Abstract
The yeast strain Pichia anomala DBVPG 3003 secretes a killer toxin (Pikt) that has antifungal activity against Brettanomyces/Dekkera sp. yeasts. Pikt interacts with β-1,6-glucan, consistent with binding to the cell wall of sensitive targets. In contrast to that of toxin K1, secreted by Saccharomyces cerevisiae, Pikt killer activity is not mediated by an increase in membrane permeability. Purification of the toxin yielded a homogeneous protein of about 8 kDa, which showed a marked similarity to ubiquitin in terms of molecular mass and N-terminal sequences. Pikt is also specifically recognized by anti-bovine ubiquitin antibodies and, similar to ubiquitin-like peptides, is not absorbed by DEAE-cellulose. However, Pikt differs from ubiquitin in its sensitivity to proteolytic enzymes. Therefore, Pikt appears to be a novel ubiquitin-like peptide that has killer activity.
After its initial discovery in Saccharomyces cerevisiae (2), the killer phenotype has been found in yeasts belonging to the genera Candida, Cryptococcus, Debaryomyces, Hanseniaspora, Hansenula, Kloeckera, Kluyveromyces, Metschnikowia, Pichia, Rhodotorula, Schwanniomyces, Torulopsis, Trichosporon, Williopsis, and Zygosaccharomyces (9, 16, 19, 25, 28, 53, 56). The distribution of this character among natural yeast isolates and in laboratory strains is accompanied by killer toxins with large biodiversity, in terms of their biochemical characteristics, genetic determinants, and spectra of action (25). In spite of this diversity, all known killer toxins are proteins (1, 31) or glycoproteins (5, 11, 28, 43, 54) that kill sensitive cells via a two-step mode of action. During the first step, the killer toxin binds a receptor site on the cell wall of its sensitive target. In the second step, which has been far less characterized, the killer toxin is assumed to interact with receptors on the cell membrane and to kill the sensitive cells via different mechanisms. These include cell membrane permeabilization, cell cycle perturbation, inhibition of DNA synthesis, and inhibition of β-1,3-glucan synthetase activity (7, 14, 15, 41, 49, 53).
As the spectrum of action of some toxins has extended to microbial pathogens of clinical interest, killer toxins and/or killer toxin-like antibodies and mimotopes are of great relevance to medicine (8, 25). Other toxins that exert a killing action on spoilage yeasts have interesting applications in the fermentative (46) and food and feed industries (10, 11, 12, 23, 32, 44), where they can be used as “natural” food antimicrobials.
In a previous study, we showed that Pichia anomala DBVPG 3003 produces a killer toxin (known as Pikt) that is active against Dekkera/Brettanomyces sp. yeasts (12). These yeasts can develop in white and red wines, resulting in unpleasant odors and tastes (18). Thus, they represent a major problem in the wine industry. Pikt is stable at acidic pH and in a range of temperatures between 20°C and 40°C (12), as are other toxins produced by different strains of P. anomala (25, 26, 37). Moreover, Pikt maintains its killing activity for at least 10 days in wine. Thus, it shows potential applications as a natural antimicrobial in the wine industry (12).
The present study investigates the mode of action of Pikt and the biochemical properties of the purified protein.
MATERIALS AND METHODS
Yeast strains, media, and growth conditions.
The yeast strains used in this study are listed in Table 1. The media were YEPD (1% yeast extract, 2% glucose, 2% peptone, and 1.8% agar when required) for medium-term storage at 4°C, YEPDpH 4.4 (YEPD with 100 mM citrate-phosphate buffer, pH 4.4, for Pikt production), YEPDpH 4.5 (YEPD with 100 mM citrate-phosphate buffer, pH 4.5, for K1 production), YEPDpH 6 (YEPD with 50 mM Na-phosphate buffer, pH 6, for the production of the K. lactis killer toxin), GYNBpH 4.4 (2% glucose, 0.67% yeast nitrogen base, with 100 mM citrate-phosphate buffer, pH 4.4, for Pikt purification), and malt agar (Difco, Voigt Global Distribution Inc., Lawrence, KS) buffered as indicated above for the well test assay. P. anomala DBVPG 3003 and S. cerevisiae DBVPG 6497 were grown at 20°C with shaking (150 rpm). Kluyveromyces lactis DBVPG 6727 was grown at 25°C with shaking (200 rpm).
TABLE 1.
Strains used in the present study
| Species | Strain designation(s)a |
|---|---|
| Pichia anomala | DBVPG 3003 |
| Saccharomyces cerevisiae | DBVPG 6497 (NCYC 232), DBVPG 6500 (NCYC 1006) |
| Kluyveromyces lactis | DBVPG 6727 (ATCC 8585) |
Alternative strain designations are in parentheses. DBVPG, Industrial Yeast Collection, Dipartimento di Biologia Vegetale, Università di Perugia, Italy; ATCC, American Type Culture Collection, Manassas, VA; NCYC, National Collection of Yeast Cultures, Institute of Food Research, Norwich, United Kingdom.
Killing assays.
