Abstract
Lactococcus lactis IL1403 was used as an experimental strain to develop a chemically defined medium for study of the physiology and metabolic pathways of lactococci. An experimental leave-one-out technique was employed to determine the necessity of each of the 57 chemical components used in medium development. A statistical experimental design approach including three fractional factorial designs and a central composite design was used to optimize the fermentation process with 21 variables composed of 19 nutritional factors grouped from the 57 components and two environmental factors (initial pH and temperature). For L. lactis IL1403, the maximum biomass concentrations obtained with the two optimal chemically defined media developed in this study (ZMB1 and ZMB2) were generally 3.5- to 4-fold higher than the maximum biomass concentrations obtained with the previously described best synthetic media (SA) and 50% to 68% higher than the maximum biomass concentrations obtained with M17, a complex medium commonly used for lactococci. The new chemically defined media support high-cell-density growth of numerous strains of L. lactis, Enterococcus faecalis, and Streptococcus thermophilus.
Lactococcus lactis is a lactic acid bacterium and has two main subspecies, L. lactis subsp. lactis and L. lactis subsp. cremoris (16). L. lactis has been granted generally recognized as safe status by the FDA and is used extensively in the dairy industry (2), among other industries (6, 8, 12). Recently, determination of the genome sequences of several L. lactis strains (2) has allowed development of metabolic models and enabled novel “omics” strategies to examine cellular metabolism during fermentation (1, 8, 11). All of these strategies are greatly facilitated by use of chemically defined media (CDMs), in which metabolism is more easily defined.
Due to a lack of various biosynthetic pathways, L. lactis generally requires media rich in nutrients. A rich nutrient environment can be provided by complex media comprised mostly of complex components (such as yeast extract, peptone, or tryptone), by semidefined media formulated mostly with defined chemicals (except for one or two complex nutrients), and by CDMs (synthetic media) containing no complex components (19). Although in most cases complex and semidefined media provide greater biomass yields than CDMs, using these types of media in physiological studies focusing on metabolism and regulation makes data more difficult to interpret (3). For this reason, a CDM that supports reasonable cell growth can be very useful in studies of gene regulation, protein expression, and metabolic fluxes. By systematically adding or removing components from a CDM formulation, the specific nutritional and regulatory requirements for growth and targeted metabolic pathways can be determined. The uncertainty due to the complicated interactions among complex components can be minimized or at least more easily understood, and the culture environment is more reproducible (16, 19).
To understand the metabolism of L. lactis, many attempts have been made to design specific CDMs, such as MS10, MS14, MS15, and SA, for particular L. lactis strains (3, 7, 11). In general, the initial formulations of these CDMs imitated the typical cellular composition of the microorganism of interest, were developed using previously described CDMs for related microorganisms, or simulated the nutrient composition of an existing complex medium (e.g., M17 or MRS) (5, 14). The level of growth on these defined media, however, has typically been far less than the level of growth obtained with complex media, which limits their utility.
Since L. lactis strains have been isolated from several environments (e.g., vegetable or dairy sources) and genetic variation may have been introduced naturally or through genetic manipulation, the specific nutrient requirements of L. lactis strains could be very different (3, 16). This makes development of a CDM generally applicable for L. lactis a particularly difficult problem. Currently no generally applicable CDMs that support high-cell-density growth of L. lactis are available. The objective of this study was to develop and optimize a CDM that can be used for a wide range of L. lactis strains and which supports better growth than existing defined media and the same or better growth than the complex medium M17.
Medium optimization generally involves determining the appropriate nutrients and establishing the concentrations of these nutrients that result in maximum levels of biomass or targeted microbial products (e.g., a recombinant protein). Traditionally, CDM optimization has been informed by biochemical studies or by using a single-factor approach, also called a one-dimensional search, which examines one nutritional factor at a time (3, 7). In spite of its experimental simplicity, this type of approach is not efficient for multiple medium components and is, in fact, likely to miss important interactions between components, resulting in a suboptimal medium. An alternative optimization strategy is a statistical design-of-experiment (DOE) method, which systematically evaluates more than one independent factor at a time (10). In practice, this method often starts with at least one fractional factorial design (FFD) (e.g., resolution III or the Plackett-Burman method with a minimum number of experiments) to screen all the potential factors to determine which ones have a significant effect on the desired response (13). With the significant factors identified, a response surface method, such as a central composite design (CCD) or D-optimal design method, is then used to search for the combination of factors supporting a near-optimal output (or the best acceptable output) in a timely manner (10, 18, 19). In this study, we employed a statistical DOE method to optimize a CDM, which resulted in two media that support growth of lactococci, enterococci, and streptococci that is comparable to or better than the growth achieved on the complex medium M17.
MATERIALS AND METHODS
Medium preparation.
