Abstract
Two α-glucosidase-encoding genes (agl1 and agl2) from Bifidobacterium breve UCC2003 were identified and characterized. Based on their similarity to characterized carbohydrate hydrolases, the Agl1 and Agl2 enzymes are both assigned to a subgroup of the glycosyl hydrolase family 13, the α-1,6-glucosidases (EC 3.2.1.10). Recombinant Agl1 and Agl2 into which a His12 sequence was incorporated (Agl1His and Agl2His, respectively) exhibited hydrolytic activity towards panose, isomaltose, isomaltotriose, and four sucrose isomers—palatinose, trehalulose, turanose, and maltulose—while also degrading trehalose and, to a lesser extent, nigerose. The preferred substrates for both enzymes were panose, isomaltose, and trehalulose. Furthermore, the pH and temperature optima for both enzymes were determined, showing that Agl1His exhibits higher thermo and pH optima than Agl2His. The two purified α-1,6-glucosidases were also shown to have transglycosylation activity, synthesizing oligosaccharides from palatinose, trehalulose, trehalose, panose, and isomaltotriose.
The gastrointestinal tract is inhabited by a complex community of microorganisms, also referred to as the microbiota, which are believed to play an important role in human health and disease (39). This concept has been driving extensive attempts to positively influence the composition and/or activity of the intestinal microbiota through the use of so-called probiotics and/or prebiotics. A probiotic has been defined as “a preparation or a product containing viable, defined microorganisms in sufficient numbers, which alter the microflora (by implantation or colonization) in a compartment of the host and by that exert beneficial health effect in this host” (48). A prebiotic has recently been (re)defined as “a selectively fermented ingredient that allows specific changes, both in the composition and/or activity in the gastrointestinal microbiota that confers benefits upon host well-being and health” (42). Finally, a synbiotic is the combination of a probiotic and a prebiotic (16).
One of the dominant bacteria of the intestinal microbiota of humans and animals (51) is bifidobacteria. These are gram-positive, pleomorphic, and anaerobic bacteria that have received increasing scientific attention in recent years due to their perceived probiotic activity (15, 27). The growth of gut-derived bifidobacteria has been shown to be selectively stimulated by various dietary carbohydrates that can thus be considered as prebiotics (30). In this context, it is interesting to note that more than 8% of the identified genes on bifidobacterial genomes are predicted to be involved in sugar metabolism, thus indicative of extensive carbon source-degrading abilities (55, 56).
Carbohydrate degradation has been extensively studied in a variety of different Bifidobacterium species (reviewed in reference 53). For example, various α- and β-galactosidases have been characterized in Bifidobacterium breve 203 (60), Bifidobacterium adolescentis DSM20083 (20), Bifidobacterium bifidum NCIMB41171 (18), and Bifidobacterium longum MB219 (43). A number of studies have also shown that Bifidobacterium spp. produce various α- and β-glucosidase activities (reviewed in reference 53), while Bifidobacterium infantis ATCC 15697 (58), Bifidobacterium lactis DSM10140(T) (13), and B. breve UCC2003 (45) have been reported to produce β-fructofuranosidases during growth on fructooligosaccharides. Additionally, starch-, amylopectin-, and pullulan-degrading activities in bifidobacteria have been investigated (36, 44). Several β-glucosidases have been biochemically characterized from a number of strains of bifidobacteria, e.g., B. adolescentis Int-57 (8), B. breve clb (35,) and Bifidobacterium sp. strain SEN (59). To date, only two α-glucosidases (AglA and AglB) have been described from B. adolescentis DSM20083 (54). AglA was shown to preferentially hydrolyze isomaltotriose, while AglB exhibits a high preference to maltose. Both AglA and AglB were also demonstrated to have transglycosylation activity. Aside from this report, little is known about the biochemical characteristics of α-glucosidase enzymes from bifidobacteria, although it is a common activity observed among these bacteria (41).
Carbohydrates other than the commercially exploited prebiotics, e.g., fructooligosaccharides (such as inulin) and trans-galactooligosaccharides (42), have received relatively little attention with regard to their possible prebiotic properties. Such potential prebiotics are, for example, honey oligosaccharides, some of which are also interesting because of their noncariogenic properties (14). One of the predominant fractions of noncariogenic sugars in honey is isomaltulose (5, 46), also called palatinose or 6-O-α-d-glucopyranosyl-d-fructose, which is a reducing disaccharide and a functional isomer of sucrose. Palatinose possesses approximately one-third of the sweetness of sucrose and is very resistant to acid and invertase hydrolysis (29, 32). The hydrolysis and adsorption of palatinose in the small intestine thus occurs at a much slower rate than does those of sucrose (17), which results in a reduction of the postprandial plasma glucose and insulin levels (3), which means that most palatinose passes through the small intestine to present a growth substrate for elements of the colonic microbiota.
In this study, we describe the identification of two genes, agl1 and agl2, present in the genome of B. breve UCC2003 and responsible for the hydrolysis of α-glycosidic linkages, such as those present in palatinose.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. B. breve UCC2003 was cultured in reinforced clostridium medium (Oxoid, Hampshire, England) or modified de Man-Rogosa-Sharpe medium (10), made from first principles, supplemented with 0.05% (wt/vol) l-cysteine HCl (Sigma-Aldrich, Steinheim, Germany) and 1% (wt/vol) of individual carbohydrate solutions as the sole carbon source. Strains were grown under anaerobic conditions in a modular atmosphere-controlled system (Davidson & Hardy Ltd., Dublin, Ireland). Lactococcus lactis strains were grown at 30°C in M17 medium supplemented with 0.5% glucose (GM17). Escherichia coli strains were grown in LB medium under aerobic conditions on a rotary shaker (150 rpm) at 37°C or plated on LB agar plates. Where appropriate, media were supplemented with 100 μg ml−1 ampicillin, 5 μg ml−1 tetracycline, and/or 5 μg ml−1 chloramphenicol for plasmid maintenance.
Bioinformatics.
Sequence data were obtained from the genome annotation of the B. breve UCC2003 sequencing project (S. C. Leahy, M. O'Connell-Motherway, J. A. Moreno Muñoz, G. F. Fitzgerald, D. G. Higgins, and D. van Sinderen, unpublished results). Database searches were performed using the nonredundant sequence database accessible at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov) using TBLASTN, BLASTX, and BLASTP (2). Sequence analysis was performed using DNASTAR MapDraw and EditSeq, and MegAlign was used to align multiple protein or DNA sequences (Madison, WI).
DNA manipulations.
Large-scale preparation of chromosomal DNA from Bifidobacterium spp. was performed as described previously (33). Plasmid DNA was obtained from L. lactis NZ9000 and E. coli XL1-Blue using the QIAprep spin plasmid miniprep kit (Qiagen GmbH, Hilden, Germany). An initial lysis step was performed using 40 mg ml−1 of lysozyme for 30 min at 37°C prior to plasmid purification from L. lactis NZ9000.
Plasmid constructions.