Well test assays were carried out as described by Woods and Bevan (52). Briefly, 70 μl of toxin samples was filter sterilized through 0.45-μm-pore-size membrane filters (Millipore Corp., Bedford, MA) and then loaded into wells (7-mm diameter) cut in malt-agar plates that had previously been seeded with 105 cells ml−1 of a strain sensitive to the toxin. The killing activity was evaluated as the diameter of the zone of inhibition around the wells after incubation for 72 h at 20°C and was defined as the mean zone of inhibition of replicate wells. The linear relationship observed between the logarithm of killer toxin concentration and the diameter of the inhibition halo from the well test was used to define Pikt activity in arbitrary units (AU). One AU was defined as the amount of toxin contained in 70 μl that generated an inhibition halo of 8 mm. One AU corresponds to about 10 ng of killer protein.
Toxin production.
The killer toxins were produced in 1.0 liter of YEPD buffered as required. After 48 h of growth, the culture broths were centrifuged (5,000 × g for 10 min at 4°C), and the supernatants were microfiltered through 0.45-μm membranes (Millipore Corp., Bedford, MA) and concentrated 100-fold by ultrafiltration (stirred ultrafiltration cells; Millipore Corp., Bradford, MA) with a 3-kDa cutoff membrane for Pikt and a 10-kDa cutoff membrane for K1 and the toxin produced by K. lactis.
Purification of Pikt.
Pikt was produced in 20 liters of GYNBpH 4.4. After 48 h of growth, the culture broth was microfiltered through a 0.2-μm membrane (Schleicher and Schuell GmbH, Germany), and the cell-free medium was concentrated about 350-fold by ultrafiltration with a 5-kDa cutoff membrane (Schleicher and Schuell GmbH, Germany). The concentrated sample was then diluted 10-fold with buffer A (10 mM citrate-phosphate, pH 4.4, 15% glycerol) and concentrated again as described above, to a final volume of about 60 ml. Microfiltration and ultrafiltration were performed using a Bioflex 50 (Schleicher and Schuell GmbH, Germany). The concentrated sample was loaded onto a DEAE-Sepharose Fast Flow (Pharmacia Biotech, Uppsala, Sweden) column (2.5 cm by 7.0 cm) equilibrated with buffer A. The void volume (60 ml) containing Pikt was applied to a Macro-Prep High S Support (Bio-Rad Laboratories, Hercules, CA) (2.5 cm by 6 cm) column equilibrated with the same buffer. After a washing with buffer A, the elution was performed with buffer A containing 1.0 M NaCl. The active fractions were pooled and dialyzed against buffer A. The dialyzed pool was applied to a fast-protein liquid chromatography column of Resource S (1 ml) (Pharmacia Biotech, Uppsala, Sweden) equilibrated with buffer A. The column was washed with the same buffer and eluted with a linear gradient of NaCl, from 0 M to 1.0 M, in buffer A.
Electrophoresis.
Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as described by Schagger and von Jagow (38). Before the gels were loaded, the samples were concentrated either by ultrafiltration on a Centricon YM3 membrane (Millipore Corp., Bedford, MA) or by precipitation as described by Wessel and Flugge (50). After being fixed with 5% glutaraldehyde for 1 h, the proteins were silver stained (Sigma Aldrich Inc., St. Louis, MO).
N-terminal amino acid sequencing.
After electrophoresis, Pikt was electroblotted onto a polyvinylidene difluoride membrane. The transfer was performed at 4°C for 1 h at 250 mA, in 10 mM CAPS (3-cyclohexylaminopropanesulfonic acid), pH 11.0, and 10% methanol. After membrane staining with Coomassie brilliant blue R-250, the Pikt band was excised and subjected to N-terminal sequencing by automated Edman degradation on a Procise model 491 sequencer (Applied Biosystems, Foster City, CA).
Immunoblotting.
After the electroblotting, the polyvinylidene difluoride membranes were incubated with antiubiquitin (whole antiserum) developed in rabbits using bovine red blood cell ubiquitin as the immunogen (Sigma Aldrich Inc., St. Louis, MO). Incubations were carried out for 2 h at a 1:100 dilution of the antibody. Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G was used as the secondary antibody. Blots were developed by chemiluminescence with the trial kit SuperSignal West Pico (Pierce, Rockford, IL).
Protease treatment.
Twenty nanograms of ubiquitin from bovine erythrocytes (Sigma Aldrich Inc., St. Louis, MO) and purified Pikt were treated with 0.04 U of protease XIV and pepsin (Sigma Aldrich Inc., St. Louis, MO) and incubated for 24 h at 20°C in 100 mM citrate phosphate buffer, pH 4.4.
Identification of the cell wall putative binding site for the P. anomala killer toxin.
The sensitive S. cerevisiae DBVPG 6500 strain (105 cells ml−1) was treated with the killer toxin (30 AU) in the absence or presence of 9 mg ml−1 of the following cell wall polysaccharides: mannan (from S. cerevisiae), glucan (from baker's yeast), laminarin (from Laminaria digitata), and pullulan (from Aureobasidium pullulans), which were all purchased from Sigma Aldrich Inc., St. Louis, MO, and pustulan (from Umbilicaria papullosa; Calbiochem, San Diego, CA). The binding of the toxin to the polysaccharides, as a function of the residual killer activity in solution, was evaluated by the well test after 24 h of incubation at 20°C.
Killer toxin mode of action.