A total of 57 components (Table 1), many from previously described CDMs for L. lactis, were investigated for development of a new optimized CDM. All chemicals were reagent grade and were purchased from Fisher Scientific (Pittsburgh, PA) or Sigma-Aldrich Co. (St. Louis, MO). More information concerning the biological function of each of the specific chemical species has been reported previously (7, 16, 19). For preparation of the CDM, concentrated stock solutions of most of the 57 chemicals were prepared and stored at −20°C; the only exception was the FeSO4 solution, which was freshly prepared. Stock solutions were autoclaved or filter sterilized (pore size, 0.2 μm; Millipore, Billerica, MA). Filter sterilization was used for heat-labile amino acids (e.g., asparagine, glutamine, and tryptophan) and water-soluble vitamins (e.g., pyridoxal HCl, thiamine HCl, riboflavin, etc.). Detailed preparation procedures for all stock solutions are shown in Table S1 in the supplemental material. The CDMs for all trials and the four previously described CDMs (MS10, MS14, MS15, and SA) were prepared using these sterile stock solutions.
TABLE 1.
Medium components for the two optimal CDMs developeda
| Group | Variable category | Chemical component | Necessity of componentb | Concn (g/liter) in:
|
|
|---|---|---|---|---|---|
| ZMB1 | ZMB2 | ||||
| 1 | Sugar | Glucose | E | 15 | 15 |
| 2 | Essential amino acid 1 | l-Histidine | E | 0.17 | 0.17 |
| 3 | Essential amino acid 2 | l-Isoleucine | E | 0.24 | 0.24 |
| 4 | Essential amino acid 3 | l-Leucine | E | 1 | 1 |
| 5 | Essential amino acid 4 | l-Methionine | E | 0.06 | 0.06 |
| 6 | Essential amino acid 5 | l-Valine | E | 0.7 | 0.7 |
| 7 | Essential amino acid 6 | l-Arginine | E | 0.72 | 0.72 |
| 8 | Vitamin | Inositol | S | 0.002 | 0.002 |
| 9 | Phosphate buffers | KH2PO4 | E | 3.1 | 3.6 |
| K2HPO4 | I | 6.4 | 7.3 | ||
| 10 | Other amino acid group | l-Glutamic acid | L | 0.6 | 0.72 |
| l-Phenylalanine | L | 0.4 | 0.48 | ||
| l-Proline | S | 0.7 | 0.84 | ||
| l-Asparagine | S | 0.5 | 0.60 | ||
| l-Aspartic acid | L | 0.05 | 0.060 | ||
| l-Glutamine | L | 0.6 | 0.72 | ||
| l-Serine | S | 0.5 | 0.60 | ||
| l-Threonine | S | 0.5 | 0.60 | ||
| l-Cysteine HCl | L | 0.2 | 0.24 | ||
| l-Alanine | S | 0.4 | 0.48 | ||
| Glycine | S | 0.3 | 0.36 | ||
| l-Lysine HCl | S | 0.5 | 0.60 | ||
| l-Tryptophan | L | 0.2 | 0.24 | ||
| l-Tyrosine | S | 0.3 | 0.36 | ||
| 11 | Important vitamin group | Biotin | S | 0.006 | 0.006 |
| Calcium pantothenate | I | 0.0012 | 0.0012 | ||
| Niacin | I | 0.0009 | 0.0009 | ||
| Pyridoxal HCl | I | 0.0048 | 0.0048 | ||
| Riboflavin | I | 0.0009 | 0.0009 | ||
| 12 | Important mineral group | MgSO4·7H2O | E | 1 | 1 |
| FeSO4·7H2O | S | 0.004 | 0.004 | ||
| ZnSO4·7H2O | I | 0.005 | 0.005 | ||
| 13 | Other vitamin group | Folic acid | S | 0.00056 | 0.00056 |
| p-Aminobenzoic acid | S | 0.000056 | 0.000056 | ||
| Thiamine HCl | S | 0.00056 | 0.00056 | ||
| 14 | Fatty acid group | Potassium acetate | I | 0.9 | 0.9 |
| Lipoic acid | S | 0.001 | 0.001 | ||
| Tween 80 | S | 0.5 | 0.5 | ||
| 15 | Nucleic acid base group | Adenine | S | 0.011 | 0.011 |
| Guanine | S | 0.0056 | 0.0056 | ||
| Uracil | S | 0.023 | 0.023 | ||
| Xanthine | S | 0.0038 | 0.0038 | ||
| 16 | Other buffer group | MOPS | I | 15 | 13.05 |
| Tricine | S | 1.5 | 1.305 | ||
| 17 | Trace mineral group | (NH4)6Mo7O24·4H2O | S | 0.00019 | 0.00019 |
| MnSO4·4H2O | S | 0.00038 | 0.00038 | ||
| CaCl2·2H2O | S | 0.04 | 0.04 | ||
| CoCl2·6H2O | S | 0.00019 | 0.00019 | ||
| CuSO4·5H2O | S | 0.00019 | 0.00019 | ||
| H3BO3 | S | 0.00075 | 0.00075 | ||
| K2SO4 | S | 0.023 | 0.023 | ||
| KI | S | 0.00011 | 0.00011 | ||
| 18 | Chelator group | EDTA | S | 0.0075 | 0.0075 |
| Nitrilotriacetic acid | S | 0.0075 | 0.0075 | ||
| 19 | Other component group | Glutathione | S | 0.015 | 0.015 |
| (NH4)2SO4 | S | 1 | 1 | ||
| NaCl | S | 3 | 3 | ||
The environmental conditions for ZMB1 and ZMB2 are as follows: starting pHs of 7.8 and 8.04, respectively; and incubation temperatures, 27.5 and 24.1°C, respectively.
