Abstract
Acid and base environmental stress responses were investigated in Bacillus subtilis. B. subtilis AG174 cultures in buffered potassium-modified Luria broth were switched from pH 8.5 to pH 6.0 and recovered growth rapidly, whereas cultures switched from pH 6.0 to pH 8.5 showed a long lag time. Log-phase cultures at pH 6.0 survived 60 to 100% at pH 4.5, whereas cells grown at pH 7.0 survived <15%. Cells grown at pH 9.0 survived 40 to 100% at pH 10, whereas cells grown at pH 7.0 survived <5%. Thus, growth in a moderate acid or base induced adaptation to a more extreme acid or base, respectively. Expression indices from Affymetrix chip hybridization were obtained for 4,095 protein-encoding open reading frames of B. subtilis grown at external pH 6, pH 7, and pH 9. Growth at pH 6 upregulated acetoin production (alsDS), dehydrogenases (adhA, ald, fdhD, and gabD), and decarboxylases (psd and speA). Acid upregulated malate metabolism (maeN), metal export (czcDO and cadA), oxidative stress (catalase katA; OYE family namA), and the SigX extracytoplasmic stress regulon. Growth at pH 9 upregulated arginine catabolism (roc), which generates organic acids, glutamate synthase (gltAB), polyamine acetylation and transport (blt), the K+/H+ antiporter (yhaTU), and cytochrome oxidoreductases (cyd, ctaACE, and qcrC). The SigH, SigL, and SigW regulons were upregulated at high pH. Overall, greater genetic adaptation was seen at pH 9 than at pH 6, which may explain the lag time required for growth shift to high pH. Low external pH favored dehydrogenases and decarboxylases that may consume acids and generate basic amines, whereas high external pH favored catabolism-generating acids.
Bacillus subtilis can grow over several log units of environmental pH while maintaining cytoplasmic pH within a relatively narrow range that preserves protein and nucleic acid stability (19, 50, 55a). Environmental pH is important for the pathogenesis of related Bacillus species, such as the food-borne pathogen Bacillus cereus, which encounters acidic environments in the gastrointestinal tract and in food products where organic acids are used as preservatives (9, 64). B. cereus shows an acid tolerance response in which vegetative growth in a moderate acid induces proteins that enable survival under extreme acid conditions (64). In Bacillus anthracis, the lethal factor toxin undergoes a low-pH-driven structural change as it passes through acidic vesicles, which allows it to translocate into the cytosol (45). The Bacillus thuringiensis toxin is activated by alkaline pH upon entering the midgut of insect larvae (13), and bacterial growth is inhibited by acids (46).
Cytoplasmic pH homeostasis has been studied extensively in B. subtilis, which maintains cytoplasmic pH within approximately pH 7.3 to pH 7.6 during vegetative growth over a range of environmental pH, from pH 6.0 to pH 9.0 (14, 50, 55a). At high external pH, cytoplasmic pH homeostasis involves Na+/H+ antiporters as well as other Na+ transport components (14, 26, 50, 67, 68). Low pH triggers spore germination and initiation of vegetative growth (35).
Maintenance of pH homeostasis in neutralophiles involves a variety of constitutive and regulated mechanisms, which have been studied most extensively in Escherichia coli (24, 56, 57, 70). A model explaining much of the E. coli pH-responsive transcriptome is that low external pH favors pathways that consume acids and generate basic amines, whereas high external pH favors metabolism-generating acids in the cytoplasm. E. coli upregulates amino acid decarboxylases, periplasmic chaperones, redox modulators, chemotaxis, and drug transporters (28, 31, 44, 60, 65), whereas exposure to high pH upregulates deaminases that generate acids, ATP synthase, and the microaerophilic cytochrome d oxidoreductase (7, 60).
We sought to test the model described above for pH-responsive metabolism in B. subtilis. While B. subtilis grows over a range of pH values similar to that of E. coli, the mechanisms B. subtilis employs in order to maintain pH homeostasis are not as well characterized and show important differences. Relatively few systems are known to be upregulated by acid; acid shock does induce components of the SigB environmental and energy stress regulon (38). Base shifting in B. subtilis is known to upregulate several systems, including the σw antibiotic resistance regulon (10, 69), the SigH sporulation regulon (17, 30), and the srf operon encoding surfactin synthetase and the competence regulator ComS (16). Spore germination is enhanced at high pH and inhibited by acids (73).
To investigate acid and base stress responses, we used DNA arrays to compare transcription profiles of B. subtilis during growth at low, neutral, and high pH. While a previous transcriptomic study addressed cultures following rapid alkalization (69), ours is the first to address B. subtilis during log-phase vegetative growth in acidic and basic conditions and at neutral pH.
MATERIALS AND METHODS
Acid and base shift growth curves.
B. subtilis AG174 (obtained from Alan Grossman) was cultured overnight in potassium-modified Luria broth (LBK; 10 g of tryptone/liter, 5 g of yeast extract/liter, 7.45 g of KCl/liter) buffered with 50 mM homopiperazine-N,N-bis-2-(ethanesulfonic acid) (HOMOPIPES; a dibasic acid; pKa, 4.55 and 8.12), adjusted to pH 8.5. Bacteria were diluted 500-fold in pH 6.0 LBK (50 mM HOMOPIPES) or 100-fold in pH 8.5 LBK (50 mM HOMOPIPES) and incubated with aeration (260 rpm) at 37°C. Bacteria cultured at pH 6 or at pH 8.5 were incubated until an optical density at 600 nm (OD600) of 0.3 and then diluted 10-fold into a prewarmed flask containing buffered LBK either at pH 8.5 or at pH 6.0 (50 mM HOMOPIPES), respectively. Dilutions were cultured for 2.5 h. For each condition, three biological replicates were tested.
Extreme acid and base resistance.
B. subtilis AG174 was tested for induction of extreme acid resistance by growth in a moderate acid. Cultures were grown from a colony inoculated in LBK medium buffered with HOMOPIPES (100 mM) at pH 7 and incubated overnight at 37°C for 16 h. The overnight culture was diluted 100-fold in LBK medium buffered with 50 mM HOMOPIPES, either at pH 7.0 or at pH 6.0. Bacteria were cultured until an OD600 of 0.3. The cultures were diluted 200-fold in LBK adjusted to pH 4.5 and incubated for 2 h at 37°C. Serial dilutions were plated on LBK and compared to plated dilutions of the original culture in medium at pH 7. Six plates from six independent cultures were scored for each condition. Error values represent the standard error of the mean (SEM) (n = 6).
B. subtilis AG174 was tested for induction of extreme base resistance by growth in a moderate base. Cultures were grown from a colony inoculated in LBK medium buffered with HOMOPIPES (100 mM) at pH 7 and incubated overnight at 37°C for 16 h. The overnight culture was diluted 100-fold in LBK buffered with 50 mM HOMOPIPES at pH 7.0 or at pH 9.0. Bacteria were cultured until an OD600 of 0.3. The cultures were diluted 200-fold in LBK adjusted to pH 10 and incubated for 2 h at 37°C. Serial dilutions were plated on LBK and compared to plated dilutions of the original culture in medium at pH 7. Six plates from six independent cultures were scored for each condition. Error values represent the SEM (n = 6).
cDNA preparation and array hybridization.
B. subtilis strain AG174 was cultured overnight at 37°C in unbuffered LBK. Bacteria were diluted 500-fold in a 125-ml baffled flask containing 12 ml of LBK buffered with 50 mM HOMOPIPES. The pH of the medium was adjusted using KOH to pH 6.0, 7.0, or 9.0. In order to maximize aeration and maintain logarithmic growth, cultures were rotated at 240 rpm at 37°C. For each pH, five biological replicate cultures were performed. Cultures were incubated to an OD600 of 0.2. The pH was tested after growth to ensure that the values were maintained at ±0.2 pH units of the pH of the original uninoculated medium. Samples were transferred to vials containing 1 ml of ice-cold 10% phenol-ethanol stop solution (6, 36) in order to stabilize the bacterial RNA. RNA was isolated as described previously (31, 36, 44) using the RNeasy kit with on-column DNA digestion (Qiagen). The RNA isolation protocol for gram-positive microorganisms was followed, in which 3 mg/ml of lysozyme is used to degrade the bacterial cell wall. The quality of the isolated RNA was examined using the Agilent Bioanalyzer 2100.