The entire coding region of agl1 (gene locus Bbr_1855) or agl2 (gene locus Bbr_0559) was amplified by PCR, with genomic DNA from B. breve UCC2003 serving as a template. Gene-specific primer pair Agl1Fw and Agl1Rv and primers Agl2Fw and Agl2Rv were used to amplify the agl1 and agl2 genes, respectively (see Table S2 in the supplemental material). These primers allowed the incorporation of a His12-encoding sequence into the 3′ ends of the agl1 and agl2 genes. PCR was performed using a PTC-200 Peltier thermal cycler (Bio-Sciences, Dublin, Ireland), Taq PCR Master Mix, and ProofStart DNA polymerase (Qiagen GmbH). The amplified 1.8-kb agl1-encompassing PCR fragment was restricted with NcoI and XbaI and ligated into plasmid pNZ8048 (25) which was cut with the same enzymes. The amplified 1.8-kb agl2 PCR fragment was restricted with SphI and BamHI restriction enzymes and ligated into similarly restricted plasmid pQE-70 (Qiagen GmbH). The ligation mixture of the agl1-containing PCR fragment and pNZ8048 was introduced into L. lactis NZ9000 by electroporation, and resulting transformants were selected based on chloramphenicol resistance. The ligation mixture of the agl2-containing PCR fragment and pQE-70 was introduced into E. coli XL1-Blue cells by electroporation, with subsequent transformant selection based on ampicillin and tetracycline resistance. Transformants were checked for plasmid content using colony PCR and restriction analysis of plasmid DNA. Several clones were obtained containing the agl1 gene downstream of the nisin-inducible promoter of pNZ8048. The plasmid insert of a number of transformants was verified by sequencing to ensure the genetic integrity of the cloned recombinant gene. One of the transformants was selected for further use and designated as pNZagl1. Similarly, several clones containing the agl2 gene downstream of the isopropyl-ß-d-thiogalactoside (IPTG)-inducible promoter of pQE-70 were identified. The plasmid insert of a number of transformants was verified by sequencing to ensure the genetic integrity of the cloned recombinant gene. One of the transformants was selected for further use and designated as pQEagl2.
Purification of Agl1His and Agl2His.
A 400-ml volume of GM17 was inoculated with 8 ml of an overnight culture of L. lactis NZ9000 cells harboring pNZagl1 and incubated at 30°C. When the A600 had reached approximately 0.5, expression of the protein was induced by the addition of 800 μl of supernatant from an overnight culture of a nisin A-producing L. lactis NZ9700 culture (11). After 2 h of incubation, cells were harvested, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole), and disrupted with glass beads in a mini-bead beater (BioSpec Products, Bartlesville, OK). Cellular debris was removed by centrifugation. The recombinant Agl1 enzyme with incorporated His12 sequence (Agl1His) was purified from the crude cell extract using a nickel-nitrilotriacetic acid column (Qiagen GmbH) according to the manufacturer's instructions (QIAexpressionist, June 2003). A 200-ml volume of LB supplemented with tetracycline and ampicillin was inoculated with 10 ml of an overnight culture of E. coli XL1-Blue cells harboring pQEagl2 and incubated at 37°C. At an A600 of 0.6, expression of agl2 was induced with 1 mM IPTG (Roche Diagnostics Ltd., West Sussex, United Kingdom). Following 5 h of incubation in the presence of IPTG, cells were harvested by centrifugation and used for the purification of Agl2His, as described above for the Agl1His purification. The Agl2His enzyme was purified using the same method as described for Agl1His.
Protein analysis was performed using 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, using a mini-Protean II system (Bio-Rad Laboratories, Richmond, CA) as described previously (26). The molecular mass of the purified enzyme was estimated by comparison with molecular mass markers (prestained protein marker, broad range [6 to 175 kDa]; New England BioLabs, Herefordshire, United Kingdom). Protein concentrations were determined using the Bradford method (7).
Biochemical characterizations of Agl1His and Agl2His.
Determination of the α-glycosidic activity of purified Agl1His and Agl2His was performed essentially as described previously (6). A 10-μl volume of crude extract (protein concentration of 1.5 mg ml−1) or purified protein (concentration of 0.5 mg ml−1) was incubated with 30 mM HEPES-KOH (pH 5.5) and 50 mM palatinose (Sigma-Aldrich) as the enzyme substrate in a final volume of 130 μl overnight at 30°C unless otherwise stated. Following incubation, the mixture was heated to 95°C for 5 min and subsequently cooled on ice. The release of equimolar amounts of glucose and fructose as detected by high-performance thin-layer chromatography (HPTLC) and/or high-performance liquid chromatography (HPLC) analyses (see below) were taken as proof for hydrolytic activity. Substrate specificity of Agl1His and Agl2His was determined by incubating purified Agl1His and Agl2His with 10 mM of a particular di-, tri-, or polysaccharide (Table 1) solution made up in 30 mM HEPES-KOH (pH 5.5) buffer for 5 min at 30°C and subsequently analyzing possible hydrolysis products by glucose release measurements using the glucose hexokinase assay kit (Sigma-Aldrich), according to the manufacturer's instructions. For the determination of the pH optimum, a pH range of 2.5 to 10.5 was used, while the temperature optimum was analyzed between 4°C and 100°C; in both cases, 50 mM palatinose was used as a substrate.
TABLE 1.
Substrate specificities of purified Agl1His and Agl2His from B. breve UCC2003
| Substrate | Linkage | Specific enzyme activity (μmol min−1 mg−1)a
|
|
|---|---|---|---|
| Agl1His | Agl2His | ||
| Disaccharides | |||
| Trehalose | O-α-d-glucosyl-(1→1)-α-d-glucose | 2.0 ± 0.01 | 1.6 ± 0.8 |
| Trehalulose | O-α-d-glucosyl-(1→1)-β-d-fructose | 10.0 ± 0.1 | 11.7 ± 0.05 |
| Sucrose | O-α-d-glucosyl-(1→2)-β-d-fructose | 0.0 | 0.0 |
| Turanose | O-α-d-glucosyl-(1→3)-d-fructose | 0.5 ± 0.1 | 1.6 ± 0.7 |
| Nigerose | O-α-d-glucosyl-(1→3)-d-glucose | 0.4 ± 0.2 | 0.7 ± 0.2 |
| Maltulose | O-α-d-glucosyl-(1→4)-d-fructose | 0.4 ± 0.1 | 1.1 ± 0.63 |
| Maltose | O-α-d-glucosyl-(1→4)-d-glucose | 0.0 | 0.0 |
| Cellobiose | O-β-d-glucosyl-(1→4)-d-glucose | 0.0 | 0.0 |
| Lactose | O-β-d-galactosyl-(1→4)-α-d-glucose | 0.0 | 0.0 |
| Leucrose | O-α-d-glucosyl-(1→5)-d-fructose | 0.0 | 0.0 |
| Palatinose | O-α-d-glucosyl-(1→6)-d-fructose | 3.2 ± 0.2 | 4.1 ± 0.11 |
| Isomaltose | O-α-d-glucosyl-(1→6)-d-glucose | 14.1 ± 0.13 | 20.4 ± 0.01 |
| Gentiobiose | O-β-d-glucosyl-(1→6)-d-glucose | 0.0 | 0.0 |
| Melibiose | O-α-d-galactosyl-(1→6)-d-glucose | 0.0 | 0.0 |
| Trisaccharides | |||
| Melezitose | O-α-d-glucosyl-(1→3)-β-d-fructosyl-(2→1)-d-glucose | 0.0 | 0.0 |
| Maltotriose | O-α-d-glucosyl-(1→4)-α-d-glucosyl-(1→4)-d-glucose | 0.0 | 0.0 |
| Raffinose | O-α-d-galactosyl-(1→6)-α-d-glucosyl-(1→2)-β-d-fructose | 0.0 | 0.0 |
| Panose | O-α-d-glucosyl-(1→6)-α-d-glucosyl-(1→4)-d-glucose | 23.0 ± 0.21 | 19.3 ± 0.1 |
| Isomaltotriose | O-α-d-glucosyl-(1→6)-α-d-glucosyl-(1→6)-d-glucose | 8.6 ± 0.12 | 7.0 ± 0.04 |
| Other substrates | |||
| Stachyose | β-d-fructosyl-O-α-d-galactosyl-(1→6)-O-α-d-galactosyl-(1→6)-α-d-glucose | 0.0 | 0.0 |
| Glycogen | (α-d-glucosyl-(1→4)-d-glucose)n, (α-d-glucosyl-(1→6)-d-glucose)n | 0.0 | 0.0 |
| Maltodextrins | (α-d-glucosyl-(1→4)-d-glucose)n | 0.0 | 0.0 |
Activity was determined with 1 μg of pure enzyme and 10 mM of each substrate at 30°C for 5 min.