The sensitive S. cerevisiae DBVPG 6500 strain was cultivated in YEPD buffered at the optimal pH of each toxin, and 105 cells ml−1 were incubated at 20°C with 46 AU of K1 or Pikt or at 25°C with 46 AU of Klkt (name given the toxin produced by K. lactis DBVPG 6727), in a final volume of 3 ml. Immediately after toxin addition (time zero) and after 4, 8, and 24 h, 100 μl of the cell suspensions was subjected to viable plate counts. Flow cytometry analyses were performed with an Epics XL (Beckman Coulter, Inc., Fullerton, CA) equipped with a 15-mW air-cooled argon-ion laser (emission, 488 nm) and five sensors for the detection of forward and side light scatter: green (525 nm, channel 1), yellow (575 nm, channel 2), and orange-red (620 nm, channel 3) fluorescence. Size calibration was performed using the flow cytometry size calibration kit (Molecular Probes, Inc., Eugene, OR). Yeast cells were harvested at various times (see Fig. 2), and ethanol-treated cells (70%, 2 h) were included as the positive control for membrane permeability. The samples were then washed and resuspended in PBS (8 mM Na2HPO4, 1.47 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl) to a final concentration of 1 × 105 to 2 × 105 cells ml−1. Fifty microliters of a propidium iodide (PI) stock solution (1 mg ml−1) in PBS was added to 500 ml of cell suspension just prior to the analysis. The fluorescence was detected in fluorescence channel 3. At least 1.5 × 104 cells were analyzed for each experiment. The data were visualized using the WinMDI flow cytometry software (Joseph Trotter, Salk Institute for Biological Studies, La Jolla, CA).
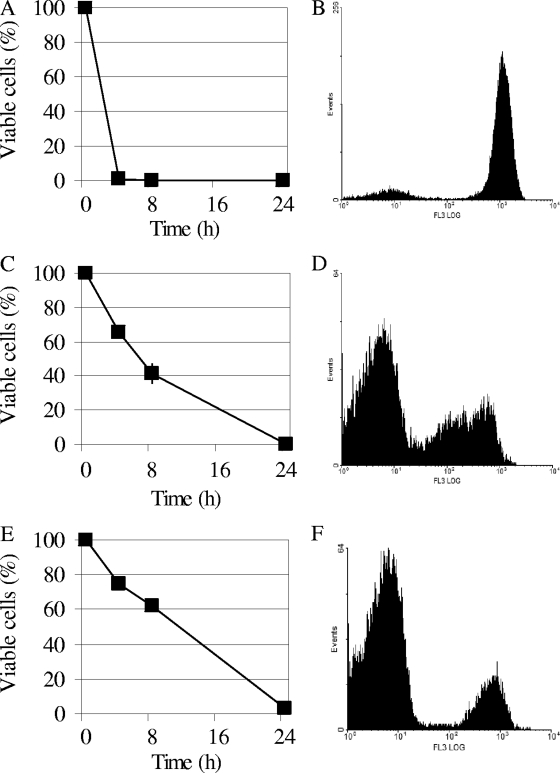
FIG. 2.
Mode of action of Pikt. DBVPG 6500 cells were treated with 46 AU of Pikt, K1, and Klkt, and the effects of each toxin were evaluated in terms of viability and membrane permeability to PI immediately after toxin addition (time zero) and at the times indicated. (A, C, and E) Viable plate counts of sensitive cells treated with K1, Klkt, and Pikt, respectively. (B, D, and F) Flow cytometric analysis of PI-stained sensitive cells after toxin treatment with K1 (8 h), Klkt (24 h), and Pikt (24 h), respectively. Abscissa, fluorescence intensity plotted on a logarithmic scale (increases from left to right); ordinate, number of events (cells). PI-stained cells are characterized by high fluorescence intensity (peaks on the right of the abscissa); unstained cells show low fluorescence intensity (peaks on the left). Data are representative of three independent experiments.
RESULTS
Pikt interacts with β-1,6-glucans.
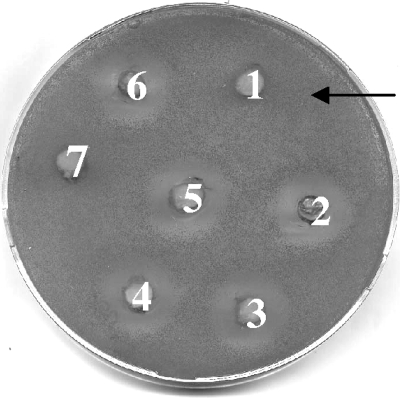
To identify the putative binding site for Pikt on the cell wall of the sensitive target, the abilities of various cell wall components to complex with, and inactivate, the killer toxin were evaluated. To achieve this, 30 AU of Pikt was mixed with glucans, pullulan, pustulan, laminarin, and mannan in citrate-phosphate buffer, pH 4.4, and the residual killer activity was evaluated by a well test on the sensitive S. cerevisiae DBVPG 6500 strain. The results obtained showed that pustulan (β-1,6-glucan) inhibited its killing activity (Fig. 1). These results were further confirmed by viable plate counts using 105 ultrafiltered cells (UFC) of the sensitive strain DBVPG 6500. The plate counts indicated that, in the presence of Pikt plus pustulan, the growth of the target cells was comparable to that of cells not treated with the toxin (positive control) (values of 30 × 105 and 36 × 105 UFC, respectively); other plate count values were as follows: Pikt alone, 0.013 × 105 UFC; Pikt plus glucan, 0.04 × 105 UFC; Pikt plus pullulan, 0.25 × 105 UFC; Pikt plus mannan, 0.3 × 105 UFC; Pikt plus laminarin, 0.38 × 105 UFC.