Standard methods were used for cDNA synthesis, fragmentation, and end-terminus biotin labeling (31, 44). Labeled cDNA samples were hybridized to Affymetrix GeneChip B. subtilis antisense genome arrays (20). Hybridized arrays were stained with streptavidin-phycoerythrin using the Affymetrix fluidic station 450. After staining, arrays were scanned with a GC3000 scanner.
Analysis of gene expression.
The GeneChip B. subtilis arrays include 4,351 probe sets that covered 4,095 protein-encoding open reading frames (ORFs) (approximately total complete genomic coverage) as well as 606 probe sets covering intergenic regions annotated to the RefSeq genome sequence, NC_000964. Duplicate probes for pH-dependent ORFs are included in Table S1A in the supplemental material (all probes show significant pH effects on expression). The intergenic regions showing pH-dependent expression are tabulated in Table S1B in the supplemental material.
To determine which probes showed pH-dependent expression ratios, a model-based expression analysis was performed on the probe-level data from Affymetrix's CEL files using dChip software (31, 44). The model relates target RNA levels to the probe signals by a linear function that weights the significance of all oligonucleotide probes for each gene. The data from different arrays were normalized and rescaled for comparison. Each array was normalized to a baseline array from a pH 7 culture, using local regression on an invariant set of probes (54). Model-based expression indices were calculated for each gene on each array using only the perfect match probes.
For each of the three pH conditions, the data set included five biological replicates (independent with respect to B. subtilis growth, RNA isolation, sample preparation, and array hybridization). Global relationships among arrays were visualized by performing a principal components analysis (31, 44) on the expression data and plotting arrays in two-dimensional space corresponding to the first two principal components. To test for significant differences in expression between the pH classes, Statistical Analysis Software (SAS) was used to perform a one-way analysis of variance (ANOVA) on the log2-transformed model-based expression indices, on a gene-by-gene basis, at a significance level of 0.001 (31, 44). Assuming a gene with average within-group variability, our sample size (five replicates for each of three conditions; 4,351 probe sets) provided a statistical power of 98% to detect a twofold difference in gene expression among pH groups. For each gene, pair-wise contrasts of pH classes were conducted using Tukey's multiple comparisons procedure to control the family-wise error rate at 0.001.
Real-time quantitative reverse transcription-PCR.
Expression of mRNA for individual genes was quantified by real-time PCR using an ABI Prism 7500 DNA analyzer (Applied Biosystems) as described in reference 36. For each growth condition, three independent biological replicate cultures were tested. Primer Express Software v2.0 (Applied Biosystems) was used for primer design. The primers chosen amplified 50 to 70 bp segments of the target genes (see Table S2 in the supplemental material). The SYBR Green PCR one-step reverse transcription-PCR protocol (Applied Biosystems) was used, in which cDNA reverse transcription and PCR amplification occur in the same well. Nucleic acid concentrations were as follows: 0.1 nM forward primer, 0.1 nM reverse primer, and 50 ng target RNA. PCR cycling conditions were as follows: reverse transcription at 48°C for 30 min and 95°C for 10 min, 40 cycles of denaturation at 92°C for 15 s, and extension at 60°C for 1 min. For detection of primer dimerization or other artifacts of amplification, a dissociation curve was run immediately after completion of the real-time PCR. Individual gene expression profiles were normalized based on measurement of the original RNA sample amplified. All expression levels are presented relative to the expression at pH 7.0.
RESULTS
Growth response to pH shift.
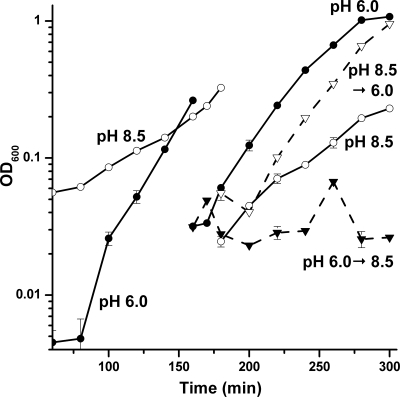
B. subtilis shows a lag in growth following a rapid pH increase (69), but its response to rapid acidification has not been reported. B. subtilis was cultured to early log phase at pH 8.5 or at pH 6.0, at which time the cultures were diluted into LBK buffered at pH 6.0 or at pH 8.5 (Fig. 1). Before dilution, the initial doubling times of the cultures at pH 8.5 and pH 6.0 were 42 min and 18 min, respectively. When the pH 8.5 cultures were transferred into pH 6.0 medium, there was a short lag in growth rate (<20 min). However, cultures that were transferred from pH 6.0 to pH 8.5 exhibited growth arrest for a period of several hours.
FIG. 1.
Acid and base shift growth curves. B. subtilis strain AG174 cultures grown overnight in unbuffered LBK were diluted 500-fold and grown at 37°C in LBK (50 mM HOMOPIPES) adjusted to pH 6.0 (•) or pH 8.5 (○) until an OD600 of 0.3. The culture at pH 6.0 was then diluted 10-fold into medium at pH 6.0 (•) and at pH 8.5 (▾). The original culture at pH 8.5 was diluted 10-fold into medium at pH 8.5 (○) and at pH 6.0 (▿). Diluted cultures were incubated for 2.5 h. The average of three biological replicates is shown for each condition. Error bars represent SEM.
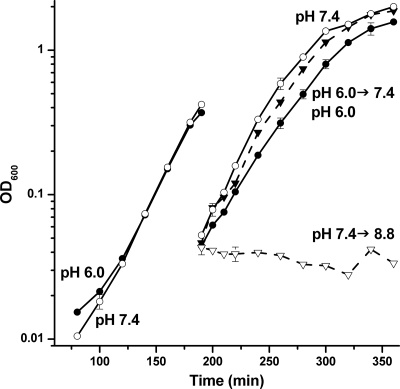
A similar lag in growth was observed when cultures were shifted from pH 7.4 to pH 8.8, although there was no lag when pH 6.0 cultures were shifted to pH 7.4 (Fig. 2). In order to control for the effect of the dilution on cell growth, cultures at pH 6.0, pH 7.4, and pH 8.5 were diluted into medium adjusted to the pH of the initial culture. There was no evidence that the dilutions without pH change significantly affected the growth rate of either the pH 6.0 or pH 7.4 cultures. Overall, B. subtilis responded to pH changes in a highly asymmetric manner: acid stress required much briefer adaptation than did base stress.
FIG. 2.
Neutral and base shift growth curves. B. subtilis strain AG174 overnight cultures in unbuffered LBK were diluted 500-fold into LBK buffered at pH 6.0 (•) and at pH 7.4 (○) and cultured until an OD600 of 0.3. The culture at pH 6.0 was then diluted 10-fold into medium at pH 7.4 (▾) and pH 6.0 (•). The culture at pH 7.4 was diluted 10-fold into medium at pH 8.8 (▿) and pH 7.4 (○). Diluted cultures were incubated for 2.5 h. The average of three biological replicates is shown; error bars represent the SEM.
Extreme acid survival and extreme base survival.
In Escherichia coli and other species, growth in a moderate acid or base enables the cell to survive under extreme acidic or basic conditions, respectively (24, 58). B. subtilis was tested for similar pH-dependent induction of acid resistance or base resistance. To maintain vegetative growth, cells were harvested in early log phase. For acid adaptation, B. subtilis was cultured at pH 6.0 to an OD600 of 0.2, and then the cultures were diluted into medium at pH 4.5. Log-phase cells grown at pH 6.0 showed 60 to 100% survival after 2 h of exposure at pH 4.5, whereas cells grown at pH 7.0 showed 5 to 15% survival. Thus, growth in moderate acid induced systems that increase survival in extreme acid.
For base resistance, B. subtilis was cultured at pH 9.0 to an OD600 of 0.2, and then cultures were diluted at pH 10.0. Log-phase cells grown at pH 9.0 showed 40 to 100% survival after 2 h at pH 10, whereas cells grown at pH 7.0 showed 1 to 5% survival. Thus, growth in a moderate base induced systems that increase survival in an extreme base.
Transcriptomic response to pH stress.
Previously, genetic responses to rapid pH shift have been reported (38, 69), but the global transcriptomic expression ratios during steady-state growth at different pH values have not been assessed. We observed differential gene expression as a function of pH for cultures following several generations of vegetative growth. B. subtilis was cultured to early log phase in buffered LBK adjusted to one of three pH values (pH 6.0, pH 7.0, and pH 9.0), a range of pHs in which cultures maintained consistent doubling times. The doubling time for pH 7.0 and pH 6.0 cultures was approximately 16 min, while the doubling time at pH 9.0 was 23 min. The cultures were grown to early log phase (OD600 = 0.2), and growth was logarithmic throughout this period.