Kinetic constants of Agl1His and Agl2His for palatinose, trehalulose, trehalose, panose, isomaltose, and isomaltotriose were determined by measuring the initial rates at various substrate concentrations, ranging from 2.5 to 25 mM. Reactions were initiated by the addition of 0.5 μg of the enzyme and stopped at different times (up to 6 min) by heat treatment (100°C for 5 min). All experiments were performed in duplicate.
Transglycosylation activities were performed with 150 mM palatinose, trehalose, isomaltose, trehalose, panose, or isomaltotriose in 30 mM HEPES-KOH (pH 5.5) and 5 μg of purified Agl1His or Agl2His in a final reaction volume of 130 μl overnight at 30°C. Reactions were terminated by boiling samples for 5 min, followed by cooling on ice. The samples were then analyzed by HPTLC and high-performance anion-exchange chromatography (HPAEC).
HPTLC analysis.
For HPTLC analysis of Agl1His and Agl2His enzyme activity, a 0.5-μl aliquot of a given reaction mixture was spotted onto a silica gel 60 plate (10 by 10 cm; Merck KGaA, Darmstadt, Germany) with a Nanomat 4 (Camag, Muttenz, Switzerland). HPTLC plates were impregnated by spraying with 0.1 M sodium bisulfate solution, followed by drying and subsequent spraying with 0.1 M citrate buffer (pH 4.8). Following solvent saturation of the HPTLC plate, the chromatogram was developed fourfold using an acetonitrile-deionized water (17:3, vol/vol) solvent system in a horizontal developing chamber. The plate was allowed to air dry in a fume hood, then sprayed evenly with 20% (vol/vol) sulfuric acid in ethanol, air dried again, and finally heated at 120°C for 10 min to visualize the carbohydrate spots.
HPLC analysis.
Glucose, fructose, and palatinose concentrations were determined by HPLC using an LKB Bromma 2150 HPLC system with a refractive index detector (Shodex RI-71) and a Highchrom heating block. An 8-μl, 8% H Rezex organic acid column (300 by 7.8 mm; Phenomenex, Torrance, CA) was used with 0.01 N H2SO4 as the elution fluid at a flow rate 0.6 ml min−1. The temperature of the column was maintained at 65°C. Substrate and end product peaks were identified by comparing their retention times with those of known standards, and concentrations were determined from standards of known concentrations (prepared in 30 mM HEPES-KOH [pH 5.5] buffer).
HPAEC-PAD.
Reaction products were quantitatively analyzed using a Dionex ICS-3000 system equipped with a CarboPac PA-100 analytical-exchange column (250 mm by 4 mm) and a pulsed electrochemical detector in the pulsed amperometric detection (PAD) mode. The elution was performed at a constant flow rate of 1 ml min−1 at 30°C using the following linear gradients of sodium acetate in 100 mM NaOH: 0 to 5 min, 0 mM; 5 to 40 min, 0 to 275 mM; and 40 to 45 min, 275 mM. The injection volume was 10 μl. Reaction products were identified and quantified by relative standard carbohydrates.
Southern hybridization analysis.
Southern hybridization analysis was performed using an enhanced chemiluminescence gene detection kit (GE Healthcare, Amersham, United Kingdom) with the following two different probes: pNZagl1 plasmid DNA representing the complete agl1 gene or a 246-bp PCR product representing the 3′ end of the agl1 gene, designated as agl1-3′. The agl1-3′ fragment was generated by PCR using the primer combination agl1550fw and agl1796rv (see Table S2 in the supplemental material). Following restriction with BamHI and separation of chromosomal DNA fragments by agarose gel electrophoresis, DNA transfer to nylon membranes (Hybond-N+; GE Healthcare) was performed as described previously (49). Probe labeling, setting up hybridization conditions, washing steps, and detection were performed according to the manufacturer's instructions (GE Healthcare).
Nucleotide sequence accession numbers.
The nucleotide sequences of agl1 and agl2 were deposited in the GenBank database under accession numbers FJ386389 and FJ386390, respectively.
RESULTS
Identification and sequence analysis of agl1 and agl2 genes from B. breve UCC2003.
To investigate the α-glucosidic activity of B. breve UCC2003, growth on palatinose as a sole carbon source was tested. B. breve UCC2003 could indeed use this disaccharide as a substrate for growth (results not shown). In order to identify possible B. breve UCC2003-encoded enzymes that are capable of palatinose hydrolysis, we performed a BLASTP analysis using the protein sequence from the characterized palatinase-encoding gene of Erwinia rhapontici (6) to search for similar proteins specified by the B. breve UCC2003 genome. This BLASTP analysis identified two open reading frames on the genome of B. breve UCC2003, designated here as agl1 and agl2, whose protein products exhibit 43% and 42% similarity/identity, respectively, to the PalQ protein of Erwinia rhapontici (see Table S3 in the supplemental material).