FIG. 1.
Identification of the cell wall binding site for the P. anomala killer toxin. Thirty AU of Pikt in citrate-phosphate buffer (pH 4.4) was incubated with different cell wall components, and the residual killer activity against the sensitive strain was evaluated using the well test. Wells 1 to 5, Pikt added with pustulan, glucan, pullulan, laminarin, and mannan, respectively; wells 6 and 7, Pikt and citrate-phosphate buffer, pH 4.4, respectively, used as the controls.
Pikt killing action differs from that of the killer toxin secreted by S. cerevisiae.
To determine the mechanism responsible for Pikt killer activity, the mode of action of this toxin was compared with those of the toxins produced by S. cerevisiae DBVPG 6497 (K1) and K. lactis DBVPG 6727 (Klkt). DBVPG 6500 cells, which are sensitive to the three toxins, were treated with 46 AU of Pikt, K1, or Klkt. The effects of each toxin were evaluated in terms of viability and membrane permeability to PI. PI penetrates cells with defective membrane integrity (13, 27). Thus, the fluorescence emitted by PI-stained cells provides a measure of membrane permeability and can serve as an indirect measure of cell death when this is mediated by alterations in the cell membrane. While the viable plate counts indicated that K1 killed the sensitive cells within 4 h, flow cytometry analysis of PI-stained cells confirmed that the K1 killing activity was mediated by a dramatic increase in membrane permeability (84.6% ± 4.3% PI-stained cells after 8 h) (Fig. 2). Klkt took a longer time to kill the sensitive cells. In particular, 65.0% ± 2.8% of the cells remained viable 4 h after toxin treatment, and complete killing was observed at 24 h. At this time, only 18.9% ± 5.71% were permeable to PI, thus indicating that Klkt killing action is not mediated by the disruption of the cell membrane. The behavior of Pikt was similar to that of Klkt with regard to the effects on both viability (75% ± 2.8% and 3.5% ± 0.21% viable cells after 4 h and 24 h, respectively) and membrane integrity of DBVPG 6500 cells (10.7% ± 0.29% of cells permeable to PI after 24 h of treatment) (Fig. 2).
Purification and N-terminal sequencing of Pikt.
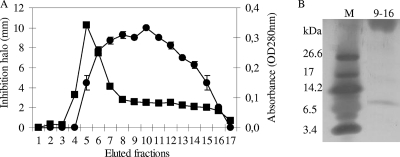
On the basis of the specific interaction between Pikt and β-1,6-glucan, toxin purification was first attempted via affinity chromatography on epoxy-Sepharose 6-B conjugated with pustulan, as already described (36). Contrary to what was expected, Pikt was found to interact very weakly with the resin under any of the conditions tested, resulting in a very low yield (data not shown). Several different chromatographic steps, including hydrophobic interactions and absorption chromatography, were attempted. The best recovery of Pikt was achieved by performing a purification procedure that consisted of two ion-exchange chromatography steps, as outlined in Materials and Methods. The anion-exchange chromatography on the DEAE resin resulted in the recovery of the toxin in the flowthrough of the column, allowing the removal of most of the protein present in the loaded sample. The column flowthrough was loaded directly onto a High S cation-exchange column, and the proteins were eluted isocratically with 1.0 M NaCl. Pikt bound more strongly than other proteins, facilitating purification of a major 8-kDa band by SDS-PAGE (Fig. 3B). The high-molecular-weight product visible in the gel shown in Fig. 3B is also present in samples from the purification procedure which do not contain the killer activity (not shown). To assess the correspondence of the 8-kDa band with the killer toxin, the High S pool was subjected to fast-protein liquid chromatography on a Resource S column, from which the bound proteins were eluted with a continuous NaCl gradient. SDS-PAGE analysis and the killer activity assays on the eluted fractions showed that active fractions again contained the protein corresponding to the 8-kDa band (data not shown). The coelution of the 8-kDa protein with the toxic activity from two different resins (High S and Resource S) and in different experimental conditions (isocratically and with a gradient) confirms the identity of the 8-kDa protein as the killer factor.
FIG. 3.
Pikt purification. (A) Elution profile from the High S cation-exchange chromatography column. Elution was performed as described in Materials and Methods. Circles indicate the diameters of the inhibition halos; squares indicate optical densities at 280 nm (OD280nm). (B) Tricine SDS-PAGE analysis of the High S pool, comprising fractions 9 to 16. Lane M, molecular mass standards (ultra-low-range molecular weight marker; Sigma).
Our purification procedure increased specific activity of the toxin 4,356-fold, with a yield of 41% (Table 2). Starting from 20 liters of culture, the final amounts of purified Pikt were extremely low and could not be quantified by the Bradford method (3). However, they were suitable for the determination of a partial N-terminal sequence by automated Edman degradation. The direct determination of the sequence demonstrated that the N terminus of Pikt was not blocked. Nine of the first 10 residues were unambiguously identified (MQIFVXTLTG). Similarity searches using BLASTP did not show any significant homology with known killer toxins. On the other hand, the sequence of Pikt was identical to that of S. cerevisiae ubiquitin (accession number CAA29196).