Analysis of expression ratios.
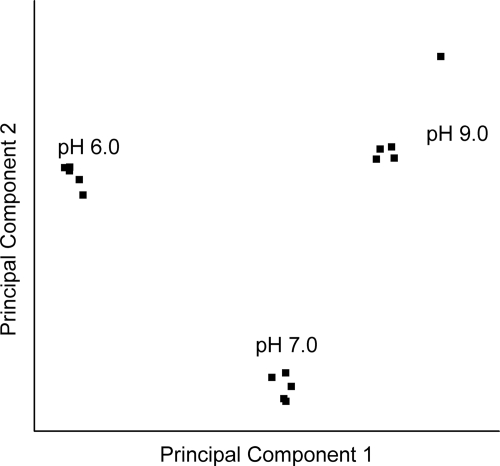
The cDNA from five independent cultures of each pH condition was hybridized to Affymetrix antisense B. subtilis arrays. Array data were deposited at the NCBI Gene Expression Omnibus (accession no. GSE11810). Global relationships among arrays were visualized by performing a principal components analysis on the expression data (Fig. 3). The array comparisons were plotted in two-dimensional space, corresponding to the first and second principal components of variation (44). The principal components of the array hybridizations from each of the three pH conditions fell into distinct groups. Within-class differences were small relative to differences among classes defined by growth pH.
FIG. 3.
Principal components analysis. The gene expression profiles of the arrays were visualized in two-dimensional Euclidian space, by using SAS software as described in Materials and Methods. The first and second principal components are shown.
To assess the statistical significance of individual gene expression ratios, we conducted a gene-by-gene ANOVA with Tukey's correction for false positives in a large data set. The expression indices for each gene among the three pH groups were compared using a one-way ANOVA at a significance level of 0.001. This means that approximately one false positive would be expected per 1,000 genes tested. Expression ratios were determined for all pairs of pH classes (pH 6.0/pH 7.0; pH 6.0/pH 9.0; pH 7.0/pH 9.0). Throughout our report, the three classes of expression ratios are presented using the quotient “acid/base,” so that the log2 value of the ratio is positive for expression increased at lower pH (decreased at higher pH) and negative for expression increased at higher pH (decreased at lower pH). Values presented are significant based on Tukey's test (P ≤ 0.001) (see Table S1A in the supplemental material). Of the approximately 4,095 known ORFs in B. subtilis, 289 showed expression ratios of twofold or greater (log2 ratio, ≥1) for one or more comparisons with decreasing pH (pH 6.0/pH 7.0, pH 7.0/pH 9.0, or pH 6.0/pH 9.0). Another 425 genes showed expression ratios of twofold or greater in the alkaline direction. All genes showing a significant expression ratio for one or more of the three pH comparisons are included in Table S1A in the supplemental material. Table S1B in the supplemental material presents probes from intergenic regions showing significant pH-dependent transcription (214 intergenic probe sets out of 606 on the array).
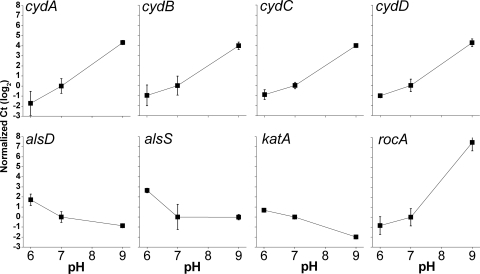
In the text (Table 1), we present the most strongly pH-dependent genes: those upregulated at least fourfold in acid or downregulated in base (pH 6.0 versus pH 9.0). The genes most strongly downregulated in acids or upregulated in bases are presented in Table 2. Tables 1 and 2 include only the promoter-proximal probe for each ORF, where duplicates were present on the GeneChip array. For eight genes of interest, expression ratios were confirmed using real-time PCR (Fig. 4). All genes tested by PCR showed expression ratios consistent with those observed in the arrays.
TABLE 1.
Genes upregulated by acid or downregulated by base (pH 6.0/pH 9.0 ratio is fourfold or greater)
| Gene | Characteristics | Log2 pH expression ratioa
|
||
|---|---|---|---|---|
| 6.0/9.0 | 6.0/7.0 | 7.0/9.0 | ||
| lrgB | Antiholin-like protein | 4.6 | 3.7 | 0.9 |
| cadA | Heavy metal cpx-type ATPase | 4.6 | 4.0 | 0.6 |
| lrgA | Antiholin-like protein | 4.5 | 3.4 | |
| yhbJ | Unknown | 4.4 | 1.5 | 3.0 |
| hxlA | 3-Hexulose-6-phosphate synthase | 4.2 | 2.3 | 1.9 |
| yhbI | MarR transcriptional regulator homolog | 4.2 | 1.5 | 2.7 |
| yhcA | Multidrug resistance protein homolog | 4.1 | 1.5 | 2.6 |
| ydeQ | NAD(P)H oxidoreductase homolog | 4.0 | 1.6 | 2.4 |
| hxlB | 6-Phospho-3-hexuloisomerase | 3.9 | 2.3 | 1.6 |
| yhcB | trp repressor binding protein homolog | 3.4 | 1.3 | 2.2 |
| alsS | Alpha-acetolactate synthase | 3.3 | 3.0 | |
| katA | Vegetative catalase 1 | 3.2 | 1.2 | 2.1 |
| ynfC | Unknown | 3.1 | 1.9 | 1.2 |
| alsD | Alpha-acetolactate decarboxylase | 3.1 | 2.5 | 0.6 |
| czcO | Potassium and divalent cation transport | 3.0 | 3.1 | |
| yhcC | Unknown | 2.8 | 1.2 | 1.6 |
| namA | Possible NADPH dehydrogenase of phenolics | 2.7 | 1.5 | 1.2 |
| czcD | Cadmium, cobalt, and zinc/H+/K+ antiporter | 2.6 | 2.5 | |
| yorE | Unknown | 2.6 | 2.4 | |
| yorF | Unknown | 2.5 | 2.2 | |
| hup2 | SPBc2 prophage-derived DNA-binding protein HU 2 | 2.3 | 1.7 | 0.6 |
| maeN | Na+/malate symporter | 2.3 | -2.6 | 4.9 |
| yrpC | Glutamate racemase homolog | 2.3 | 2.0 | |
| yorB | Unknown | 2.2 | 1.6 | 0.6 |
| maeA | Malate dehydrogenase homolog | 2.2 | 2.7 | |
| gspA | General stress protein | 2.2 | 2.6 | |
| ywjC | Unknown | 2.1 | 2.4 | |
| yrhE | Formate dehydrogenase homolog | 2.1 | −1.1 | 3.3 |
| yflT | General stress protein, 17 M | 2.1 | 2.5 | |
| yotJ | Unknown | 2.1 | 2.3 | |
| ydgA | Unknown | 2.0 | 1.0 | 1.0 |
| ybyB | Unknown | 2.0 | 2.5 | |
| ydfE | Unknown | 2.0 | 1.7 | |
Significant values are shown (P ≤ 0.001).
TABLE 2.
