Abstract
Asiatic citrus canker is a major disease worldwide, and its causal agent, Xanthomonas citri pv. citri, is listed as a quarantine organism in many countries. Analysis of the molecular epidemiology of this bacterium is hindered by a lack of molecular typing techniques suitable for surveillance and outbreak investigation. We report a comparative evaluation of three typing techniques, amplified fragment length polymorphism (AFLP) analysis, insertion sequence ligation-mediated PCR (IS-LM-PCR) typing, and multilocus variable-number tandem-repeat analysis (MLVA), with 234 strains originating from Asia, the likely center of origin of the pathogen, and reference strains of pathotypes A, A*, and Aw, which differ in host range. The typing techniques were congruent in describing the diversity of this strain collection, suggesting that the evolution pattern of the bacterium may be clonal. Based on a hierarchical analysis of molecular variance, the AFLP method best described the genetic variation found among pathotypes whereas MLVA best described the variation found among individual strains from the same countries or groups of neighboring countries. IS-LM-PCR data suggested that the transposition of insertion sequences in the genome of X. citri pv. citri occurs rarely enough not to disturb the phylogenetic signal. This technique may be useful for the global surveillance of non-epidemiologically related strains. Although pathological characteristics of strains could be most often predicted from genotyping data, we report the occurrence in the Indian peninsula of strains genetically related to pathotype A* strains but with a host range similar to that of pathotype A, which makes the classification of this bacterium even more complicated.
The definition of host range is a central parameter for the understanding and, ultimately, the control of infectious diseases in general and bacterial plant diseases in particular. In phytobacteriology, host range is an important aspect of pathogenicity. Control of diseases can be achieved with resistance genes which reduce host range (50, 64). Epidemiological characteristics are highly dependent on host range, and the emergence of new diseases is sometimes correlated with broadened host ranges (74). Xanthomonads have the particularity of an extremely narrow host range (sometimes reduced to a single plant genus), although a very large number of plant families can be hosts when all members of the genus are considered (33), which led plant pathologists to create the concept of pathovar at an infrasubspecific level. Pathovars were defined as groups of strains sharing several pathological characteristics, such as their host range and the disease facies they cause (18). Based on molecular data, strains classified as a single pathovar usually form a discrete monomorphic or weakly polymorphic cluster, suggesting that strains of a pathovar have a common ancestral origin (3, 56). Xanthomonas citri pv. citri is the causal agent of Asiatic canker, a severe disease infecting most commercial citrus cultivars and some genera in the Rutaceae family in many citrus-producing areas worldwide (6, 60, 61). This pathovar has two types of strains, which differ in their host ranges: pathotype A has a wide host range and a worldwide distribution and is a permanent threat for citriculture (29); in contrast, the more recently characterized pathotype A* causes citrus canker on Mexican lime (Citrus aurantifolia) and has a much less severe impact on citriculture (72). Strains of this pathotype were considered to belong to the pathovar citri because of their phenotypic and genetic relatedness to pathotype A. Their distribution was initially reported to include Saudi Arabia, Oman, Iran, and India and was recently found to extend to southeast Asia, with reports of these strains in Thailand (10) and Cambodia (11). Finally, strains genetically related to pathotypes A and A* but able to infect Mexican lime and Citrus macrophylla naturally were recently detected in Florida and classified as a pathotype designated Aw (68). The molecular basis of the specific interaction of X. citri pv. citri pathotypes A* and Aw with a restricted range of citrus hosts is not known (5). An interaction between a host resistance gene and an avr gene product from the pathogen inducing host-pathogen incompatibility has not yet been demonstrated for the X. citri pv. citri-citrus pathosystem, as it has been previously for other plant pathogenic bacteria (43).
No assumption can be made about whether the apparent contemporary emergence of pathotype A* strains is due to a change in virulence or to environmental or human factors. Host range shifts have sometimes been related to modifications in the repertoire of virulence genes by horizontal gene transfer or intragenomic recombinations or mutations (20, 32, 75). A clear understanding of the evolutionary relationships among pathotypes A, A*, and Aw and of the diversity among strains of each pathotype would be helpful for assessing these issues.
Due to the extreme difficulty and cost of the complete eradication of Asiatic citrus canker, several canker-threatened citrus-producing regions rely on integrated pest management strategies for control (30). Data derived from the huge effort put into the molecular typing of human bacterial pathogens (46, 63, 67, 69) suggest that an extensive knowledge of populations of plant pathogenic bacteria may improve our understanding of epidemic situations.
The tools most often used for the molecular epidemiology of citrus canker have been repetitive-element-based PCR (rep-PCR) and pulsed-field gel electrophoresis (PFGE) (13, 16, 19, 28, 68). The lack of discriminatory power of rep-PCR and the high labor requirement for PFGE make it difficult to use these techniques extensively for outbreak investigations or regional or global surveillance (67). Therefore, alternative high-resolution and high-throughput molecular typing systems for X. citri pv. citri should be developed. Amplified fragment length polymorphism (AFLP) analysis of an Iranian collection of strains causing Asiatic citrus canker suggested previously that this technique has better discriminatory power than the rep-PCR method (39). AFLP has the advantage of generating a large number of randomly located markers over the whole genome. The detected polymorphism may arise from point mutations at the targeted restriction sites or from insertions and/or deletions in the amplified region (73). The determination of the complete sequence of X. citri pv. citri strain 306 (17) should facilitate the development of molecular typing tools well-suited for deciphering taxonomy, evolution, and/or epidemiology. For instance, it gave access to specific primers associated with transposable elements present in this bacterium (45), which were used for typing DNA from herbarium specimens showing canker-like symptoms and originating from different geographical origins. This technique revealed an unexpectedly high degree of genetic diversity. However, this typing scheme requires more than 50 PCRs for the full analysis of unknown DNA. A new insertion sequence ligation-mediated PCR (IS-LM-PCR) scheme (9) also revealed considerable diversity and is less labor-intensive. This technique amplifies DNA fragments between an insertion sequence element and a selected restriction site (9). We also recently developed a multilocus variable-number tandem-repeat analysis (MLVA) approach for this bacterium, a promising technique targeting tandem repeats (minisatellite-like loci) for fine-scale epidemiology with distinctive advantages, such as high discriminatory power, maximal reproducibility of results, and portability of equipment (12). The characteristics of these newly developed techniques need to be subjected to a comparative evaluation in order to determine which methods would be most useful for global surveillance and molecular epidemiology on small spatial scales. In this study, we compared the AFLP, MLVA, and IS-LM-PCR techniques to explore the genetic diversity of a collection of pathotype A, A*, and Aw strains originating from Asia. Furthermore, we sought to determine the genetic diversity and structure of X. citri pv. citri strains, including a large collection of pathotype A* strains for which no extensive characterization study is available at the moment, from the area of origin of the pathogen.
MATERIALS AND METHODS
Bacterial strains and DNA extraction.
A total of 234 bacterial strains isolated from citrus canker lesions and collected from 26 countries in Asia were used in this study, together with reference pathotype A, A*, and Aw strains. All strains were assigned to a pathogenicity group on the basis of results from detached-leaf inoculation assays performed on Mexican lime (C. aurantifolia), C. macrophylla, and grapefruit (C. paradisi) (72). Strains producing canker-like lesions on the three host species were classified as pathotype A strains. Both pathotype A* (n = 59) and Aw (n = 6) strains produced canker-like lesions on C. aurantifolia and C. macrophylla but not on C. paradisi. Thus, the two types were pathogenically indistinguishable (see Table S1 in the supplemental material for information on each strain, including the geographical origin, host species, date of collection, and pathogenicity group). Single colonies were subcultured on plates containing YPGA (yeast extract, 7 g liter−1; peptone, 7 g liter−1; glucose, 7 g liter−1; agar, 18 g liter−1; and propiconazole, 20 mg liter−1) for 24 h at 28°C. These subcultures were used to inoculate tubes containing 4 ml of YP broth (yeast extract, 7 g liter−1; peptone, 7 g liter−1; pH 7.2), and the tubes were incubated at 28°C on an orbital shaker for 16 to 18 h. These suspensions were used for DNA extraction with the DNeasy tissue kit according to the instructions of the manufacturer (Qiagen, Courtaboeuf, France). DNA concentrations were estimated by fluorometry with a TKO 100 fluorometer (Hoefer, San Francisco, CA).
AFLP analysis.