TABLE 2.
Purification of the killer protein from Pichia anomala DBVPG 3003
| Step | Total vol (ml) | Total protein (mg) | Total activity (AU) | Sp act (AU/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Culture broth | 20,000 | 32 | 763 | 23.8 | 100 | |
| Ultrafiltration | 60 | 8.4 | 572 | 68.0 | 2.8 | 75 |
| DEAE | 60 | 2.5 | 515 | 206.0 | 8.6 | 67 |
| High S | 41 | 0.003a | 311 | 103,667 | 4,356 | 41 |
Estimate based on the amount of the first amino acid residue, determined by N-terminal sequencing.
Pikt is a ubiquitin-like protein.
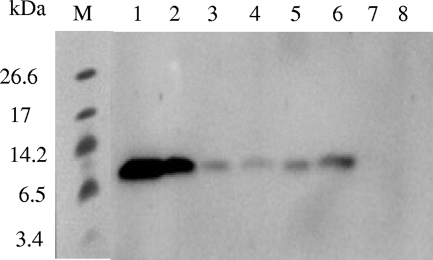
The similarity of the N-terminal sequence of Pikt with that of ubiquitin prompted us to determine whether purified Pikt was recognized by an antiubiquitin antibody. Figure 4 shows an immunoblot obtained by incubating different fractions from the purification steps with such an antibody. A positive signal was evident in all toxin samples, and the corresponding band migrated exactly like the bovine ubiquitin used as a positive control. On the other hand, none of the fractions lacking killer activity cross-hybridized with the antiubiquitin antibody (Fig. 4).
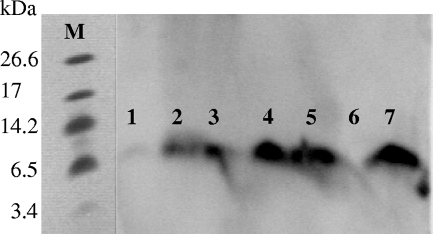
FIG. 4.
Immunoblot of samples from the different steps of Pikt purification, performed with antiserum against bovine ubiquitin. Lanes 1 and 2, 50 ng and 25 ng of bovine ubiquitin, respectively; lane 3, 350-fold-concentrated cell-free medium; lanes 4 and 5, fractions 15 and 10, respectively, eluted from the High S cation column and showing killer activity; lane 6, DEAE column flowthrough, showing killer activity; lane 7, High S cation column flowthrough, not showing killer activity; lane 8, fraction 5 eluted from the High S cation column, not showing killer activity; lane M, molecular mass standards (ultra-low-range molecular weight marker; Sigma).
To further investigate the similarity between Pikt and bovine ubiquitin, both proteins were treated with pepsin and protease XIV. After proteolytic digestion, the samples were both analyzed for residual Pikt killer activity and subjected to immunoblot analysis with the antiubiquitin antibody. As shown in Fig. 5, only protease XIV completely degraded the toxin. This result was in agreement with the well test, which highlighted a total loss of Pikt toxic activity following proteolytic digestion with protease XIV (12). Pepsin treatment caused a decrease in the signal produced by the Pikt-antibody complex, in agreement with the slight reduction in the activity of the toxin according to the well test (12). In contrast, and in agreement with results described by Schlesinger et al. (39), neither of the two proteolytic enzymes degraded bovine ubiquitin.
FIG. 5.
Immunoblot of bovine ubiquitin and Pikt after proteolytic digestion performed with antiserum against bovine ubiquitin. Lanes 1 and 2, Pikt treated with protease XIV and pepsin, respectively; lanes 3 and 7, untreated Pikt and ubiquitin, respectively; lanes 4 and 5, ubiquitin treated with protease XIV and pepsin, respectively; lane 6, empty; lane M, molecular mass standards (ultra-low-range molecular weight marker; Sigma).
DISCUSSION
Since Pikt is a secreted toxin, it is possible that it first acts on the outside of the sensitive cells by interacting with the cell wall. Accordingly, competitive inhibition of killer toxin activity by pustulan showed that the toxin selectively interacts with β-1,6-glucan. Therefore, as has been seen for the K1 and K2 S. cerevisiae killer toxins (17) and the toxins produced by Hanseniaspora uvarum (34), Debaryomyces hansenii (36), and Pichia membranifaciens (35), β-1,6-glucans provide a putative binding site for Pikt on the cell wall of the sensitive target. As suggested for other toxins, this first interaction may be necessary to concentrate the toxin on the cell wall of the sensitive target and facilitate the contact between the toxin and the plasma membrane (40).
To examine the mode of action of Pikt, comparisons were made to K1 and Klkt. K1 is an ionophoric toxin that disrupts membrane integrity (4, 6, 14, 42), while Klkt causes a permanent arrest of the progression of the cell cycle at the G1 phase (7, 24, 51). The behavior of Pikt appeared distinct from that of K1, for both the death kinetics and the membrane permeability. In short, the Pikt killing action is not mediated through the disruption of cell membrane integrity (Fig. 2).
