands and 3 independent genes) and 20 genes from four other strains. The characteristics of these 60 genes are summarized in Table 2.
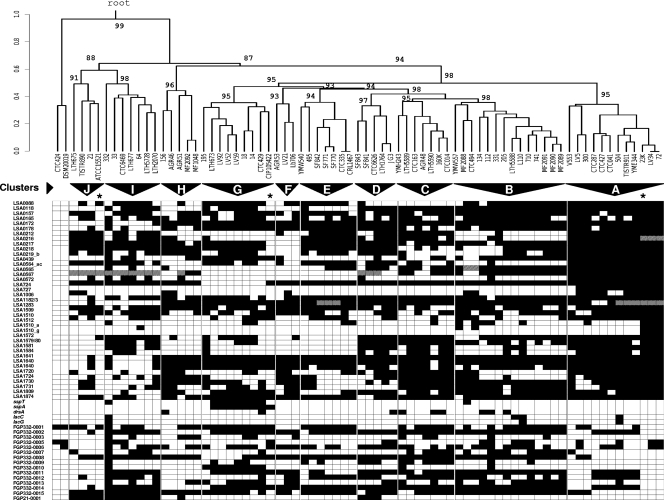
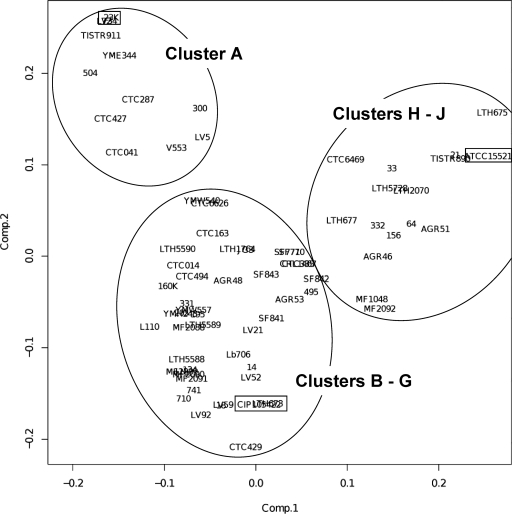
Based on the PCR detection profile of the 60 genes, we attempted to classify the L. sakei natural isolates by using an unsupervised average-linkage hierarchical clustering algorithm and by estimating P values via multiscale bootstrap resampling to assess the uncertainty of the clustering analysis (Fig. 1). From the resulting dendrogram, we could clearly identify at least 10 bootstrap-supported clusters of strains. These clusters have unequal sizes ranging from 14 strains to only 3 strains. Cluster A includes the reference strain 23K, cluster G includes the L. sakei subsp. carnosus type strain CIP 105422, and cluster J includes the most distantly related strains to cluster A, including the L. sakei subsp. sakei type strain, ATCC 15521. It should be noted that although the external branch separating clusters A to G from clusters H to J was statistically supported (P = 94%), the estimated P values of the main branches above clusters H to J were lower than 90%, suggesting that the hierarchical order of these three clusters between them is not supported. To confirm the overall grouping, a multivariate PCA was carried out (Fig. 2). This analysis confirmed the different grouping of clusters H to J from clusters A to G and also the external position of cluster A from the remaining clusters B to G.
FIG. 1.
L. sakei genomic diversity. Shown is the distribution of the 60 genetic markers among the 73 L. sakei isolates and the 2 L. curvatus isolates taken as an outgroup reference. Genes are ordered by their position in the 23K chromosome. White, absent; black, present; gray, IS-truncated gene or gene cluster. The dendrogram showing estimates of genomic relationships of the strains was constructed by average-linkage hierarchical analysis. The scale represents the distance at each node. A coefficient of 1 denotes complete independence, and zero indicates absolute identity. P values at main nodes indicate confidence of the clustering by multiscale bootstrap resampling using the PVCLUST program (see Materials and Methods). Strains were grouped on the basis of cluster branches where confidence was above 90% and with a maximum distance of 0.5 between isolates. Strain clusters (genotypes) are indicated by triangles. Clusters were named by starting with cluster A containing reference strain L. sakei 23K and were incremented hierarchically to cluster J, the most distantly related to cluster A. The subspecies type strains and the reference strain L. sakei 23K are indicated by asterisks.
FIG. 2.