The agl1 gene is 1,824 bp in length, encoding a protein of 607 amino acids (molecular mass, ∼68.22 kDa), while the agl2 gene is 1,815 bp in length, encoding a protein of 604 amino acids (molecular mass, ∼68.32 kDa). BLASTP and multiple-sequence alignments showed that Agl1 and Agl2 are similar to putative and characterized α-1,6-glucosidases encoded by various different microorganisms (see Table S3 in the supplemental material), while also exhibiting 73% identity to each other, with the highest level of identity in the N terminus of the protein. Additionally, the deduced amino acid sequences of Agl1 and Agl2 were shown to display 75 and 77% identity to the characterized α-1,6-glucosidase AglA and 29 and 31% identity to the α-glucosidase AglB, both of which are from B. adolescentis DSM20083, respectively (54). All these α-glucosidases are members of glycosyl hydrolase family 13 (GH13) (19), and Agl1 and Agl2 can therefore be assigned as putative α-glucosidases belonging to this GH13 family. Furthermore, Agl1 and Agl2 possess six conserved sequence regions, including the highly conserved QPDLN (conserved sequence region V), which are typical of the oligo-1,6-glucosidase subfamily (EC 3.2.1.10) (see Fig. S1 in the supplemental material) (38), indicating that Agl1 and Agl2 belong to this GH13 subfamily.
Purification and characterization of purified Agl1His and Agl2His.
In order to characterize the putative α-glucosidases Agl1 and Agl2 and their possible involvement in palatinose metabolism, we purified and biochemically characterized Agl1His and Agl2His (see Materials and Methods). The molecular masses of the purified enzymes were estimated in both cases by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis to be approximately 70 kDa, which is in agreement with the calculated size of His-tagged proteins (68.87 and 69.96 kDa for Agl1His and Agl2His, respectively).
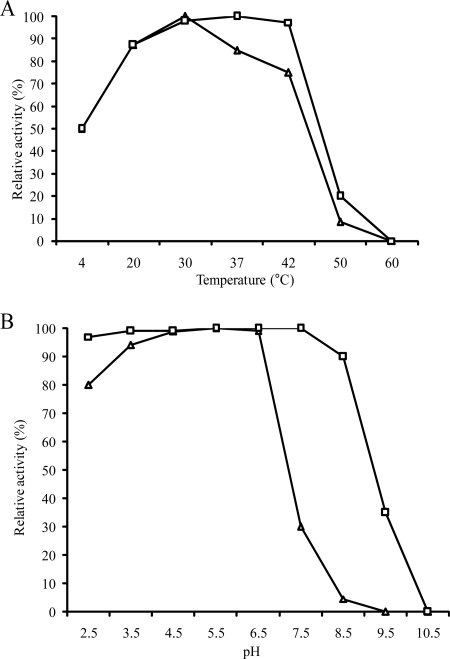
The suspected α-glucosidic activities of the agl1 and agl2 gene products were indeed confirmed by HPTLC and HPLC analyses, using palatinose as a substrate (see Materials and Methods). Optimal temperatures of Agl1His and Agl2His were determined to be 37 and 30°C, respectively. Agl1His appeared to be more thermostable than Agl2His (Fig. 1); the former was shown to still exhibit 100% activity at 42°C (relative to 30°C), but Agl2His activity dropped to 80% at this temperature. Neither enzyme exhibited any activity at incubation temperatures of 60°C or higher (Fig. 1A). The optimum pH for both Agl1His and Agl2His activities was estimated to be 5.5. Remarkably, both enzymes were shown to be active at rather low pH values, retaining more than 90% relative activity at pH 2.5. Agl1His activity dropped to 86% at pH 8.5 and to 40% at pH 9.5. Agl2His activity was not stable at a higher pH and was shown to lose 98% of its activity at pH 8.5, under the conditions used (Fig. 1B).
FIG. 1.
Effects of temperature (A) and pH (B) on the activity of Agl1His (open squares) and Agl2His (open triangles). The optimum pH and temperature conditions for catalytic activity, using palatinose as a substrate, were determined to be pH 5.5 and 30°C and 37°C for Agl1His and Agl2His, respectively.
Substrate specificity of the recombinant α-glucosidases from B. breve UCC2003.
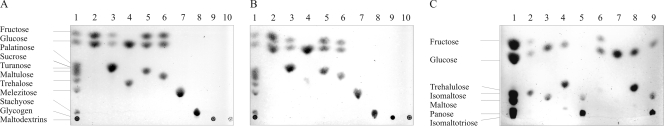
The two identified α-glucosidases from B. breve UCC2003 displayed a rather broad substrate range not limited to palatinose (Fig. 2). Both Agl1His and Agl2His were capable of degrading α-1,6 and α-1,1 linkages between two glucose molecules and between glucose and fructose molecules in sucrose isomers (Table 1). Agl1His hydrolyzed the α-1,6 glucosidic bond of the trisaccharides panose (relative activity, 100%) and isomaltotriose (37.4%) and the glucosidic bond within the disaccharides isomaltose (61.3%), trehalulose (43.5%), palatinose (13.9%), trehalose (8.7%), turanose (2.2%), nigerose (1.7%), and maltulose (1.7%). Similar preferred substrates were observed for α-glucosidase Agl2His (Table 1). No release of hydrolysis products for either enzyme was observed for the disaccharides maltose, cellobiose, lactose, gentiobiose, leucose, and melibiose; the trisaccharides raffinose, melezitose, and maltotriose; the oligosaccharide stachyose; or the polysaccharides glycogen and maltodextrins (Table 1). Both AglHis enzymes showed very low activity against sucrose and only when high concentrations of enzyme and substrate were used (5 μg enzyme and 50 mM sucrose) (Fig. 2). To summarize, the characterized Agl1His and Agl2His enzymes were shown to exhibit α-glycosidic activity with a preference for the hydrolysis of the α-1,6-glycosidic linkage present in panose, isomaltose, and isomaltotriose and the α-1,1-glycosidic linkage in trehalulose.
FIG. 2.
Substrate specificities of Agl1His and Agl2His as determined by HPTLC. Reaction conditions were as described in Materials and Methods. Lane 1, carbohydrate standards listed on the left of panels A and C. Lanes 2 to 10 contained hydrolysis products obtained from (24 h [A]; 5 min [B]) the incubation of purified proteins (Agl1His [A] or Agl2His [B]) with palatinose (lane 2), sucrose (lane 3), trehalose (lane 4), turanose (lane 5), maltulose (lane 6), melezitose (lane 7), stachyose (lane 8), glycogen (lane 9), and maltodextrins (lane 10). (C) Lanes 2 to 9 contained hydrolysis products obtained from the incubation of Agl1His (lanes 2 to 5) and Agl2His (lanes 6 to 9) with trehalulose (lanes 2 and 6), isomaltose (lanes 3 and 7), panose (lanes 4 and 8), and isomaltotriose (lanes 5 and 9).
Kinetic studies of the two recombinant α-glucosidases allowed for the calculation of Vmax and Km values for the substrates palatinose, isomaltose, trehalulose, trehalose, panose, and isomaltose as well as the rate constants (kcat) and catalytic efficiencies (kcat/Km). The obtained data confirmed the findings described above and showed that panose, isomaltose, and trehalulose were the preferred substrates for the Agl1His and Agl2His enzymes under the conditions used in this kinetic study (Table 2).
TABLE 2.