AFLP fingerprinting was performed mainly according to the original protocol by Vos et al. (73) as described previously (2). In brief, 25 ng of DNA was digested with MspI and SacI restriction enzymes as recommended by the manufacturer (New England Biolabs/Ozyme, Saint Quentin en Yvelines, France). Then 2.5-μl aliquots of the digested products were added to 22.5-μl ligation mixes containing 2 μM MspI adaptor (see Table S2 in the supplemental material), 0.2 μM SacI adaptor (Applied Biosystems, Courtaboeuf, France) (see Table S2 in the supplemental material), and 2 U of T4 DNA ligase (New England Biolabs/Ozyme, Saint Quentin en Yvelines, France) in 1× T4 DNA ligation buffer. Ligations were performed for 3 h at 37°C before enzyme inactivation at 65°C for 10 min. For preselective PCR, 10-fold-diluted ligation products were used as a template in a mix containing 5 mM MgCl2, 0.23 μM (each) MspI and SacI primers (see Table S2 in the supplemental material), 0.45 mM (each) deoxynucleoside triphosphates (New England Biolabs/Ozyme, Saint Quentin en Yvelines, France), and 0.5 U of Taq DNA polymerase (Goldstar red; Eurogentec, Seraing, Belgium) in 1× Goldstar buffer. The following PCR conditions were used: initial extension to ligate the second strand of the adaptors at 72°C for 2 min; a denaturation step at 94°C for 2 min; 25 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 2 min; and a final extension step at 72°C for 10 min. Tenfold-dilutions of PCR products were used as templates for selective amplifications. The selective amplifications using the unlabeled MspI+A, MspI+C, MspI+T, or MspI+G primer and the SacI+C primer labeled with one of four different fluorochromes (Applied Biosystems, Courtaboeuf, France) (see Table S2 in the supplemental material) were performed under the same conditions as the preselective PCR, except that the SacI+C primer concentration was 0.12 μM. The following PCR conditions were used: initial denaturation at 94°C for 2 min; 37 cycles of 94°C for 30 s, annealing for 30 s at 65°C in the first cycle, at temperatures decreasing by 0.7°C per cycle for the next 12 cycles, and then at 56°C for the last 24 cycles, and extension at 72°C for 2 min; and a final extension step at 72°C for 10 min. Samples were then prepared for capillary electrophoresis by adding 1 μl of the final PCR product to a mixture of 18.7 μl of formamide and 0.3 μl of a GeneScan 500 LIZ DNA ladder (Applied Biosystems, Courtaboeuf, France) as an internal standard. The samples were then denatured for 5 min at 95°C and placed on ice for at least 5 min. Electrophoresis was performed in an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Courtaboeuf, France) using a performance-optimized polymer, POP-4, at 15,000 V for about 20 min at 60°C, with an initial injection of 66 s. The AFLP fingerprints were analyzed visually using the software GeneScan 3.7 (Applied Biosystems, Courtaboeuf, France). To test the reproducibility of the results from the AFLP technique, two independent DNA extractions were used for all strains and strain 306 of X. citri pv. citri (17) was used as a control in each AFLP experiment.
IS-LM-PCR analysis.
IS-LM-PCR fingerprinting was performed as described previously (9). In brief, aliquots of bacterial genomic DNA were subjected to restriction enzyme and ligated to the adaptor by incubation for 3 h at 37°C in total volumes of 20 μl containing 2 ng of DNA, 9 U of MspI (New England Biolabs/Ozyme, Saint Quentin en Yvelines, France), 50 U of T4 DNA ligase (New England Biolabs/Ozyme, Saint Quentin en Yvelines, France), 50 mM NaCl, 1 μM MspI adaptor (Applied Biosystems, Courtaboeuf, France) (see Table S3 in the supplemental material), and 1× bovine serum albumin in 1× T4 DNA ligation buffer, followed by enzyme inactivation at 65°C for 10 min. Tenfold dilutions were used as template DNA in 20 μl of a PCR mix which contained 1 mM (each) deoxynucleoside triphosphates, 5 mM MgCl2, 0.25 μM unlabeled MspI primer (Applied Biosystems, Courtaboeuf, France) (see Table S3 in the supplemental material), 0.25 μM 5′-end-labeled insertion sequence-specific primer (Applied Biosystems, Courtaboeuf, France) (see Table S3 in the supplemental material), and 0.5 U of Taq DNA polymerase (Goldstar red; Eurogentec, Seraing, Belgium) in 1× Taq Goldstar buffer. The following PCR conditions were used: initial extension to ligate the second strand of the adaptors at 72°C for 2 min; a denaturation step at 94°C for 2 min; 35 cycles at 94°C for 45 s, 60°C for 60 s, and 72°C for 60 s; and a final extension step at 72°C for 10 min. Samples were prepared and subjected to capillary electrophoresis as explained above. To test the reproducibility of results from the IS-LM-PCR technique, two independent DNA extractions were used for all strains and strain 306 of X. citri pv. citri (17) was used as a control in each experiment.
MLVA scheme.
Fourteen primer pairs targeting single-locus alleles designed from the full sequence of X. citri pv. citri strain 306 (17) were used in a multiplex PCR format with a PCR kit from Qiagen (Courtaboeuf, France) (12). Briefly, 2 to 5 ng of genomic DNA was used as a template in mixes containing 0.2 μM (each) primers (one of which was marked with one of the fluorescent dyes 6-carboxyfluorescein, NED, PET, and VIC [Applied Biosystems]), 1× Qiagen multiplex mastermix (containing a hot-start Taq DNA polymerase), 0.5× Q-solution (Qiagen, Courtaboeuf, France), and RNase-free water to yield a volume of 15 μl. PCR amplifications were performed in a GeneAmp PCR system 9700 thermocycler (Applied Biosystems) under the following conditions: 15 min at 95°C for hot-start activation; 25 cycles of 94°C for 30 s, annealing at temperatures ranging from 64 to 70°C for 90 s, and 72°C for 90 s; and a final extension step at 72°C for 30 min (12). Aliquots of 1 μl of amplified products diluted 1/50 to 1/200 were mixed with 10.7 μl of Hi-Di formamide and 0.3 μl of a GeneScan 500 LIZ internal lane size standard (Applied Biosystems). Capillary electrophoresis was performed in an ABI PRISM 3130xl genetic analyzer (Applied Biosystems). To test the reproducibility of results from the MLVA technique, two independent DNA extractions were used for all strains and strain 306 of X. citri pv. citri (17) was used as a control in each experiment.
Data scoring and exploration.
For the AFLP and IS-LM-PCR techniques, the presence and absence of fragments were scored as a binary matrix and analyzed with the software R (version 2.6.1; R Development Core Team, Vienna, Austria). The size of each fragment in the range of 50 to 500 bp was determined. Fragments with fluorescence above a threshold set to 500 relative fluorescence units were scored. This threshold was found to be suitable for minimizing scoring discrepancies among DNA replicates in earlier studies (2, 53). Only fragments detected for both DNA replicates were scored as positive in the data matrix. Dice dissimilarities were used as distances to construct a weighted neighbor-joining (NJ) tree (26, 59) with the software R. The robustness of the tree was assessed by bootstrap analysis (1,000 resamplings). Metric multidimensional scaling (MDS) was used to represent distances between strains based on a Dice dissimilarity matrix. MDS transforms a distance matrix (which cannot be analyzed by eigendecomposition) into a cross-product matrix and then solves the eigenvector problem to find the coordinates of individuals so that distortions in the distance matrix are minimized. As in principal component analysis, individuals are projected into n dimensions (1). MDS was performed using the cmd-scale function in the R software.
For MLVA, integer numbers of tandem repeats were used as input data. Manhattan distances were calculated and used to build NJ trees with the R software (version 2.6.1; R Development Core Team, Vienna, Austria) using “cluster” and “ape” packages. The robustness of trees was assessed by bootstrap analysis (1,000 resamplings). MDS was also performed as described above, based on the Manhattan distance matrix. The identification of MLVA types (i.e., groups of strains differing by one to three variable-number tandem-repeat [VNTR] loci) was performed with eBURST, version 3 (25), available at http://eburst.mlst.net/. Whether MLVA loci evolve following a stepwise mutation model (SMM), i.e., preferentially by the addition or loss of a single repeat, was explored separately for each locus. For this purpose, the difference in the number of repeats for each pair of haplotypes along the evolutionary path inferred by eBURST analysis was calculated. The occurrence of each value of repeat difference was recorded for each group (defined as a collection of strains each with a maximum of three allelic mismatches with at least one other member of the collection), and values from all eBURST groups were pooled. This analysis was performed using multilocus analyzer software (S. Brisse, unpublished data), which is an independent implementation (coded in Python) of the eBURST algorithm, to which the SMM test function was added.
Predictive in silico analyses of AFLP, IS-LM-PCR, and MLVA techniques.
A predictive analysis of the AFLP, IS-LM-PCR, and MLVA methods was performed with the genome sequence of X. citri pv. citri strain 306 (17) to determine the accuracy and reproducibility of the results from each system (7). In the AFLP analysis, the lengths of predicted fragments corresponded to the lengths of the restriction fragments produced by simulating digestion with SacI and MspI and then selecting restriction fragments based on selective nucleotides present on selective AFLP primers, plus 24 bp corresponding to the length of adaptors. In the IS-LM-PCR analysis, the lengths of the predicted fragments were calculated as the size of the fragment bordered by each primer pair from a selection of MspI restriction fragments containing the targeted insertion sequences. The predicted fragment size for MLVA corresponded to the length of the PCR product for each primer pair. A total of 31, 41, and 82 fingerprints using strain 306 were analyzed for the AFLP, IS-LM-PCR, and MLVA techniques, respectively.
Genetic diversity and population structure.