PCA of L. sakei isolates based on the presence or absence of 60 genetic markers. The main groups are indicated within ellipsoids, and the subspecies type strains and the reference strain L. sakei 23K are denoted by rectangles.
Global proteomic variability between L. sakei isolates from the different genotypic clusters and subspecies affiliation of the clusters.
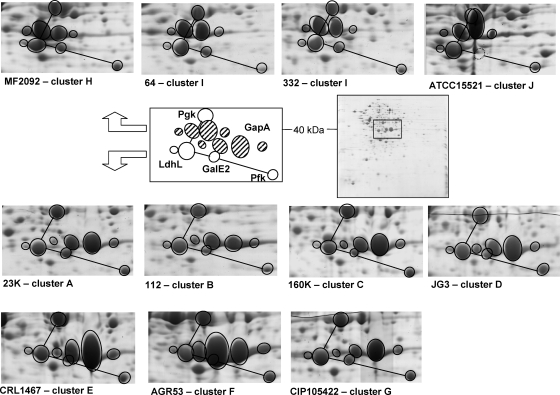
In previous studies, the prevailing L. sakei subspecies were defined by patterns of total soluble cell protein patterns obtained on native polyacrylamide gel electrophoresis gels (26, 38). The two subspecies patterns show a noticeable variation of an abundant protein around 40 kDa (see reference 26 for gel examples between the two subspecies type strains). We then used 2D electrophoresis to investigate this pattern in more detail and to compare the proteomes of a selection of 10 strains chosen from the various genotypic clusters including the two subspecies type strains. Although an average of ∼400 spots were commonly observed in the pI range of 4 to 7, we noticed a marked variation (>20%) around the average in the number of spots detected between strains (data not shown). Spots representing major differences (absence/presence of spots between strains) were identified by using matrix-assisted laser desorption ionization-time of flight mass spectroscopy. Most of the differences were shown to be related to proteins potentially encoded by genes not present in L. sakei 23K because they could not be identified in the protein database from this reference genome and were usually strain-specific spots (data not shown but in agreement with previous observations [21]). Hence, these variations, mainly strain dependent, could not be used as a criterion to differentiate the isolates to the subspecies level. On the other hand, a striking difference was observed in the 40-kDa region of the 2D gels containing spots of high intensity and corresponding to several glycolytic enzymes. In particular, we observed that the four isoforms (with different pIs) of the GapA protein (glyceraldehyde-3P dehydrogenase) displayed a size variation between the two subspecies type strains (Fig. 3). We found that this difference was not due to sampling issues or growth conditions, since four analyses were conducted (see Materials and Methods) and revealed no variation of this phenotype. We have suspected that this migration difference could be due to variations in the amino acid sequence of the GapA protein. However, the determination of the gapA gene sequence in strain 332 revealed only few modifications to that of strain 23K, most variations leading to silent mutations, suggesting that the pI and molecular weight modifications observed in the two types of GapA isoforms result most likely from posttranslational modifications of the protein (data not shown). This difference between strains was in agreement with the main difference observed in total cell protein pattern analysis previously used to distinguish the subspecies level (26, 38). The GapA variation was confirmed for a second isolate in each cluster and was then used to affiliate the genotypic clusters to the prevailing subspecies. In further agreement with the PCA clustering analysis shown in Fig. 2, clusters A to G (57 isolates representing 78% of the population) were affiliated with L. sakei subsp. carnosus and clusters H to J (16 isolates representing 22% of the population) were affiliated with L. sakei subsp. sakei. Our data also indicate that L. sakei 23K, although formally described as belonging to L. sakei subsp. sakei, should be reaffiliated with L. sakei subsp. carnosus.
FIG. 3.
Comparative analysis of the GapA isoforms between one representative isolate from each of the 10 genotypic clusters. For cluster I, two strains are shown: strain 332, used as the source of many genes for clustering analysis, and strain 64. A typical L. sakei 2D electrophoresis gel in the pI range 4 to 7 is shown on the right (L. sakei 23K). A window has been drawn in the middle area showing a closeup of the GapA region, which is schematically superimposed onto each strain tested to illustrate the variations in GapA isoforms. The open circles connected with a black line indicate a protein pattern highly conserved between strains, both on the basis of migration position and on the basis of level of expression. The striped ellipsoids represent the four GapA isoforms. Based on the 23K genome, the theoretical molecular masses of the corresponding proteins shown in this figure are as follows: GapA, 35.5 kDa; LdhL, 35.4 kDa; PgK, 42.7 kDa.
