Abstract
Cydia pomonella granulovirus (CpGV) has been used for 15 years as a bioinsecticide in codling moth (Cydia pomonella) control. In 2004, some insect populations with low susceptibility to the virus were detected for the first time in southeast France. RGV, a laboratory colony of codling moths resistant to the CpGV-M isolate used in the field, was established with collection of resistant insects in the field followed by an introgression of the resistant trait into a susceptible colony (Sv). The resistance level (based on the 50% lethal concentrations [LC50s]) of the RGV colony to the CpGV-M isolate, the active ingredient in all commercial virus formulations in Europe, appeared to be over 60,000-fold compared to the Sv colony. The efficiency of CpGV isolates from various other regions was tested on RGV. Among them, two isolates (I12 and NPP-R1) presented an increased pathogenicity on RGV. I12 had already been identified as effective against a resistant C. pomonella colony in Germany and was observed to partially overcome the resistance in the RGV colony. The recently identified isolate NPP-R1 showed an even higher pathogenicity on RGV than other isolates, with an LC50 of 166 occlusion bodies (OBs)/μl, compared to 1.36 × 106 OBs/μl for CpGV-M. Genetic characterization showed that NPP-R1 is a mixture of at least two genotypes, one of which is similar to CpGV-M. The 2016-r4 isolate obtained from four successive passages of NPP-R1 in RGV larvae had a sharply reduced proportion of the CpGV-M-like genotype and an increased pathogenicity against insects from the RGV colony.
The development of resistance has become a major concern arising from the use of chemical insecticides. This problem is not exclusive to chemical compounds; it can also affect biological products. In 1965, the development of resistance to Bacillus thuringiensis Berliner toxins in a laboratory colony of house flies was reported (18). In the early 1990s, the first reports of field populations of the diamondback moth resistant to B. thuringiensis were published (12, 20, 33). In contrast, the development of resistance to insect baculoviruses has been observed in the laboratory in several species of insects (2, 4, 11, 16, 19) but had not been detected in the field until recently.
The codling moth, Cydia pomonella (L.), is a major pest causing severe economic damage in apple and pear orchards throughout most of the temperate world (9). Intensive chemical control led to resistance to most classes of insecticides (27, 31). Since the registration of the Cydia pomonella granulovirus (CpGV) (Baculoviridae), the use of this biological product in codling moth control has increased continuously due to both the increasing interest toward organic farming and the spread of resistance to synthetic chemical insecticides. Various baculoviruses are used in biological control. Most belong to the nucleopolyhedrovirus genus. Among those belonging to the second genus of the baculovirus family, the granulovirus, CpGV is one of the most commonly used, both in organic farming and in conventionally managed orchards.
In 2003, suspicions of field resistance to the virus were raised first in Germany (14) and then in France. Resistance was confirmed for several C. pomonella populations collected in organic orchards where CpGV treatments had failed (3, 30). This resistance to CpGV is the first case of resistance to a virus observed in field populations of an insect. As control failures permit an increase in the density of the insect population, the risk of dispersal of the resistance trait is high. Development of resistance against CpGV is, therefore, a major issue for apple production.
The first CpGV isolate was found in Mexico and was described by Tanada in 1964 (34). In Europe, all commercial formulations of CpGV are derived from this Mexican isolate (CpGV-M). The genome of CpGV-M1, a genotype isolated from the CpGV-M population by an in vivo limiting dilution method, was first characterized by restriction enzyme fragment length polymorphism (7). Then, it was completely sequenced (25), and this has become the reference genotype. Two additional CpGV genotypic variants have been described and classified according to their geographical origins: the R type, from Russia, and the E-type, from England (8, 17). Usually, the characterization of a new isolate is made by comparison with these three genotypes, M, R, and E. This classification is based on modifications in some restriction fragments and is not directly related to the biological properties of the variants.
Among the principal differences between chemical and biological control agents are the genetic diversity of the latter and their ability to evolve when conditions allow adaptation of the pathogen to the host. Success in adaptation to homologous or heterologous hosts by serial passages has been reported for baculoviruses (21, 26). The approach developed in this study was to survey existing isolates for their efficacy against the resistant codling moth and then to examine the possible adaptation of the most pathogenic candidate to the resistant host by serial passages through a laboratory colony of C. pomonella originating from field-collected resistant insects.
MATERIALS AND METHODS
Insects.
The susceptible strain Sv of C. pomonella was established in INRA (Avignon) from a codling moth population collected in field in the southeast of France (Les Vignières, Vaucluse, France). It has been mass reared on an artificial diet (15) without selection for 14 years.