Genes downregulated by acid or upregulated by base (pH 6.0/pH 9.0 ratio is down by fourfold or greater)
| Gene | Characteristics | Log2 pH expression ratioa
|
||
|---|---|---|---|---|
| 6.0/9.0 | 6.0/7.0 | 7.0/9.0 | ||
| rocA | Arginine catabolism | −6.0 | −1.4 | −4.6 |
| cydA | Cytochrome bd ubiquinol oxidase | −5.9 | −5.6 | |
| pstS | Phosphate ABC transporter | −5.1 | −4.7 | |
| cydB | Cytochrome bd ubiquinol oxidase | −4.9 | −4.8 | |
| yrkO | Unknown | −4.9 | −4.7 | |
| pstBA | Phosphate ABC transporter | −4.7 | −4.5 | |
| cydD | ABC transporter | −4.7 | −4.5 | |
| ctaO | Heme O synthase activity | −4.4 | −1.3 | −3.1 |
| ldh | l-Lactate dehydrogenase | −4.3 | −1.0 | −3.3 |
| cydC | ABC transporter | −4.2 | −4.0 | |
| pstBB | Phosphate ABC transporter | −4.1 | −3.8 | |
| yhaT | Unknown | −4.1 | −0.6 | −3.5 |
| yrkN | Unknown | −4.1 | −3.9 | |
| yrkA | Hemolysin-like homolog | −4.1 | −1.5 | −2.6 |
| ykuO | Unknown | −4.0 | −1.8 | −2.3 |
| ykuN | Flavodoxin homolog | −3.9 | −1.7 | −2.2 |
| rocG | Glutamate dehydrogenase | −3.8 | −1.2 | −2.6 |
| ykuP | Flavodoxin homolog | −3.8 | −1.5 | −2.3 |
| fruB | Fructose 1-phosphate kinase | −3.8 | ||
| yhaS | Unknown | −3.8 | −0.9 | −2.9 |
| pstA | Phosphate ABC transporter | −3.7 | −3.4 | |
| yhaU | Na+/H+ antiporter homolog | −3.7 | −0.3 | −3.4 |
| rocB | Arginine catabolism | −3.6 | −3.3 | |
| nasD | Assimilatory nitrite reductase | −3.6 | −0.6 | −3.0 |
| fruA | PTS fructose-specific IIABC component | −3.6 | −2.2 | |
| pstC | Phosphate ABC transporter | −3.5 | −3.1 | |
| ywcJ | Nitrite transporter homolog | −3.4 | −0.9 | −2.5 |
| ycnI | Unknown | −3.4 | −1.0 | −2.4 |
| ycnK | DeoR transcription regulator homolog | −3.4 | −1.2 | −2.2 |
| ycnJ | Copper export protein homolog | −3.4 | −1.1 | −2.2 |
| fruR | Transcriptional repressor of the fructose operon | −3.3 | −1.8 | |
| ydbN | Unknown | −3.3 | −1.5 | −1.8 |
| psiE | Predicted phosphate starvation-inducible protein | −3.2 | −3.6 | |
| dhbC | Isochorismate synthase | −3.1 | −1.3 | −1.8 |
| fnr | Transcriptional regulator of anaerobic genes | −3.0 | −1.2 | −1.9 |
| lctP | l-Lactate permease | −3.0 | −0.9 | −2.2 |
| ysfC | Glycolate oxidase subunit homolog | −3.0 | −0.4 | −2.6 |
| yrkP | Two-component response regulator (YrkQ) homolog | −2.9 | −3.0 | |
| ykoM | MarR transcriptional regulator homolog | −2.9 | −1.0 | −1.9 |
| dhbF | Involved in 2,3-dihydroxybenzoate biosynthesis | −2.9 | −0.7 | −2.2 |
| yerA | Adenine deaminase homolog | −2.8 | −2.3 | |
| rapK | Response regulator aspartate phosphatase | −2.8 | −2.9 | |
| dhbE | 2,3-Dihydroxybenzoate-AMP ligase | −2.8 | −1.0 | −1.8 |
| bglH | Beta-glucosidase | −2.7 | −2.1 | |
| dhbA | 2,3-Dihydro-2,3-dihydroxybenzoate dehydrogenase | −2.7 | −1.3 | −1.5 |
| bglP | PTS beta-glucoside-specific enzyme IIBCA component | −2.7 | −2.0 | |
| gltA | Glutamate synthase | −2.6 | −1.4 | −1.3 |
| yrkQ | Two-component sensor histidine kinase (YrkP) homolog | −2.6 | 0.3 | −2.9 |
| ysfD | Glycolate oxidase subunit homolog | −2.6 | −2.6 | |
| liaI | Putative membrane protein | −2.6 | −0.9 | −1.8 |
| ycdA | Unknown | −2.6 | −2.8 | |
| ytkA | Unknown | −2.6 | −1.3 | −1.3 |
| ywrJ | Unknown | −2.6 | −0.5 | −2.1 |
| dhbB | Isochorismatase | −2.6 | −1.1 | −1.5 |
| gltB | Glutamate synthase | −2.6 | −1.3 | −1.3 |
| yrhP | Efflux protein homolog | −2.6 | −0.4 | −2.1 |
| yybP | Unknown | −2.5 | −2.6 | |
| cwlS | Cell wall lytic enzyme associated with cell separation | −2.5 | −0.4 | −2.1 |
| yxeL | Unknown | −2.5 | −2.2 | |
| liaH | Phage shock protein A homolog | −2.5 | −0.7 | −1.8 |
| htpX | Probable protease homolog | −2.5 | −1.1 | −1.4 |
| yqgY | Unknown | −2.5 | −2.0 | −0.5 |
| ywoF | Unknown | −2.5 | −1.0 | −1.4 |
| xkdN | PBSX prophage | −2.4 | −2.8 | |
| blt | Multidrug-efflux transporter | −2.4 | 0.4 | −2.9 |
| yqaT | Phage-related terminase homolog | −2.4 | −0.6 | −1.8 |
| bltD | Spermine/spermidine acetyltransferase | −2.4 | 0.7 | −3.1 |
| qcrA | Menaquinol:cytochrome c oxidoreductase | −2.4 | −1.5 | −0.9 |
| qcrC | Menaquinol:cytochrome c oxidoreductase (bc subunit) | −2.4 | −1.5 | −0.9 |
| yuiI | Unknown | −2.3 | −1.7 | −0.6 |
| xkdM | PBSX prophage | −2.3 | −2.8 | |
| pspA | Phage shock protein A homolog | −2.3 | −2.2 | |
| spoVG | Required for spore cortex synthesis | −2.3 | −0.5 | −1.8 |
| ylqB | Unknown | −2.3 | −0.9 | −1.4 |
| ykoY | Similar to toxic anion resistance protein | −2.3 | −0.5 | −1.8 |
| rocD | Ornithine aminotransferase | −2.3 | 1.1 | −3.3 |
| yfmQ | Unknown | −2.2 | −0.8 | −1.4 |
| ybbA | Unknown | −2.2 | −1.0 | −1.2 |
| bdbA | Thiol-disulfide oxidoreductase | −2.2 | −1.2 | −0.9 |
| sdpB | Sporulation-delaying protein | −2.2 | −2.0 | |
| sboA | Subtilisin A | −2.1 | −1.9 | |
| ctaE | Cytochrome caa3 oxidase | −2.1 | −0.8 | −1.3 |
| xhlB | Hydrolysis of 5-bromo 4-chloroindolyl phosphate | −2.1 | −2.5 | |
| xhlA | Involved in cell lysis upon induction of PBSX | −2.1 | −2.6 | |
| ctaC | Cytochrome caa3 oxidase | −2.1 | −1.0 | −1.1 |
| fosB | Metallothiol transferase | −2.1 | −1.8 | −0.3 |
| phrK | Phosphatase (RapK) regulator | −2.1 | −2.1 | |
| feuB | Iron-uptake system | −2.1 | −1.2 | −0.9 |
| yxeK | Monooxygenase homolog | −2.1 | −1.8 | |
| ydjP | Chloroperoxidase homolog | −2.0 | −1.9 | |
| yxiE | Unknown | −2.0 | −1.1 | −1.0 |
| yvdS | Molecular chaperone homolog | −2.0 | −0.6 | −1.5 |
| tcyP | l-Cysteine uptake | −2.0 | −0.5 | −1.5 |
| feuC | Iron-uptake system | −2.0 | −1.2 | −0.8 |
| yosU | Unknown | −2.0 | −1.4 | |
| yydI | ABC transporter homolog | −2.0 | −0.7 | −1.3 |
| feuA | Iron-uptake system | −2.0 | −1.1 | −0.9 |
| ykoX | Alkaline phosphatase homolog | −2.0 | −1.0 | −0.9 |
| xkdF | PBSX prophage | −2.0 | −2.5 | |
| yuaF | Unknown | −2.0 | −1.1 | −0.9 |
Significant values are shown (P ≤ 0.001).
FIG. 4.
Real-time PCR expression ratios of selected genes as a function of external pH. B. subtilis strain AG174 overnight cultures in unbuffered LBK were diluted 500-fold into LBK buffered with 50 mM HOMOPIPES at pH 6.0, at pH 7.0, and at pH 9.0. RNA was isolated, and mRNA expression for individual genes was quantified by real-time PCR using an ABI Prism 7500 DNA analyzer (Applied Biosystems) with SYBR Green one-step protocol. All expression levels are presented relative to the expression at pH 7.0. Error bars represent the SEM (n = 3).
Catabolism and respiration.