The discriminatory power of each typing system was calculated using Hunter's single numerical index of discrimination (D) (34). This analysis was performed on our collection (n = 234) typed by the AFLP, IS-LM-PCR, and MLVA methods and on a subcollection (n = 34) including strains studied by Cubero and Graham (16). The correlations between distance matrices were tested pairwise using the Mantel test (48). All Mantel tests were performed using GenAlEx, version 6.1, with 9,999 permutations (52). Nei's unbiased estimates of genetic diversity (HE) for MLVA data were calculated using FSTAT 2.9.3 (http://www.unil.ch/popgen/softwares/fstat.htm). For biallelic data (AFLP and IS-LM-PCR results), AFLP-SURV software, version 1.0 (71) (http://www.ulb.ac.be/sciences/lagev/aflp-surv.html), was used for computing (i) allelic frequencies from observed frequencies of fragments according to the method of Lynch and Milligan for haploid species (47) and (ii) HE.
The allelic richness of our two sample sets composed of strains of pathotypes A and A*/Aw was estimated by a rarefaction method (36) producing unbiased estimates due to uneven sample sizes. The rarefaction method was performed with HP-RARE, version 1.0 (37). The degree of linkage disequilibrium was determined using the index of association (IA) (49) with the software GenAlEx, version 6.1 (52). IA is calculated by comparing the observed variance (VO) in the distribution of allelic mismatches in all pairwise comparisons of the allelic profiles with the expected variance (VE) in the freely recombining population, as follows: IA = (VO/VE) − 1. Significant linkage disequilibrium is established if the variance observed in the MLVA allele profiles is greater than the maximum variance observed in 1,000 randomized allele profiles (P < 0.001) (49).
We described the structure of the strain collection by using different approaches. Populations were defined at the geographical level when at least 10 strains of pathotype A or A*/Aw originated from the same country. When fewer than 10 strains per pathotype-country combination were present, we grouped strains from neighboring countries with no natural geographical barriers and/or with a common past history, e.g., those of the Indian peninsula; otherwise, such countries were not included in the analyses. A total of eight (10- to 27-strain) and three (10- to 24-strain) populations of pathotype A and A* strains, respectively, were defined (see Table S1 in the supplemental material). Genetic differentiation among populations was examined using different approaches. For MLVA data, pairwise population differentiation was assessed by Fisher exact tests (57) and tested for significance by the Markov chain Monte Carlo method using Arlequin, version 3.1 (22). For biallelic data, the results of exact tests were computed using the software TFPGA (Tools for Population Genetic Analyses), version 1.3 (42).
A hierarchical analysis of molecular variance (AMOVA) was conducted with Arlequin software, version 3.1 (22). Significance was tested using a nonparametric approach. Partitioning between data sets for pathotypes A and A* and among data sets for strains of each type from different countries and from the same country was conducted to evaluate the different contributions of these sources of variation for each genotyping technique. Estimates for Wright's fixation index of genetic differentiation (FST) and Slatkin's FST analogue RST, which takes into account the size difference among alleles and is more appropriate than FST when the loci studied evolve under the basic SMM (66), were obtained from the three sets of data (AFLP, IS-LM-PCR, and MLVA results) and from MLVA data, respectively. Estimates of FST for MLVA data and their respective P values for population pairs and all populations were calculated by using FSTAT 2.9.3. In the same way, RST estimates were calculated using the software RST CALC, version 2.2 (27). For AFLP and IS-LM-PCR data, FST was calculated using the software AFLP-SURV, version 1.0.
Isolation by distance among the eight populations of pathotype A was tested (58). The correlation between the logarithm of the geographical distances, retaining a central point for each population area, as defined above, and FST/(1 − FST) was evaluated with a Mantel test using the software GenAIEx 6.1 for each data set. Statistical testing was conducted using random permutation (n = 9,999).
The Bayesian clustering approach implemented in the software STRUCTURE, version 2.2.3, was used to infer population structure and assign individuals to groups characterized by distinct allele frequencies (54). The method estimates a probability of ancestry for each individual from each of the groups. Individuals are assigned to one of the groups or populations or jointly to two or more populations if their genotypes indicate that they are admixed. Twenty independent runs of STRUCTURE were performed by setting the number of subpopulations or groups (K) from 1 to 10, with 20,000 burn-in replicates and a run length of 105 replicates to decide which value of K best fit the data. The selection of K was done by examining the estimates of the posterior probability of the data for a given value of K, Pr(X K) (where X represents the number of genotypes in the sample), as a guide and by estimating the modal value of the distribution of ΔK [calculated from the STRUCTURE output Pr(X K)], which is a good indicator of the real K (21). We examined the clustering of strains of X. citri pv. citri for the inferred number of groups. We used different ancestry models in STRUCTURE according to the data sets. The linkage model (24) was used for MLVA data. For the biallelic markers (AFLP and IS-LM-PCR data), we used the admixture model and performed clustering without using prior population information under the F model, which assumes that the allele frequencies in the populations are correlated (23).
RESULTS
Predictive in silico AFLP, IS-LM-PCR, and MLVA typing.
In the AFLP analysis, 111 of 147 predicted fragments (76%) for the sequenced X. citri pv. citri strain 306 were always recorded and 8 predicted fragments were amplified in an irreproducible way. AFLP markers were scattered throughout the X. citri pv. citri genome. In the IS-LM-PCR analysis, 34 of 39 predicted fragments (87%) were always recorded, 3 predicted fragments were occasionally amplified, and 4 unpredicted fragments were reproducibly recorded. In MLVA, all predicted fragments were observed.
AFLP analysis.
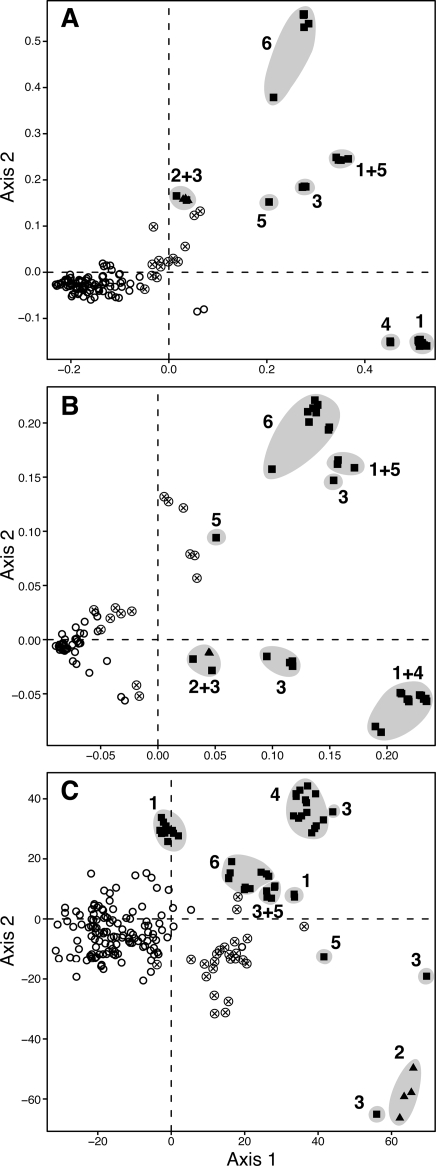
Although fingerprints obtained for DNA replicates sometimes differed in fragment intensity, the scoring system allowed overall good reproducibility of AFLP results, with >95% of the corresponding fragments in replicates being identically assigned. A total of 182 fragments, 100 (55%) of which were polymorphic, were scored. Ninety-four haplotypes were identified within the strain collection (n = 234) (Table 1). Based on the NJ tree (see Fig. S1A in the supplemental material) and the MDS plot (Fig. 1A), a clear-cut separation between pathotype A and A*/Aw strains was most often observed. A noticeable exception concerned a set of pathotype A strains from Bangladesh and India that were genetically close to pathotype A* strains but produced canker-like lesions on all assayed Citrus species (data not shown). The two graphical representations also suggested greater polymorphism within the A*/Aw group, an indication supported by the calculation of Nei's unbiased total gene diversity index (HT; 0.13 for pathotype A*/Aw strains versus 0.03 for pathotype A strains) (Table 1). Although pathotype Aw strains formed a subclade supported by a maximal bootstrap value, these strains were closely related to pathotype A* strains from India, with which they formed a cluster with a bootstrap value of 97%. The A*/Aw group formed a total of eight robust clusters supported by bootstrap values of ≥84% (see Fig. S1A in the supplemental material). Some A*/Aw clusters contained strains from a single origin (e.g., India or Thailand), whereas other clusters contained strains from more than one country (e.g., Saudi Arabia and Iran or Saudi Arabia, Oman, and India). In contrast, pathotype A strains did not generally form robust clusters (see Fig. S1A in the supplemental material). In rare cases, pathotype A strains grouped according to their countries of origin (e.g., strains from the Philippines and Bangladesh), but most often, closely related strains originated from different countries. Some haplotypes of pathotype A strains were identified in several countries. For example, haplotype 1 (24 strains) was detected in China, South Korea, Japan, the Philippines, Taiwan, and Thailand. Similarly, haplotype 5 (19 strains) was detected in China, Japan, Malaysia, the Philippines, and Taiwan (see Table S1 in the supplemental material).
TABLE 1.