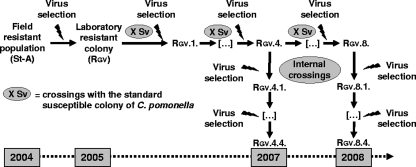
Diapausing larvae collected during the 2004 season in an organic orchard (St. Andiol, Bouches du Rhône, France) in which CpGV-based control had failed were used to start a virus-resistant laboratory colony. Their offspring were challenged with the commercial CpGV-M formulation to select the resistant individuals. This resistant laboratory colony was named RGV. To allow precise comparisons between Sv and RGV, it was important to homogenize their genetic composition. For that, an introgression of the resistance trait was performed by successive backcrosses with the Sv colony. At each generation, the resistant larvae were selected by exposing the population to a discriminating concentration of virus defined previously (30). After four successive backcrosses (RGV.4), the RGV colony was maintained for four additional generations under continuous virus selection. This colony was used for the bioassays in 2007. The introgression of the resistance trait was then continued until the eighth backcross (RGV.8), also followed by four generations of selection (Fig. 1). This colony, named RGV.8, was used in 2008.
FIG. 1.
Introgression of the resistance trait into the Sv-associated susceptible genetic background.
Virus isolates.
The Mexican isolate used in this study (CpGV-M) is the inoculum used for the production of Carpovirusine (NPP, Arysta LifeScience, Pau, France), one of the three CpGV commercial products in Europe. The same stock of CpGV-M (lot E06) was used in all bioassays. The virus used in 2007 was named CpGV-M07, and that used in 2008 was named CpGV-M08. These two isolates differed only in their durations of storage at −20°C, 1 year after production for CpGV-M07 and 2 years for CpGV-M08.
The I12 isolate was provided by M. Rezapanah from PPDRI (Pests and Deseases Research Institute, Teheran, Iran) and by J. Jehle from DLR (Dienstleistungszentrum Ländlicher Raum, Neustadt, Germany). This isolate, originally from Iran, was recently shown to overcome CpGV resistance in a German population of C. pomonella (10).
The NPP-R1 isolate was provided by NPP. This isolate comes from the virus collection of the firm. The original stock was amplified using the susceptible colony reared at the NPP facility prior to use in bioassays. The 2016-r4 isolate is the result of four cycles of multiplication of NPP-R1 on RGV larvae.
These four isolates (CpGV-M, I12, NPP-R1, and 2016-r4) were used for restriction analysis of the DNA and for bioassays. The occlusion body (OB) concentration of each viral stock was determined using a light microscope (Olympus BX41TF) with dark field optics (×600) and a Petroff-Hauser counting chamber (depth, 0.01 mm).
Virus multiplication on RGV larvae.
A series of viral passages on RGV larvae were carried out, starting from the NPP-R1 isolate. Third-instar RGV larvae (7 days old) reared on a virus-free diet were infected with OBs produced in the previous passage. A 50-μl suspension comprising between 1 × 104 and 2 × 104 OBs/μl was deposited on the surface of a formaldehyde-free insect diet (Heliothis Diet; Stonefly Industries, TX) in 24-well plates. Approximately 50 larvae per passage were inoculated and incubated at 25°C with a 16:8-h (light/dark) photoperiod. Diseased larvae were extracted from the rearing diet at early signs of viral infection, usually from the fourth to the seventh day postinoculation, and stored at 25°C until death. The dead larvae were homogenized in distilled water, and the resulting suspension was filtered through nylon to remove insect debris. The filtrate was centrifuged twice at 10,000 × g for 30 min, and the pellet was resuspended in distilled water. This suspension constituted the inoculum for the following viral passage.
DNA extraction and restriction endonuclease (REN) analysis of CpGV isolates.
To isolate viral DNA, OBs were dissolved in 0.1 M Na2CO3. Viral DNA was extracted twice with an equal volume of phenol and once with chloroform. DNA was then precipitated in cold ethanol (70%, vol/vol) in the presence of 0.2 M sodium acetate and incubated overnight at −20°C. Samples were centrifuged for 30 min at 10,000 × g to collect the nucleic acid precipitate. Resulting pellets were washed with 70% cold ethanol, dried, and resuspended into distilled water.
Approximately 500 ng of purified virus DNA was digested with EcoRI, BamHI, PstI, SalI, or XhoI (Fermentas) in the supplied buffer at 37°C for 3 h. The DNA restriction fragments were separated by electrophoresis in 1% agarose gels in Tris-acetate-EDTA buffer at 80 V for approximately 4 h. λ-phage DNA/HindIII fragments were used as standards for size determination. Fragments were visualized under UV light by ethidium bromide staining. The fragment sizes and DNA quantity were estimated using the Kodak 1D image system (Eastman Kodak Company).
PCR amplification and REN analysis of the PCR product.
PCR was carried out using primer sets CPGV3F (5′-AAA CGA GGA CCG TCA AAA TG-3′) and CPGV3R (5′-CTC GGA CAA TGA ACG TGT TG-3′), located at nucleotides (nt) 54513 and 58482 on the CpGV-M1 sequence (GB-U53466). These primers generate a PCR product of about 4,000 bp. The amplified DNA was tested for the presence of an EcoRI restriction site by treatment with the endonuclease followed by electrophoresis as described above.
Bioassays.