The biochemical characterization of purified Pikt highlighted the similarity to ubiquitin in terms of molecular masses and N-terminal sequences. Moreover, Pikt was specifically recognized by an anti-bovine ubiquitin antibody and, as with ubiquitin and ubiquitin-like peptides, Pikt was not absorbed onto DEAE-cellulose (30). In aggregate, these results support the “ubiquitin-like” nature of Pikt.
Ubiquitin is localized mainly in the cytoplasm. However its presence in the extracellular environment has already been reported. In particular, Kieffer et al. (20) observed that bovine chromaffin cells secrete ubiquitin upon nicotinic stimulation and Vad et al. (45) showed that a Pichia pastoris strain engineered to produce human parathyroid hormone cosecreted ubiquitin in the culture medium.
Interestingly, even though the biological role of secreted ubiquitin needs further investigation, it has been reported that ubiquitin has antimicrobial activity. In particular, Kieffer et al. (20) showed that secreted ubiquitin exerts antibacterial and antifungal activities by destabilizing the fungal cell wall and plasma membrane. Likewise, Yoshio et al. (55) reported that the ubiquitin present in Vernix caseosa, together with other antimicrobial polypeptides, is involved in defense against microbial invasion at birth. Kutty et al. (22) reported that exogenously added ubiquitin inhibits Schizosaccharomyces pombe growth by interfering with cell cycle progression. Kieffer et al. (20) also demonstrated that antimicrobial activity is retained by a short ubiquitin fragment. Accordingly, Purdy and Russel (33) and Kim et al. (21) showed that ubiquitin-derived peptides show antibacterial and antifungal activities in the μM range. Interestingly, Pikt also appears to possess a high specific activity. Indeed, the very small amount of Pikt recovered from 20 liters of cultural broth was characterized by a strong killing action.
One difference between ubiquitin and Pikt is seen in resistance to proteolytic enzymes. According to Schlesinger et al. (39), ubiquitin has a marked resistance to the proteolytic enzymes used in the present study. In contrast, Pikt was completely hydrolyzed by protease XIV and partially hydrolyzed by pepsin.
Thus, Pikt appears to be a ubiquitin-like peptide that has antimicrobial activity. Ubiquitin-like peptides from the fruiting bodies of edible mushrooms, including Pleurotus sajor-caju, Pleurotus ostreatus, and Cantharellus cibarius, have been characterized so far (29, 47, 48). These peptides show an N-terminal sequence identical to that of ubiquitin, and all have RNase- and translation-inhibiting activities. Studies are in progress to determine whether Pikt exhibits similar activities.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Ashida, S., T. Shimazaki, K. Kitano, and S. Hara. 1983. New killer toxin of Hansenula mrakii. Agric. Biol. Chem. 47:2953-2955. [Google Scholar]
- 2.Bevan, E. A., and M. Makower. 1963. The physiological basis of the killer character in yeast. Proc. Int. Congr. Genet. 1:202-203. [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Breinig, F., D. J. Tipper, and M. J. Schmitt. 2002. Kre1p, the plasma membrane receptor for the yeast K1 viral toxin. Cell 108:395-405. [DOI] [PubMed] [Google Scholar]
- 5.Bussey, H., and N. Skipper. 1975. Membrane-mediated killing of Saccharomyces cerevisiae by glycoproteins from Torolopsis glabrata. J. Bacteriol. 124:476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bussey, H. 1991. K1 killer toxin, a pore-forming protein from yeast. Mol. Microbiol. 5:2339-2343. [DOI] [PubMed] [Google Scholar]
- 7.Butler, A. R., J. H. White, and M. J. R. Stark. 1991. Analysis of the response of Saccharomyces cerevisiae cells to Kluyveromyces lactis toxin. J. Gen. Microbiol. 137:1749-1757. [DOI] [PubMed] [Google Scholar]
- 8.Buzzini, P., and A. Martini. 2001. Large scale screening of selected Candida maltosa, Debaryomyces hansenii and Pichia anomala killer toxin activity against pathogenic yeasts. Med. Mycol. 39:479-482. [DOI] [PubMed] [Google Scholar]
- 9.Chen, W. B., Y. F. Han, S. C. Jong, and S. C. Chang. 2000. Isolation, purification, and characterization of a killer protein from Schwanniomyces occidentalis. Appl. Environ. Microbiol. 66:5348-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciani, M., and F. Fatichenti. 2001. Killer toxin of Kluyveromyces phaffii DBVPG 6076 as a biopreservative agent to control apiculate wine yeast. Appl. Environ. Microbiol. 67:3058-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comitini, F., N. Di Pietro, L. Zacchi, I. Mannazzu, and M. Ciani. 2004. Kluyveromyces phaffii killer toxin active against wine spoilage yeasts: purification and characterization. Microbiology 150:2535-2541. [DOI] [PubMed] [Google Scholar]