Genetic diversity parameters for X. citri pv. citri strains estimated by using MLVA (14 VNTR loci), AFLP (4 conditions), and IS-LM-PCR (4 conditions)a
| Method | Locus(i) or primer combination(s) | No. of allelesb | Total no. of fragments | No. of haplotypes | HT | HWc | FSTd | RSTe |
|---|---|---|---|---|---|---|---|---|
| MLVA | XL1 | 19 | 0.92 | 0.84 | 0.11*** | 0.23NS | ||
| XL2 | 29 | 0.93 | 0.58 | 0.38*** | 0.84*** | |||
| XL3 | 11 | 0.87 | 0.70 | 0.25*** | 0.37NS | |||
| XL4 | 22 | 0.93 | 0.77 | 0.21*** | 0.78*** | |||
| XL5f | 15 | 0.89 | 0.73 | 0.19*** | 0.49** | |||
| XL6 | 26 | 0.93 | 0.83 | 0.15*** | 0.56*** | |||
| XL7 | 18 | 0.89 | 0.60 | 0.27*** | 0.17NS | |||
| XL8 | 10 | 0.73 | 0.44 | 0.33*** | 0.22NS | |||
| XL9 | 14 | 0.74 | 0.49 | 0.33*** | 0.60NS | |||
| XL10 | 10 | 0.83 | 0.47 | 0.43*** | 0.50NS | |||
| XL11 | 6 | 0.33 | 0.23 | 0.49*** | 0.53NS | |||
| XL13 | 11 | 0.47 | 0.31 | 0.39*** | 0.80* | |||
| XL14 | 23 | 0.94 | 0.62 | 0.35*** | 0.86*** | |||
| XL15 | 18 | 0.86 | 0.70 | 0.19*** | 0.51** | |||
| All X. citri loci | 232 | 0.80 | 0.59 | 0.27*** | 0.62*** | |||
| Loci from pathotype A strains (167 strains) | 172.6 | 0.77 | 0.64 | 0.13*** | 0.38*** | |||
| Loci from pathotype A* strains (65 strains)g | 137.6 | 0.79 | 0.48 | 0.54*** | 0.92*** | |||
| Biallelic-marker methods | ||||||||
| AFLP analysis | SacI+C/MspI+A | 26 | 31 | 0.07 | 0.02 | 0.71** | ||
| SacI+C/MspI+T | 39 | 29 | 0.09 | 0.03 | 0.76** | |||
| SacI+C/MspI+C | 66 | 45 | 0.07 | 0.02 | 0.80** | |||
| SacI+C/MspI+G | 51 | 44 | 0.06 | 0.03 | 0.55** | |||
| All combinations | 182 | 94 | 0.07 | 0.02 | 0.73** | |||
| Combinations for pathotype A strains (167 strains) | 154 | 59 | 0.03 | 0.02 | 0.28** | |||
| Combinations for pathotype A* strains (65 strains)g | 158 | 36 | 0.13 | 0.02 | 0.78** | |||
| IS-LM-PCR analysis | pMsp/ISXac1-D3 | 80 | 88 | 0.13 | 0.03 | 0.78** | ||
| pMsP/ISXac1-GF | 95 | 92 | 0.12 | 0.03 | 0.76** | |||
| pMsP/ISXac2-D3 | 96 | 80 | 0.06 | 0.03 | 0.63** | |||
| pMsp/ISXac3-IRF | 65 | 32 | 0.08 | 0.02 | 0.83** | |||
| All combinations | 336 | 146 | 0.10 | 0.03 | 0.78** | |||
| Combinations for pathotype A strains (167 strains) | 216 | 110 | 0.05 | 0.03 | 0.39** | |||
| Combinations for pathotype A* strains (65 strains)g | 249 | 35 | 0.20 | 0.03 | 0.87** |
***, P < 0.001; **, P < 0.01; *, P < 0;05; NS, not significant (P > 0.05).
Number of alleles observed at each locus. Allelic richness for both pathotypes was calculated using the rarefaction method (n [corresponding to the number of individuals in the smallest group] = 65).
HW, Nei's mean index of gene diversity within populations.
FST estimates were calculated for eight and three geographical groups (with 10 or more members) composed of pathotype A and A* strains, respectively.
RST estimates from VNTR data were calculated for eight and three geographical groups (with 10 or more members) composed of pathotype A and A* strains, respectively.
Parameters were calculated for 225 strains, as 7 pathotype A*/Aw strains did not produce any amplicon for the XL5 locus.
Strains originally described as pathotype Aw (68) were included in the pathotype A* group for data derived from all three genotyping techniques.
FIG. 1.
MDS representation of the distances among 234 strains of X. citri pv. citri based on AFLP (A), IS-LM-PCR (B), and MLVA (C) data. X. citri pv. citri pathotypes are represented by different symbols, including symbols indicating the geographical origins of pathotype A strains, as follows: ○, pathotype A from eastern and southern Asia; ⊗, pathotype A from western Asia; ▪, pathotype A*; and ▴, pathotype Aw. Numbers indicate the geographical origins of pathotype A* strains, as follows: 1, Saudi Arabia; 2, Florida; 3, the Indian peninsula; 4, Iran; 5, Oman; and 6, Thailand or Cambodia. The two first MDS axes described 86.2% (A), 85.8% (B), and 83.4% (C) of the total variation.
IS-LM-PCR analysis.
The overall reproducibility of IS-LM-PCR results was similar to that described for AFLP results. The global diversity (HT) ranged from 0.06 (for insertion element ISXac2) to 0.13 (for ISXac1) (Table 1). Based on Mantel test results, all distance matrices derived from a single primer pair were significantly correlated (P < 0.001). Pooling data ensured that the NJ tree had a robust structure, as indicated by bootstrap values (data not shown). When data derived from the four insertion sequence primer pairs were pooled, a total of 336 markers, 329 (98%) of which were polymorphic, were scored. The relationships among the 146 haplotypes that were identified within the strain collection (n = 234) are shown by an NJ tree (see Fig. S1B in the supplemental material) and the MDS plot in Fig. 1B. The two first MDS axes described 85.8% of the total variation. A clear-cut separation between pathotype A and A*/Aw strains was observed, which is consistent with AFLP results (Fig. 1A and B). Pathotype Aw strains formed a subclade supported by a bootstrap value of 87% but were closely related to pathotype A* strains from India, forming a cluster with a maximal bootstrap value. The A*/Aw group formed a total of 10 robust clusters supported by maximal bootstrap values (see Fig. S1B in the supplemental material). This high level of diversity was confirmed by calculating the genotypic diversity. The gene diversity (Nei's unbiased gene diversity index [HT]) calculated for pathotype A*/Aw strains (HT = 0.20) was greater than that calculated for pathotype A strains (HT = 0.05). The strain compositions of A*/Aw clusters derived from AFLP and IS-LM-PCR techniques were highly consistent. Pathotype A strains did not generally form robust clusters (see Fig. S1 in the supplemental material), which is consistent with AFLP data. Due to the greater discriminatory power of IS-LM-PCR, haplotypes that included a large number of strains were rare. Nevertheless, as in the AFLP analysis, it was possible to identify a few haplotypes of pathotype A strains that were found in several countries. For example, haplotype 10 (seven strains) was detected in China, Japan, and Taiwan. Pathotype A strains that showed the greatest genetic relatedness to pathotype A* strains originated from western Asia (Bangladesh, India, and the Maldives), which is consistent with AFLP data.
MLVA scheme.
A total of 209 haplotypes among the 234 strains were detected when data from the 14 loci were pooled, with allele numbers per locus ranging from 6 (for locus XL11) to 29 (for locus XL2). Five pathotype Aw strains and two pathotype A* strains from India did not produce amplicons for the XL5 locus. The mean numbers of observed alleles for types A and A* were 12.3 and 9.8, respectively, revealing a slightly higher degree of allelic richness in pathotype A strains (Table 1). Strains with the same allelic profile were primarily those isolated from the same site during the same year. In a few cases, strains sharing the same allelic profile were isolated from sites several miles apart in the same year or from the same site over two years. Similarly, each MLVA type originated from a single country. The relationships among the haplotypes that were identified within the strain collection are shown by an NJ tree (see Fig. S1C in the supplemental material) and the MDS plot in Fig. 1C. An eBURST analysis of the 209 MLVA types yielded 32 groups and 108 singletons, which is consistent with AFLP and IS-LM-PCR data. In order to determine whether tandem repeats evolve by following an SMM, we computed all the differences in the number of repeats along the evolutionary path deduced by the eBURST analysis within each eBURST group. Generally, the distribution of occurrences showed a single mode centered around zero. For most loci where at least four changes occurred in total (e.g., loci XL1, XL3, XL7, and XL11 to XL14), the most frequent change was either −1 or +1 repeat unit and the symmetric change was generally the second most frequent. Two noticeable exceptions were locus XL10, with more −2 changes (n = 6) than −1 (n = 3) and +1 (n = 3) changes, and locus XL9, with more +2 changes (n = 4) than −1 (n = 3) or +1 (n = 1) changes. However, the numbers were small in these two cases.
The IA calculated for the whole strain collection or for each pathotype separately showed that there was significant linkage disequilibrium (P < 0.001) between loci.
Genetic diversity and population structure.