Bioassays against neonate larvae (0 to 12 h old) were carried out using a diet surface contamination method in 96-well plates containing about 200 μl of a formaldehyde-free artificial diet (Heliothis Diet; Stonefly Industries, TX). A 6-μl volume of an OB suspension was spread over the diet surface of each well (well surface area = 28 mm2). The same volume of distilled water was used in control wells. Bioassays were performed using five or six CpGV concentrations, ranging from 3 to 729 OBs/μl for the most efficient isolates (corresponding to 0.643 to 156.2 OBs/mm2 of diet surface) and up to 3.125 × 106 OBs/μl for the RGV colony for the least efficient isolate (6.696 × 105 OBs/mm2). One larva was placed in each well. The wells were sealed with parafilm, and the microplates were incubated in a growth chamber at 25°C with a 16:8-h (light/dark) photoperiod. The larvae that died within the first day postinoculation were excluded from the test. Mortality was recorded at 7 days postinfection. Larvae that did not react to physical stimuli were considered dead.
To avoid any bias in the results due to a reduction in pathogenicity of the virus or a variation in susceptibility of the hosts, the data were analyzed independently each year. Mortality data were subjected to probit analysis (13) after correction for control mortality (1).
RESULTS
REN analysis. (i) Analysis of NPP-R1.
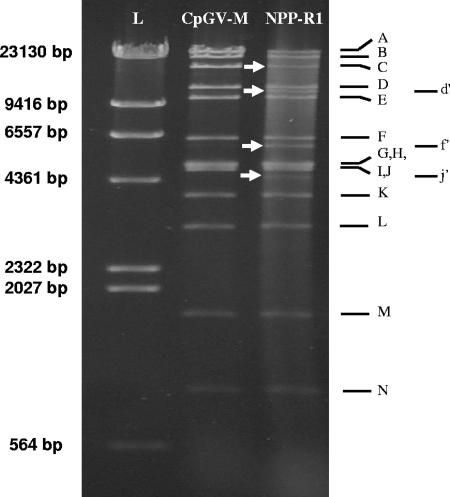
DNA from NPP-R1 was compared to that from CpGV-M by using five RENs (EcoRI, BamHI, SalI, PstI, and XhoI). The CpGV-M isolate showed the same REN profiles those as published by Crook et al. in 1997 (Fig. 2 and 3). NPP-R1 showed genotypic differences after digestion with all the restriction enzymes tested (data not shown). Three additional fragments were detected in the EcoRI restriction pattern of NPP-R1, compared to that of CpGV-M. The apparent sizes of these fragments were estimated at 11.2 kb (d′), 5.9 kb (f′), and 4.4 kb (i′) (Fig. 2). Moreover, the intensity of EcoRI fragment C (16.8 kb) was lower than expected for the NPP-R1 isolate, suggesting that an additional EcoRI site is located within this fragment. PCR amplification of an internal 4-kbp fragment (nt 54513 to 58482) followed by an EcoRI digestion resulted in a partial digestion (data not shown). The submolar bands d′ and f′ belong to the same genotype and are the result of an additional EcoRI site at a position close to nt 58000. Thus, at least two distinguishable genotypes are present in the NPP-R1 isolate, one with the profile of the CpGV-M and one called R1. According to the intensity of the additional bands, the ratio of the two major genotypes appeared to be around one-third of CpGV-M and two-thirds of R1. The submolar band of 4.4 kb was not identified. Its ratio was estimated at about 10%.
FIG. 2.
Agarose gel electrophoresis of CpGV-M DNA and NPP-R1 DNA digested with EcoRI. Arrows indicate additional or modified fragments. Lambda-DNA digested with HindIII is included for molecular size standards (L).
FIG. 3.
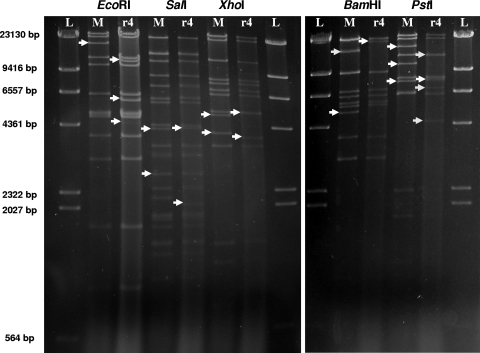
DNA restriction profiles for EcoRI, SalI, XhoI, BamHI, and PstI. Lanes M represent the CpGV-M DNA, and lane r4 represents the 2016-r4 DNA. Arrows indicate additional or modified fragments. Lambda-DNA digested with HindIII is included for molecular size standards (L).
(ii) Analysis of 2016-r4.