Variations of chromosome size and geometry between L. sakei genotypic clusters.
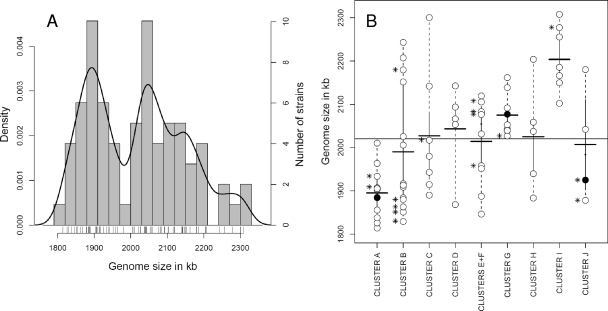
Next, we investigated the extent of genome size variation between L. sakei isolates by PFGE analysis of I-CeuI-digested fragments. I-CeuI mapping of the L. sakei chromosome reveals seven DNA fragments of various sizes and is an efficient tool for resolving the overall size and geometry of L. sakei genome (13). Genome size data are shown in Table S1 in the supplemental material. This analysis revealed important differences in genome size between the L. sakei strains. The mean chromosome size is 2,020 kb for the species, ranging from 1,814 ± 30 kb (strain CTC427, cluster A) to 2,309 ± 79 kb (strain LTH677, cluster I), which represents about 25% (∼500 kb) genome variation. A Gaussian probability distribution of the genome size data indicated that the chromosome size of L. sakei strains was not homogeneously distributed across this range (Fig. 4A).
FIG. 4.
(A) Histogram distribution of genome sizes among L. sakei isolates. Strains are represented by bars above the genome size axis. Histogram bars represent the number of strains within a genome size window of 30 kb (average standard deviation of PFGE measurements). The Gaussian probability distribution of genome size in the population (estimated density on the left axis) is shown by the black smooth line. (B) Plot showing genome size distribution of L. sakei isolates according to their genotypic clustering. The thin horizontal black line denotes the average genome size of 2,020 kb. Thick black horizontal lines indicate the average genome size in each cluster. Cluster F, containing few isolates, was pooled with cluster E for the analysis. The genome size of each isolate is shown by an open circle. The subspecies type strains and the reference 23K strain are shown by filled circles. Asterisks on the left side of some circles denote the positions of strains lacking one copy of the rrnAB doublet.
However, genome size cannot be used to distinguish between the two subspecies, since we noticed only a small difference between their average genome sizes (2,000 kb for L. sakei subsp. carnosus versus 2,100 kb for L. sakei subsp. sakei). Furthermore, analysis of the genome size distributions across the 10 genotypic clusters (Fig. 4B) revealed an important heterogeneity between them, explaining thereby the subpopulations observed in Fig. 4A. Indeed some clusters show uniform intraspecies genome sizes but a marked difference between them (e.g., average genome sizes of 1,895 kb for cluster A, 2,075 kb for cluster G, and 2,205 kb for cluster I). Other clusters (e.g., clusters B and C) display a large heterogeneity, suggesting the possibility of further subpopulations. Clusters F and J contain too few isolates to draw conclusions about the genome size trend in these clusters.
Finally, the absence of fragment C7 corresponding to the rrnAB doublet was noticed in 15 strains (∼20% of the whole population).
Many genotypes can be isolated from various meat/fish products.
We observed that each of the 10 clusters comprised strains from at least three different laboratory collections. Strains sourced from large collections (e.g., INRA, Jouy-en-Josas, France, and IRTA, Monells, Spain) did not cluster to themselves, but were distributed across the genotypic groups and clusters (average affiliation with seven clusters for each set of 10 strains), indicating that the results were not influenced by local sampling bias (collection bias or geographical bias). Similarly, the genotypic clusters (at least those with more than five isolates) did not show significant bias from the types of food products from which they were isolated (ecological bias). Finally, strains isolated from either raw or fermented products (process bias) were also evenly distributed between the groups. Therefore, our results suggest that the natural diversity of L. sakei strains may be identified within each type of food product.