- 12.Comitini, F., J. De Ingeniis, L. Pepe, I. Mannazzu, and M. Ciani. 2004. Pichia anomala and Kluyveromyces wickeramii killer toxins as new tools against Dekkera/Brettanomyces spoilage yeasts. FEMS Microbiol. Lett. 238:235-240. [DOI] [PubMed] [Google Scholar]
- 13.Deere, D., J. Shen, G. Vesey, P. Bell, P. Bissinger, and D. Veal. 1998. Flow cytometry and cell sorting for yeast viability assessment and cell selection. Yeast 14:147-160. [DOI] [PubMed] [Google Scholar]
- 14.de la Pena, P., F. Barros, S. Gascon, P. S. Lazo, and S. Ramos. 1981. The effect of yeast killer toxin on sensitive cells of Saccharomyces cerevisiae. J. Biol. Chem. 256:10420-10425. [PubMed] [Google Scholar]
- 15.Eisfeld, K., F. Riffer, J. Mentges, and M. J. Schmitt. 2000. Endocytotic uptake and retrograde transport of a virally encoded killer toxin in yeast. Mol. Microbiol. 37:926-940. [DOI] [PubMed] [Google Scholar]
- 16.Hodgson, V. J., D. Button, and G. M. Walker. 1995. Anti-Candida activity of a novel killer toxin from the yeast Williopsis mrakii. Microbiology 141:2003-2012. [DOI] [PubMed] [Google Scholar]
- 17.Hutchins, K., and H. Bussey. 1983. Cell wall receptor for yeast killer toxin: involvement of (1-6)-β-d-glucan. J. Bacteriol. 154:161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibeas, J. I., I. Lozano, F. Perdigones, and I. Jimenez. 1996. Detection of Dekkera/Brettanomyces strains in sherry by nested PCR method. Appl. Environ. Microbiol. 62:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandel, J. S., and T. A. Stern. 1979. Killer phenomenon in pathogenic yeast. Antimicrob. Agents Chemother. 15:568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieffer, A. E., Y. Goumon, O. Ruh, S. Chasserot-Golaz, G. Nullans, C. Gasnier, D. Aunis, and M. H. Metz-Boutigue. 2003. The N- and C-terminal fragments of ubiquitin are important for the antimicrobial activities. FASEB J. 17:776-788. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. Y., S. Y. Lee, S. C. Park, S. Y. Shin, S. J. Choi, Y. Park, and K. S. Hahm. 2007. Purification and antimicrobial activity studies of the N-terminal fragment of ubiquitin from human amniotic fluid. Biochim. Biophys. Acta 1774:1221-1226. [DOI] [PubMed] [Google Scholar]
- 22.Kutty, B. C., K. Pasupathy, and K. P. Mishra. 2005. Effects of exogenous ubiquitin on cell division cycle mutants of Schizosaccharomyces pombe. FEMS Microbiol. Lett. 244:187-191. [DOI] [PubMed] [Google Scholar]
- 23.Lowes, K. F., C. A. Shearman, D. Mackenzie, D. B. Archer, R. J. Merry, and M. J. Gasson. 2000. Prevention of yeast spoilage in feed and food by the yeast mycocin HMK. Appl. Environ. Microbiol. 66:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, J., B. Huang, A. Esberg, M. Johansson, and A. S. Byström. 2005. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 11:1648-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magliani, W., S. Conti, M. Gerloni, D. Bertolotti, and L. Polonelli. 1997. Yeast killer systems. Clin. Microbiol. Rev. 10:369-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquina, D., A. Santos, and J. M. Peinado. 2002. Biology of killer yeasts. Int. Microbiol. 5:65-71. [DOI] [PubMed] [Google Scholar]
- 27.Marza, E., N. Camougrand, and S. Manon. 2002. Bax expression protects yeast plasma membrane against ethanol-induced permeabilization. FEBS Lett. 521:47-52. [DOI] [PubMed] [Google Scholar]
- 28.Middelbeek, E. J., J. M. H. Hermans, and C. Stumm. 1979. Production, purification and properties of a Pichia klujveri killer toxin. Antonie van Leeuwenhoek 45:437-450. [DOI] [PubMed] [Google Scholar]
- 29.Ng, T. B., S. K. Lam, and S. Y. Chan. 2002. A ubiquitin-like peptide from the mushroom Pleurotus sajor-caju exhibits relatively potent translation-inhibitory and ribonuclease activities. Peptides 23:1361-1365. [DOI] [PubMed] [Google Scholar]
- 30.Ng, T. B. 2004. Peptides and proteins from fungi. Peptides 25:1055-1073. [DOI] [PubMed] [Google Scholar]
- 31.Palfree, R. G., and H. Bussey. 1979. Yeast killer toxin: purification and characterization of the protein toxin from Saccharomyces cerevisiae. Eur. J. Biochem. 93:487-493. [DOI] [PubMed] [Google Scholar]
- 32.Palpacelli, V., M. Ciani, and G. Rosini. 1991. Activity of different 'killer' yeasts on strains of yeast species undesirable in the food industry. FEMS Microbiol. Lett. 68:75-78. [DOI] [PubMed] [Google Scholar]
- 33.Purdy, G. E., and D. G. Russel. 2007. Lysosomal ubiquitin and the demise of Mycobacterium tuberculosis. Cell. Microbiol. 9:2768-2774. [DOI] [PubMed] [Google Scholar]