The discriminatory abilities of the AFLP, IS-LM-PCR, and MLVA techniques was determined and compared by calculating D for 234 strains typed by all three methods. MLVA differentiated 209 strains and showed the best level of discrimination, with a D value of 0.998. IS-LM-PCR and AFLP markers distinguished 146 strains (D = 0.991) and 94 strains (D = 0.970), respectively (Table 2). The combination of MLVA and IS-LM-PCR data improved the discrimination among the strains to a D value of 0.999 (218 of 234 strains were discriminated). The three methods were more discriminative than rep-PCR based on the data from a collection of 34 strains (Table 2).
TABLE 2.
Discriminatory powers of AFLP, IS-LM-PCR, and MLVA methods based on results for a collection of 234 strains of X. citri pv. citri and a subset of 34 strains typed by rep-PCRa
| Genotyping method | No. of haplotypes identified (n = 234) | D | No. of haplotypes identified (n = 34) | D |
|---|---|---|---|---|
| AFLP | 94 | 0.970 | 19 | 0.932 |
| IS-LM-PCR | 146 | 0.991 | 31 | 0.994 |
| MLVA | 209 | 0.998 | 33 | 0.998 |
| Rep-PCR | NDb | ND | 12 | 0.854 |
Rep-PCR data are from Cubero and Graham (16).
ND, not determined.
Mantel test results suggested that data derived from the three typing techniques were congruent (P < 0.001) (Table 3). The highest values of correlation between individual genetic distances were observed for AFLP and IS-LM-PCR data (r = 0.866; P < 0.001). Lower but highly significant values of correlation between MLVA and IS-LM-PCR data (r = 0.618, P < 0.001) and MLVA and AFLP data (r = 0.599; P < 0.001) were found. Correlations for individual pairs of genetic distances were also highly significant when determined by considering pathotype A and A*/Aw strains separately (Table 3).
TABLE 3.
Mantel test results for pairwise correlations between genetic distances among individual strains for the different typing methods and pairwise population genetic differentiationa
| Population(s) | rA | rB | rC |
|---|---|---|---|
| Pathotype A strains | 0.472 | 0.430 | 0.673 |
| Pathotype A* strains | 0.818 | 0.801 | 0.759 |
| All populations | 0.866 | 0.599 | 0.618 |
rA, AFLP versus IS-LM-PCR data; rB, AFLP versus MLVA data; and rC, IS-LM-PCR versus MLVA data. P, <0.001 for all values.
Hierarchical AMOVA revealed highly significant genetic variation between pathotypes (P < 0.01), as well as between populations or within populations, whatever the molecular markers used (Table 4). The patterns of partitioning of the total variance differed among the different data sets. For AFLP, most of the total genetic variation (66.5%) was found between pathotypes and the remainder was distributed evenly among or within populations. Most of the genetic variation indicated by IS-LM-PCR was also between pathotypes, but to a lesser extent (52.5%), and the second source of variation was found at the population level. In contrast, analysis of the MLVA data set revealed that most of the total genetic variance was found among individual strains within populations (65.5%) and that the genetic variation between pathotypes accounted for only 12.2% of the total variation (Table 4).
TABLE 4.
Hierarchical AMOVA of X. citri pv. citri populations (eight pathotype A and three pathotype A* groups) by using AFLP, IS-LM-PCR, and VNTR markers
| Level at which variation occurred | dfa | Variance by:
|
% Total variation by:
|
Φ statisticb for:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AFLP analysis | IS-LM-PCR analysis | MLVA | AFLP analysis | IS-LM-PCR analysis | MLVA | AFLP analysis | IS-LM-PCR analysis | MLVA | ||
| Between pathotypes | 1 | 7.96 | 14.61 | 0.77 | 66.56 | 52.53 | 12.27 | 0.66** | 0.52** | 0.12** |
| Among populations within pathotypes | 9 | 1.75 | 8.02 | 1.37 | 14.67 | 28.83 | 22.19 | 0.44*** | 0.61*** | 0.27*** |
| Within populations | 173 | 2.24 | 5.19 | 4.04 | 18.77 | 18.64 | 65.54 | 0.81*** | 0.81*** | 0.34*** |
| Total | 183 | 11.95 | 27.83 | 6.17 | ||||||
df, degrees of freedom.
**, P < 0.01; ***, P < 0.001. Populations/geographical groups with 10 or more strains were considered.
For the eight defined X. citri pv. citri pathotype A populations, significant correlation was found between the logarithmic geographical distances and FST/(1 − FST), as estimated from IS-LM-PCR and MLVA data sets (P < 0.05) but not from the AFLP data set, with correlation values of 0.536, 0.507, and 0.371, respectively.
Fisher exact tests revealed little differentiation among the eight groups of pathotype A strains. The Indian peninsula group was differentiated from the group defined for Thailand (P < 0.05; Fisher exact test) and, with some significance, from those defined for China and the Philippines (P < 0.10), whatever the genotyping method (Table 5). Genetic differentiation between pathotype A strains from Thailand and the groups from China and the Philippines (P < 0.05) was also observed, but only for the MLVA data set. Unlike pathotype A strains, the three defined groups of pathotype A* strains, i.e., those from Saudi Arabia, Iran, and Thailand, were found to be genetically differentiated, whatever the typing technique (P ≤ 0.01; Fisher exact test). Based on FST analogues, when pathotypes were analyzed separately, larger differentiation estimates were obtained for pathotype A* strains than for pathotype A strains, whatever the typing technique (Table 1). Furthermore, genetic differentiation was observed in pairwise population analyses performed with biallelic markers, with AFLP-based and IS-LM-PCR-based FST estimates showing 27 of 28 and 28 of 28 pairs, respectively, to be significantly different (P < 0.01). FST estimates, when significant, varied from 0.07 (Taiwan/Thailand) to 0.62 (South Korea/the Maldives) and from 0.059 (Japan/Taiwan) to 0.67 (Japan/the Maldives) for AFLP and IS-LM-PCR, respectively. The level of genetic differentiation revealed by the MLVA-based FST values and RST values was lower, with 24 of 28 and 26 of 28 pairs, respectively, being significantly different (P < 0.01). Differentiation estimates varied from 0.07 (Philippines/Thailand) to 0.31 (Japan/the Maldives) and from 0.07 (Philippines/Thailand) to 0.81 (China/the Maldives) for MLVA-based FST and RST estimates, respectively. All the analyzed pairs of pathotype A* strains (n = 3) revealed genetic differentiation (P < 0.01) when FST and RST were estimated from biallelic markers or MLVA data. FST estimates were between 0.47 and 0.87, 0.78 and 0.93, and 0.46 and 0.63 for AFLP, IS-LM-PCR, and MLVA data, respectively. MLVA-based RST estimates for comparisons among pathogen A* groups varied from 0.90 to 0.95.
TABLE 5.
Genetic differentiation between X. citri pv. citri pathotype A subpopulations as estimated by Fisher exact testsa
| Population origin |
P value for comparison with population from:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| China | Indian peninsula | Japan | South Korea | The Maldives | The Philippines | Taiwan | Thailand | |
| China | 0.016 | 0.295 | 0.362 | 0.344 | 0.062 | 0.326 | 0.013 | |
| Indian peninsula | 0.0001/0.113 | 0.062 | 0.080 | 0.080 | 0.014 | 0.065 | 0.002 | |
| Japan | 1/1 | 0.458/1 | 1.000 | 1.000 | 0.265 | 1.000 | 0.069 | |
| South Korea | 1/1 | 1/1 | 1/1 | 1.000 | 0.282 | 1.000 | 0.097 | |
| Maldives | 0.999/1 | 1/1 | 0.998/1 | 1/1 | 0.288 | 1.000 | 0.096 | |
| Philippines | 1/1 | 0.006/0.811 | 1/1 | 1/1 | 0.999/1 | 0.277 | 0.013 | |
| Taiwan | 1/1 | 0.996/1 | 1/1 | 1/1 | 1/1 | 1/1 | ||
| Thailand | 1/1 | 0.024/0.563 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | |
P values for the AFLP/IS-LM-PCR data sets (below the diagonal) and for the MLVA data set (above the diagonal) are given.
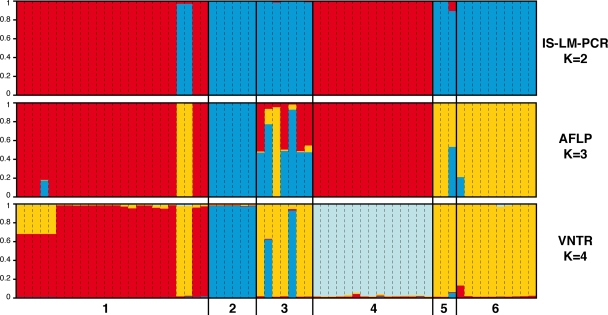
Bayesian clustering was used to analyze multilocus haplotypes to infer the genetic ancestry of the individual strains from both pathotypes separately. When pathotype A strains (n = 167) were considered, the analyses showed a pattern typical of an unstructured population, whatever the typing technique. No plateau in the estimates of the log likelihoods was reached, and no consistency among typing techniques for the compositions of putative populations was found. On the contrary, STRUCTURE identified two to four ancestral groups within the collection of pathotype A* strains, depending on the typing technique. Observations of plateaus in the estimates of ln [Pr(X K)] and a clear modal value in the distribution of ΔK indicated values of K of 2, 3, and 4 for the IS-LM-PCR, AFLP, and MLVA data sets, respectively (Fig. 2). The assignments of individual pathotype A* strains to ancestral clusters were consistent among the different data sets and correlated primarily with the strains' geographical origins. Strains from Florida, together with two strains from the Indian peninsula, were identified as a single population, whatever the typing technique. Two populations of strains from Saudi Arabia were consistently identified. One of the populations identified shared a common ancestor with strains from Iran, based on AFLP and IS-LM-PCR results, but the latter strains were unique based on MLVA data (Fig. 2). The second identified population of strains from Saudi Arabia shared a common ancestor with strains from India, Oman, Thailand, and Cambodia, whatever the genotyping technique (Fig. 2).