Kinetic parameters of purified Agl1His and Agl2Hisa
| Substrate | Agl1His
|
Agl2His
|
||||||
|---|---|---|---|---|---|---|---|---|
| Vmax | Km | kcat | kcat/Km | Vmax | Km | kcat | kcat/Km | |
| Palatinose | 14.4 ± 2.78 | 27.2 ± 1.99 | 12.4 ± 2.38 | 0.5 ± 0.12 | 16.2 ± 2.0 | 7.1 ± 0.07 | 13.0 ± 4.53 | 1.8 ± 0.65 |
| Isomaltose | 35.9 ± 3.01 | 8.3 ± 0.02 | 30.8 ± 2.58 | 3.7 ± 0.32 | 42.3 ± 2.7 | 9.0 ± 0.8 | 36.3 ± 2.28 | 4.0 ± 0.09 |
| Trehalulose | 41.2 ± 5.2 | 30.2 ± 3.7 | 35.4 ± 7.76 | 1.2 ± 0.03 | 45.1 ± 5.1 | 22.9 ± 3.5 | 38.7 ± 5.93 | 1.7 ± 0.24 |
| Trehalose | 8.5 ± 1.28 | 33.6 ± 4.25 | 7.3 ± 1.8 | 0.2 ± 0.08 | 6.45 ± 0.54 | 31.15 ± 3.45 | 5.8 ± 0.33 | 0.2 ± 0.01 |
| Panose | 30.8 ± 3.05 | 10.4 ± 1.21 | 28.7 ± 3.52 | 2.8 ± 1.02 | 33.0 ± 4.42 | 15.0 ± 3.56 | 30.0 ± 3.94 | 2.0 ± 0.07 |
| Isomaltotriose | 14.5 ± 0.86 | 16.7 ± 1.89 | 12.5 ± 0.74 | 0.8 ± 0.13 | 14.0 ± 1.83 | 5.5 ± 0.99 | 12.0 ± 1.57 | 2.5 ± 1.06 |
Vmax values are expressed as μmol min−1 mg−1, Km values are expressed as mM, kcat values are expressed as s−1, and kcat/Km values are expressed as mM−1s−1. All values are means from two experiments ± standard errors.
Growth profiles of B. breve UCC2003.
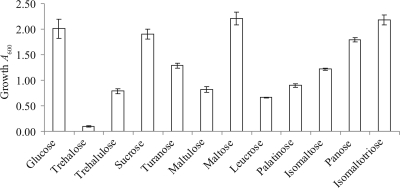
In order to investigate whether B. breve UCC2003 is capable of metabolizing the carbohydrates identified in the substrate specificity study, growth abilities of this strain on these sugars were determined by measuring the A600 after 24 h of anaerobic growth at 37°C (Fig. 3). B. breve UCC2003 was shown to grow on all substrates as its sole carbon source except trehalose.
FIG. 3.
Growth profile analysis of B. breve UCC2003 in modified Rogosa broth supplemented with different carbohydrates.
Prevalence of agl1 and agl2 in bifidobacterial genomes.
In order to investigate whether homologs of the agl1 and agl2 genes are present in other B. breve strains, genomic DNA was extracted from eight B. breve strains, which following BamHI restriction were subjected to Southern hybridization analysis using probes specific for agl1 (see Fig. S2 in the supplemental material). All tested strains were shown to harbor at least one copy of the agl gene. Southern hybridization analysis with B. breve UCC2003 genomic DNA and pNZagl1 plasmid DNA as a probe generated, as expected, two hybridization products corresponding to agl1 and agl2 which exhibit high levels of sequence identity (79% overall). Therefore, to avoid cross-hybridization of pNZagl1 with agl2, we performed a Southern hybridization analysis with another probe, agl1-3′, specific only to the 3′ end of agl1. This probe generated a single hybridization signal for the genomes of five B. breve strains (UCC2003, JCM7017, JCM7019, NCFB2258, and NCIMB8815), indicating that these strains are likely to contain a single copy of the agl1 gene (see Fig. S2B in the supplemental material) and a single copy of the agl2 gene (see Fig. S2A in the supplemental material). Genomic DNA of B. breve UCC2005 generated two hybridization signals when pNZagl1 plasmid DNA was used as a probe, but did not give any signal with the agl1-specific probe (see Fig. S2A and B in the supplemental material), indicating that this strain contains at least one copy of the agl2 gene. Genomic DNA of B. breve CCUG43878 only hybridized to the pNZagl1 probe, resulting in a single hybridization product, thereby indicating the genomic presence of a single agl2 copy.
In addition, other strains such as B. adolescentis CIP64.13, B. adolescentis DSM20083, B. adolescentis NCFB2229, B. adolescentis NCFB2204, B. adolescentis LMG10502, B. adolescentis CIP64.61, Bifidobacterium globosum JCM5820, B. globosum JCM7092, Bifidobacterium dentium NCFB2843, B. longum JCM7052, and Bifidobacterium thermophilum JCM7027 were also shown to produce hybridization signals using pNZagl1 as a probe, which indicates that these strains also carry homologs of the B. breve UCC2003 agl genes (data not shown).
Thus, the Southern hybridization analysis showed that the presence of an α-1,6-glucosidase-encoding gene(s) is a common feature of bifidobacterial genomes.
Glycosyltransferase activity of purified Agl1His and Agl2His.
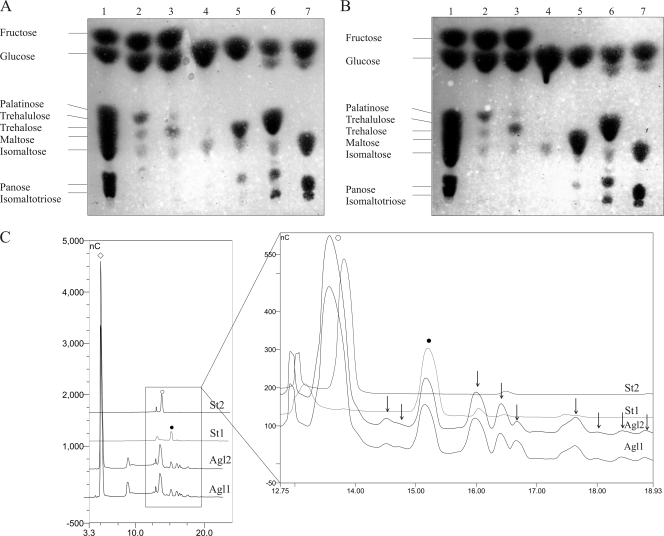
Glycosyltransferase activity has been shown previously for α-glucosidases (54). In order to investigate whether Agl1His and Agl2His possess the ability to produce higher-molecular-weight oligosaccharides, the glycosyltransferase activity of these enzymes was tested (see Materials and Methods). In addition to the expected hydrolysis products, the purified Agl1His and Agl2His indeed synthesized higher-molecular-weight oligosaccharides when incubated with high (150 mM) concentrations of palatinose, trehalulose, trehalose, panose, or isomaltotriose, as was obvious from thin-layer chromatography and HPAEC-PAD analyses (Fig. 4). Interestingly, under the conditions used, it was found that isomaltose did not act as a substrate for glycosyltransferase activity. HPAEC analysis of the produced carbohydrate profiles showed that the concentration of fructose (in case of palatinose or trehalulose) was approximately twofold higher than the glucose concentration in these samples, which suggests, as would be expected, that glucose residues are transferred to a carbohydrate acceptor molecule for this transglycosylation process.