DISCUSSION
This study provides an evaluation of intraspecies genomic variation of L. sakei and generates, for the first time, a comprehensive classification of natural isolates.
Our results show that the L. sakei species displays extensive (up to 500 kb) chromosome size variation between isolates. The difference covers ∼25% of the average species genome size of 2,020 kb. Many comparative genomic studies have revealed that intraspecies genomic diversity may vary widely between species from zero to more than 20% (for reviews, see references 9, 10, 12, and 25 and references cited therein). The extent of genomic variation within a species is believed to contribute to the ecological and phenotypic potentials bacteria require for survival in and exploitation of different ecological niches and the ability to respond to fluctuations in their natural environment (1). Therefore, the broad L. sakei intraspecies diversity observed in meat strains is likely to be a consequence of meat providing a range of complex ecological niches for microbial populations. Although a link between genotypic clusters and possible ecotypes could not yet be established using our analyses, we cannot discount the possibility that the number of isolates examined (n = 73) may have been too small to fully appreciate the wide natural diversity of the species. However, the strains analyzed were specifically chosen from a wide range of laboratory collections and care was taken wherever possible to include isolation sources representing the range of L. sakei natural habitats in food products. We have also shown that these two parameters (geographical and ecological) were evenly distributed among the genotypic clusters. Consequently, our data suggest that many meat/fish products could be the source of multiple L. sakei genotypes, possibly in combinations of yet-to-be-described ecotypes. It is possible that strains from several genotypes could successively dominate the ecological niche during meat storage as a result of the dynamic variations in microbial competition, fluctuation of nutrient availability, and changing redox conditions.
In this study, we provide a first insight into the possible number of molecular subtypes within the L. sakei species. The natural population can be seen as comprising two main groups of strains: (i) L. sakei subsp. carnosus, the more diverse, comprised of 7 clusters; and (ii) L. sakei subsp. sakei, comprised of 3 clusters. In both subspecies, we observed a trend toward a substantial difference in genome size between the various clusters. Therefore, our results favor the hypothesis that the major L. sakei genotypic groups have evolved sufficiently away from each other to yield populations with discernible genome sizes and this difference could be due to a differential adaptation to a specific environmental pressure (microhabitat). Nevertheless, we noticed some intracluster heterogeneity. For instance, some isolates show evidence of genomic rearrangements (loss of one rrn copy) and some other isolates have a genome size not fully representative of the L. sakei population to which they belong (e.g., strain CTC163 in cluster C, which appears to have evolved by acquiring a large amount of genetic material that distinguishes it from its close relative). Such a “leap” may result from the integration of large DNA segments like prophages or conjugative plasmids, which are often associated with genome evolution in bacteria (14, 34). These observations strongly suggest that deletion and integration events dynamically contribute to the evolution of the L. sakei species.
It remains possible that our clustering approach could result in a small degree of aberrant clustering for some strains or a biased estimation of the real number of molecular subtypes. For instance, PCR-based detection of genetic markers can only confirm the presence of particular genes but not their absence. PCR amplification may fail due to high nucleotidic polymorphism. Confirmation of the absence of genes could be achieved using DNA microarray technology. However, we consider that a high nucleotidic polymorphism between strains will be reflected by phylogenetic distance and would therefore have value for strain clustering. We have focused on the variable genes of strain L. sakei 23K only because currently it is the only strain with a sequenced genome. We have tried to address bias issues by introducing genes from other strains (especially from strain 332F from L. sakei subsp. sakei and distantly related from strain 23K) and by removing mobile elements from the analysis because they are known to skew clustering methods and to mask the lineages. However, the reason for the seemingly strong bias observed in the subspecies ratio (78%/22%) remains unclear. These findings could suggest a more successful adaptation of L. sakei subsp. carnosus to meat environments or that other sampling sources (food or environmental) should be included to improve the recovery of L. sakei subsp. sakei isolates. In this regard, we note that although the historical origin of L. sakei has yet to be determined, the species tend to be isolated from sources almost exclusively related to meat microbial ecosystems. It is also possible that the genotypic and ecological boundaries between the two subspecies may not be so sharp. It could also be of interest to determine whether microbial community membership and specific pressures in the natural environmental samples could influence the isolation of a given genotype.