FIG. 2.
Population structure among the collection of X. citri pv. citri pathotype Aw strains and pathotype A* strains from the six geographical origins as inferred by STRUCTURE and assignment of individuals to the K ancestral groups. Numbers on the x axis indicate the geographical origins of strains, as follows: 1, Saudi Arabia; 2, Florida; 3, the Indian peninsula; 4, Iran; 5, Oman; and 6, Thailand or Cambodia. Each strain is represented by a vertical segment. Inside each segment, the length of each color section (y axis) indicates the estimated probability of assignment to each of the defined K ancestral groups. The use of the same color suggests that strains have a common ancestral group.
DISCUSSION
The availability of the complete genome of a Brazilian strain of X. citri pv. citri with a wide host range among citrus plants (pathotype A) (17) made it easier to design appropriate markers and predict the DNA fragments produced by different PCR-based techniques (9, 12, 45). This genome is relatively rich in insertion sequence elements and tandem repeats (17, 51). We evaluated the potentials of AFLP, IS-LM-PCR, and MLVA techniques as genotyping methods for molecular epidemiology studies of X. citri pv. citri. The analysis of the genetic diversity of large collections or populations requires discriminative, high-throughput techniques able to identify strain types and provide reproducible results. Despite the worldwide distribution and the major economic importance of this quarantine organism, no comparative evaluation of different molecular typing techniques for population structure analyses based on large strain collections has been conducted previously. Earlier studies considered primarily the usefulness of PFGE, AFLP, or rep-PCR markers in terms of their abilities to determine genetic relationships between Xanthomonas pathovars that are pathogenic to citrus and X. citri pv. citri pathological variants (16, 19, 39, 44, 68).
All three techniques used in this study typed all strains, except in the case of the XL5 VNTR locus, for which no amplicon was obtained from a limited number of strains within pathotype A*. MLVA revealed the greatest genetic diversity (based on Nei's and Hunter's indices), followed by IS-LM-PCR and AFLP analyses. For instance, pathotype A* strains from Saudi Arabia and Iran were not differentiated using AFLP, while IS-LM-PCR analysis and MLVA clearly separated strains from the two origins. Furthermore, MLVA identified each Iranian strain as a haplotype. The discriminatory powers of these methods were markedly greater than that of rep-PCR, based on results for a subset of 34 strains used in our study and a previous study by Cubero and Graham (16). VNTRs are known to exhibit much higher levels of mutation than other parts of the genome (35). The discriminatory power of MLVA was indeed found to be higher than or similar to those of insertion sequence-based typing techniques in population studies of Mycobacterium bovis and M. tuberculosis (4, 40, 41).
The comparison of experimental data with in silico data derived from the complete sequence of X. citri pv. citri strain 306 (17) indicated that the MLVA scheme was 100% accurate, whereas not all predicted fragments were amplified in the IS-LM-PCR and AFLP analyses, leading to accuracy values of 87 and 75%, respectively. This finding suggests that digestion and/or ligation may be deficient at some sites. The level of accuracy obtained by the AFLP method for X. citri pv. citri strain 306 was between those obtained previously for a strain of Escherichia coli (92%) and two strains of M. tuberculosis (55 and 66%) (7, 65). Although the overall intralaboratory reproducibility of AFLP and IS-LM-PCR data was good (95 to 97%), it did not reach that obtained for MLVA results (100%). Consistent with our data, previous interlaboratory comparisons of DNA typing methods for M. tuberculosis indicated better reproducibility of MLVA results than IS-LM-PCR and AFLP results (40).
Based on results from Mantel tests of dissimilarities, AFLP, IS-LM-PCR, and MLVA data were significantly congruent. Congruency among the data from the different methods was further observed for the parameters of genetic diversity. Pathotype A* strains were always clearly separated from the large majority of pathotype A strains. However, a few pathotype A strains were consistently shown to be related to pathotype A* strains. These strains originated from Bangladesh and India. Interestingly, they were all isolated from lime and displayed pathogenicity patterns typical of pathotype A. These observations suggest that variations in host range may have occurred in populations of X. citri pv. citri that originated from the Indian peninsula. Populations of the pathogen from this region display the greatest genetic diversity revealed to date, with the presence of two groups of pathotype A* strains and with pathotype A strains that were genetically related either to pathotype A strains present in southeast Asia or to pathotype A* strains. It is likely that undescribed genetic and/or pathological variants of the pathogen are yet to be discovered.
Clusters identified in relation to the geographical origins of the strains were consistently reproduced among X. citri pv. citri pathotype A* strains, whatever the typing technique. A pathotype A* strain was detected in Cambodia in 2007 (11), and this strain was closely related to pathotype A* strains from Thailand (10). Similarly, pathotype Aw strains originating from Florida were closely related to some pathotype A* strains isolated in India: these strains belonged to a single cluster with high bootstrap values, whatever the typing technique, and none of them could be typed using the XL5 VNTR locus. In addition, STRUCTURE suggested a common ancestry for pathotype Aw strains and the pathotype A* strains isolated in India. Thus, our molecular data fully support the results of investigations in Florida, which revealed that pathotype Aw strains had been isolated from Mexican lime in the yard of a family that had recently arrived from India and did not reveal the source of the diseased plant (62). The distribution area of pathotype A* strains in Asia is larger than previously reported and confirms the epidemiological significance of this narrow-host-range pathotype.
Interestingly, the level of genetic diversity among pathotype A* strains was higher than that among pathotype A strains, whatever the typing technique. This finding suggests that pathotype A* may have a longer evolutionary history. Li et al. (45) analyzed a few lime herbarium specimens collected in western Asia during the first half of the 20th century, and no conclusion could be drawn. Genetic differentiation analyses indicated that pathotype A* strain populations are well-structured. The number of ancestral populations computed by STRUCTURE was genotyping technique dependent, but pathotype A* strains could originate from at least two ancestral populations. Pathotype A* strains from Saudi Arabia were consistently classified into two populations, whatever the typing technique, which may suggest at least two different incidents involving the introduction of the pathogen into this country.
Based on NJ trees and MDS, no clear geographically inferred structure among the pathotype A strains was revealed. This result is consistent with those of previous studies targeting diversity by using rep-PCR or transposable elements (16, 45). Significant patterns of isolation by distance were observed at the level of the Asian continent. The low degree of global genetic diversity and the significant isolation by distance suggest a spatially limited gene flow for pathotype A strains and/or a rather short evolutionary history.
The different levels of structuring were also reflected in the different values of differentiation estimates for subpopulations of each pathotype. The elevated RST values compared to FST estimates calculated from the MLVA data set suggested that the geographical subpopulations of X. citri pv. citri or of each pathotype differed not only in allele frequencies but also significantly in the evolutionary distance between alleles. Different estimates of FST from each molecular marker data set revealed lower values for MLVA than for the two biallelic marker systems. MLVA revealed the highest discriminatory power, as shown in this study by the identification of 209 different haplotypes among 234 strains. Estimations of FST depend partly on the level of within-population diversity, and estimates derived from highly mutable loci should be considered with caution (14). Gene flow between populations homogenizes allele frequencies and tends to decrease genetic differentiation. Furthermore, the differentiation level is not affected strictly by the mutation potential of the observed loci but also by the ratio between mutation and migration (8). This situation makes FST sensitive to the mutation rates for pathogens such as X. citri pv. citri with limited long-distance migration (31). The RST analogue values from the MLVA data set are independent of the mutation rate and are more appropriate estimates than FST values, given that tandem repeats in our scheme can be considered as evolving by the progressive gain or loss of single repeat units. RST values were in accordance with the estimates of genetic differentiation obtained from the two biallelic data sets. It is likely that the significance of the long-distance movement of infected plant material, although globally limited, is pathotype dependent. Pathotype A strains can be transported by most of the citrus species, primarily through propagative material, providing much greater potential for the exchange of pathotype A strains than for that of pathotype A* strains, which are hosted mainly by limes and, to a lesser extent, C. macrophylla. In many Asiatic countries, lime trees are produced from seed and C. macrophylla is used mainly as rootstock, and the pathogen is not seed borne (15). These host characteristics drastically decrease the movement of budwood (a major source of long-distance spread). The low level of migration of pathotype A* strains, together with their narrow host range, minimizes gene flow, which can explain the greater genetic differentiation observed within A* subpopulations. This observation may also be the result of the greater genetic diversity observed within X. citri pv. citri pathotype A*.