FIG. 4.
HPTLC identification of the transferase activities of Agl1His (A) and Agl2His (B). Lane 1, carbohydrate standards listed on the left of both images. Lanes 2 to 7 contained hydrolysis and transglycosylation products obtained from the incubation of purified proteins (Agl1His [A] or Agl2His [B]) with the following 150 mM substrates: palatinose (lane 2), trehalulose (lane 3), isomaltose (lane 4), trehalose (lane 5), panose (lane 6), and isomaltotriose (lane 7). (C) HPAEC analysis of reaction products formed after the transglycosylation of panose by Agl1His and Agl2His. Open diamond, glucose; open circle, maltose; filled circle, panose (as identified by standards St2 and St1, respectively). Black arrows point to the newly synthesized oligosaccharides.
DISCUSSION
In this study, we identified and characterized two novel α-glucosidases, encoded by agl1 and agl2 of B. breve UCC2003, that preferentially cleave α-1,6 glycosidic linkages (in panose, isomaltose, isomaltotriose, and palatinose) but also hydrolyze α-1,1 (in trehalulose and trehalose), and under certain conditions and with lower efficacy, α-1,3 (in turanose and nigerose), α-1,4 (in maltulose), and α-1,2 (in sucrose) glycosyl linkages. Even though both characterized α-glucosidases from B. breve UCC2003 were capable of hydrolyzing palatinose, this sugar was not the preferred substrate. Instead, certain sucrose isomers, isomaltose, trehalose, and other oligosaccharides used were shown to be good substrates for the B. breve UCC2003 Agl enzymes.
Interestingly, these sugars, including others such as isomaltotriose, raffinose, melezitose (4, 5, 46), and theanderose, which can be synthesized by an α-glucosidase isolated from Bacillus sp. strain SAM1606 (34), are found naturally in honey (5, 46). Only a small number of studies have reported results on the growth-promoting and prebiotic activities of honey on bifidobacteria (22, 47, 52). It has been claimed that growth of commercial strains, such as those belonging to the species B. longum, B. breve, and B. infantis, was enhanced more by honey than by the commercially available prebiotics inulin and fructooligosaccharides (22). Testing B. breve UCC2003 for its capability to ferment various carbohydrates that are also present in honey showed that it is able to metabolize most of these sugars, with the exception of trehalose (Fig. 3), which may be due to the absence of a transport system specific for this carbohydrate.
Both Agl protein sequences exhibit approximately 70% identity to the protein sequence of a putative α-glucosidase-encoding gene present on the genome of a B. asteroides strain that was isolated from the stomach of a honeybee (M. Ventura, C. Canchaya, and D. van Sinderen, unpublished results). Honey is produced by honeybees from the nectar they gather. It has been shown previously that nectar is composed of the disaccharide sucrose and the monosaccharides glucose and fructose (24), whereas honey is known to contain fructose and glucose, which make up approximately 60% of the sugars present, and sucrose isomers, isomaltose, panose, nigerose, and other oligosaccharides, which together constitute around 15 to 20% of the available carbohydrates (5, 46). In a recent study on the bacterial flora of the honeybee Apis mellifera, B. asteroides was found to be the most dominant Bifidobacterium sp. which can metabolize the nectar-derived sugars in the honeybee stomach (37). Considering the enzymatic activities of Agl1 and Agl2 and the observed similarity of these identified α-glucosidases to a homolog encoded by a recently sequenced B. asteroides genome, it is tempting to speculate that the agl gene products may be involved in the fermentation and perhaps the transglycosylation of honey oligosaccharides, with the latter activity made possible due to the high concentration of its substrates being present in honey.
The presence of α-glucosidase activity in bifidobacteria has been reported previously (40, 54). However, the particular α-glucosidase activities described in this paper have not been reported for other characterized (bifido)bacterial α-glucosidases (1, 6, 9, 21, 28, 34, 54). For example, the food-borne pathogen Enterobacter sakazakii produces an α-glucosidase with activity toward palatinose (28), while an α-glucosidase isolated from Erwinia rhapontici showed strict substrate specificity toward palatinose. A recently identified α-glucosidase from Thermus thermophilus HB27 (AglHHB27) was shown to preferentially hydrolyze α,α-trehalose (α-1,1); however, panose, isomaltose, isomaltotriose, palatinose, turanose, nigerose, sucrose, maltose, maltotriose, and leucrose were also substrates for this enzyme (1).
In B. adolescentis DSM20083, two α-glucosidases were previously characterized, AglA (oligo-1,6-glucosidase; EC 3.2.1.10) and AglB (α-glucosidase; EC 3.2.1.20) (54). AglA from B. adolescentis DSM20083 also belongs to the GH13 family and was shown to preferentially hydrolyze isomaltose, isomaltotriose, and trehalose, whereas AglB exhibited a preference for maltose. Although, in particular, AglA exhibits enzymatic properties that resemble those of Agl1 and Agl2, differences in primary structure and substrate specificities as well as differences in pH and temperature optima between these enzymes indicate that all three proteins are distinctive enzymes.
Agl1His and Agl2His can both catalyze the following two types of reactions: hydrolysis and transglycosylation (Fig. 4). Further studies are required to analyze the latter transglycosylation process and the precise carbohydrate products formed. Also, AglA and AglB α-glucosidases from B. adolescentis DSM20083 are known to synthesize different oligosaccharides using specific substrates (54). This is consistent with two studies that also showed that α-glucosidases from other bacterial strains possess the ability to synthesize oligosaccharides using disaccharides maltose and sucrose as acceptors (31, 34). Interestingly, it has been reported previously that transglycosylated oligosaccharides can stimulate the growth of Bifidobacterium spp. (23). Further research will be required to investigate the exact catalytic activities of Agl1 and Agl2 to synthesize oligosaccharides and their potential usage as an enzymatic method to synthesize novel oligosaccharides with potential prebiotic activities.
All bifidobacterial strains tested for the presence of the ag11 and agl2 genes appeared to have at least one homolog of this type of α-glucosidase-encoding gene in their genome. Previous studies have demonstrated that all examined bifidobacteria produce α-glucosidase activity, although limited information is available on the precise characteristics of such α-glucosidases (12, 53, 57).
The results obtained in this study provide new insights into the carbohydrate metabolism of bifidobacteria. We identified and characterized two novel α-glucosidases from B. breve UCC2003 with biochemical properties that have not previously been found in bifidobacteria. Our results also show that oligosaccharides, which are commonly present in honey, support growth of B. breve UCC2003. The requirements of the agl1 and/or agl2 genes for the ability of this strain to grow on such carbohydrates awaits experimental confirmation, although it is tempting to make this correlation. These honey-derived carbohydrates may be used in functional foods as prebiotic components, but such an application will depend on further research that is needed to demonstrate their specific growth-promoting effect on (probiotic) bifidobacteria present in the human gut microbiota.
Supplementary Material
Acknowledgments
This work was financially supported by the Science Foundation Ireland through the Alimentary Pharmabiotic Centre located at University College Cork.