The NPP-R1 isolate had been submitted to four successive passages on the RGV colony. The obtained isolate was named 2016-r4. After this process, it became impossible to detect the characteristic CpGV-M EcoRI restriction fragment of 16.8 kb. All five enzymes tested gave restriction profiles which differed from those of CpGV-M and allowed a specific and reliable identification of the R1 genotype (Fig. 3). The specific EcoRI and BamHI fragments were unambiguously located in the genome. The additional EcoRI site in fragment C previously mentioned to occur in NPP-R1 was present, and the submolar band of 4.4 kb was also present, albeit weaker in appearance. The BamHI site located at position 17729 on the CpGV-M1 genome was absent in 2016-r4, resulting in a new fragment of 20.5 kb. The SalI and XhoI profiles showed small differences in size, indicating some insertions and deletions 50 to 200 nt in length in the 2016-r4 genome. Digestion with PstI revealed the greatest number of differences in restriction profile in comparison with CpGV-M; among them, 2016-r4 seemed to have an additional PstI site in CpGV-M fragment C.
Bioassays. (i) Resistance ratio of the RGV colony.
Under our experimental conditions, the CpGV-M presented a 50% lethal concentration (LC50) of 25 to 30 OBs/μl on Sv (Table 1). In 2007, the LC50 obtained for RGV.4 by using CpGV-M07 was 1.38 × 106 OBs/μl, which represents a resistance factor of 60,000-fold between Sv and RGV colonies. In 2008, upon treatment with CpGV-M08, the LC50 for RGV.8 was 2.37 × 106 OBs/μl, which was not significantly different from the value estimated in 2007 (χ2 = 0.844; df = 3; P = 0.8389). Moreover, considering the LC50 of Sv treated with CpGV-M08 (29.22 OBs/μl), the resistance factor of 80,000-fold in 2008 was considered to be equivalent to that for 2007.
TABLE 1.
Pathogenicities, measured by LC50 and LC90, of four virus isolates on C. pomonella laboratory colonies susceptible (Sv) and resistant (RGV) to CpGV-Ma
| Strain (yr) | Virus | No. of insects tested | No. of OBs/μl (95% CI)
|
Slope ± SE | χ2 | Resistance factor (fold)
|
||
|---|---|---|---|---|---|---|---|---|
| LC50 | LC90 | LC50 | LC90 | |||||
| Sv (2007) | CpGV-M07 | 771 | 24.00 (14.10-35.86) | 275.45 (176.83-512.48) | 1.21 ± 0.12 | 1.11 | 1 | 1 |
| I12 | 486 | 26.10 (13.8-41.8) | 261.82 (149.04-652.39) | 1.28 ± 0.19 | 4.70 | 1 | 1 | |
| NPP-R1 | 689 | 25.80 (14.48-39.93) | 328.55 (196.93-702.51) | 1.16 ± 0.13 | 1.28 | 1 | 1 | |
| Sv (2008) | CpGV-M08 | 593 | 29.22 (13.87-49.27) | 377.37 (231.96-725.56) | 1.15 ± 0.14 | 3.07 | 1 | 1 |
| 2016-r4 | 999 | 39.65 (6.40-133.91) | 805.85 (260.20-1.36 × 103) | 0.98 ± 0.11 | 13.6 | 1 | 2 | |
| RGV.4 (2007) | CpGV-M07 | 416 | 1.38 × 106 (3.17 × 105-4.91 × 106) | NAb | 0.62 ± 0.12 | 1.61 | 60,000 | NA |
| I12 | 656 | 5.01 × 103 (1.85 × 103-1.03 × 104) | 2.74 × 105 (1.27 × 105-8.35 × 105) | 0.63 ± 0.08 | 4.42 | 200 | 1,000 | |
| NPP-R1 | 578 | 166.31 (91.21-278.27) | 1.28 × 104 (5.95 × 103-3.80 × 104) | 0.70 ± 0.08 | 4.81 | 7 | 46 | |
| RGV.8 (2008) | CpGV-M08 | 698 | 2.37 × 106 (8.53 × 105-7.61 × 106) | NA | 0.50 ± 0.10 | 4.62 | 80,000 | NA |
| 2016-r4 | 1,201 | 102.31 (63.20-146.91) | 1.57 × 103 (1.01 × 103-2.97 × 103) | 1.10 ± 0.10 | 6.21 | 3.5 | 4 | |
The pathogenicity of CpGV-M for Sv larvae is used as a reference level. Bioassays were carried out by diet surface contamination with neonate larvae. Mortality was scored 7 days after infection.
NA, >50% mortality could not be achieved on RGV larvae inoculated with CpGV-M isolates.
(ii) Biological activity of the I12 and NPP-R1 isolates.
In 7-day bioassays with Sv larvae, no significant difference was observed in the biological activities of CpGV-M07 and I12 isolates (LC50s of 24.0 OBs/μl and 26.1 OBs/μl, respectively; χ2 = 0.178; df = 3; P = 0.9811) (Table 1). With an estimated LC50 of 5.01 × 103 OBs/μl on RGV larvae, I12 presented a significantly higher pathogenicity than CpGV-M (χ2 = 118.5; df = 3; P < 0.0001). The resistance factor at the LC50 between the Sv and RGV colonies was reduced from 60,000-fold (the level for inoculation with CpGV-M) to 200-fold (upon inoculation with I12).