E, essential component (OD < 0.1); I, important component (0.1 ≤ OD < 0.4); S, somewhat important component (0.4 ≤ OD < 0.6); L, least important (or even detrimental) component (OD ≥ 0.6). All classifications were determined using the results of the LOO experiments (see Table S1 in the supplemental material).
In this study, the complex medium that was used for inoculum preparation and comparison with new CDMs was modified M17, in which 5 g/liter lactose was replaced by 20 g/liter glucose to support a higher level of growth (8). This complex medium (37.25 g/liter Difco M17 broth) was autoclaved for 20 min at 121°C before it was mixed with glucose that was sterilized separately.
Bacterial strains and inoculum preparation.
A completely sequenced strain, L. lactis IL1403 of dairy origin (2, 3), was used as an experimental strain throughout the optimization and scale-up studies. Frozen stock cultures (1 ml of culture and 0.75 ml of 60% glycerol in a 2-ml cryovial) were prepared and stored at −80°C. For each batch of fermentations, 1 ml of frozen stock culture was used to inoculate 50 ml of M17 in a 125-ml flask, which was then incubated for 9 h at 30°C. The seed culture was centrifuged at 6,000 × g for 5 min. To minimize the carryover of M17 components to the fermentation, the cell pellet was washed twice in the same amount of 0.01 M phosphate-buffered saline, and the pellet was then resuspended in the same amount of sterile water immediately before inoculation.
In addition to L. lactis IL1403, eight other lactococcal strains (L. lactis subsp. lactis ML3 and LM0230, L. lactis subsp. lactis bv. diacetylactis 18-16, UCD172, and DCR 3, and L. lactis subsp. cremoris MG1363, FG2, and SK11), two streptococcal strains (Streptococcus thermophilus MTC330 and MTC360), and two enterococcal strains (Enterococcus faecalis OG1RF and KA177) were used to test the new optimal CDMs and to compare the performance of the new CDMs with that of M17. An inoculum for each of these strains was prepared as described above.
Culture preparation and conditions.
A series of CDM optimization experiments was carried out in standard 96-well microtiter plates (Costar* clear polystyrene 96-well plates; Fisher Scientific, Pittsburgh, PA) to simplify biomass measurement. At the development stage for each CDM, 4.95 ml of CDM (including the desired amounts of stock solutions and distilled water) was pipetted into a 5-ml volumetric flask. The starting pH of the CDM was adjusted to the desired value using an Orion microcombination pH/sodium electrode (Thermo Scientific, Beverly, MA) by addition of 10 N NaOH and 10 N H2SO4. The inoculum (50 μl) was transferred into the 4.95 ml of CDM. Inoculated CDM (200 μl) was then pipetted into a well of a microtiter plate, covered with 40 μl of sterile mineral oil to prevent evaporation and maintain an anaerobic environment, and incubated without shaking for 12 to 24 h at the desired temperature. For each set of growth conditions four replicates were used. Cultures of the eight other lactococcal strains were prepared as described above. The procedure used for the two streptococcal strains and the two enterococcal strains was similar, except that 150 μl of inoculated CDM was used in each well of the microtiter plate without mineral oil as incubation was carried out in an anaerobic chamber.
For test-tube-scale fermentations, 25 ml of a CDM was prepared and transferred into a 50-ml sterile centrifuge tube. The starting pH of the CDM was adjusted to the desired value using the pH probe described above. After inoculation with 250 μl of inoculum, the CDM was incubated without shaking for 20 h at the desired temperature. All experimental conditions were examined in triplicate.
Biomass measurement.
The response examined for this experimental optimization procedure was cell concentration. The optical density (OD) of each culture in the microtiter plates was determined by using an Emax precision microplate reader (Molecular Devices, Sunnyvale, CA) at a preset wavelength of 650 nm. To prevent cell aggregation, the cultures were stirred with a stainless steel, 48-pin replicator before the OD was determined. Each measurement was normalized by subtracting the background value for the original medium from the final OD.