Significant levels of linkage disequilibrium among the VNTR loci were observed using a multilocus estimate, which revealed the absence of frequent genetic exchanges. Given our strain collection, however, this finding should be considered with caution because specific ecological niche and/or geographical barriers may explain the limited DNA exchange observed (49). Nevertheless, these results, together with the significant congruence among data derived from the three independent genotyping techniques, suggested that both pathotypes of X. citri pv. citri are clonal. Testing for nonrandom association of loci on small scales should confirm the level of clonality, which may vary among populations. Extensive sampling of pathotype A* strains in the Indian peninsula, for which admixtures within some individual strains suggest different origins, may reveal a population structure which differs from that in other geographical areas.
The various discriminative powers of the three genotyping techniques made it possible to conduct molecular epidemiology analyses on different spatial scales and for different purposes. Highly variable markers, such as VNTRs, are not adapted for international long-term strain screening (70). Although the concomitant use of several typing techniques strengthens analyses, the increased discriminatory power of the IS-LM-PCR method compared to those of rep-PCR (16) and AFLP (this study) analyses makes this technique most appropriate for the global surveillance of non-epidemiologically related strains of X. citri pv. citri. IS-LM-PCR targets transposable elements, similar to the technique developed by Li et al. (45). IS-LM-PCR seems superior because of the smaller number of PCR amplifications required. Furthermore, in contrast with the method used by Li et al. (45), IS-LM-PCR was proved to provide results highly congruent with those of the AFLP method, a technique well-suited for analyzing phylogenetic relationships among xanthomonads (55). These findings suggest that the transposition of insertion sequences in the genome of X. citri pv. citri occurs so rarely that it does not disturb the phylogenetic signal. In previous interlaboratory tests in which nine typing techniques were compared, two IS-LM-PCR-based techniques were the best alternative to MLVA for M. tuberculosis in terms of reproducibility (40). This observation suggests that our IS-LM-PCR protocol may be amenable to an efficient interlaboratory typing system.
VNTR loci display a high degree of polymorphism and are well-adapted for the typing of epidemiologically related isolates. Among the three typing techniques used in this study, MLVA best described the X. citri pv. citri intrapopulation genetic variation. MLVA may be useful for tracing haplotypes during epidemics on small spatial scales and for investigating inoculum sources associated with outbreaks. MLVA was proved previously to be very useful for the discrimination of anthrax-causing bacterial populations with a low level of genetic diversity (38). We are currently evaluating MLVA for the molecular epidemiology of X. citri on different spatial scales and in different epidemiological contexts (such as integrated management and eradication).
Supplementary Material
Acknowledgments
We thank E. L. Civerolo for helpful discussion and K. Vital, C. Boyer, and V. Ledoux for their technical expertise.
The European Union (FEOGA and FEDER), Conseil Régional de La Réunion, and CIRAD provided financial support. The platform “Genotyping of Pathogens and Public Health” acknowledges financial support from the Institut Pasteur (Paris, France) and the Institut de Veille Sanitaire (Saint-Maurice, France).
Footnotes
Published ahead of print on 16 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abdi, H. 2007. Metric multidimensional scaling (MDS): analyzing distance matrices, p. 1-13. In N. Salkind (ed.), Encyclopedia of measurement and statistics. Sage, Thousand Oaks, CA.
- 2.Ah-You, N., L. Gagnevin, F. Chiroleu, E. Jouen, J. R. Neto, and O. Pruvost. 2007. Pathological variations within Xanthomonas campestris pv. mangiferaeindicae support its separation into three distinct pathovars that can be distinguished by amplified fragment length polymorphism. Phytopathology 97:1568-1577. [DOI] [PubMed] [Google Scholar]
- 3.Ah-You, N., L. Gagnevin, P. A. D. Grimont, S. Brisse, X. Nesme, F. Chiroleu, L. Bui Thi Ngoc, E. Jouen, P. Lefeuvre, C. Vernière, and O. Pruvost. 2009. Polyphasic characterization of xanthomonads pathogenic to Anacardiaceae and their relatedness to different Xanthomonas species. Int. J. Syst. Evol. Microbiol. 59:306-318. [DOI] [PubMed] [Google Scholar]
- 4.Allix, C., K. Walravens, C. Saegerman, J. Godfroid, P. Supply, and M. Fauville-Dufaux. 2006. Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping. J. Clin. Microbiol. 44:1951-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Saadi, A., J. D. Reddy, Y. P. Duan, A. M. Brunings, Q. Yuan, and D. W. Gabriel. 2007. All five host-range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host-range variation. Mol. Plant-Microbe Interact. 20:934-943. [DOI] [PubMed] [Google Scholar]
- 6.Anonymous. 2007. List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 57:893-897. [DOI] [PubMed] [Google Scholar]
- 7.Arnold, C., L. Metherell, J. P. Clewley, and J. Stanley. 1999. Predictive modelling of fluorescent AFLP: a new approach to the molecular epidemiology of E. coli. Res. Microbiol. 150:33-44. [DOI] [PubMed] [Google Scholar]
- 8.Balloux, F., and N. Lugon-Moulin. 2002. The estimation of population differentiation with microsatellite markers. Mol. Ecol. 11:155-165. [DOI] [PubMed] [Google Scholar]
- 9.Bui Thi Ngoc, L., C. Vernière, J. Belasque, K. Vital, S. Boutry, L. Gagnevin, and O. Pruvost. 2008. Ligation-mediated PCR, a fast and reliable technique for insertion sequence-based typing of Xanthomonas citri pv. citri. FEMS Microbiol. Lett. 288:33-39. [DOI] [PubMed] [Google Scholar]
- 10.Bui Thi Ngoc, L., C. Vernière, O. Pruvost, N. Kositcharoenkul, and S. Phawichit. 2007. First report in Thailand of Xanthomonas axonopodis pv. citri-A* causing citrus canker on lime. Plant Dis. 91:771. [DOI] [PubMed] [Google Scholar]
- 11.Bui Thi Ngoc, L., C. Vernière, O. Pruvost, S. Thavrith, and G. I. Johnson. 2008. First report of Xanthomonas axonopodis pv. citri-A* causing citrus canker on lime in Cambodia. Plant Dis. 92:1588. [DOI] [PubMed] [Google Scholar]
- 12.Bui Thi Ngoc, L., C. Vernière, K. Vital, F. Guérin, L. Gagnevin, S. Brisse, N. Ah-You, and O. Pruvost. 2009. Fourteen minisatellite markers for population studies of the citrus canker bacterium, Xanthomonas citri pv. citri. Mol. Ecol. Res. 9:125-127. [DOI] [PubMed] [Google Scholar]
- 13.Carvalho, F. M. S., L. P. C. Caramori, and R. P. Leite. 2005. Genetic diversity of Xanthomonas axonopodis pv. citri based on plasmid profile and pulsed field gel electrophoresis. Genet. Mol. Biol. 28:446-451. [Google Scholar]
- 14.Charlesworth, B. 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol. Biol. Evol. 15:538-543. [DOI] [PubMed] [Google Scholar]
- 15.Civerolo, E. L. 1984. Bacterial canker disease of citrus. J. Rio Grande Valley Hortic. Soc. 37:127-145. [Google Scholar]
- 16.Cubero, J., and J. H. Graham. 2002. Genetic relationship among worldwide strains of Xanthomonas causing canker in citrus species and design of new primers for their identification by PCR. Appl. Environ. Microbiol. 68:1257-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. Do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. De Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 18.Dye, D. W., J. F. Bradbury, M. Goto, A. C. Hayward, R. A. Lelliott, and M. N. Schroth. 1980. International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Rev. Plant Pathol. 59:153-168. [Google Scholar]
- 19.Egel, D. S., J. H. Graham, and R. E. Stall. 1991. Genomic relatedness of Xanthomonas campestris strains causing diseases of citrus. Appl. Environ. Microbiol. 57:2724-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Yacoubi, B., A. M. Brunings, Q. Yuan, S. Shankar, and D. W. Gabriel. 2007. In planta horizontal transfer of a major pathogenicity effector gene. Appl. Environ. Microbiol. 73:1612-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evanno, G., S. Regnaut, and J. Goudet. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14:2611-2620. [DOI] [PubMed] [Google Scholar]
- 22.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 23.Falush, D., M. Stephens, and J. K. Pritchard. 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7:574-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falush, D., M. Stephens, and J. K. Pritchard. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gascuel, O. 1997. Concerning the NJ algorithm and its unweighted version UNJ, p. 149-171. In B. Mirkin, F. R. McMorris, F. Roberts, and A. Rzhetsky (ed.), Mathematical hierarchies and biology, vol. 37. American Mathematical Society, Providence, RI. [Google Scholar]
- 27.Goodman, S. J. 1997. RST Calc: a collection of computer programs for calculating estimates of genetic differentiation from microsatellite data and determining their significance. Mol. Ecol. 6:881-885. [Google Scholar]
- 28.Gottwald, T. R., J. H. Graham, and D. S. Egel. 1992. Analysis of foci of Asiatic citrus canker in a Florida citrus orchard. Plant Dis. 76:389-396. [Google Scholar]
- 29.Gottwald, T. R., J. H. Graham, and T. S. Schubert. 12 August 2002, posting date. Citrus canker: the pathogen and its impact. Plant Health Prog. doi: 10.1094/PHP-2002-0812-01-RV. [DOI]
- 30.Gottwald, T. R., and M. Irey. 23 May 2007, posting date. Post-hurricane analysis of citrus canker. II. Predictive model estimation of disease spread and area potentially impacted by various eradication protocols following catastrophic weather events. Plant Health Prog. doi: 10.1094/PHP-2007-0405-01-RS. [DOI]
- 31.Graham, J. H., T. R. Gottwald, J. Cubero, and D. S. Achor. 2004. Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 5:1-15. [DOI] [PubMed] [Google Scholar]
- 32.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 33.Hayward, A. C. 1993. The hosts of Xanthomonas, p. 1-119. In J. G. Swings and E. L. Civerolo (ed.), Xanthomonas. Chapman & Hall, London, United Kingdom.