In E. coli and other bacteria, pH regulates catabolism and respiration to consume acids at low pH and produce acids at high pH (44, 48, 57). We sought to determine whether a similar pattern of metabolism appears in B. subtilis. Table 3 summarizes enzymes involved in catabolism or respiration that showed pH-dependent expression ratios of twofold or greater (pH 6.0 versus pH 9.0). Growth at pH 6 upregulated acetoin production (alsDS), a pathway known to shunt fermentation into neutral products, minimizing acid production (1, 32, 37, 55). Upregulation of alsSD at low external pH was confirmed by real-time PCR (Fig. 4). Acid also upregulated a large number of NAD(P)-dependent dehydrogenases: alcohol dehydrogenase (adhA), alanine dehydrogenase (ald), succinate-semialdehyde dehydrogenase (gabD), and several putative formate dehydrogenases (fdhD, yjgC, yrhE, and yrhG). These enzymes are capable of removing acidity through NAD(P)H which transfers electrons to the electron transport system (ETS) and pumps protons out of the cell. In addition, acid upregulated phosphatidylserine decarboxylase (psd) and arginine decarboxylase (speA) as well as decarboxylase homologs (gcvPA and maeA). In other organisms, CO2 production removes acid from the cytoplasm (48, 57).
TABLE 3.
Catabolism-related genes showing twofold or greater pH dependence (pH 6.0/pH 9.0 expression ratios)
| Gene(s) | Characteristics | Reaction or pathway (known or putative) |
|---|---|---|
| Acid-upregulated | ||
| adhA | Alcohol dehydrogenase | Alcohol + NADP+ ⇌ aldehyde + NADPH |
| ald | l-Alanine dehydrogenase | Alanine + NAD+ ⇌ pyruvate + NADH + NH4+ |
| alsDS, ilvH | 2-Acetolactate synthase, 2-acetolactate decarboxylase | Pyruvate → acetolactate + CO2 → acetoin + 2CO2 |
| cypE | NADPH-cytochrome P450 reductase | ETS |
| fdhD, yjgC, yrhE, and yrhG | Formate dehydrogenase homologs | HCO2− + NAD+ → CO2 + NADH |
| gabD | Succinate-semialdehyde dehydrogenase | Succinic semialdehyde + glutamate → succinate |
| gcvPA | Probable glycine decarboxylase | Glycine + H-protein-lipoyllysine → H-protein-S-aminomethyldihydrolipoyllysine + CO2 |
| hxlA, hxlB, hxlR | 3-Hexulose 6-phosphate synthase | d-Ribulose 5-phosphate + formaldehyde ⇌ d-arabino-3-hexulose 6-phosphate |
| maeA | Malate decarboxylase homolog | Malate + NAD+ → pyruvate + NADH + CO2 |
| psd, pssA | Phosphatidylserine decarboxylase | CDP-diacylglycerol + l-serine → l-1-phosphatidyl-ethanolamine + CO2 + CMP |
| speA | Arginine decarboxylase | Arginine → agmatine + CO2 |
| ycdF, ycdG | Glucose 1-dehydrogenase and oligo-1,6-glucosidase homologs | d-Glucose + NAD(P)+ → d-glucono-1,5-lactone + NAD(P)H |
| ydaD | Alcohol dehydrogenase homolog | Alcohol + NAD(P)+ → aldehyde + NAD(P)H + H+ |
| ytbE | Plant metabolite dehydrogenase homolog | |
| yvfV | Glycolate oxidase homolog | Glycolate → glyoxylate |
| Base-upregulated | ||
| acuABC | Acetoin dehydrogenase | 2-Acetolactate → acetoin + CO2 |
| amyX, bglS | Pullulanase and glucanase | Glucan catabolism |
| atpA, atpC, atpD, atpI | ATP synthase | H+(out) + P + ADP = H+(in) + H2O + ATP |
| azoR2 | NAD(P)H dehydrogenase (quinone) homolog | ETS |
| ctaA, ctaC, ctaD, ctaE, ctaF | Cytochrome caa3 oxidase | ETS |
| cydABCD | Cytochrome bd ubiquinol oxidase | ETS |
| fruA, fruB | Fructose l-phosphate kinase | Glycolysis |
| gltAB | Glutamate synthase | l-Glutamine + α-ketoglutarate → 2 l-glutamate |
| idh, iolB, iolD, iolH, iolI | myo-Inositol catabolism | myo-Inositol → dihydroxyacetone phosphate + acetyl-coenzyme A + CO2 |
| lacA | Beta-galactosidase | Lactose → β-d-glucose + β-d-galactose |
| ldh, lctP | l-Lactate dehydrogenase, permease | Pyruvate → l-lactate |
| mccA, mccB | Methionine catabolism to cysteine | Methionine → cysteine → pyruvate |
| mmgD | Propionyl-coenzyme A metabolism | Propionyl-coenzyme A → succinate (trichloroacetic acid cycle) |
| narG, narH, narI | Nitrate reductase | 2 Nitrate → 2 nitrite → N2 |
| rocA, rocB, rocDEF, rocG | Arginine catabolism | l-Arginine → l-glutamate + 2NH4+ |
| treA, treP | Trehalose-6-phosphate hydrolase | Trehalose catabolism |
| xlyA, xhlA, xhlB | N-Acetylmuramoyl-l-alanine amidase; cell wall lysis | Peptidoglycan catabolism |
| ybcF | Carbonic anhydrase homolog | CO2 + H2O → HCO3− + H+ |
| ydbM | Butyryl-CoA dehydrogenase homolog | Butyrate catabolism |
| yisK | Pentene tricarboxylic acid decarboxylase homolog | |
| yrbE | Oxidoreductase homolog |
High external pH upregulated the entire roc operon (Tables 2 and 3), which encodes enzymes that are part of a three-step arginase pathway responsible for converting arginine to glutamic acid and trichloroacetic acids (2, 11). Confirmed by real-time PCR, the upregulation of rocA at high pH was over 200-fold (Fig. 4). High pH also upregulated gltAB (Table 2), which encodes glutamate synthase (5). Several enzymes that could generate acids were upregulated, including l-lactate dehydrogenase (ldh), myo-inositol catabolism (icd), and sugar chain catabolism (amyX, bglS, and fruAB).
High pH upregulated several membrane-bound complexes involved in electron transport and proton transfer across the membrane. The cytochrome d oxidase subunits (cydABCD) (39, 71) were among the most highly base-dependent genes (Table 2), and all upregulated greater than 16-fold (log2 values, >4), confirmed by real-time PCR (Fig. 4). The cytochrome c oxidase genes (ctaACE) (53) were upregulated in bases to a lesser extent, less than twofold in most cases (see Table S1 in the supplemental material). The cytochrome d complex, with greater oxygen affinity, minimizes proton transport out of the cell. High pH also upregulated the cytochrome bc complex (menaquinone:cytochrome c reductase) qcrC (75) (Table 2).
Oxidative stress and metal transport.
In E. coli, several acid stress genes overlap with those involved in oxidative stress (44). In B. subtilis, 87 genes that are known to be induced by oxidative stress were pH dependent, and 75% of those genes were upregulated in acidic conditions compared to those in both pH 7 and pH 9 (see Table S1 in the supplemental material). For example, the genes katA and namA (yqjM) were highly upregulated in acidic conditions (Table 1). The gene katA encodes the enzyme catalase (25), which protects the cell from oxidative stress by catalyzing the decomposition of hydrogen peroxide to water and oxygen. The gene namA encodes an antioxidant enzyme of the “old yellow enzyme” (OYE) family, induced by oxidative stress (23).
Low pH increases the solubility of various metals, allowing those metals to cross the bacterial membrane and resulting in undesirable redox reactions (27, 34, 66). The metal transporters cadA and czcDO were strongly induced by acids (Table 2). The gene cadA encodes a cpx-type ATPase, which exports toxic metals such as Cd(II), Zn(II), and Co(II) (27, 59), and the czcDO operon encodes an antiporter that exchanges heavy metals for K+ (47).
Transport of inorganic ions and polyamines.
The proton-translocating ATP synthase operon showed many genes upregulated in basic conditions; although the amount of increase was less than twofold, the values were significant and observed for virtually the entire operon (see Table S1A in the supplemental material). High-pH upregulation of ATPase has been shown for several other species (see Discussion).
In many bacteria, pH homeostasis depends on ion transport, particularly Na+ and K+. In B. subtilis, at high pH, pH homeostasis involves cation/proton antiporters (50). These antiporters allow for the acidification of the cytoplasm by exchanging cations such as Na+ and K+ for protons. After alkaline shock, B. subtilis induces the K+/H+ antiporter yhaTU (26, 69). In our arrays, yhaTU showed similar upregulation during log-phase growth at pH 9.0 (Table 2). Another mediator of Na+ flux is tetL, whose deletion decreases alkaline tolerance (14, 68). Mutants containing the tetL deletion repress maeN (62), which encodes a sodium-coupled malate transporter (68). We found maeN strongly upregulated in acids and downregulated in bases.