The OD of all test-tube-scale fermentations were determined using a UV 1201 spectrophotometer with sipper mode (Shimadzu, Columbia, MD) at 600 nm, a wavelength more commonly used for OD measurement, after the cultures were stirred using a vortex mixer to break any cell aggregates. If the OD was greater than 0.7, the medium sample was diluted with water (generally between two- and sevenfold) and the resulting absorbance value was multiplied by the corresponding dilution factor to calculate the final OD. The background value for the original medium was subtracted from the final OD to normalize the measurements.
Software used for experimental design and analysis.
Design-Expert version 7.1.1 (Stat-Ease, Inc., Minneapolis, MN) was used to design three sets of Resolution III designs and one set of CCD. For the CCD, a quadratic, polynomial model that best fit the CCD data for a given response (e.g., OD) was automatically suggested by the Design-Expert program. This polynomial had the form:
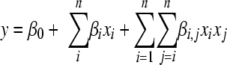 |
(1) |
where β is a regression coefficient, n is the dimensionality of an input space, and x is a variable in the CCD design. A statistically significant model was selected by minimizing the sum-squared errors between the measured OD and the predicted OD (detailed procedures for model fitting have been described previously [10]). Data analysis was performed using Design-Expert, MATLAB version 7.0.1 (The Math Works, Natick, MA), and Microsoft Office Excel 2003 (SP2).
RESULTS
Evaluating the importance of the 57 medium components.
A leave-one-out (LOO) technique, in which single nutrients are removed to design a minimal defined medium (3), was used with the 57 chemicals as the first stage in the development of the L. lactis IL1403 CDM. This technique was used to determine the necessity of different nutritional factors and to reduce the number of experimental factors. One component at a time was eliminated from the starting CDM that contained all 57 components. All cultures in the inoculated CDM were grown in 96-well microtiter plates for 24 h with an initial pH of 6.6 at 30°C. The resulting maximum OD in each of the 57 LOO experiments was compared to the maximum OD of the starting CDM (0.587 ± 0.026). As mentioned above, Table S1 in the supplemental material shows the concentrations of the components in the starting CDM and the maximum biomass for each of the 57 LOO experiments. Based on the observations made in these experiments, the 57 chemical species were separated into four classes (Table 1): essential components (OD, <0.1), important components (0.1 ≤ OD < 0.4), somewhat important components (0.4 ≤ OD < 0.6), and least important components (or even detrimental components, since more growth was observed without the component) (OD, ≥0.6). To reduce the dimensions that might be needed for medium optimization, the 57 components were divided into 19 variable groups (Table 1) depending on the classification of importance in the LOO experiments, as well as the biological functions of the components. For instance, each of the essential amino acids was used as a separate factor, while some of the vitamins and trace minerals were grouped.
Optimizing the fermentation in CDM using the statistical DOE method.
To minimize the number of experiments required to optimize the CDM, a statistical DOE approach was employed to identify the most significant variables and the optimal ranges for them. Besides the 19 grouped nutritional variables, two environmental parameters (temperature and starting pH) were also investigated. The original ranges of these two environmental variables were determined using data from previous studies (3, 8, 16).
Three batches of experiments using a two-level Resolution III FFD were designed and performed. Each of these batches of experiments included 32 experiments (in which each combination of levels for a pair of factors appeared the same number of times and the frequency of each level of a factor was also the same) plus five extra center points (halfway between the high and low levels for each of the 21 factors) in order to evaluate the effect curvature and experimental variability. For each of the experiments, a culture was grown in a 96-well microtiter plate, and the resulting cell concentration was determined after 12 h. This time was chosen based on LOO experiments in which the maximum OD was observed after 12 h in most experiments.
For each factor, two levels were used in the experiments, a high level and a low level, which defined the range of the factor. The difference between the average response in the experiments with the high level of the factor (Rh) and the average response in the experiments with the low level of the factor (R1) could be calculated using equation 2, in which N is the total number of experiments (excluding the center point experiments).
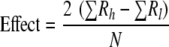 |
(2) |
Depending on whether the effect determined in equation 2 was positive or negative, the range was shifted in the direction of the more likely optimum. The progression of the variable ranges for the three sets of FFDs is shown in Table S2 in the supplemental material. Figure 1a to c show the corresponding Pareto charts for the three sets of FFDs, in which the absolute effects of the 21 variables for each batch of experiments are shown in descending order. To determine the overall significance of each independent variable, the rank of each variable for each of the batches of experiments was summed and plotted (Fig. 1d). The significance of each variable should be inversely proportional to its cumulative rank.
FIG. 1.