- 34.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarne, P., and P. J. L. Lagoda. 1996. Microsatellites, from molecules to populations and back. Trends Ecol. Evol. 11:424-429. [DOI] [PubMed] [Google Scholar]
- 36.Kalinowski, S. T. 2004. Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv. Genetics 5:539-543. [Google Scholar]
- 37.Kalinowski, S. T. 2005. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 5:187-189. [Google Scholar]
- 38.Keim, P., M. N. Van Ert, T. Pearson, A. J. Vogler, L. Y. Huynh, and D. M. Wagner. 2004. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect. Genet. Evol. 4:205-213. [DOI] [PubMed] [Google Scholar]
- 39.Khodakaramian, G., and J. Swings. 2002. AFLP fingerprinting of the strains of Xanthomonas axonopodis inducing citrus canker disease in southern Iran. J. Phytopathol. 150:227-231. [Google Scholar]
- 40.Kremer, K., C. Arnold, A. Cataldi, M. C. Gutierrez, W. H. Haas, S. Panaiotov, R. A. Skuce, P. Supply, A. G. M. Van der Zanden, and D. Van Soolingen. 2005. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. J. Clin. Microbiol. 43:5628-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kremer, K., B. K. Y. Au, P. C. W. Yip, R. Skuce, P. Supply, K. M. Kam, and D. Van Soolingen. 2005. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J. Clin. Microbiol. 43:314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labate, J. A. 2000. Software for population genetic analyses of molecular marker data. Crop Sci. 40:1521-1528. [Google Scholar]
- 43.Leach, J. E., and F. F. White. 1996. Bacterial avirulence genes. Annu. Rev. Phytopathol. 34:153-179. [DOI] [PubMed] [Google Scholar]
- 44.Lee, Y. H., S. Lee, D. H. Lee, S. H. Ji, H. Y. Chang, S. Heu, J. W. Hyun, D. S. Ra, and E. W. Park. 2008. Differentiation of citrus bacterial canker strains in Korea by host range, rep-PCR fingerprinting and 16S rDNA analysis. Eur. J. Plant Pathol. 121:97-102. [Google Scholar]
- 45.Li, W., Q. Song, R. H. Brlansky, and J. S. Hartung. 2007. Genetic diversity of citrus bacterial canker pathogens preserved in herbarium specimens. Proc. Natl. Acad. Sci. USA 104:18427-18432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindstedt, B. A. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 26:2567-2582. [DOI] [PubMed] [Google Scholar]
- 47.Lynch, M., and G. Milligan. 1994. Analysis of population genetic structure with RAPD markers. Mol. Ecol. 3:91-99. [DOI] [PubMed] [Google Scholar]
- 48.Mantel, N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209-220. [PubMed] [Google Scholar]
- 49.Maynard-Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minsavage, G. V., D. Dahlbeck, M. C. Whalen, B. Kearney, U. Bonas, B. J. Staskawicz, and R. E. Stall. 1990. Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria-pepper interactions. Mol. Plant-Microbe Interact. 3:41-47. [Google Scholar]
- 51.Mrázek, J., X. Guo, and A. Shah. 2007. Simple sequence repeats in prokaryotic genomes. Proc. Natl. Acad. Sci. USA 104:8472-8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peakall, R., and P. E. Smouse. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6:288-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picard, Y., P. Roumagnac, D. Legrand, L. Humeau, I. Robène-Soustrade, F. Chiroleu, L. Gagnevin, and O. Pruvost. 2008. Polyphasic characterization of Xanthomonas axonopodis pv. allii associated with outbreaks of bacterial blight on three Allium species in the Mascarene archipelago. Phytopathology 98:919-925. [DOI] [PubMed] [Google Scholar]
- 54.Pritchard, J. K., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rademaker, J. L. W., B. Hoste, F. J. Louws, K. Kersters, J. Swings, L. Vauterin, P. Vauterin, and F. J. De Bruijn. 2000. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int. J. Syst. Evol. Microbiol. 50:665-677. [DOI] [PubMed] [Google Scholar]
- 56.Rademaker, J. L. W., F. J. Louws, M. H. Schultz, U. Rossbach, L. Vauterin, J. Swings, and F. J. De Bruijn. 2005. A comprehensive species to strain taxonomic framework for Xanthomonas. Phytopathology 95:1098-1111. [DOI] [PubMed] [Google Scholar]
- 57.Raymond, M., and F. Rousset. 1995. An exact test for population differentiation. Evolution 49:1280-1283. [DOI] [PubMed] [Google Scholar]
- 58.Rousset, F. 1997. Genetic differentiation and estimation of gene flow from F statistics under isolation by distance. Genetics 145:1219-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 60.Schaad, N. W., E. Postnikova, G. H. Lacy, A. Sechler, I. Agarkova, P. E. Stromberg, V. K. Stromberg, and A. K. Vidaver. 2006. Emended classification of xanthomonad pathogens on citrus. Syst. Appl. Microbiol. 29:690-695. [DOI] [PubMed] [Google Scholar]
- 61.Schaad, N. W., E. Postnikova, G. H. Lacy, A. Sechler, I. Agarkova, P. E. Stromberg, V. K. Stromberg, and A. K. Vidaver. 2005. Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans subsp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv. malvacearum (ex Smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) Dye 1978 as X. alfalfae subsp. alfalfae (ex Riker et al., 1935) sp. nov. nom. rev.; and “var. fuscans” of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Syst. Appl. Microbiol. 28:494-518. [DOI] [PubMed] [Google Scholar]
- 62.Schubert, T., S. Rizvi, X. Sun, T. Gottwald, J. Graham, and W. Dixon. 2001. Meeting the challenge of eradicating citrus canker in Florida—again. Plant Dis. 85:341-356. [DOI] [PubMed] [Google Scholar]
- 63.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen, Y. W., and P. Ronald. 2002. Molecular determinants of disease and resistance in interactions of Xanthomonas oryzae pv. oryzae and rice. Microbes Infect. 4:1361-1367. [DOI] [PubMed] [Google Scholar]
- 65.Sims, E. J., M. Goyal, and C. Arnold. 2002. Experimental versus in silico fluorescent amplified fragment length polymorphism analysis of Mycobacterium tuberculosis: improved typing with an extended fragment range. J. Clin. Microbiol. 40:4072-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Slatkin, M. 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Struelens, M. J., Y. De Gheldre, and A. Deplano. 1998. Comparative and library epidemiologic typing systems: outbreak investigations versus surveillance systems. Infect. Control Hosp. Epidemiol. 19:565-569. [DOI] [PubMed] [Google Scholar]
- 68.Sun, X. A., R. E. Stall, J. B. Jones, J. Cubero, T. R. Gottwald, J. H. Graham, W. N. Dixon, T. S. Schubert, P. H. Chaloux, V. K. Stromberg, G. H. Lacy, and B. D. Sutton. 2004. Detection and characterization of a new strain of citrus canker bacteria from key Mexican lime and Alemow in South Florida. Plant Dis. 88:1179-1188. [DOI] [PubMed] [Google Scholar]
- 69.Van Belkum, A. 2002. Molecular typing of micro-organisms: at the centre of diagnostics, genomics and pathogenesis of infectious diseases? J. Med. Microbiol. 51:7-10. [DOI] [PubMed] [Google Scholar]
- 70.Van Belkum, A., M. Struelens, A. De Visser, H. Verbrugh, and M. Tibayrenc. 2001. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin. Microbiol. Rev. 14:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vekemans, X., T. Beauwens, M. Lemaire, and I. Roldan-Ruiz. 2002. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol. Ecol. 11:139-151. [DOI] [PubMed] [Google Scholar]
- 72.Vernière, C., J. S. Hartung, O. P. Pruvost, E. L. Civerolo, A. M. Alvarez, P. Maestri, and J. Luisetti. 1998. Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from Southwest Asia. Eur. J. Plant Pathol. 104:477-487. [Google Scholar]
- 73.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Vandelee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wicker, E., L. Grassart, R. Coranson-Beaudu, D. Mian, C. Guilbaud, M. Fegan, and P. Prior. 2007. Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Appl. Environ. Microbiol. 73:6790-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woolhouse, M. E. J., D. T. Haydon, and R. Antia. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20:238-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.