Given these considerations, the next task will be to establish if the genotypic clusters identified in our present study correspond to phylogenetic lineages. This task could be achieved using multilocus sequence typing, and our genotypic clustering could be used as a basis for selecting strains for such studies. Similarly, comparative microarray-based genomic hybridization analysis may help to refine the genotypic clustering, especially for L. sakei subsp. sakei isolates for which our study may have lacked some discriminating power. This will require the characterization of the variable gene pools in a wider pool of strains to produce new and meaningful information from this type of analysis. To this effect, we are currently establishing a project that aims to characterize the flexible gene pool in strains covering the whole genotypic diversity of the L. sakei species (http://genome.jouy.inra.fr/sakei/biodiversity/html). We also believe that a sampling of new “undomesticated” strains from various traditional food products and from geographical areas underrepresented in our study (Asia, America, and Africa) would benefit further studies. Meat and fish products currently represent the major ecological source of L. sakei isolates. However, such products have only existed for a relatively short period of time. It is therefore possible that before meat products first became available for adaptive colonization by L. sakei, the species originated from sources such as the gastrointestinal tract of animals or the environment (e.g., pasture), where it may still survive as a minor component of the overall microbial population. Isolating and analyzing strains from such nonmeat environments might therefore reveal a greater and/or possibly different intraspecies diversity than currently appreciated.
Supplementary Material
Acknowledgments
Matrix-assisted laser desorption ionization-time of flight experiments were performed on the PAPPS platform of the INRA Center of Jouy-en-Josas. Statistical analyses were run on the LINUX server for bioinformatics (http://genome.jouy.inra.fr) of the INRA Center of Jouy-en-Josas. SSH analyses of L. sakei 332 were carried out with the financial support of the ANR (Agence Nationale de la Recherche) under the “Programme National de Recherche en Alimentation et Nutrition Humaine” (project ANR-05-PNRA-020.
We thank C. Hennequet-Jacquemard for useful advice on hierachical clustering analysis. We are also indebted to T. Aymerich, M. Garriga, L. Axelson, R. Valyasevi, Y. Morishita, B. G. Shaw, C. Hertel, G. Vignolo, F. Leroy, and J.-J. Joffreau, who provided us with L. sakei strains from their laboratory collections.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abby, S., and V. Daubin. 2007. Comparative genomics and the evolution of prokaryotes. Trends Microbiol. 15:135-141. [DOI] [PubMed] [Google Scholar]
- 2.Alpert, C.-A., A.-M. Crutz-Le Coq, C. Malleret, and M. Zagorec. 2003. Characterization of a theta-type plasmid from Lactobacillus sakei: a potential basis for low-copy-number vectors in lactobacilli. Appl. Environ. Microbiol. 69:5574-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthier, F., M. Zagorec, M. C. Champomier-Vergès, S. D. Ehrlich, and F. Morel-Deville. 1996. Efficient transformation of Lactobacillus sake by electroporation. Microbiology 142:1273-1279. [DOI] [PubMed] [Google Scholar]
- 5.Berthier, F., and S. D. Ehrlich. 1999. Genetic diversity within Lactobacillus sakei and Lactobacillus curvatus and design of PCR primers for its detection using randomly amplified polymorphic DNA. Int. J. Syst. Bacteriol. 49:997-1007. [DOI] [PubMed] [Google Scholar]
- 6.Chaillou, S., M. C. Champomier-Vergès, A. M. Crutz Le Coq, M. Cornet, A. M. Dudez, V. Martin, S. Beaufils, E. Darbon-Rongère, V. Loux, R. Bossy, and M. Zagorec. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23:1527-1533. [DOI] [PubMed] [Google Scholar]