Another source of base stress in E. coli is that polyamines can be deprotonated (neutralized) at high pH, enabling them to cross the cytoplasmic membrane as permeant weak bases, which results in growth inhibition (74). Thus, at high external pH, the upregulation of polyamine efflux pumps decreases polyamine toxicity. In B. subtilis, the export of spermine and spermidine is initiated by polyamine acetylation (72). We found the spermine/spermidine transporter and acetyltransferase (blt and bltD) to be upregulated strongly at high pH (Table 2).
Stress regulons.
Several global regulons governed by alternate sigma factors showed pH dependence. The general stress regulon SigB (51) is known to be induced after a mild acid shock (38). In our arrays, we found that 61% of the genes under SigB regulation were pH dependent but that various genes showed divergent regulation by acidic or basic conditions (see Table S1C in the supplemental material).
Base shock is known to induce the SigW extracytoplasmic function regulon, controlling functions involving the secretion or uptake of macromolecules or ions (33, 69). The SigW regulon is activated by proteolytic destruction of the membrane-bound anti-SigW in response to antimicrobial peptides and other agents that damage the cell envelope (22). We found the SigW regulon to be upregulated during steady-state log-phase growth at pH 9.0 compared to that at low pH (see Table S1D in the supplemental material).
Components of the SigH (17) and SigL (15) regulons showed increasing expression at higher pH (see Table S1E in the supplemental material). SigH is essential for sporulation (30) and is known to be downregulated by acids (17, 41). On the other hand, acid upregulated the SigX regulon, which responds to extracytoplasmic stress, conferring resistance to positively charged antimicrobial peptides (12). Several other sigma regulons (SigF, -G, -M, and -Y) showed no consistent pattern of pH response.
The transcriptional regulator Fnr is an integral part of the regulatory cascade required for the adaption of B. subtilis to low oxygen tension (52). Of the 44 genes in the Fnr regulon, 74% were found to be pH dependent. Fnr controls four groups of differentially expressed genes (52), of which genes in groups 1, 2, and 3 were found to be strongly pH dependent (see Table S1F in the supplemental material). With the exception of the alsSD operon and ywcJ, virtually all of the genes under Fnr regulation were induced by base.
The flagellar motility and chemotaxis regulon, including the fla-che operon and flg and fli genes, is under the control of the DegS-DegU two-component regulator, which induces motility in early starvation phase (3). We found most of the motility genes to be upregulated significantly at pH 9 (see Table S1A in the supplemental material).
DISCUSSION
Growth and survival.
In log-phase cultures of B. subtilis, a shift in pH from pH 7.5 to pH 8.8 caused a prolonged lag in growth, consistent with a previous report of base shock (69). We observed little or no growth lag after an acid shift of comparable magnitude (Fig. 1). Thus, for the most part, genes expressed at high external pH allow the ability to grow as well at low pH. The lag period after a base shift, however, suggests that a greater number of genes upregulated at high pH are essential for growth than those upregulated at low pH. In fact, a greater number of genes was found to be upregulated by a base compared to that with an acid.
Near either end of the pH range, growth may induce components that allow survival at more extreme pHs. When grown in moderate acid, the food-borne pathogen B. cereus induces an acid tolerance response; that is, enhanced survival of nongrowing cells in an extreme acid (64), a response requiring protein neosynthesis. We observed a comparable acid adaptation in B. subtilis. In addition, we observed a comparable response of base adaptation. We expected growth in a mild acid or base to upregulate genes needed for extreme-pH survival.
Catabolism and respiration.
Our array analysis showed pH-dependent expression of numerous components of catabolism and respiration that could be consuming acids at low pH or generating acids at high pH (Table 3). The NAD(P)-dependent dehydrogenases upregulated by acid could increase the transfer of electrons through the electron transport chain, pumping protons out of the cell to help maintain internal pH homeostasis. Certain decarboxylases such as ilvH (acetolactate decarboxylase) may also contribute by consuming acids in conjunction with CO2 export. Previously, oxalate decarboxylase (oxdC) was found to be upregulated by acids (43, 63). Our arrays show that oxdC is upregulated by acids relative to that at pH 7 but also show high-pH upregulation compared to that at pH 7 (see Table S1A in the supplemental material).
The overall acetoin biosynthesis pathway (alsSD) was strongly upregulated at low pH. The alsSD pathway is induced by acetate (55), whose accumulation acidifies the growth medium. Acetoin production may in fact be triggered by low pH, independent of the presence of acetate.
Arginine catabolism (roc) is upregulated in the alkaline-sensitive B. subtilis strain JC112 (Δ tetL) (68), and in our array analysis, roc was strongly upregulated at high pH. The conversion of arginine to glutamate with the export of ammonia, facilitated by arginine permeases (rocC and rocE), provides a means to acidify the cytoplasm. Thus, arginine uptake and breakdown to acids could help counteract base stress (68). The increase of arginine catabolism at high pH differs from the acid upregulation of arginine catabolism in other species, such as the arc operon of Streptococcus spp. (42) and the adi arginine decarboxylase of E. coli (57). Both the arc and adi pathways, however, produce ornithine rather than acids; thus, they generate alkaline products within the cytoplasm, which would counteract acidity.
In B. subtilis, several cytochrome oxidase complexes were upregulated at higher pHs. The most strongly base-dependent complex (over 16-fold upregulation at pH 9 compared to that at pH 6) was cyd, which minimizes proton export. For a comparison, E. coli minimizes proton export at high external pH by upregulating the terminal cytochrome bd oxidase cyd (44), which exports half as many protons per electron as does the downregulated cyo oxidase (18). In B. subtilis, base also upregulated both ctaACE and qcrC oxidases, which translocate additional protons across the cytoplasmic membrane. However, the upregulation of these pumps was only about twofold, compared to 16-fold for cyd. Thus, B. subtilis did show substantial preference for cyd over the proton-pumping oxidase complexes at high pHs.
The ATP synthase operon (atp) as a whole was upregulated about twofold at pH 9 compared to that at pH 6. This finding is consistent with the acid upregulation (or base downregulation) of ATP synthase observed in other respiratory bacteria (see Tables S1G and H in the supplemental material), including E. coli (44), Mycobacterium tuberculosis (29), Staphylococcus aureus (8), and Desulfovibrio vulgaris (61). The increased ATP synthase production has been proposed to compensate for the decreased proton motive force at high external pH, where the ΔpH is inverted (44).
Metals and oxidative stress.
The acidification of the external environment increases the solubility of metals, which allows the metals to be transported into the cytoplasm where they may accumulate to toxic levels. For example, excess metals can inhibit transport systems and can disrupt cellular membrane integrity (49). In order to resist metal toxicity, bacteria efflux the toxic metal out of the cell (21). We found that two such metal transport systems, cadA and czcDO, were highly upregulated under acidic conditions (Table 1). Both systems transport metals out of the cytoplasm, although cadA is a cpx-type ATPase, whereas czc acts as a K+/metal antiporter.
The upregulation of czcDO at low pH may have other benefits independent of metal efflux. In acidophiles, K+ plays a role in pH homeostasis by producing an inside-positive electrochemical potential, which would inhibit the influx of protons using a chemiosmotic barrier against the proton gradient (4). K+ transporters are also upregulated by acid in E. coli (36).
Several of the genes we found upregulated at low pH, such as katA and namA, are known to respond to oxidative stress (23, 25). A similar overlap between low pH and oxidative stress is found in E. coli (44). Low pH may increase the production rates of reactive oxygen species, such as those of the iron-dependent Fenton reaction (34, 66).
Motility and chemotaxis.
The flagellar motility and chemotaxis regulon fla-che, under the control of DegS-DegU (3), was upregulated at pH 9 (see Table S1A in the supplemental material). Enhancement of motility at high pH could be part of the DegU-dependent motility peak during late-log-phase growth. The upregulation of motility genes at high pH is not seen in other gram-positive bacteria (see Table S1G in the supplemental material) and contrasts with the low-pH enhancement of motility in E. coli (44).
Comparison with other species.