As for the I12 isolate, the biological activities of the CpGV-M07 and NPP-R1 isolates on Sv larvae were similar (LC50s of 24.0 OBs/μl and 25.8 OBs/μl, respectively; χ2 = 0.228; df = 3; P = 0.9730) (Table 1). In contrast, the biological activities of NPP-R1 and CpGV-M07 on RGV larvae differed significantly (LC50s of 166 OBs/μl and 1.38 × 106 OBs/μl, respectively; χ2 = 229; df = 3; P < 0.0001). The RGV resistance factor of 60,000-fold in CpGV-M07 was thus reduced to 7-fold in NPP-R1. Accordingly, the potency of NPP-R1 was close to 10,000-fold higher than that of CpGV-M on resistant larvae.
The slopes of concentration-mortality regression were not significantly different among the three viral isolates CpGV-M07, I12, and NPP-R1 for the same C. pomonella colony (Sv [χ2 = 0.278; df = 2; P = 0.870] and RGV [χ2 = 1.395; df = 2; P = 0.498]). Conversely, significant differences were observed for the slopes with same isolate between the Sv and RGV colonies (CpGV-M07 [χ2 = 25.5; df = 1; P < 0.0001], I12 [χ2 = 7.749; df = 1; P = 0.0054], and NPP-R1 [χ2 = 9.78; df = 1; P = 0.0018]). The slope for resistant larvae was shallower than that for susceptible insects. The resistance ratio of the RGV colony treated with NPP-R1 was thus higher at the LC90 than at the LC50, reaching a ratio of 46-fold instead of 7-fold (Table 1). Similarly, the resistance factor of the RGV colony to I12 was higher at the LC90 than at the LC50, reaching 1,000-fold instead of a 200-fold ratio.
(iii) Biological activity of 2016-r4.
To determine if the amplification of NPP-R1 on RGV larvae resulted in a modification of the virus efficiency, the biological activity of 2016-r4 was evaluated using bioassays on both Sv and RGV colonies. On the Sv colony, the LC50s of 2016-r4 and CpGV-M08 on Sv larvae did not differ significantly (χ2 = 4.0; df = 3; P = 0.261) (Table 1). In contrast, the LC50 of RGV larvae treated with 2016-r4 decreased to 102 OBs/μl, which represented a reduction in resistance ratio from 7-fold for NPP-R1 to 3.5-fold for 2016-r4. The most relevant improvement of efficiency was observed at the LC90, which was reduced from 1.28 × 104 OBs/μl (NPP-R1) to 1.57 × 103 OBs/μl (2016-r4). As a result, the slopes of the concentration-mortality regressions of 2016-r4 on Sv and RGV were no longer significantly different (χ2 = 0.603; df = 1; P = 0.437). Since the concentration-mortality lines could be considered parallel, the value for the relative potency between CpGV-M on Sv larvae and 2016-r4 on RGV was estimated to be 4.0.
DISCUSSION
The RGV colony presents a level of resistance against CpGV-M of about 60,000-fold, compared to the level for the reference susceptible colony Sv. This level is higher than the 100-fold resistance described by Eberle and coworkers (10) for the CpR colony. These two C. pomonella colonies have different geographic origins (France and Germany, respectively). A virus isolate from Iran, CpGV-I12, was recently identified as overcoming the resistance in CpR (10). To test if the mechanisms of resistance involved in both colonies are different, the I12 virus isolate was also tested in RGV. I12 also showed a higher efficiency against RGV than did CpGV-M. The major resistance mechanisms of CpR and RGV probably have the same basis, and the difference in resistance level might be related to a difference in level of expression.
As the improvement in the control of RGV larvae by I12 did not reach appropriate levels for its commercial use (Table 1), the search for new isolates with greater efficiency against RGV larvae has been pursued. Among the various original isolates that were tested on RGV (data not shown), a new isolate, NPP-R1, showed the highest efficacy, with a reduction in resistance ratio from 60,000- to 7-fold at the LC50. Field trials with NPP-R1 were performed in 2007 in fields with confirmed resistance in France and Germany, but no significant reduction of pest infestation was achieved by applications of NPP-R1, compared to levels for applications involving CpGV-M (data not shown). There are various possible explanations for these results. First, the LC90 probably represents a better estimation of the field efficiency of an insecticide than the LC50, as high dosages are required for effective pest suppression. Therefore, the high LC90 value of the NPP-R1 isolate against RGV (1.28 × 104 OBs/μl) may result in low efficacy for a field application. Second, the failure to control C. pomonella populations in the field with the NPP-R1 isolate could be explained by a RGV colony that is unrepresentative of the field populations (for example, coexistence of more than one resistant genotype, lost during the RGV selection process). Finally, the CpGV-R1 isolate might not resist field conditions as well as CpGV-M, resulting in its rapid inactivation. The efficiency of NPP-R1, and of any new isolate, should be systematically verified using both laboratory bioassays and field trials on representative insect populations that differ in susceptibility to CpGV-M.