- 7.Champomier-Vergès, M. C., M. C. Montel, F. Grimont, and P. A. Grimont. 1987. Genomic identification of meat lactobacilli as Lactobacillus sake. Ann. Inst. Pasteur Microbiol. 138:751-758. [DOI] [PubMed] [Google Scholar]
- 8.Champomier-Vergès, M. C., S. Chaillou, M. Cornet, and M. Zagorec. 2002. Lactobacillus sakei: recent developments and future prospects. Res. Microbiol. 152:115-123. [DOI] [PubMed] [Google Scholar]
- 9.Coenye, T., D. Gevers, Y. Van de Peer, P. Vandamme, and J. Swings. 2005. Towards a prokaryotic genomic taxonomy. FEMS Microbiol. Rev. 29:147-167. [DOI] [PubMed] [Google Scholar]
- 10.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 11.De man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:133-135. [Google Scholar]
- 12.Doolittle, W. F., and R. T. Papke. 2006. Genomics and the bacterial species problem. Genome Biol. 7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudez, A. M., S. Chaillou, L. Hissler, R. Stentz, M. C. Champomier-Vergès C. A. Alpert, and M. Zagorec. 2002. Physical and genetic map of the Lactobacillus sakei 23K chromosome. Microbiology 148:421-431. [DOI] [PubMed] [Google Scholar]
- 14.Eisen, J. A. 2000. Horizontal gene transfer among microbial genomes: new insights from complete genome analysis. Curr. Opin. Genet. Dev. 10:606-611. [DOI] [PubMed] [Google Scholar]
- 15.Fontana, C., G. Vignolo, and P. S. Cocconcelli. 2005. PCR-DGGE analysis for the identification of microbial populations from Argentinean dry fermented sausages. J. Microbiol. Methods 63:254-263. [DOI] [PubMed] [Google Scholar]
- 16.Guillot, A., C. Gitton, P. Anglade, and M. Y. Mistou. 2000. Proteomic analysis of Lactococcus lactis, a lactic acid bacterium. Proteomics 3:337-354. [DOI] [PubMed] [Google Scholar]
- 17.Hugas, M., M. Garriga, T. Aymerich, and J. M. Monfort. 1993. Biochemical characterization of lactobacilli from dry fermented sausages. Int. J. Food Microbiol. 18:107-113. [DOI] [PubMed] [Google Scholar]
- 18.Hugas, M., M. Garriga, T. Aymerich, and J. M. Monfort. 1995. Inhibition of Listeria in dry fermented sausages by the bacteriogenic Lactobacillus sake CTC494. J. Appl. Bacteriol. 79:322-330. [Google Scholar]
- 19.Jacquard, A. 1974. The genetic structure of populations. Springer-Verlag, Berlin, Germany.
- 20.Joffraud, J. J., M. Cardinal, J. Cornet, J. S. Chasles, S. Léon, F. Gigout, and F. Leroi. 2006. Effect of bacterial interactions on the spoilage of cold-smoked salmon. Int. J. Food Microbiol. 112:51-61. [DOI] [PubMed] [Google Scholar]
- 21.Jofré, A., M. C. Champomier-Vergès, P. Anglade, F. Baraige, B. Martín, M. Garriga, M. Zagorec, and T. Aymerich. 2007. Protein synthesis in lactic acid and pathogenic bacteria during recovery from a high pressure treatment. Res. Microbiol. 158:512-520. [DOI] [PubMed] [Google Scholar]
- 22.Katagiri, H., K. Kitahara, and K. Fukami. 1934. The characteristics of the lactic acid bacteria isolated from moto, yeast mashes for saké manufacture. IV. Classification of the lactic acid bacteria. Bull. Agr. Chem. Soc. Jpn. 10:156-157. [Google Scholar]
- 23.Klein, G., L. M. Dicks, A. Pack, B. Hack, K. Zimmerman, F. Dellaglio, and G. Reuter. 1996. Emended descriptions of Lactobacillus sake (Katagiri, Kitahara, and Fukami) and Lactobacillus curvatus (Abo-Elnaga and Kandler): numerical classification revealed by protein fingerprinting and identification based on biochemical patterns and DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 46:367-376. [Google Scholar]
- 24.Klein, G., and International Committee on Systematic Bacteriology. 2001. Subcommittee on the taxonomy of Bifidobacterium, Lactobacillus and related organisms. Int. J. Syst. Evol. Microbiol. 51:259-261. [DOI] [PubMed] [Google Scholar]