Ranking the importance of 21 variables based on the Pareto charts for three sets of FFD. (a to c) Pareto charts for (a) FFD set I, (b) FFD set II, and (c) FFD set III. (d) Plot of the cumulative rank of the 21 variables. OAAG, other amino acid group; T, temperature; Met, l-methionine; Gluc, glucose; PB, phosphate buffers; IMG, important mineral group; OBG, other buffer group; FAG, fatty acid group; Leu, l-leucine; Arg, l-arginine; CG, chelator group; Ile, l-isoleucine; IVG, important vitamin group; NABG, nucleic acid base group; OCG, other component group; Val, l-valine; Ino, inositol; OVG, other vitamin group; His, l-histidine; TMG, trace mineral group.
To minimize the number of experiments necessary in the final optimization procedure, only the five variables with the most significant effects (initial pH, other amino acid group, phosphate buffers, other buffer group, and temperature) identified in Fig. 1d were selected for inclusion in the final response surface set of experiments. The response surface method chosen was a CCD (rotatable α = 2.378), which included 32 two-level full factorial experiments, two star points for each of the five factors, and eight center points. As shown in Table S2 in the supplemental material, the concentrations of the other 16 variables were set to be constant based on the previous trials that yielded the highest level of biomass. Table S2 in the supplemental material also shows the concentrations or concentration ranges of the variables in this final design. The experimental results obtained with the CCD show that the medium conditions associated with the maximum OD (ZMB1) (Table 1) resulted in a maximum OD of 0.92, with a standard deviation of 0.02. The improvement in the 12-h biomass accumulation with progression of the statistical optimization method is shown in Fig. S1 in the supplemental material. Based on the maximum OD distribution for all fermentations in a batch, it is clear that the average biomass of a batch generally increased with each batch of experiments, as did the highest level of biomass.
In order to assess whether growth could be improved further using a combination of the five factors not tested experimentally, a polynomial function was fitted to the CCD data. A numerical optimization technique was then used with this polynomial function. To increase the chance of locating the “best” local optimum, the optimization procedure started from m random points in the CCD space (including the axial points) and proceeded up the steepest slopes to find the m local maximum. The maximum OD predicted was 0.998, using the set of variable values of the CCD optimum (ZMB2 in Table 1). Four fermentation replicates with the predicted optimal operating conditions were grown at the microplate scale to verify that the predicted set of conditions actually further improved the process. These fermentations resulted in a new optimal medium, ZMB2, in which the maximum OD was 1.04 ± 0.02.
Comparing the new CDMs with previously described media at optimization scale.
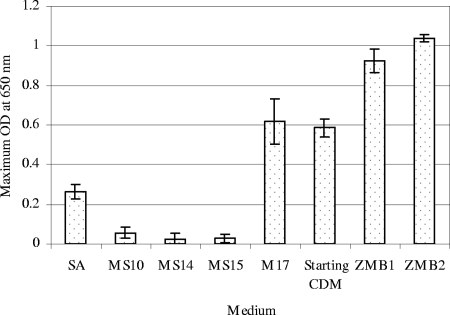
To evaluate the effectiveness of the two new CDMs (ZMB1 and ZMB2), the growth of L. lactis IL1403 in these two media was compared with the growth in the four previously described CDMs, the starting CDM prior to optimization, and the complex medium M17. Most inoculated media were incubated under the standard process conditions (30°C with a starting pH of 6.6 in a microtiter plate); the exceptions were the new CDMs, whose optimized environmental conditions are shown in Table 1. In this way, we were able to compare standard media and standard operating conditions with the new media and optimized conditions. Figure 2 shows the growth of L. lactis IL1403 in these media. As can be seen, poor growth was observed for three minimal media (MS10, MS14, and MS15, which contain only nutritional factors that satisfy no more than the minimal requirement for cell growth [19]). At the microtiter plate scale, the maximum OD obtained with ZMB1 and ZMB2 were generally 3.5- to 4-fold higher than the maximum OD obtained with the best previously described synthetic medium (SA) and 50 to 68% higher than the maximum OD obtained with M17 and the starting CDM, respectively.
FIG. 2.
Comparison of the optimal CDMs (ZMB1 and ZMB2) with four previously described CDMs, the starting CDM, and a complex medium (M17). Most cultures were grown in 96-well microtiter plates with a working volume of 200 μl for 12 h at 30°C; the exceptions were the ZMB1 and ZMB2 cultures, which were grown at 27.5 and 24.1°C, respectively. ODs were determined at a preset wavelength of 650 nm.
While a temperature of 30°C is commonly used for L. lactis growth in the complex medium M17 and thus was used for comparison to the new conditions, this temperature may not be the optimum temperature for all strains in M17. Therefore, we examined the growth of L. lactis IL1403 in M17 at various temperatures, including temperatures in the range from 30 to 40°C, all at the 200-μl scale (data not shown). In this range, maximum growth was observed at 37°C. The mean OD at 37°C was approximately 15% higher than the mean OD at 30°C. Therefore, even compared to M17 at the optimal temperature, the new defined media, ZMB1 and ZMB2, supported significantly more growth.