Restriction fragment enzyme length polymorphism analysis revealed that NPP-R1 is a mixture of genotypes. PCR amplification followed by EcoRI digestion allowed us to determine that two major genotypes constitute the NPP-R1 isolate. Of these genotypes, a CpGV-M1-like genotype was present at a prevalence of ca. 30%. The unidentified submolar band of 4.4 kb may belong to a third genotype, similar to the R1 genotype. The analysis of CpGV-M (7) did not reveal any genetic variability in this isolate. However, a recent analysis of natural virus isolates from Iran revealed a high proportion of genotypic mixtures (29). It seems that GV populations are composed of distinct genotypes, as are those of the best-studied nuclear polyhedrosis virus (32).
The passages on RGV resulted in the adaptation of the NPP-R1 isolate to the new, more uniform, more restrictive host conditions. This adaptation is likely to have affected the original genetic composition of the isolate. The CpGV-M genotype that is inefficient against RGV larvae was reduced to an undetectable level in fewer than four passages. The selection of the 2016-r4 isolate free of CpGV-M resulted in a slight improvement in the LC50 in RGV larvae and an eightfold improvement in LC90 (from 1.28 × 104 OBs/μl to 1.57 × 103 OBs/μl). This reduction in LC90 should have an important impact on the level of control of the insect population. In a Spodoptera frugiperda nucleopolyhedrovirus isolate, an increase of pathogenicity was observed only when various genotypes coexisted in the same larvae. Among them, one genotype was not able to infect alone (24). For NPP-R1, our results confirmed that the R1 genotype alone was indeed responsible for overcoming the resistance in the insect colony; the M type present in NPP-R1 was not required for R1 multiplication. The submolar band of 4.4 kb resulting from the EcoRI digestion of NPP-R1 DNA was still present in 2016-r4, although at a reduced intensity. 2016-r4 is probably not a single genotype but a group of closely related genotypes, indistinguishable with the five enzymes used. The 2016-r4 restriction profile clearly indicates that the R1 genotype is a CpGV variant. Three previously described CpGV variants, the Mexican (M), the English (E), and the Russian (R) variants, present some differential genotypic characteristics. The R type has a single deletion of about 2.4 kbp in the total genome, and the E type is modified in one area, resulting in an additional EcoRI site, a shift in a BamHI, site and a total insertion of 0.8 kbp (deletion of 0.3 kbp and insertion of 1.1 kbp) (8). The R1 genotype is different from all three variants. R1 also differs from the CpGV-I12 isolate described by Eberle et al. (10), which overcomes the CpGV-M resistance in German populations of C. pomonella. To date, after only a few years of investigation, two different isolates have been identified as active against resistant larvae, with different levels of efficiency.
The concentration-mortality lines of 2016-r4 on RGV and CpGV-M on Sv were estimated to be parallel, suggesting that the pathogenicities of both virus populations in these alternative hosts follow similar dynamics. Continuous selection on RGV larvae might lead to a virus isolate even more efficient against this larval phenotype. However, increasing virus pathogenicity could reduce the isolate's pathogenicity against susceptible larvae. Some studies have shown that serial passages could favor the proliferation of defective genotypes (6, 23). No statistically significant differences were detected in the pathogenicities of 2016-r4 and CpGV-M on Sv larvae after four cycles of selection. It seems that the increased performance of 2016-r4 on RGV is not counterbalanced by a decrease in pathogenicity in the other host. Alternatively, this adaptation could influence other biological properties of the virus, like OB production or speed of kill, as observed for other insect viruses (5). It has been shown that increasing the speed of kill results in a lower OB yield (22, 28), but to our knowledge, no direct relationship with pathogenicity (LC50) or other biological traits has been reported.
The rapid response to the selection of NPP-R1 indicates that the virus isolate already contained a genotype able to control the RGV-associated resistant phenotype. However, the 88% decrease in the 2016-r4 LC90 value is higher than would be expected following the elimination of the M genotype, since it represents only ∼30% of the total genotypic composition of the NPP-R1 isolate. In-depth analysis would be required to determine whether there is interference between the M and R1 genotypes or additional changes in the R1 genome that have occurred during the adaption to the conditions of the laboratory host colony.
The final aim of this work was to assist in finding a solution to CpGV resistance for European apple producers. The efficiency of 2016-r4 on RGV did not reach the same level as that on Sv. This difference in efficiency between susceptible and resistant larvae might be considered minor in comparison with the 60,000-fold resistance level observed with CpGV-M. However, field trials with 2016-r4 are required to determine its efficacy as a pest control agent in the field. Three different categories of C. pomonella populations can be found in orchards: populations only composed of susceptible phenotypes, populations mainly composed of resistant phenotypes, and mixed populations. Each type of population may require different pest control strategies. When C. pomonella populations are susceptible to CpGV-M, the use of the 2016-r4 isolate for control is appropriate only when its efficiency is similar to or greater than that of CpGV-M. In contrast, when the C. pomonella population is heterogeneous for the resistance trait, the use of 2016-r4 should be recommended to avoid the proliferation and dispersal of resistant individuals.