Now that pH-dependent transcriptomes have been compiled for various bacteria, both gram negative and gram positive, some broader comparisons may be made. We tabulated shared orthologs from our study of B. subtilis with pH-dependent transcriptomes reported for E. coli (44) (see Table S1A in the supplemental material), acid stress studies of M. tuberculosis (29), S. aureus (8), and Agrobacterium tumefaciens (76) (see Table S1G in the supplemental material), and base stress studies of Desulfovibrio vulgaris (61) and Shewanella oneidensis (40) (see Table S1H in the supplemental material). The most widespread feature shared by B. subtilis with four other species was the increased expression of ATP synthase with increasing pH. Thus, the upregulation of ATP synthase appears to be a widespread adaptation of neutralophiles to growth at higher pH, where the proton motive force may be low. Another recurring preference at higher pH appears to be the non-proton-pumping cytochrome bd oxidase, seen in B. subtilis, E. coli, and A. tumefaciens.
In comparison with that in E. coli (44), catabolism and proton transport in both species are regulated so as to minimize acid production at low pH or maximize acid production at high pH, but the specific components differ between the two species. At low pH, E. coli expresses several amine-generating amino acid decarboxylases (gad, cad, and adi). In B. subtilis, acids upregulated fewer decarboxylases but a greater number of dehydrogenases than in E. coli. At high pH, E. coli expresses deaminases for tryptophan (tna) and serine (sda), which generate fermentation acids. In B. subtilis, sda was upregulated, but the strongest response was the roc operon for arginine catabolism at high pH.
With S. aureus (8), B. subtilis shared acid upregulation of several metabolic enzymes and oxidative stress proteins, such as isocitrate dehydrogenase (icd), and catalase (katA). With S. oneidensis (40), B. subtilis shared base upregulation of cysteine synthase A (cysK) and superoxide dismutase (sodA). Overall, our study of the pH-dependent transcriptome in B. subtilis adds a dimension to the growing picture of pH response in bacteria.
Supplementary Material
Acknowledgments
We thank Melanie Berkmen for providing the strains and Alan Grossman and Terry Krulwich for helpful discussions.
This work was supported by NIH AREA award no. R15 GM079731-01.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ali, N. O., J. Bignon, G. Rapoport, and M. Debarbouille. 2001. Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis. J. Bacteriol. 183:2497-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, N. O., J. Jeusset, E. Larquet, E. L. Cam, B. Belitsky, A. L. Sonenshein, T. Msadek, and M. Debarbouille. 2003. Specificity of the interaction of RocR with the rocG-rocA intergenic region in Bacillus subtilis. Microbiology 149:739-750. [DOI] [PubMed] [Google Scholar]
- 3.Amati, G., P. Bisicchia, and A. Galizzi. 2004. DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J. Bacteriol. 186:6003-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker-Austin, C., and M. Dopson. 2007. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 15:165-171. [DOI] [PubMed] [Google Scholar]
- 5.Belitsky, B. R., and A. L. Sonenshein. 2004. Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase. J. Bacteriol. 186:3399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhagwat, A. A. 2003. Regulation of the glutamate-dependent acid-resistance system of diarrheagenic Escherichia coli strains. FEMS Microbiol. Lett. 227:39-45. [DOI] [PubMed] [Google Scholar]
- 7.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bore, E., S. Langsrud, Ø. Langsrud, T. M. Rode, and A. Holck. 2007. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153:2289-2303. [DOI] [PubMed] [Google Scholar]
- 9.Browne, N., and B. C. A. Dowds. 2002. Acid stress in the food pathogen Bacillus cereus. J. Appl. Microbiol. 92:404-414. [DOI] [PubMed] [Google Scholar]
- 10.Butcher, B. G., and J. D. Helmann. 2006. Identification of Bacillus subtilis σW-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol. Microbiol. 60:765-782. [DOI] [PubMed] [Google Scholar]
- 11.Calogero, S., R. Gardan, P. Glaser, J. Schweizer, G. Rapoport, and M. Debarbouille. 1994. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J. Bacteriol. 176:1234-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao, M., and J. D. Helmann. 2004. The Bacillus subtilis extracytoplasmic-function σx regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186:1136-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, L., R. Grant, and A. Aronson. 2001. Regulation of the packaging of Bacillus thuringiensis δ-endotoxins into inclusions. Appl. Environ. Microbiol. 67:5032-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng, J., A. A. Guffanti, W. Wang, T. A. Krulwich, and D. H. Bechhofer. 1996. Chromosomal tetA(L) gene of Bacillus subtilis: regulation of expression and physiology of a tetA(L) deletion strain. J. Bacteriol. 178:2853-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi, S. K., and M. H. Saier. 2005. Regulation of sigL expression by the catabolite control protein CcpA involves roadblock mechanism in Bacillus subtilis: potential connection between carbon and nitrogen metabolism. J. Bacteriol. 187:6856-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cosby, W. M., D. Vollenbroich, O. H. Lee, and P. Zuber. 1998. Altered srf expression in Bacillus subtilis resulting from changes in culture pH is dependent on the Spo0K oligopeptide permease and the ComQX system of extracellular control. J. Bacteriol. 180:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosby, W. M., and P. Zuber. 1997. Regulation of Bacillus subtilis σH (spo0H) and AbrB in response to changes in external pH. J. Bacteriol. 179:6778-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotter, P. A., V. Chepuri, R. B. Gennis, and R. P. Gunsalus. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalma-Weiszhausz, D. D., J. Warrington, E. Y. Tanimoto, and C. G. Miyada. 2006. The Affymetrix GeneChip platform: an overview. Methods Enzymol. 410:3-28. [DOI] [PubMed] [Google Scholar]
- 21.Dopson, M., C. Baker-Austin, P. R. Koppineedi, and P. L. Bond. 2003. Growth in sulfidic mineral environments: metal resistance mechanisms in acidophilic micro-organisms. Microbiology 149:1959-1970. [DOI] [PubMed] [Google Scholar]
- 22.Ellermeier, C. D., and R. Losick. 2006. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 20:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzpatrick, T. B., N. Amrhein, and P. Macheroux. 2003. Characterization of yqjM, an old yellow enzyme homolog from Bacillus subtilis involved in the oxidative stress response. J. Biol. Chem. 278:19891-19897. [DOI] [PubMed] [Google Scholar]
- 24.Foster, J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898-907. [DOI] [PubMed] [Google Scholar]
- 25.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and per genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujisawa, M., M. Ito, and T. A. Krulwich. 2007. Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity. Proc. Natl. Acad. Sci. USA 104:13289-13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaballa, A., and J. D. Helmann. 2003. Bacillus subtilis cpx-type ATPases: characterization of Cd, Zn, Co and Cu efflux systems. Biometals 16:497-505. [DOI] [PubMed] [Google Scholar]
- 28.Gajiwala, K. S., and S. K. Burley. 2000. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605-612. [DOI] [PubMed] [Google Scholar]
- 29.Golby, P., K. A. Hatch, J. Bacon, R. Cooney, P. Riley, J. Allnutt, J. Hinds, J. Nunez, P. D. Marsh, R. G. Hewinson, and S. V. Gordon. 2007. Comparative transcriptomics reveals key gene expression differences between the human and bovine pathogens of the Mycobacterium tuberculosis complex. Microbiology 153:3323-3336. [DOI] [PubMed] [Google Scholar]
- 30.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayes, E. T., J. C. Wilks, P. Sanfilippo, E. Yohannes, D. P. Tate, B. D. Jones, M. D. Radmacher, S. S. BonDurant, and J. L. Slonczewski. 2006. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 6:89-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hornbaek, T., M. Jakobsen, J. Dynesen, and A. K. Nielsen. 2004. Global transcription profiles and intracellular pH regulation measured in Bacillus licheniformis upon external pH upshifts. Arch. Microbiol. 182:467-474. [DOI] [PubMed] [Google Scholar]
- 33.Huang, X., A. Gaballa, M. Cao, and J. D. Helmann. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function σ factor, σW. Mol. Microbiol. 31:361-371. [DOI] [PubMed] [Google Scholar]
- 34.Hug, S. J., and O. Leupin. 2003. Iron-catalyzed oxidation of arsenic(III) by oxygen and by hydrogen peroxide: pH-dependent formation of oxidants in the Fenton reaction. Environ. Sci. Technol. 37:2734-2742. [DOI] [PubMed] [Google Scholar]