Scale-up fermentations.
Practically, increasing the scale of fermentation to the scale commonly used in laboratory experiments could affect the yield of cells or the target product. To investigate the influence of scale on CDM fermentation performance, L. lactis IL1403 was incubated in 25 ml of each medium (including the two new CDMs, SA, and M17) for 20 h. The growth curves in Fig. 3 show that the final cell growth in these test-tube-scale fermentations was similar to that in the microtiter-plate-scale fermentations. L. lactis IL1403 grew much faster in the new CDMs than in SA and M17, and the maximum OD at 600 nm was higher. The maximum OD at 600 nm for ZMB2 (at 24.2°C) was slightly higher than that for ZMB1 (at 27.5°C), while the growth rate in ZMB2 was much lower than the growth rate in ZMB1, likely due to the incubation temperature used for ZMB2, which was not optimal for growth.
FIG. 3.
Comparison of the growth curves for L. lactis IL1403 incubated in four media at the test tube scale. Cultures were grown in 50-ml test tubes with a working volume of 25 ml for 20 h. Symbols: ▴, ZMB1 incubated at 27.5°C; ×, ZMB2 incubated at 24.1°C; ▪, M17 incubated at 30°C; •, SA incubated at 30°C. Optical densities were determined at a wavelength of 600 nm.
Performance of new CDMs with other strains of lactococci.
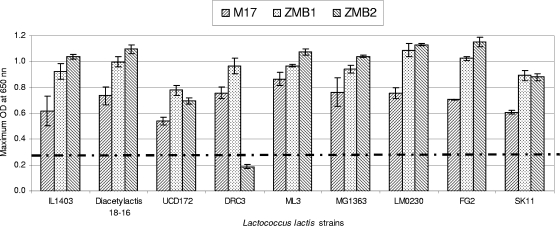
To determine if the composition of the optimal CDMs is specific for strain IL1403, eight other L. lactis strains, including two other completely sequenced strains, MG1363 (17) and SK11 (9), were examined. The performance of the new CDMs and M17 as the control medium is shown in Fig. 4. In view of the results presented here, it is obvious that all nine strains grew very well in the new CDMs, and the maximum cell growth of most strains in the new CDMs was greater than that in M17; the only exception was L. lactis DCR3, which grew poorly in ZMB2. Moreover, the growth rate of most of the strains tested in ZMB2 was slightly higher than in ZMB1; the exceptions were L. lactis strains UCD172, DRC 3, and SK11.
FIG. 4.
Evaluation of the maximum OD of nine L. lactis strains cultivated in three media. The dashed and dotted line indicates the maximum biomass of L. lactis IL1403 growing in the best previously described CDM, SA. Cultures were grown in 96-well plates with a working volume of 200 μl for 20 h at set incubation temperatures (ZMB1 at 27.5°C, ZMB2 at 24.1°C, M17 at 30°C, and SA at 30°C). Optical densities were determined at a wavelength of 650 nm.
General applicability of new CDMs for growth of streptococci and enterococci.
To investigate the general applicability of the new CDMs, two streptococcal strains (S. thermophilus MTC330 and MTC360) and two enterococcal strains (E. faecalis OG1RF and KA177) were cultivated in 150 μl of the new CDMs at 37°C for 24 h, with M17 as the control medium. The maximum cell growth of each of the strains tested is shown in Fig. 5. As Fig. 5 shows, all streptococcal and enterococcal strains tested grew very well in the new CDMs. The maximum cell growth values for all strains tested in ZMB1 and ZMB2 were generally 61 to 94% and 82 to 129% higher than those in M17, respectively; the only exception was S. thermophilus MTC360, for which similar biomass production was observed with all media.
FIG. 5.
Analysis of the general applicability of the new media using strains of streptococci and enterococci, with M17 as the control medium. Cultures were grown anaerobically in 96-well plates with a working volume of 150 μl for 24 h at 37°C. Optical densities were determined at a wavelength of 650 nm.
DISCUSSION
Although the optimal CDM formulation for a fermentation process is generally strain specific, starting with previously described defined media to develop a CDM for related species can be a great help. Based on existing media for L. lactis and previous knowledge of lactococci (3, 7, 11, 16), two new CDMs (ZMB1 and ZMB2) that support high-cell-density growth of L. lactis IL1403 were developed in this study. We confirmed that these new CDMs support significantly more growth than four previously described CDMs (3, 7, 11) and a commonly used complex medium (M17) in microtiter-plate- and test-tube-scale fermentations. This is due to the variables (nutritional species, concentrations, and environmental parameters) optimized in the fermentation system used. It is also likely that the new CDMs can be used for lactococcal strains in general because they were neither designed as minimal defined media nor tailor-made for a particular strain. Defined media MS10, MS14, and MS15, which were designed as minimal media, are not as useful for a broad range of organisms, as shown by the fact that the experimental strain IL1403 could not be cultivated in three of the four previously described CDMs. Compared to cultivation in the complex M17 medium, cultivation of L. lactis in the new CDMs might make it easier to interpret metabolic flux data in physiological studies of lactococci.