We are grateful to Dan Walsh from the Department of Microbiology at University College Cork in Ireland for his help in performing HPLC analysis. We thank S. Leahy, J. A. Moreno Muñoz, D. G. Higgins, M. Ventura, and C. Canchaya for sharing unpublished sequencing data with us. We also sincerely thank J. Thompson (National Institutes of Health, Bethesda, MD) and N. Emphadinhas (Universidade de Coimbra, Coimbra, Portugal) for supplying various sucrose isomers.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alarico, S., M. S. da Costa, and N. Empadinhas. 2008. Molecular and physiological role of the trehalose-hydrolyzing alpha-glucosidase from Thermus thermophilus HB27. J. Bacteriol. 190:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai, H., A. Mizuno, M. Sakuma, M. Fukaya, K. Matsuo, K. Muto, H. Sasaki, M. Matsuura, H. Okumura, H. Yamamoto, Y. Taketani, T. Doi, and E. Takeda. 2007. Effects of a palatinose-based liquid diet (Inslow) on glycemic control and the second-meal effect in healthy men. Metabolism 56:115-121. [DOI] [PubMed] [Google Scholar]
- 4.Arias, V. C., R. C. Castells, N. Malacalza, C. E. Lupano, and C. B. Castells. 2003. Determination of oligosaccharide patterns in honey by solid-phase extraction and high-performance liquid chromatography. Chromatographia 58:797-801. [Google Scholar]
- 5.Astwood, K., B. Lee, and M. Manley-Harris. 1998. Oligosaccharides in New Zealand honeydew honey. J. Agric. Food Chem. 46:4958-4962. [Google Scholar]
- 6.Börnke, F., M. Hajirezaei, and U. Sonnewald. 2001. Cloning and characterization of the gene cluster for palatinose metabolism from the phytopathogenic bacterium Erwinia rhapontici. J. Bacteriol. 183:2425-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Choi, Y. J., C. J. Kim, and G. E. Ji. 1996. A partially purified beta-glucosidase from Bifidobacterium adolescentis converts cycasin to a mutagenic compound. Lett. Appl. Microbiol. 22:145-148. [DOI] [PubMed] [Google Scholar]
- 9.Degnan, B. A., and G. T. Macfarlane. 1994. Synthesis and activity of a-glucosidase produced by Bifidobacterium pseudolongum. Curr. Microbiol. 29:43-47. [Google Scholar]
- 10.de Man, J. C., A. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 11.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desjardins, M.-L., D. Roy, and J. Goulet. 1990. Growth of bifidobacteria and their enzyme profiles. J. Dairy Sci. 73:299-307. [Google Scholar]
- 13.Ehrmann, M. A., M. Korakli, and R. F. Vogel. 2003. Identification of the gene for beta-fructofuranosidase of Bifidobacterium lactis DSM10140(T) and characterization of the enzyme expressed in Escherichia coli. Curr. Microbiol. 46:391-397. [DOI] [PubMed] [Google Scholar]
- 14.English, H. K., A. R. Pack, and P. C. Molan. 2004. The effects of manuka honey on plaque and gingivitis: a pilot study. J. Int. Acad. Periodontol. 6:63-67. [PubMed] [Google Scholar]
- 15.Gibson, G. R. 2008. Prebiotics as gut microflora management tools. J. Clin. Gastroenterol. 42(Suppl. 2):S75-S79. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, G. R., H. M. Probert, J. Van Loo, R. A. Rastall, and M. B. Roberfroid. 2004. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr. Res. Rev. 17:259-275. [DOI] [PubMed] [Google Scholar]
- 17.Goda, T., and N. Hosoya. 1983. Hydrolysis of palatinose by rat intestinal sucrase-isomaltase complex. J. Jpn. Soc. Nutr. Food Sci. 36:169-173. [Google Scholar]
- 18.Goulas, T. K., A. K. Goulas, G. Tzortzis, and G. R. Gibson. 2007. Molecular cloning and comparative analysis of four beta-galactosidase genes from Bifidobacterium bifidum NCIMB41171. Appl. Microbiol. Biotechnol. 76:1365-1372. [DOI] [PubMed] [Google Scholar]
- 19.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280(Pt. 2):309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinz, S. W., L. A. van den Brock, G. Beldman, J. P. Vincken, and A. G. Voragen. 2004. β-Galactosidase from Bifidobacterium adolescentis DSM20083 prefers beta(1,4)-galactosides over lactose. Appl. Microbiol. Biotechnol. 66:276-284. [DOI] [PubMed] [Google Scholar]
- 21.Jorge, C. D., M. M. Sampaio, G. O. Hreggvidsson, J. K. Kristjanson, and H. Santos. 2007. A highly thermostable trehalase from the thermophilic bacterium Rhodothermus marinus. Extremophiles 11:115-122. [DOI] [PubMed] [Google Scholar]
- 22.Kajiwara, S., H. Gandhi, and Z. Ustunol. 2002. Effect of honey on the growth of and acid production by human intestinal Bifidobacterium spp.: an in vitro comparison with commercial oligosaccharides and inulin. J. Food Prot. 65:214-218. [DOI] [PubMed] [Google Scholar]
- 23.Kim, T.-K., D.-C. Park, and Y.-H. Lee. 1998. Synthesis of transglucosylated xylitol using cyclodextrin glucanotransferase and its stimulating effect on the growth of Bifidobacterium. San'oeb Misaengmul Haghoeji 26:442-448. [Google Scholar]
- 24.Kromer, T., M. Kessler, G. Lohaus, and A. N. Schmidt-Lebuhn. 2008. Nectar sugar composition and concentration in relation to pollination syndromes in Bromeliaceae. Plant Biol. (Stuttgart) 10:502-511. [DOI] [PubMed] [Google Scholar]
- 25.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Leahy, S. C., D. G. Higgins, G. F. Fitzgerald, and D. van Sinderen. 2005. Getting better with bifidobacteria. J. Appl. Microbiol. 98:1303-1315. [DOI] [PubMed] [Google Scholar]
- 28.Lehner, A., K. Riedel, T. Rattei, A. Ruepp, D. Frishman, P. Breeuwer, B. Diep, L. Eberl, and R. Stephan. 2006. Molecular characterization of the alpha-glucosidase activity in Enterobacter sakazakii reveals the presence of a putative gene cluster for palatinose metabolism. Syst. Appl. Microbiol. 29:609-625. [DOI] [PubMed] [Google Scholar]
- 29.Lina, B. A., D. Jonker, and G. Kozianowski. 2002. Isomaltulose (palatinose): a review of biological and toxicological studies. Food Chem. Toxicol. 40:1375-1381. [DOI] [PubMed] [Google Scholar]
- 30.Macfarlane, G. T., H. Steed, and S. Macfarlane. 2008. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J. Appl. Microbiol. 104:305-344. [DOI] [PubMed] [Google Scholar]