Acknowledgments
We thank Ghislain Cabrol (NPP, Arysta LifeScience, France) for OB counting, Mohammadreza Rezapanah (PPDRI, Iran) and Johannes Jehle (DLR, Germany) for providing the CpGV isolates within the framework of the EU project SustainCpGV (032857), and Trevor Williams (Instituto de Ecología A.C., Mexico) for useful comments on the manuscript.
This work was supported by the French Research Agency (ANR-06-RIB-003-02) and by NPP. M.B. received a fellowship from the Ecole des Mines d'Alès.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Abbott, W. S. 1925. A method of computing the effectiveness of an insecticide. J. Econ. Entomol. 18:265-267. [Google Scholar]
- 2.Abot, A. R., F. Moscardi, J. R. Fuxa, D. R. Sosa-Gomez, and A. R. Richter. 1996. Development of resistance by Anticarsia gemmatalis from Brazil and the United States to a nuclear polyhedrosis virus under laboratory selection pressure. Biol. Control 7:126-130. [Google Scholar]
- 3.Asser-Kaiser, S., E. Fritsch, K. Undorf-Spahn, J. Kienzle, K. E. Eberle, N. A. Gund, A. Reineke, C. P. Zebitz, D. G. Heckel, J. Huber, and J. A. Jehle. 2007. Rapid emergence of baculovirus resistance in codling moth due to dominant, sex-linked inheritance. Science 317:1916-1918. [DOI] [PubMed] [Google Scholar]
- 4.Briese, D. T., and H. A. Mende. 1983. Selection for increased resistance to a granulosis virus in the potato moth, Phthorimaea operculella (Zeller) (Lepidoptera: Gelechiidae). Bull. Entomol. Res. 73:1-9. [Google Scholar]
- 5.Chakraborty, S., and S. Reid. 1999. Serial passage of a Helicoverpa armigera nucleopolyhedrovirus in Helicoverpa zea cell cultures. J. Invertebr. Pathol. 73:303-308. [DOI] [PubMed] [Google Scholar]
- 6.Croizier, G., L. Croizier, G. Biache, and J. Chaufaux. 1985. Evolution de la composition génétique et du pouvoir infectieux du baculovirus de Mamestra brassicae L. au cours de 25 multiplications successives sur les larves de la noctuelle du chou. Entomophaga 30:365-374. [Google Scholar]
- 7.Crook, N. E., J. D. James, I. R. Smith, and D. Winstanley. 1997. Comprehensive physical map of the Cydia pomonella granulovirus genome and sequence analysis of the granulin gene region. J. Gen. Virol. 78:965-974. [DOI] [PubMed] [Google Scholar]
- 8.Crook, N. E., R. A. Spencer, C. C. Payne, and D. J. Leisy. 1985. Variation in Cydia pomonella granulosis virus isolates and physical maps of the DNA from three variants. J. Gen. Virol. 66:2423-2430. [Google Scholar]
- 9.Cross, J. V., M. G. Solomon, D. Chandler, P. Jarrett, P. N. Richardson, D. Winstanley, H. Bathon, J. Huber, B. Keller, G. A. Langenbruch, and G. Zimmermann. 1999. Biocontrol of pests of apples and pears in Northern and Central Europe: 1. Microbial agents and nematodes. Biocontrol Sci. Technol. 9:125-149. [Google Scholar]
- 10.Eberle, K. E., S. Asser-Kaiser, S. M. Sayed, H. T. Nguyen, and J. A. Jehle. 2008. Overcoming the resistance of codling moth against conventional Cydia pomonella granulovirus (CpGV-M) by a new isolate CpGV-I12. J. Invertebr. Pathol. 98:293-298. [DOI] [PubMed] [Google Scholar]
- 11.Engelhard, E. K., and L. E. Volkman. 1995. Developmental resistance in fourth instar Trichoplusia ni orally inoculated with Autographa californica M nuclear polyhedrosis virus. Virology 209:384-389. [DOI] [PubMed] [Google Scholar]
- 12.Ferre, J., M. D. Real, J. Van Rie, S. Jansens, and M. Peferoen. 1991. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc. Natl. Acad. Sci. USA 88:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finney, D. J. 1971. Probit analysis. Cambridge University Press, Cambridge, United Kingdom.