The LOO technique that we employed can be a useful method for classification of components based on whether nutrients are necessary for cell growth. Even though this technique provides a good indication of how critical each nutritional factor is for growth, there is no guarantee that a minimal CDM can be designed simply with the essential components identified by this approach. Indeed, a minimal CDM composed of only the nine essential components identified in this screen did not support growth of L. lactis IL1403 (data not shown). This could have been because the degree to which a nutrient is truly essential may be dependent on other nutritional factors present in a CDM. Although the LOO technique has this limitation, it was still useful for grouping the nutritional factors, which reduced the high dimensionality of the optimization problem at the first step and subsequent steps in CDM development.
It is clear from the data shown here that pH is a key factor in the success of a microtiter-plate- or test-tube-scale fermentation (as confirmed by the significant variables selected from the cumulative rank of 21 variables shown in Fig. 1d). This has also been observed by other workers (3, 16). This is likely because L. lactis can regulate its internal pH between 7 and 7.5 only when the external pH is not too extreme (e.g., from pH 4.5 to pH 8.5). While complex media such as M17 have many components with natural buffering capacity in addition to specific buffer components (e.g., disodium-β-glycerophosphate), addition of specific buffer components to CDM is absolutely required to balance the lactic acid produced in the absence of this natural buffering. In this work, the buffering was supplied mainly by both phosphate and morpholinepropanesulfonic acid (MOPS) buffer systems. In addition, we showed that a higher initial pH is also beneficial for the yield. However, higher concentrations of buffers might inhibit the formation of certain desired products (19), and a higher initial pH can also delay exponential growth and cause precipitation of some components prior to initiation of fermentation. While most of the precipitates dissolve during fermentation due to acidification of the CDM (data not shown), this phenomenon may still prohibit use of key medium ingredients, such as trace metals, early in cell growth. In our experience, growth ceases when the pH decreases to a certain value (the lowest final pH observed was around 4.3 ± 0.1 in this study), suggesting that the pH limits the final cell density in a CDM, which has been observed previously for other L. lactis strains (1, 3, 16).
In this study, the ratios of concentrations within each of the 19 nutritional families were chosen based on previous reports (3, 11, 16). This poses the question of whether the ratios of chemical concentrations chosen are optimal for cell growth. For instance, some amino acids can be synthesized from metabolic intermediates derived from other nitrogenous components of a CDM (3). It is likely that the ratios could, in fact, be further optimized with further experimentation. However, making ratio assumptions dramatically reduced the number of fermentations required for the statistical DOE method and still significantly improved the CDMs. Additional investigation could be carried out to further improve the CDMs using a mixture design or a simplex design method with certain nutritional groups.
Since there is great strain variation among the lactococci (15), a useful CDM would ideally allow growth of many strains to increase the reliability of comparative interspecies studies. In general, dairy-origin strains are more demanding than plant-origin strains (3), although the absolute nutrient requirements of most L. lactis strains tested here were quite similar. Because our LOO experiments did not account for the possibility of interactions between components or the impact of the concentration ranges chosen for study, some of the nutrients identified as insignificant factors for L. lactis IL1403 in our LOO experiment were not removed from the CDM formulation. We took this action with the understanding that truly insignificant variables would be removed by the optimization process. Leaving all 57 components in the optimization likely contributed to the general applicability of the media developed. Compared with the CDM described by other investigators, our optimal CDMs included more components, which may be one reason why they supported the growth of eight other L. lactis strains in this study. One exception to this is that L. lactis DRC3 grew rather poorly in ZMB2. It is not clear whether this was due to unbalanced nutritional concentrations for this particular strain or detrimental changes in the pH, temperature, or buffer composition, although experimental replicates and other data for the same inoculum and same media indicated that this was a real effect. Incorporating genomic information for specific strains may help us understand the observed behavior and lead to further improved, albeit less general, media. Besides the lactococcal strains tested, several strains of enterococci and streptococci, for which M17 is a common growth medium, can also be cultivated to high cell densities in the new CDMs. This further expands the scope of this work and suggests that these new media could also be used for other organisms that grow in M17.