- 34.Radler, F., M. J. Shmitt, and B. Meyer. 1990. Killer toxin of Hanseniaspora uvarum. Arch. Microbiol. 154:175-178. [DOI] [PubMed] [Google Scholar]
- 35.Santos, A., D. Marquina, J. A. Leal, and J. M. Peinado. 2000. (1-6)-β-d-Glucan as cell wall receptor for Pichia membranifaciens killer toxin. Appl. Environ. Microbiol. 66:1809-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos, A., D. Marquina, J. Barroso, and J. M. Peinado. 2002. (1-6)-β-D-glucan as the cell wall binding site for Debaryomyces hansenii killer toxin. Lett. Appl. Microbiol. 34:95-99. [DOI] [PubMed] [Google Scholar]
- 37.Sawant, A. D., A. T. Abdelal, and D. G. Ahearn. 1989. Purification and characterization of the anti-Candida toxin of Pichia anomala WC 65. Antimicrob. Agents Chemother. 33:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulphate polyacrilamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 39.Schlesinger, D. H., G. Goldstein, and H. D. Niall. 1975. The complete amino acid sequence of ubiquitin, on adenylate cyclase stimulating polypeptide probably universal in living cells. Biochemistry 14:2214-2218. [DOI] [PubMed] [Google Scholar]
- 40.Schmitt, M. J., and F. Radler. 1987. Mannoprotein of the yeast cell wall as the primary receptor of the killer toxin of Saccharomyces cerevisiae strain 28. J. Gen. Microbiol. 133:3347-3354. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt, M. J., P. Klavehn, J. Wang, I. Shoning, and D. J. Tipper. 1996. Cell cycle studies on the mode of action of yeast K28 killer toxin. Microbiology 142:2655-2662. [DOI] [PubMed] [Google Scholar]
- 42.Skipper, N., and H. Bussey. 1977. Mode of action of yeast toxin: energy requirement for Saccharomyces cerevisiae killer toxin. J. Bacteriol. 129:667-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugisaki, Y., N. Gunge, K. Sakaguchi, M. Yamasaky, and G. Tamura. 1984. Characterization of a novel killer toxin encoded by a double-stranded linear DNA plasmid of Kluyveromyces lactis. Eur. J. Biochem. 141:241-245. [DOI] [PubMed] [Google Scholar]
- 44.Todd, B. E. N., G. H. Fleet, and P. A. Henscke. 2000. Promotion of autolysis through the interaction of killer and sensitive yeasts: potential application in sparkling wine production. Am. J. Enol. Vitic. 51:65-72. [Google Scholar]
- 45.Vad, R., E. Nafstad, L. A. Dahl, and O. S. Gabrielsen. 2005. Engineering of a Picha pastoris expression system for secretion of high amounts of intact human parathyroid hormone. J. Biotechnol. 116:251-260. [DOI] [PubMed] [Google Scholar]
- 46.Van Vuuren, H. J. J., and C. J. Jacobs. 1992. Killer yeast in the wine industry: a review. Am. J. Enol. Vitic. 43:119-128. [Google Scholar]
- 47.Wang, H. X., and T. B. Ng. 2000. Isolation of a novel ubiquitin-like protein from Pleurotus ostreatus mushroom with anti-human immunodeficiency virus, translation-inhibitory, and ribonuclease activities. Biochem. Biophys. Res. Commun. 276:587-593. [DOI] [PubMed] [Google Scholar]
- 48.Wang, H. X., H. K. Ngai, and T. B. Ng. 2003. A ubiquitin-like peptide with ribonuclease activity against various polyhomoribonucleotides from the yellow mushroom Cantharellus cibarius. Peptides 24:509-513. [DOI] [PubMed] [Google Scholar]
- 49.Weiler, F., and M. J. Schmitt. 2003. Zygocin, a secreted antifungal toxin of the yeast Zygosaccharomyces bailii, and its effect on sensitive fungal cells. FEMS Yeast Res. 3:69-76. [DOI] [PubMed] [Google Scholar]
- 50.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]
- 51.White, J. H., A. R. Butler, and M. J. Stark. 1989. Kluyveromyces lactis toxin does not inhibit adenylyl cyclase. Nature 341:666-668. [Google Scholar]
- 52.Woods, D. R., and E. A. Bevan. 1968. Studies on the nature of the killer factor produced by Saccharomyces cerevisiae. J. Gen. Microbiol. 51:115-126. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto, T., T. Hiratani, H. Hirata, M. Imani, and H. Yamaguchi. 1986. Killer toxin from Hansenula mrakii selectively inhibits cell wall synthesis in a sensitive yeast. FEBS Lett. 197:50-54. [DOI] [PubMed] [Google Scholar]
- 54.Yokomori, Y., H. Akiyama, and K. Shimizu. 1988. Toxin of wild Candida killer yeast with a novel killer property. Agric. Biol. Chem. 52:2797-2801. [Google Scholar]
- 55.Yoshio, H., M. Tollin, G. H. Gudmundsson, H. Lagercrantz, H. J.örnvall, G. Marchini, and B. Agerberth. 2003. Antimicrobial polypeptides of human vernix caseosa and amniotic fluid: implications for newborn innate defense. Pediatr. Res. 53:211-216. [DOI] [PubMed] [Google Scholar]
- 56.Young, T. W., and M. Yagiu. 1978. A comparison of the killer character in different yeasts and its classification. Antonie van Leeuwenhoek 44:59-77. [DOI] [PubMed] [Google Scholar]