- 35.Illades-Aguiar, B., and P. Setlow. 1994. Autoprocessing of the protease that degrades small, acid-soluable proteins of spores of Bacillus species is triggered by low pH, dehydration, and dipicolinic acid. J. Bacteriol. 176:7032-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kannan, G., J. C. Wilks, D. M. Fitzgerald, B. D. Jones, S. S. BonDurant, and J. L. Slonczewski. 2008. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol. 8:37-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinsinger, R. F., D. B. Kearns, M. Hale, and R. Fall. 2005. Genetic requirements for potassium ion-dependent colony spreading in Bacillus subtilis. J. Bacteriol. 187:8462-8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovács, T., A. Hargitai, K. L. Kovacs, and I. Mecs. 1998. pH-dependent activation of the alternative transcriptional factor σB in Bacillus subtilis. FEMS Microbiol. Lett. 165:323-328. [DOI] [PubMed] [Google Scholar]
- 39.Larsson, J. T., A. Rogstam, and C. von Wachenfeldt. 2005. Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis. Microbiology 151:3323-3335. [DOI] [PubMed] [Google Scholar]
- 40.Leaphart, A. B., D. K. Thompson, K. Huang, E. Alm, X.-F. Wan, A. Arkin, S. D. Brown, L. Wu, T. Yan, X. Liu, G. S. Wickham, and J.-Z. Zhou. 2006. Transcriptome profiling of Shewanella oneidensis gene expression following exposure to acidic and alkaline pH. J. Bacteriol. 188:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, J., W. M. Cosby, and P. Zuber. 1999. Role of lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol. Microbiol. 33:415-428. [DOI] [PubMed] [Google Scholar]
- 42.Liu, Y., Y. Dong, Y.-Y. M. Chen, and R. A. Burne. 2008. Environmental and growth phase regulation of the Streptococcus gordonii arginine deiminase genes. Appl. Environ. Microbiol. 74:5023-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacLellan, S. R., T. Wecke, and J. D. Helmann. 2008. A previously unidentified sigma factor and two accessory proteins regulate oxalate decarboxylase expression in Bacillus subtilis. Mol. Microbiol. 69:954-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurer, L. M., E. Yohannes, S. S. Bondurant, M. Radmacher, and J. L. Slonczewski. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187:304-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ménard, A., K. Altendorf, D. Breves, M. Mock, and C. Montecucco. 1996. The vacuolar ATPase proton pump is required for the cytotoxicity of Bacillus anthracis lethal toxin. FEBS Lett. 386:161-164. [DOI] [PubMed] [Google Scholar]
- 46.Montiel, M. D., R. D. Tyagi, and J. R. Valero. 2001. Wastewater treatment sludge as a raw material for the production of Bacillus thuringiensis based biopesticides. Water Res. 35:3807-3816. [DOI] [PubMed] [Google Scholar]
- 47.Moore, C. M., A. Gaballa, M. Hui, R. W. Ye, and J. D. Helmann. 2005. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol. Microbiol. 57:27-40. [DOI] [PubMed] [Google Scholar]
- 48.Neely, M. N., C. L. Dell, and E. R. Olson. 1994. Roles of LysP and CadC in mediating the lysine requirement for acid induction of the Escherichia coli cad operon. J. Bacteriol. 176:3278-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 50.Padan, E., E. Bibi, M. Ito, and T. A. Krulwich. 2005. Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta 1717:67-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 52.Reents, H., R. Munch, T. Dammeyer, D. Jahn, and E. Hartig. 2006. The Fnr regulon of Bacillus subtilis. J. Bacteriol. 188:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saraste, M., T. Metso, T. Nakari, T. Jalli, M. Lauraeus, and J. Van der Oost. 1991. The Bacillus subtilis cytochrome c oxidase. Variations on a conserved protein theme. Eur. J. Biochem. 195:517-525. [DOI] [PubMed] [Google Scholar]
- 54.Schadt, E. E., C. Li, B. Ellis, and W. Wong. 2001. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J. Cell. Biochem. 84:120-125. [DOI] [PubMed] [Google Scholar]
- 55.Schilling, O., O. Frick, C. Herzberg, A. Ehrenreich, E. Heinzle, C. Wittmann, and J. Stulke. 2007. Transcriptional and metabolic responses of Bacillus subtilis to the availability of organic acids: transcription regulation is important but not sufficient to account for metabolic adaptation. Appl. Environ. Microbiol. 73:499-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a.Shioi, J. I., S. Matsuura, and Y. Imae. 1980. Quantitative measurements of proton motive force and motility in Bacillus subtilis. J. Bacteriol. 144:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slonczewski, J. L., B. P. Rosen, J. R. Alger, and R. M. Macnab. 1981. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc. Natl. Acad. Sci. USA 78:6271-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slonczewski, J. L., and J. W. Foster. 1996. pH-related genes and survival at extreme pH, p. 1539-1552. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 58.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solovieva, I. M., and K. D. Entian. 2002. Investigation of the yvgW Bacillus subtilis chromosomal gene involved in Cd2+ ion resistance. FEMS Microbiol. Lett. 208:105-109. [DOI] [PubMed] [Google Scholar]
- 60.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stolyar, S., Q. He, M. P. Joachimiak, Z. He, Z. K. Yang, S. E. Borglin, D. C. Joyner, K. Huang, E. Alm, T. C. Hazen, J.-Z. Zhou, J. D. Wall, A. P. Arkin, and D. A. Stahl. 2007. Response of Desulfovibrio vulgaris to alkaline stress. J. Bacteriol. 189:8944-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka, K., K. Kobayashi, and N. Ogasawara. 2003. The Bacillus subtilis YufLM two-component system regulates the expression of the malate transporters MaeN (YufR) and YflS, and is essential for utilization of malate in minimal medium. Microbiology 149:2317-2329. [DOI] [PubMed] [Google Scholar]
- 63.Tanner, A., and S. Bornemann. 2000. Bacillus subtilis YvrK is an acid-induced oxalate decarboxylase. J. Bacteriol. 182:5271-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomassin, S., M. P. Jobin, and P. Schmitt. 2006. The acid tolerance response of Bacillus cereus ATCC14579 is dependent on culture pH, growth rate, and intracellular pH. Arch. Microbiol. 186:229-239. [DOI] [PubMed] [Google Scholar]
- 65.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of pH responses in Escherichia coli. J. Bacteriol. 184:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varela, E., and M. Tien. 2003. Effect of pH and oxalate on hydroquinone-derived hydroxyl radical formation during brown rot wood degradation. Appl. Environ. Microbiol. 69:6025-6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei, Y., A. A. Guffanti, M. Ito, and T. A. Krulwich. 2000. Bacillus subtilis YqkI is a novel Malic/Na+-lactate antiporter that enhances growth on malate at low protonmotive force. J. Biol. Chem. 275:30287-30292. [DOI] [PubMed] [Google Scholar]
- 68.Wei, Y., G. Deikus, B. Powers, V. Shelden, T. A. Krulwich, and D. H. Bechhofer. 2006. Adaptive gene expression in Bacillus subtilis strains deleted for tetL. J. Bacteriol. 188:7090-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiegert, T., G. Homuth, S. Versteeg, and W. Schumann. 2001. Alkaline shock induces the Bacillus subtilis σW regulon. Mol. Microbiol. 41:59-71. [DOI] [PubMed] [Google Scholar]
- 70.Wilks, J. C., and J. L. Slonczewski. 2007. pH of cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 189:5601-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winstedt, L., K.-I. Yoshida, Y. Fujita, and C. von Wachenfeldt. 1998. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J. Bacteriol. 180:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woolridge, D. P., J. D. Martinez, D. E. Stringer, and E. W. Gerner. 1999. Characterization of a novel spermidine/spermine acetyltransferase, bltD, from Bacillus subtilis. Biochem. J. 340:753-758. [PMC free article] [PubMed] [Google Scholar]
- 73.Wuytack, E. Y., and C. W. Michiels. 2001. A study of the effects of high pressure and heat on Bacillus subtilis spores at low pH. Int. J. Food Microbiol. 64:333-341. [DOI] [PubMed] [Google Scholar]
- 74.Yohannes, E., A. E. Thurber, J. C. Wilks, D. P. Tate, and J. L. Slonczewski. 2005. Polyamine stress at high pH in Escherichia coli K-12. BMC Microbiol. 5:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu, J., L. Hederstedt, and P. J. Piggot. 1995. The cytochrome bc complex (menaquinone:cytochrome c reductase) in Bacillus subtilis has a nontraditional subunit organization. J. Bacteriol. 177:6751-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuan, Z.-C., P. Liu, P. Saenkham, K. Kerr, and E. W. Nester. 2008. Transcriptome profiling and functional analysis of Agrobacterium tumefaciens reveals a general conserved response to acidic conditions (pH 5.5) and a complex acid-mediated signaling involved in Agrobacterium-plant interactions. J. Bacteriol. 190:494-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.