- 25.Konstantinidis, K. T., A. Ramette, and J. M. Tiedje. 2006. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B 361:1929-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koort, J., P. Vandamme, U. Schillinger, W. Holzapfel, and J. Bjorkroth. 2004. Lactobacillus curvatus subsp. melibiosus is a later synonym of Lactobacillus sakei subsp. carnosus. Int. J. Syst. Evol. Microbiol. 54:1621-1626. [DOI] [PubMed] [Google Scholar]
- 27.Labadie, J. 1999. Consequences of packaging on bacterial growth. Meat is an ecological niche. Meat Sci. 52:299-305. [DOI] [PubMed] [Google Scholar]
- 28.Lauret, R., F. Morel-Deville, F. Berthier, M. C. Champomier-Vergès, P. Postma, S. D. Ehrlich, and M. Zagorec. 1996. Carbohydrate utilization in Lactobacillus sake. Appl. Environ. Microbiol. 62:1922-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marceau, A., T. Méra, M. Zagorec, and M. C. Champomier-Vergès. 2001. Protein expression under uracil privation in Lactobacillus sakei. FEMS Microbiol. Lett. 200:49-52. [DOI] [PubMed] [Google Scholar]
- 30.Marceau, A., M. Zagorec, S. Chaillou, T. Méra, and M. C. Champomier-Vergès. 2004. Evidence for involvement of at least six proteins in adaptation of Lactobacillus sakei to cold temperature and addition of NaCl. Appl. Eviron. Microbiol. 70:7260-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montel, M. C., R. Talon, J. Fournaud, and M. C. Champomier. 1991. A simplified key for identifying homofermentative Lactobacillus and Carnobacterium spp. from meat. J. Appl. Bacteriol. 70:469-472. [DOI] [PubMed] [Google Scholar]
- 32.Morishita, Y., and K. Shiromizu. 1986. Characterization of lactobacilli from meats and meat products. Int. J. Food Microbiol. 3:19-29. [Google Scholar]
- 33.Noonpakdee, W., K. Phucharoen, P. Teerawattanamontri, T. Valyasevi, and S. Panyim. 1996. Molecular cloning, DNA sequencing and expression of catalase gene of Lactobacillus sake TISTR911 in Escherichia coli UM2. Asia Pac. J. Mol. Biol. Biotechnol. 4:229-235. [Google Scholar]
- 34.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 35.Schillinger, U., and F. K. Lucke. 1987. Identification of lactobacilli from meat and meat products. Food Microbiol. 4:199-208. [Google Scholar]
- 36.Shaw, B., and C. Harding. 1984. A numerical taxonomic study of lactic acid bacteria from vacuum-packed beef, pork, lamb and bacon. J. Appl. Bacteriol. 56:25-40. [DOI] [PubMed] [Google Scholar]
- 37.Tanasupawat, S., and W. Daengsubha. 1983. Pediococcus species and related bacteria found in fermented foods and related materials in Thailand. J. Gen. Appl. Microbiol. 29:487-506. [Google Scholar]
- 38.Torriani, S., C. A. Van Reenen, G. Klein, G. Reuter, F. Dellaglio, and L. M. T. Dicks. 1996. Lactobacillus curvatus subsp. curvatus nov. and Lactobacillus curvatus subsp. melibiosus nov. and Lactobacillus sake subsp. sake subsp. nov. and Lactobacillus sake subsp. carnosus subsp. nov., new subspecies of Lactobacillus curvatus Abo-Elnaga and Kandler 1965 and Lactobacillus sake Katagiri, Kitahara, and Fukami 1934 (Klein et al. 1996, emended descriptions), respectively. Int. J. Syst. Bacteriol. 46:1158-1163. [DOI] [PubMed] [Google Scholar]
- 39.Vermeiren, L., F. Devlieghere, and J. Debevere. 2004. Evaluation of meat born lactic acid bacteria as protective cultures for the biopreservation of cooked meat products. Int. J. Food Microbiol. 96:149-164. [DOI] [PubMed] [Google Scholar]
- 40.Vogel, R. F., M. Lohmann, A. N. Weller, M. Hugas, and W. P. Hammes. 1991. Structural similarity and distribution of small cryptic plasmids of Lactobacillus curvatus and L. sake. FEMS Microbiol. Lett. 84:183-190. [DOI] [PubMed] [Google Scholar]
- 41.Vogel, R. F., M. Lohmann, M. Nguyen, A. N. Weller, and W. P. Hammes. 1993. Molecular characterization of Lactobacillus curvatus and L. sake isolated from sauerkraut and their application in sausage fermentations. J. Appl. Bacteriol. 74:295-300. [DOI] [PubMed] [Google Scholar]
- 42.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.