- 31.Mala, S., H. Dvorakova, R. Hrabal, and B. Kralova. 1999. Towards regioselective synthesis of oligosaccharides by use of alpha-glucosidases with different substrate specificity. Carbohydr. Res. 322:209-218. [DOI] [PubMed] [Google Scholar]
- 32.Matsukubo, T., and I. Takazoe. 2006. Sucrose substitutes and their role in caries prevention. Int. Dent. J. 56:119-130. [DOI] [PubMed] [Google Scholar]
- 33.Mazé, A., M. O'Connell-Motherway, G. F. Fitzgerald, J. Deutscher, and D. van Sinderen. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakao, M., T. Nakayama, M. Harada, A. Kakudo, H. Ikemoto, S. Kobayashi, and Y. Shibano. 1994. Purification and characterization of a Bacillus sp. SAM1606 thermostable alpha-glucosidase with transglucosylation activity. Appl. Microbiol. Biotechnol. 41:337-343. [DOI] [PubMed] [Google Scholar]
- 35.Nunoura, N., K. Ohdan, T. Yano, K. Yamamoto, and H. Kumagai. 1996. Purification and characterization of beta-d-glucosidase (beta-d-fucosidase) from Bifidobacterium breve clb acclimated to cellobiose. Biosci. Biotechnol. Biochem. 60:188-193. [DOI] [PubMed] [Google Scholar]
- 36.O'Connell Motherway, M., G. F. Fitzgerald, S. Neirynck, S. Ryan, L. Steidler, and D. van Sinderen. 2008. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 74:6271-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olofsson, T. C., and A. Vasquez. 2008. Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr. Microbiol. 57:356-363. [DOI] [PubMed] [Google Scholar]
- 38.Oslancova, A., and S. Janecek. 2002. Oligo-1,6-glucosidase and neopullulanase enzyme subfamilies from the alpha-amylase family defined by the fifth conserved sequence region. Cell. Mol. Life Sci. 59:1945-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parracho, H., A. L. McCartney, and G. R. Gibson. 2007. Probiotics and prebiotics in infant nutrition. Proc. Nutr. Soc. 66:405-411. [DOI] [PubMed] [Google Scholar]
- 40.Rada, V. 1997. Detection of Bifidobacterium species by enzymatic methods and antimicrobial susceptibility testing. Biotechnol. Tech. 11:909-912. [Google Scholar]
- 41.Rada, V., E. Vlkova, J. Nevoral, and I. Trojanova. 2006. Comparison of bacterial flora and enzymatic activity in faeces of infants and calves. FEMS Microbiol. Lett. 258:25-28. [DOI] [PubMed] [Google Scholar]
- 42.Roberfroid, M. 2007. Prebiotics: the concept revisited. J. Nutr. 137:830S-837S. [DOI] [PubMed] [Google Scholar]
- 43.Rossi, M., L. Altomare, A. Gonzalez Vara y Rodriguez, P. Brigidi, and D. Matteuzzi. 2000. Nucleotide sequence, expression and transcriptional analysis of the Bifidobacterium longum MB 219 lacZ gene. Arch. Microbiol. 174:74-80. [DOI] [PubMed] [Google Scholar]
- 44.Ryan, S. M., G. F. Fitzgerald, and D. van Sinderen. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 72:5289-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryan, S. M., G. F. Fitzgerald, and D. van Sinderen. 2005. Transcriptional regulation and characterization of a novel beta-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 71:3475-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanz, M. L., M. Gonzalez, C. de Lorenzo, J. Sanz, and I. Martinez-Castro. 2004. Carbohydrate composition and physico chemical properties of artisanal honeys from Madrid (Spain): occurrence of Echium sp. honey. J. Sci. Food Agric. 84:1577-1584. [Google Scholar]
- 47.Sanz, M. L., N. Polemis, V. Morales, N. Corzo, A. Drakoularakou, G. R. Gibson, and R. A. Rastall. 2005. In vitro investigation into the potential prebiotic activity of honey oligosaccharides. J. Agric. Food Chem. 53:2914-2921. [DOI] [PubMed] [Google Scholar]
- 48.Schrezenmeir, J., and M. de Vrese. 2001. Probiotics, prebiotics, and synbiotics—approaching a definition. Am. J. Clin. Nutr. 73:361S-364S. [DOI] [PubMed] [Google Scholar]
- 49.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 50.Reference deleted.
- 51.Turroni, F., A. Ribbera, E. Foroni, D. van Sinderen, and M. Ventura. 2008. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie van Leeuwenhoek 94:35-50. [DOI] [PubMed] [Google Scholar]
- 52.Ustunol, Z., and H. Gandhi. 2001. Growth and viability of commercial Bifidobacterium spp. in honey-sweetened skim milk. J. Food Prot. 64:1775-1779. [DOI] [PubMed] [Google Scholar]
- 53.van den Broek, L. A., S. W. Hinz, G. Beldman, J. P. Vincken, and A. G. Voragen. 2008. Bifidobacterium carbohydrases—their role in breakdown and synthesis of (potential) prebiotics. Mol. Nutr. Food Res. 52:146-163. [DOI] [PubMed] [Google Scholar]
- 54.van den Broek, L. A., K. Struijs, J. C. Verdoes, G. Beldman, and A. G. Voragen. 2003. Cloning and characterization of two alpha-glucosidases from Bifidobacterium adolescentis DSM20083. Appl. Microbiol. Biotechnol. 61:55-60. [DOI] [PubMed] [Google Scholar]
- 55.Ventura, M., C. Canchaya, A. Tauch, G. Chandra, G. F. Fitzgerald, K. F. Chater, and D. van Sinderen. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ventura, M., M. O'Connell-Motherway, S. Leahy, J. A. Moreno-Munoz, G. F. Fitzgerald, and D. van Sinderen. 2007. From bacterial genome to functionality; case bifidobacteria. Int. J. Food Microbiol. 120:2-12. [DOI] [PubMed] [Google Scholar]
- 57.Vlkova, E., J. Nevoral, B. Jencikova, J. Kopecny, J. Godefrooij, I. Trojanova, and V. Rada. 2005. Detection of infant faecal bifidobacteria by enzymatic methods. J. Microbiol. Methods. 60:365-373. [DOI] [PubMed] [Google Scholar]
- 58.Warchol, M., S. Perrin, J. P. Grill, and F. Schneider. 2002. Characterization of a purified beta-fructofuranosidase from Bifidobacterium infantis ATCC 15697. Lett. Appl. Microbiol. 35:462-467. [DOI] [PubMed] [Google Scholar]
- 59.Yang, L., T. Akao, K. Kobashi, and M. Hattori. 1996. Purification and characterization of a novel sennoside-hydrolyzing beta-glucosidase from Bifidobacterium sp. strain SEN, a human intestinal anaerobe. Biol. Pharm. Bull. 19:705-709. [DOI] [PubMed] [Google Scholar]
- 60.Zhao, H., L. Lu, M. Xiao, Q. Wang, Y. Lu, C. Liu, P. Wang, H. Kumagai, and K. Yamamoto. 2008. Cloning and characterization of a novel alpha-galactosidase from Bifidobacterium breve 203 capable of synthesizing Gal-alpha-1,4 linkage. FEMS Microbiol. Lett. 285:278-283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.