- 14.Fritsch, E., K. Undorf-Spahn, J. Kienzle, C. Zebitz, and J. Hüber. 2005. Codling moth granulovirus: first indication of variations in the susceptibility of local codling moth populations. Nachrbl. Dtsch. Pflanzenschutzd. 57:29-34. (In German.) [Google Scholar]
- 15.Guennelon, G., H. Audemard, J. C. Fremond, and A. M. Idrissi Ammari. 1981. Progrès réalisés dans l'élevage du carpocapse (Laspeyresia pomonella L.) sur milieu artificiel. Agronomie 1:59-64. [Google Scholar]
- 16.Haas-Stapleton, E. J., J. O. Washburn, and L. E. Volkman. 2005. Spodoptera frugiperda resistance to oral infection by Autographa californica multiple nucleopolyhedrovirus linked to aberrant occlusion-derived virus binding in the midgut. J. Gen. Virol. 86:1349-1355. [DOI] [PubMed] [Google Scholar]
- 17.Harvey, J. P., and L. E. Volkman. 1983. Biochemical and biological variation of Cydia pomonella (codling moth) granulosis virus. Virology 124:21-34. [DOI] [PubMed] [Google Scholar]
- 18.Harvey, T. L., and D. E. Howell. 1965. Resistance of the house fly to Bacillus thuringiensis Berliner. J. Invertebr. Pathol. 20:92-100. [DOI] [PubMed] [Google Scholar]
- 19.Hoover, K., J. O. Washburn, and L. E. Volkman. 2000. Midgut-based resistance of Heliothis virescens to baculovirus infection mediated by phytochemicals in cotton. J. Insect Physiol. 46:999-1007. [DOI] [PubMed] [Google Scholar]
- 20.Kirsch, K., and H. Schmutterer. 1988. Low efficacy of a Bacillus thuringiensis (Berl.) formulation in controlling the diamondback moth, Plutella xylostella (L.), in the Philippines. Z. Angew. Entomol. 105:249-255. [Google Scholar]
- 21.Kolodny-Hirsch, D. M., and N. A. M. Van Beek. 1997. Selection of a morphological variant of Autographa californica nuclear polyhedrosis virus with increased virulence following serial passage in Plutella xylostella. J. Invertebr. Pathol. 69:205-211. [Google Scholar]
- 22.Kumar, S., and L. K. Miller. 1987. Effects of serial passage of Autographa californica nuclear polyhedrosis virus in cell culture. Virus Res. 7:335-349. [DOI] [PubMed] [Google Scholar]
- 23.Lee, H. Y., and P. J. Krell. 1992. Generation and analysis of defective genomes of Autographa californica nuclear polyhedrosis virus. J. Virol. 66:4339-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.López-Ferber, M., O. Simón, T. Williams, and P. Caballero. 2003. Defective or effective? Mutualistic interactions between virus genotypes. Proc. Biol. Sci. 270:2249-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luque, T., R. Finch, N. Crook, D. R. O'Reilly, and D. Winstanley. 2001. The complete sequence of the Cydia pomonella granulovirus genome. J. Gen. Virol. 82:2531-2547. [DOI] [PubMed] [Google Scholar]
- 26.Maleki-Milani, H. 1978. Influence de passages répétés du virus de la polyédrose nucléaire de Autographa californica chez Spodoptera littoralis. Entomophaga 23:217-224. [Google Scholar]
- 27.Moffitt, H. R., P. H. Westigard, K. D. Mantey, and H. E. Van de Baan. 1988. Resistance to diflubenzuron in the codling moth (Lepidoptera: Tortricidae). J. Econ. Entomol. 81:1511-1515. [Google Scholar]
- 28.O'Reilly, D. R., and L. K. Miller. 1991. Improvement of a baculovirus pesticide by deletion of the EGT gene. Bio/Technology 9:1086-1089. [Google Scholar]
- 29.Rezapanah, M., S. Shojai-Estabragh, J. Huber, and J. Jehle. 2008. Molecular and biological characterization of new isolates of Cydia pomonella granulovirus from Iran. J. Pest Sci. 81:187-191. [Google Scholar]
- 30.Sauphanor, B., M. Berling, J.-F. Toubon, M. Reyes, J. Delnatte, and P. Allemoz. 2006. Carpocapse des pommes: cas de résistance au virus de la granulose en vergers biologiques. Phytoma Def. Veg. 590:24-27. [Google Scholar]
- 31.Sauphanor, B., J. C. Bouvier, and V. Brosse. 1998. Spectrum of insecticide resistance in Cydia pomonella (Lepidoptera: Tortricidae) in southeastern France. J. Econ. Entomol. 91:1225-1231. [Google Scholar]
- 32.Simón, O., T. Williams, M. López-Ferber, and P. Caballero. 2005. Functional importance of deletion mutant genotypes in an insect nucleopolyhedrovirus population. Appl. Environ. Microbiol. 71:4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabashnik, B. E., N. L. Cushing, N. Finson, and M. W. Johnson. 1990. Field development of resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera: Plutellidae). J. Econ. Entomol. 83:1671-1676. [Google Scholar]
- 34.Tanada, Y. 1964. A granulosis virus of the codling moth, Carpocapsa pomonella (Linnaeus) (olethreutidae, Lepidoptera). J. Insect Pathol. 6:378-380. [Google Scholar]