The statistical DOE method employed in this study proved to be very useful for identification of significant variables and development of a CDM that supports a high level of growth. A total of 161 experiments were performed, which significantly reduced the number of trials that might have been involved in optimizing a fermentation system with 57 components and two environmental factors if it were done in a less efficient manner (e.g., one component at a time). However, there are still limitations when the statistical DOE method is used. The ranking of 21 variables in each Pareto chart is greatly affected by the variable ranges chosen in each set of FFD. All variables were kept until the final set of optimization design in order to avoid exclusion of some important chemicals that might be considered insignificant due to the concentration ranges selected. In addition, the CCD approach is fundamentally a gradient-based approach which is very likely to get trapped in a local optimum if it is used for a highly nonlinear and multidimensional system. In this case, a more efficient nonlinear optimization method (e.g., a hybrid radial basis function neural network-truncated genetic algorithm-based experimental design method [18] or Bayesian regularized neural networks based on information theoretic criterion [4]) are likely to be attractive alternatives for developing better CDMs with fewer experiments. This is an active area of research for us (4, 18), and these new methods have shown promise for the same optimization problem described here.
Overall, we developed two generally applicable CDMs that support much greater growth of lactococci than the four previously described CDMs and greater growth than the complex M17 medium. The new CDMs are generally applicable to lactococci, enterococci, and streptococci and should allow more reliable metabolic analysis than has been possible previously. In this work, we designed optimal CDMs only for L. lactis IL1403; nevertheless, the approaches and computational techniques used to develop the rich CDMs here may be applicable to other organisms as well.
Supplementary Material
Acknowledgments
This work was supported by the California Dairy Research Foundation and by the UC Discovery Grants Program.
We acknowledge Angela Marcobal for her help conducting the fermentation experiments using streptococcal and enterococcal strains. We thank Lucy Joseph for expert technical assistance. We thank Bart Weimer, Larry McKay, and Gary Dunny for providing strains used in this study.
Footnotes
Published ahead of print on 12 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akerberg, C., K. Hofvendahl, G. Zacchi, and B. Hahn-Hagerdal. 1998. Modelling the influence of pH, temperature, glucose and lactic acid concentrations on the kinetics of lactic acid production by Lactococcus lactis ssp. lactis ATCC 19435 in whole-wheat flour. Microbiol. Biotechnol. 49:682-690. [Google Scholar]
- 2.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cocaign-Bousquet, M., C. Garrigues, L. Novak, N. D. Lindley, and P. Loubiere. 1995. Rational development of a simple synthetic medium for the sustained growth of Lactococcus lactis. J. Appl. Bacteriol. 79:108-116. [Google Scholar]
- 4.Coleman, M. C., and D. E. Block. 2007. Nonlinear experimental design using Bayesian regularized neural networks. AIChE J. 53:1496-1509. [Google Scholar]
- 5.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 23:130-135. [Google Scholar]
- 6.Fernandez de Palencia, P., C. Nieto, P. Acebo, M. Espinosa, and P. Lopez. 2000. Expression of green fluorescent protein in Lactococcus lactis. FEMS Microbiol. Lett. 183:229-234. [DOI] [PubMed] [Google Scholar]
- 7.Jensen, P. R., and K. Hammer. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 59:4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan, C. Q., G. Oddone, D. A. Mills, and D. E. Block. 2006. Kinetics of Lactococcus lactis growth and metabolite formation under aerobic and anaerobic conditions in the presence or absence of hemin. Biotechnol. Bioeng. 95:1070-1080. [DOI] [PubMed] [Google Scholar]
- 9.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers, R. H., and D. C. Montgomery. 2002. Response surface methodology: process and product optimization using designed experiments, 2nd ed. J. Wiley, New York, NY.
- 11.Novak, L., M. Cocaign-Bousquet, N. D. Lindley, and P. Loubiere. 1997. Metabolism and energetics of Lactococcus lactis during growth in complex or synthetic media. Appl. Environ. Microbiol. 63:2665-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piard, J. 1998. Lactic acid bacteria in live vaccine development: designing tools to target antigens to a defined cell compartment. Res. Immunol. 149:101-102. [Google Scholar]
- 13.Stanbury, P. F., A. Whitaker, and S. J. Hall. 1995. Principles of fermentation technology, 2nd ed. Pergammon Press, Oxford, United Kingdom.
- 14.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teuber, M., and A. Geis. 2006. The genus Lactococcus, p. 205-228. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, vol. 4. Springer, New York, NY. [Google Scholar]
- 16.van Nie, E. W. J., and B. Hahn-Hagerdal. 1999. Nutrient requirements of lactococci in defined growth media. Appl. Microbiol. Biotechnol. 52:617-627. [Google Scholar]
- 17.Wegmann, U., M. O'Connell-Motherwy, A. Zomer, G. Buist, C. Shearman, C. Canchaya, M. Ventura, A. Goesmann, M. J. Gasson, O. P. Kuipers, D. van Sinderen, and J. Kok. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, G., M. M. Olsen, and D. E. Block. 2007. New experimental design method for highly nonlinear and dimensional processes. AIChE J. 53:2013-2025. [Google Scholar]
- 19.Zhang, J., and R. Greasham. 1999. Chemically defined media for commercial fermentations. Appl. Microbiol. Biotechnol. 51:407-421. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.